Abstract
Lipids are vital biological molecules and play multiple roles in cellular function of mammalian organisms such as cellular membrane anchoring, signal transduction, material trafficking and energy storage. Driven by the biological significance of lipids, lipidomics has become an emerging science in the field of omics. Lipidome in biological systems consists of hundreds of thousands of individual lipid molecules that possess complex structures, multiple categories, and diverse physicochemical properties assembled by different combinations of polar headgroups and hydrophobic fatty acyl chains. Such structural complexity poses a huge challenge for comprehensive lipidome analysis. Thanks to the great innovations in chromatographic separation techniques and the continuous advances in mass spectrometric detection tools, analytical strategies for lipidomics have been highly diversified so that the depth and breadth of lipidomics have been greatly enhanced. This review will present the current state of mass spectrometry-based analytical strategies including untargeted, targeted and pseudotargeted lipidomics. Recent typical applications of lipidomics in biomarker discovery, pathogenic mechanism and therapeutic strategy are summarized, and the challenges facing to the field of lipidomics are also discussed.
Keywords: Liquid chromatography, High-resolution mass spectrometry, Targeted lipidomics, Untargeted lipidomics, Pseudotargeted lipidomics, Biomarker discovery
Graphical abstract
Highlights
-
•
Recent advances in analytical techniques allow rapid, reliable and sensitive detection of hundreds to thousands of lipids
-
•
The diversified MS-based analytical strategies have greatly enhanced the development and application of lipidomics.
-
•
The integration of multi-omics has a bright prospect in studying the pathogenesis and potentially targetable therapy.
-
•
Comprehensive analysis of the lipidome still faces considerable challenges in analytical techniques.
1. Introduction
Lipids are crucial small biomolecules and play vital roles in a variety of physio-pathological events by serving as constituents of cellular membranes, cellular barriers, signal transduction, energy sources, and intermediates in signaling pathways [1]. Maintenance of lipid metabolic homeostasis is an essential feature of a normal organism. An increasing number of studies have demonstrated that lipid metabolism imbalance is linked to many diseases such as obesity, hypertension, metabolic syndrome, diabetes, cardiovascular diseases, Parkinson and cancer [2].
The biological significance of lipids has impelled the development of lipid research as a discipline, i.e., lipidomics. In analogy to metabolomics, lipidomics aims at the panoramic analysis of the lipidome existing in biological systems and the concomitant detection of the subtle changes in individual lipids response to internal and/or external stimuli, such as environmental stress, diseases, drug intervention and genetic mutation. It is obvious that the analytical technology is the core for lipidomics.
The key elements facing to lipidomic analytical methods are lipid coverage, sensitivity, identification and quantification as well as throughput. Recent advances in mass spectrometry (MS) together with the highly efficient separation techniques allow the stable, rapid and sensitive detection of a large number of individual lipids present in different biological samples. It has greatly enhanced our understanding of the metabolic characteristics and biological activities of individual lipids and/or lipid (sub)classes in different patho-physiological processes. Although it is an emerging science, lipidomics has displayed great potential in different fields related to disease, drug R&D, food and plant.
The present review will first describe the structural diversity of lipids and its resultant challenges, and then summarize the recent advances in MS-based lipidomics in terms of untargeted, targeted and pseudotargeted analytical strategies. On this basis, the applications of lipidomics reported in recent 5 years in disease biomarker discovery as well as pathogenic mechanism and therapeutic strategy will be presented, and future perspectives of lipidomics is also given.
2. Structural diversity and analytical challenge of lipids
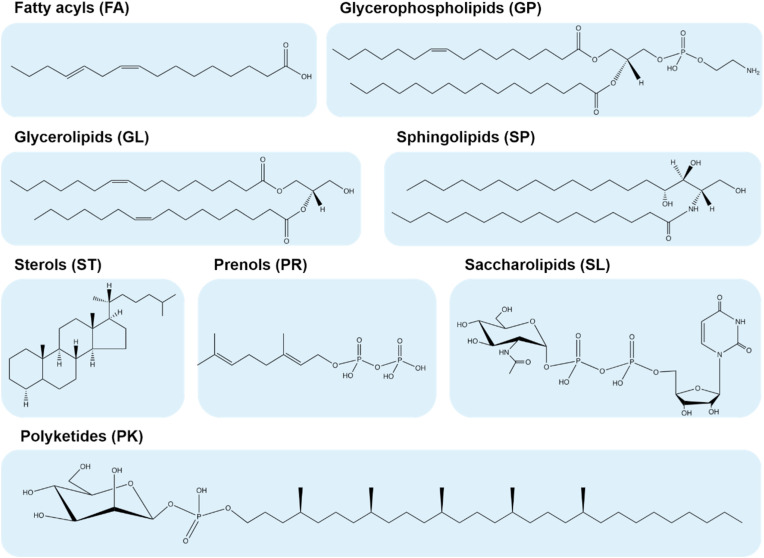
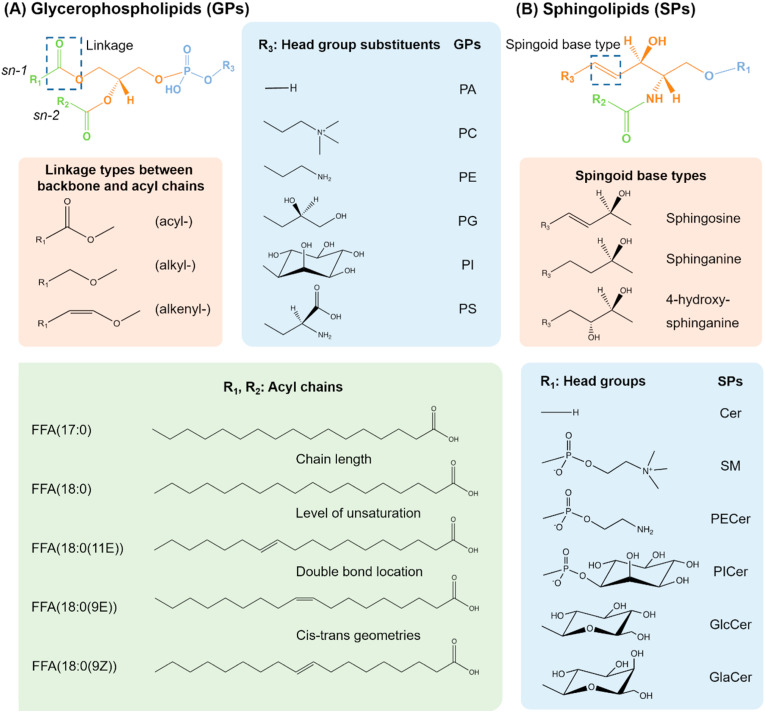
Lipids in biological systems consist of tens to hundreds of thousands distinct chemical entities with wide diversities in structures and physiochemical properties [10]. Even a single mammalian cell encompasses hundreds of thousands of lipid species. Currently, the LIPID MAPS Structure Database (LMSD, http://www.lipidmaps.org/resources/databases) has enrolled 44,701 unique lipid structures dispersed in eight categories including fatty acyls (FA), glycerolipids (GL), glycerophospholipids (GP), sphingolipids (SP), sterol lipids (ST), prenol lipids (PR), saccharolipids (SL), and polyketides (PK). Each category can be finely divided into several classes and/or subclasses [[3], [4], [5], [6]], Fig. 1 summarizes the basic structural information of eight lipid categories. The majority of natural lipids are various combinations of hydrophobic fatty acyl chains and polar head groups attached to different lipid backbone structures (e.g., glycerol and sphingoid bases). The structural diversity of the lipidome arises via variations in the type of the head groups, the fatty acyl chain length, the level of unsaturation, double bond location, cis-trans geometric isomerism, branched functional groups in the fatty acyl chains, the type of the covalent bond, i.e., ester (acyl-), ether (alkyl-) and vinyl-ether (alkenyl-), linked to the head groups. Taking GPs and SPs as examples, Fig. 2 illustrates the diversity and complexity of lipid structures.
Fig. 1.
Basic structural information of eight lipid categories classified by Fahy et al. [3].
Fig. 2.
The illustration of the diversity and complexity of lipid structures by taking GPs and SPs as examples. GPs structure diversity lies in the variety of head group, fatty acyl chains at the sn-1 and sn-2 positions as well as the linkage between glycerol backbone and sn-1 fatty acids. The head group generally represents phosphate group, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol and phosphatidylserine, which respectively defines the classes of GPs as PA, PC, PE, PG, PI and PS, etc. Another aspect of diversity stems from three different linkage: ester (acyl-), ether (alkyl-) and vinyl-ether (alkenyl-). Besides, the fatty acyl chains also vary in chain length, level of unsaturation, double bond location and cis-trans geometries. Similarly, combinations of different structures of head group, sphingoid based type and N-acyl chain constitute tens of thousands of SPs.
The structural diversity of lipids endows lipid molecular species with diversified physiological functions. For instance, triacylglycerols (TAGs) are the main energy storage substances in cells. The long-chain fatty acids (LCFAs) play critical roles in regulating energy metabolism [7]. Lipid species such as eicosanoids, lyso-phospholipids (LPLs), and phosphoinositides (PIs) serve as signaling messengers in the cellular biosynthetic pathways [8]. Besides, lipid composition is highly associated with the cellular membrane physicochemical properties. For example, the relative size of the head group and the length of FA chain have an effect on cellular membrane curvature and fission [9], which may further affect the activity and localization of the membrane proteins [10,11].
Given the variety and importance of the physiological functions of lipids, it is indispensable to analyze the entire lipidome for fully understanding the functional consequences of lipids with different structures. However, the high diversity in physicochemical properties of lipids makes it a huge challenge for qualitative and quantitative analysis of the lipidome comprehensively. The challenges are mainly reflected in two points: (1) how to expand the scope of analysis with the view to achieving full and unbiased coverage of all lipid species and (2) how to quantitate individual lipid molecules accurately.
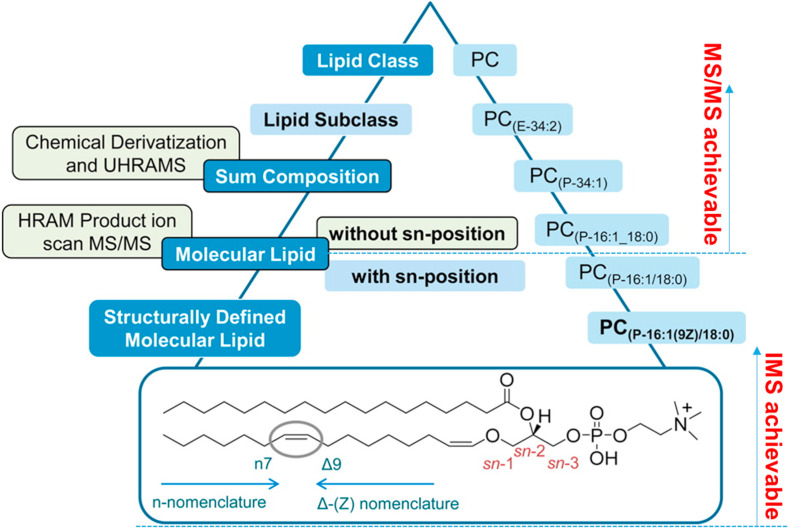
The first challenge is determined by the huge number of the biological lipids and the intrinsic limitations of the existing separation techniques. The complexity and diversity of lipids in biological matrices make it impossible to analyze all lipids in a single method. On the one hand, the existing chromatographic methods separate lipids based on either classes or fatty acyl chain length and unsaturation level, often leading to one chromatographic peak containing a certain number of isomeric lipids. Even a two-dimensional (2D) orthogonal chromatographic system is not able to separate lipids one by one. On the other hand, MS has its own limitations on elucidating the lipid structures by full scan and MS/MS fragments, it usually can only provide the structural information on the head groups, the link types of the covalent bond and the fatty acyl compositions. The finer structural information of the fatty acyl position, the unsaturated double bond position and the cis-trans geometries, requires other auxiliary facilities (e.g., ion mobility spectroscopy, IMS). Taking PC (P-16:1(9Z)/18:0) as an example, Fig. 3 displays its annotation hierarchy by MS and IMS [12]. Another intractable problem for high coverage lipidome analysis is that some bioactive lipids are often easily obscured on account of their low abundance, instability or low ionization efficiency. For example, lipids serving as intermediates of lipid metabolism and signaling molecules are usually present at a very low abundance in biological systems [13,14]. Some highly bioactive lipids are not only at low level but also unstable, and easily attacked by free radicals to form oxidized lipids [15]. The sterol lipids, having vital biological functions, cannot be detected in the regular lipid extracts due to their low ionization efficiency in MS. All of these factors impede the comprehensive analysis of lipids [16].
Fig. 3.
Annotation hierarchy of PC (P-16:1(9Z)/18:0) by high resolution mass spectrometry and ion mobility spectroscopy (HRAM: high resolution and accurate mass; UHRAMS: ultrahigh resolution and accurate mass spectrometry). Reproduced with permission from literature [12].
The second challenge is mainly attributed to that there is no explicit quantitative relationship between the ion intensity and the concentration. It is well recognized that the ion intensity of a peak is influenced by the sample preparation, the ionization efficiency, the ion transmission efficiency and the detector response [17]. Therefore, the ideal way to fulfill the accurate quantitation of lipids is to use stable isotope labelled standards corresponding to the lipids to be measured for intensity correction. However, the commercial availability of isotope labelled lipids is limited. It is also not realistic for researchers to buy a huge number of isotope labelled lipid standards. As an alternative, a novel technique called lipidome isotope labelling of yeast (LILY) has recently been proposed for a relatively accurate quantitation via growing Pichia pastoris on 13C-labelled glucose. This technique showed some potential in accurate quantification with the labelling degree of 13C-labelled yeast lipids up to 99.5% [18]. However, the complicated experimental procedures of LILY may restrict its wide usage. In practice, multiple external or internal standards from different lipid classes are often used for the (semi-)quantitation of the detected lipids. As it is hardly guaranteed the same experimental conditions for the samples as for the external standards, lipid internal standards are routinely used in most cases. Detailed information on the type, the number and the amounts of internal standards for lipid quantification was well described in a review by Wang et al. [19].
Facing above challenges, lipidomics is developing at a rapid pace because of innovations in chromatographic techniques and the advances in mass spectrometric tools. So far, three analytical strategies including untargeted, targeted and pseudotargeted lipidomics, have been applied in lipidomics. Each has its own merits and demerits. Lipid investigators can choose an appropriate one according to their research requirements. The following section will highlight the recent advances in these analytical strategies.
3. Advances in analytical strategies for mass spectrometry-based lipidomics
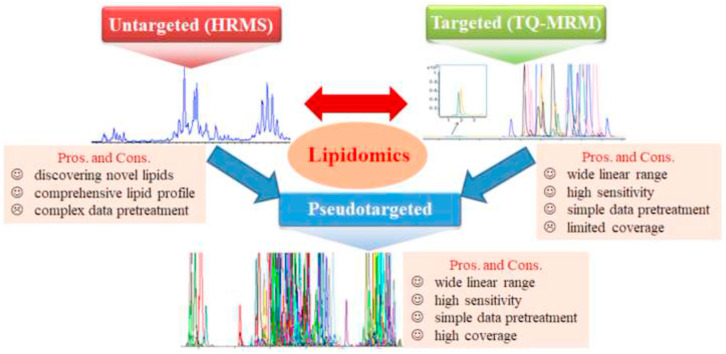
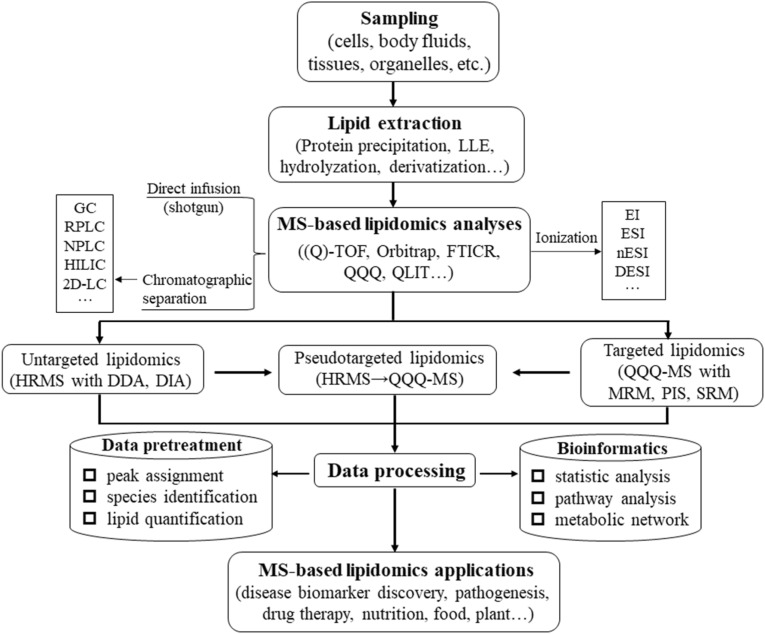
Recent advances in detection techniques including nuclear magnetic resonance (NMR) and MS have greatly expanded the depth and breadth of lipid analysis. MS is superior to NMR in virtue of its extremely high sensitivity, high resolution and molecular specificity and thereby has been a mainstream technique in current lipidomics. Fig. 4 illustrates the workflow of MS-based untargeted, targeted and pseudotargeted analytical strategies in the field of lipidomics. Due to the space limitation, techniques of NMR, gas chromatography (GC)-MS and SFC-based lipidomics will not be included in this review.
Fig. 4.
A workflow of MS-based analytical strategies for untargeted, targeted and pseudotargeted lipidomics.
3.1. Untargeted lipidomics
Untargeted lipidomics, also called global lipidomics, aims at comprehensive and unbiased analysis of the lipidome in a biological matrix. High-resolution MS (HRMS), that is particularly favorable for the elucidation of lipid structural composition due to its powerful mass resolution and high mass accuracy, is preferred for untargeted lipidomics analysis [17]. (Quadruple) time-of-flight MS (Q-TOF MS), Orbitrap MS, and Fourier Transform Ion Cyclotron Resonance MS (FTICR MS) are the outstanding representatives of HRMS.
Untargeted lipidomics is eximious for discovering novel lipids and defining differential lipids. Therefore, it is well suited for screening novel lipid biomarkers in relation to diseases. The most popular platforms in untargeted lipidomics include direct injection-HRMS (DI-HRMS) and liquid chromatography-HRMS (LC-HRMS), depending on whether separation techniques are needed prior to HRMS detection.
3.1.1. DI-HRMS-based untargeted lipidomics
DI-MS-based lipidomics is usually called shotgun lipidomics, in which lipids extracted from biological samples are directly introduced into the ion source at a constant concentration either by an LC autosampler or by a chip-based automated syringe (e.g. Advion TriVersa NanoMate) [20]. Such technique avoids the time-consuming separation step and enables high quality data acquisition of hundreds of thousands of lipids only in minutes. It has shown a high-throughput analysis ability with great potential in untargeted lipidomics [21].
Nevertheless, the ion suppression effect caused by the injection of complex lipid extracts into the ion source simultaneously triggered the signal of low abundance or low ionization level lipids to be undetectable [22]. Diluting the sample appropriately is helpful to reduce the ion suppression to some extent and also to prevent the lipid aggregation from impairing the ionization efficiency. It is recommended that serial analysis should be performed according to the lipid type in the extracts and the resuspending solvents to obtain optimal concentration with the maximum peak number [23,24]. Utilizing the differential charge properties of various lipid categories under specific experimental conditions could achieve selective ionization of specific lipids. Therefore, adding acidic, basic modifiers or chemical derivatization reagents to the multiple extracts of lipids for positive and negative ionization modes, respectively, helps to identify more lipids, such as anionic, weakly anionic, and electrically neutral lipids [16,25]. In addition, “spectral stitching” technique in which data are collected as a serial continuously overlapping narrow windows instead of a wide m/z window to cover the entire mass range, can effectively improve the detection sensitivity. Southam et al. described a complete spectral-stitching nano-electrospray ionization (nESI) DI-HRMS workflow for untargeted metabolomics and lipidomics analysis. A fivefold increase in the number of the detected peaks obtained by this spectral-stitching nESI DIMS method as compared with that by standard wide-scan DIMS made it efficient to detect low-abundance lipids, thereby achieving extended coverage of lipids [23].
Lacking of effective identification of isomers and isobars was once considered as a limitation for DI-MS techniques. However, HRMS-based MS/MS data acquisition exploiting specific fragmentation patterns of different lipid species could achieve in-depth structural identification of isomers and isobars with high reliability and credibility [16]. Additionally, combining ion mobility MS (IM-MS) with DI-HRMS provided an additional separation dimension for untargeted lipidomics, thereby simplifying the complexity of lipid extracts and facilitating more explicit isomer and isobar structural elucidation [[26], [27], [28]].
3.1.2. LC-HRMS-based untargeted lipidomics
LC-HRMS, benefiting accurate identification of individual lipid molecular species, has been extensively used in untargeted lipidomics. Owing to LC separation prior to MS detection, LC-HRMS can alleviate the ion suppression and provide explicit identification of isomeric or isobaric lipids to a great extent. Normal-phase LC (NPLC), hydrophilic interaction liquid chromatography (HILIC) and reversed-phase LC (RPLC) are the three most frequently used chromatographic techniques in untargeted lipidomics. RPLC is the most applicable LC due to its high separation efficiency, high reproducibility and high robustness. The elution order of lipids on RPLC is related to carbon-chain length and unsaturation degree. NPLC and HILIC both utilize polar stationary phases and are well suitable for the separation of the lipid classes with different headgroups. However, NPLC requires nonaqueous eluting solvents (e.g., n-hexane and ethyl acetate) and is limited in separating polar and hydrophilic compounds, while HILIC uses the mobile phase consisting of high polar organic solvents with small portion of water and allows highly efficient separation of polar compounds which usually have poor retention on the RPLC system.
Recent efforts in LC-HRMS-based untargeted lipidomic analytical strategies are mainly in two aspects: 1) shortening the analysis time as much as possible to achieve a high-throughput analysis of lipids, and 2) improving the peak capacity and resolution to achieve a high coverage of lipids. A faster flow rate [29,30], a higher column tolerant temperature [31] and a bi- or ternary gradient with high solvent strength [31,32] have positive effects on shortening the analysis time and thus improving the throughput. Lorenzen et al. developed a fast RPLC-HRMS untargeted lipidomics method and detected 166 lipids in Myxocoxxus xanthus within 14 min [30]. Xuan et al. shortened a 20 min binary gradient for lipidomics profiling to 13 min and detected nearly 500 serum lipids covering 20 lipid (sub)classes. This UHPLC-HRMS-based untargeted lipidomic method enhanced the analysis throughput on the premise of ensuring the separation performance [31]. A rapid untargeted lipidomic method based on Q Exactive HRMS developed by Lagerborg et al. realized the relative quantitation of 350 identified lipids in plasma only in 8.5 min [32].
In addition, multi-dimensional LC (MDLC) with orthogonal separation properties is the optimal choice for the increase of peak capacity, in which the complex lipids can be separated by different mechanisms. The separation of TAG lipid species in biological matrices is rather difficult due to their complex structural compositions and very similar physicochemical properties. In a pioneering study, Dugo et al. developed a silver-ion reversed-phase comprehensive two-dimensional liquid chromatography - mass spectrometric method for the triacylglycerol analysis in a rice oil sample [33]. Our group also developed several MDLC separation techniques for untargeted lipidomics. Initially, a comprehensive Ag-LC × RPLC 2D method was used for the analysis of TAGs, in which the first and second dimensions were separated according to the unsaturation and the fatty acyl chain length, respectively. However, the differences in the 2D flow velocity caused a loss of sensitivity [34]. Subsequently, a stop-flow HILIC × RPLC 2D system was developed to reduce the dilution effect [35]. Further, a parallel-column switching 2D system coupled with pre-fractionation was proposed to further extend the separation window and increase the coverage. It enabled to achieve a simultaneous untargeted analysis of metabolome and lipidome within a single run [36]. Other recent advances in untargeted lipidomics analysis based on MDLC techniques can be referred to a newly published review [37].
The integration of IM-MS into conventional LC-MS workflow, can provide a third dimensional separation satisfactorily orthogonal to LC-MS, and improve peak capacity by separating isomeric and isobaric lipids [[38], [39], [40]]. The innovative IM-MS design [41], optimized instrument parameters [42] and gas-phase ion reaction (e.g. ozonolysis) [43] help to further improve lipid separation and increase the specificity of lipid identification in complex samples. Baglai et al. evaluated LC-IMS-MS and LC LC-MS-based untargeted lipidomic performances in terms of the number of molecular features, analysis time and structural characterization. From this study, one can see that although a higher number of identified lipids were obtained in LC LC-MS through improving the separation of co-eluting lipids, LC-IMS-MS is overperformed in the faster analysis and accurate lipid identification [44]. It is recommended to complement the advantages of both techniques for untargeted lipidomics of large-scale complex samples, i.e., conducting comprehensive lipids profiling on the LC LC-MS platform followed by IM-MS for detailed structural characterization [44].
3.1.3. MS/MS scan modes in untargeted lipidomics
Data-dependent acquisition (DDA) and data-independent acquisition (DIA) modes are the two major MS/MS scan modes in untargeted lipidomics. As a classic MS data acquisition technique for untargeted analysis, DDA only obtains fragment information of specific precursor ions above ion intensity threshold. Although the narrow isolation window for precursor ion selection contributed to a high-quality MS2 spectrum [45], the low MS/MS information coverage was not conductive to structural annotation. Many efforts have been devoted to providing more MS2 information. Targeted-directed DDA with pre-selected precursor ions filtered by 80% rule and ion fusion have been proposed to trigger MS2 targeted fragmentation [46,47]. Iterative exclusion (IE) strategy, where precursor ions selected previously would be iteratively excluded during each MS2 analysis [48], has shown an increase in lipid coverage of over 50% [49] and great benefits for detecting trace lipids [49,50]. Various ionization mode and activation energy would be another choice for increasing the coverage of MS2 data in untargeted lipidomics. The integration of ionization mode, such as collision-induced dissociation (CID) and high-energy collisions dissociation (HCD), provided more than 60% of metabolite complementary information. More possibilities to obtain abundant precursor ion fragments could be further achieved through the comprehensive use of low, medium and high activation energy [51].
As an alternative MS/MS acquisition, DIA is enabled to provide a comprehensive secondary spectrum, thereby improving the coverage of precursor ions. All-ion fragmentation (AIF) scanning represented by MSE [52] or AIF [53] and segmentation scanning represented by sequential windowed acquisition of all theoretical fragment ion (SWATH) are two different acquisition modes using DIA strategies [54]. Since comprehensive MS/MS data covering the entire mass range is generated through continuous isolation windows instead of a single wide window, SWATH overperforms all-ion fragmentation method in terms of spectral quality and annotation coverage [45,55]. Therefore, SWATH-based DIA technique has gained widespread popularity in untargeted lipidomics [[56], [57], [58], [59]]. However, SWATH encounters a big challenge, i.e., the complexity of MS2 spectra destructs the direct association between precursor ions and fragments [60]. Deconvolution of MS2 spectrum is needed to reconstruct the correlation between precursor ions and product ions. At present, deconvolution-based pseudo MS2 spectra reconstruction strategy represented by MS-DIAL [61] as well as DecoMetDIA [62] and MS2 spectra library-based targeted extraction strategy represented by MetDIA [63] as well as MetaboDIA [64] have been utilized to reconstruct precursor ions and fragments connection. We firmly believe that as data complexity alleviates, DIA technique will surely have wider applications in untargeted lipidome analysis.
Notably, accurate identification and quantification of lipids are the key to untargeted lipidomics. Efforts should be made to avoid/eliminate some artefacts potentially included in the analyses, such as lipid aggregation, ion suppression, and in-source fragmentation. In high-concentration and highly polar lipid solution, especially in the presence of salt, lipids tend to form aggregations, which cannot be well ionized and detected. High-concentration lipid solutions can also trigger space and charge competition during the ionization process, which significantly reduces the ionization efficiency. Diluting the sample appropriately or reducing the injection volume is helpful to reduce the ion suppression to a certain extent. With the increasing ionization temperature and voltage, the fragmentation reaction in the ion source possibly occur, for example, some phosphatidylserine species become phosphatidic acid species. Fine-tuning the instrument using lipid standard mixture to define appropriate ionization conditions can help eliminate in-source fragmentation. Readers can refer to recent reviews to have more related information [[65], [66], [67]].
3.2. Targeted lipidomics
Targeted lipidomics focuses on the accurate quantification of specific lipid species usually involved in one or several lipid pathways. Lipids to be measured in targeted lipidomics are often those prescreened or predefined to be key targeted molecules and/or potential biomarkers via an untargeted lipidomics analysis. MS-based multiple reaction monitoring (MRM) and parallel-reaction monitoring (PRM) acquisition modes are the most widely used techniques in both shotgun and LC-MS-based targeted lipidomics.
3.2.1. MRM MS-based targeted lipidomics
Triple quadrupole (QQQ) MS or quadrupole linear-ion trap (QLIT) MS with MRM acquisition are the two predominant techniques applied in targeted lipidomics owing to their superiorities in wide linear range, high sensitivity and great stability [68], in which Q1 is used to acquire MS data of predefined precursor ions based on known or assumed information and Q3 monitors the characteristic fragments generated by CID in Q2 [4,17]. In MRM-MS strategies, selective monitoring of precursor ions and corresponding product ions that are closely related to the structure would aid in isomers discrimination. For example, PC (16:0_20:4) and PC (18:2_18:2) were successfully distinguished based on fatty acyl-derived transition s [69]. In addition, chemically specific transitions were also effective in discriminating co-eluting lipids [70]. Moreover, the quantitation of low-abundance analytes benefits from the high sensitivity of MRM. In a 5 min UPLC-MS/MS targeted lipidomics study, over 120 eicosanoids have been identified in 20 μL plasma sample, showing a linear range of multiple orders of magnitude [71].
In particular, QQQ-MRM-based MDMS shotgun lipidomics (MDMS SL) platform equipped with nESI is very powerful in targeted analysis of the majority of lipid classes in biological samples owing to its high sensitivity, resolution and efficiency as well as its wide coverage. It enables lipid investigators to make good use of the special advantages inherited in MS for lipid analysis and maximally exploit the unique physicochemical properties of lipid species to obtain the maximal separation and ionization, and the minimal ion suppression [72].
However, the inherent disadvantages of MRM restrict its success in some specific fields due to the unit resolution of the quadrupole, resulting in false positive discoveries and incorrect quantitation [73]. The measurement of at least two MRM transitions for each analyte improved the credibility of analyte confirmation in targeted lipidomics [74]. Restricted by the dwell time, there is an upper limit on the number of detectable ion pairs in each MRM data acquisition. The current technological advancements are capable of hundreds of MRM transitions [75]. A further increase in the number of transitions is still expected, which means large-scale and high-throughput quantitation of lipids in a single experiment, thereby placing higher demands on MRM technology. Ultra-fast scanning QQQ MS, such as Shimadzu LCMS-8060 and Waters Xevo TQ-S micro, makes it possible to conduct wide MRM transitions with minimum dwell time (1 ms) and maximum polar switching speed (5–15 ms) [69]. Besides, more efficient MRM acquisition method has been developed, namely, dynamic MRM (dMRM) or time-scheduled MRM, which is used to monitor specific transitions only within the preset retention time (t R) windows. This technique, on the one hand, avoids unnecessary scans realizing wider MRM transition coverage in a single run; on the other hand, reduces the number of simultaneous ion pairs achieving higher sensitivity and accuracy [[76], [77], [78]]. A targeted lipidomics approach has demonstrated that dMRM allowed the quantification of 483 ceramides with a tenfold signal response higher than MRM [79]. In another example of targeted lipidomics, large-scale quantification of 413 lipid species belonging to 22 lipid classes has been conducted under optimized collision energy and transition settings [69].
3.2.2. PRM MS-based targeted lipidomics
Although QQQ-based MRM has been recognized as a gold standard for lipid quantification, PRM implemented on HRMS represents an alternative technique. PRM (also referred to as HR MRM or MRMHR) can achieve selection of interested precursor ions in the quadrupole and detection of characteristic fragments generated in the HRMS including Orbitrap and TOF, which has been widely used in the field of proteomics for large-scale quantification of peptides [[80], [81], [82], [83]]. Recently, several PRM-based targeted lipidomics platforms have been applied for lipid discovery and quantification in human serum [84], zebrafish [85], barley root [86], yeast [87] and enterococcus faecalis [88]. For instance, Yu et al. developed a HR-HPLC-QqTOF-based targeted lipidomics platform using PRM and realized the analysis of more than 600 lipids including FAs, STs and long chain base in the barley root [86]. HRMS-based PRM method has also been used to detect and quantify cardiolipins with low ionization efficiency, realizing an increase in lipidomics coverage [88].
Thanks to the HRMS, the acquisition of high-quality precision m/z enabled more accurate identification of precursor ions and fragments to eliminate false positives, which was impossible for MRM performed on the QQQ MS with unit mass resolution. Besides, the entire MS2 information corresponding to specific precursor ions could be collected without preselection of ion pairs, instead of providing precursor ions-product ions list under optimized collision energies before conducting MRM experiments, reflecting the flexibility of PRM [86]. Nevertheless, the disadvantage of PRM is relatively low scan rate which might be time-consuming for large-scale MS2 experiments when compared with MRM. To overcome this deficiency, it is needed to upgrade the scan speed of HRMS. Fortunately, current QTOF are capable of up to 100 PRM experiments per cycle achieving large-scale precursor and product ions monitoring [86]. Besides, each lipid was monitored only within the predetermined t R window, which allows richer MS2 information in a single experiment [89]. However, this was based on the premise of maintaining stable reproducibility of the chromatographic system [81]. Adding internal standards to the samples to establish a reference elution time and compensate for time drift contributed to achieving real-time collection of t R of the predetermined lipids [81,82]. For extremely complex sample systems, “multiplex mode” could be enabled to scan co-eluting precursor ions within the same t R window. Up to ten precursor ions were sequentially isolated and fragmented, and all product ions were simultaneously transferred into the Orbitrap to generate a complex MS2 spectrum [81]. However, deconvolution was required to establish a correlation between precursor and product ions in the later date processing.
Taking the respective advantages of these two techniques into consideration, some researchers have proposed that it would be wise to complement the superiorities of high-quality precision in PRM and high scan speed in MRM. Once the MS2 spectra have been identified by PRM, we can transfer the ion pairs list to MRM at fast scanning speed to achieve large-scale quantitative determination [89].
3.3. Pseudotargeted lipidomics
Pseudotargeted lipidomics integrates the untargeted and targeted lipidomics strategies, i.e., using the targeted techniques based on the information from untargeted methods to achieve a high coverage lipidomics data acquisition. The complete pseudotargeted lipidomics analytical procedures include: 1) HRMS-based untargeted lipidomics techniques are used to analyze mixed biological matrices of interest in full scan or IDA-based auto MS/MS scan to collect rich MS/MS fragment ions; 2) lipid ion pairs are defined based on the characteristic fragment ions and the corresponding parent ions generated from untargeted lipidomics analyses; and 3) QQQ-MS is used to realize high-coverage dynamic MRM analysis of the constructed lipid ion pairs. Pseudotargeted lipidomics possesses the advantages of both untargeted and targeted lipidomics, showing a high sensitivity, good reliability, wide linear range, higher coverage, and easy data treatment.
The conception of pseudotargeted metabolomics was first proposed by our group in 2012 [90], a GC-MS-based untargeted metabolomics approach was established by combining an optimized algorithm to select ions from all of the detected analytes. This method enables to measure the abundance of both known and unknown metabolites in biological samples using the t R locking GC-MS-selected ion monitoring (RTL-GC/MS-SIM). In 2013, this strategy was further extended to LC-MS via obtaining ion pairs of known and unknown metabolites by LC-HRMS and measuring their abundances by targeted LC-MRM MS [91]. In the same year, a widely targeted metabolomics strategy was also proposed by Chen et al. in which the authors used a strategy named step-wise multiple ion monitoring-enhanced product ions (stepwise MIM-EPI) to construct a MS/MS spectral tag library comprising 698 non-redundant metabolite signals from rice leaf tissues [92]. To raise the efficiency of selecting the ion pairs for the subsequent MRM acquisition, an in-house software called MRM-ion pair picking software (i.e., MRM-Ion Pair Finder) was developed, and can be used to execute precursor ion alignment, MS/MS spectrum extraction and reduction, characteristic product ion selection and ion fusion automatically [93], which greatly reduces the time to establish a pseudotargeted method. Meanwhile, a novel correction strategy for large-scale pseudotargeted metabolomics studies was developed for GC-MS, which enabled to integrate metabolomics data from multiple batches and different instruments by calibrating gross and systemic errors, to make the metabolomics data from different batches and different instruments practicable [94,95]. Furthermore, a novel pseudotargeted metabolomics approach was developed based on SWATH MS acquisition to increase the coverage of the ion pairs to a maximum extent [96].
All these pseudotargeted analytical strategies for metabolomics are also suitable for lipidomics. In a recent study, we developed a pseudotargeted lipidomics approach and fulfilled a high coverage lipidomic analysis. Untargeted UHPLC-HRMS lipidomics analyses of mixed matrices were firstly performed to obtain as much information on molecular structures and t R of lipids as possible. Secondly, the quantitative structure and the chromatographic retention relationship was constructed for each lipid (sub)class, and used to i) confirm the entity of the detected lipids; ii) recognize the identity of the lipids that showed no MSMS fragments but could be obtained by EIC; and iii) extend the structures of the lipids that were not detected by the untargeted method but could theoretically exist in the biological matrix. A total of 3377 targeted lipid ion pairs containing over 7000 lipid molecular structures were defined for the MRM data acquisition [97].
MDLC-MS is an effective way to extend the separation window of HPLC. A pseudotargeted method based on parallel column 2D LC-MS has recently been established for broad coverage of metabolome and lipidome [98]. In total, 1294 and 687 ion pairs could be detected in positive and negative ion modes, respectively.
The importance of pseudotargeted metabolomics/lipidomics has been more and more recognized by researchers for it can provide high quality data and high coverage of analytes. To make it be applicable in more laboratories, taking the standard reference material (SRM) 1950 plasma as an example our group has recently released an entire protocol for LC-MS-based pseudotargeted metabolomics method in which some related software and tools are provided [99]. It is hoped that this protocol can provide reference for readers interested in pseudotargeted metabolomics/lipidomics.
Pseudotargeted metabolomics approach has also some limitations. For instance, i) some metabolites detected in the pseudotargeted method are not identified although the MRM transitions were known, their identification is to be performed by UHPLC-HRMS/MS; and ii) the number of metabolites that can be detected in a pseudotargeted metabolomics approach is limited by the TQMS scan rate; and iii) two instruments and special data treatment are needed to define the ion pairs and perform subsequent MRM data acquisition.
4. Applications of MS-based lipidomics in diseases
The essential biological functions of lipids and the advances in MS-based lipidomics have greatly impelled the research progress of many fields such as disease, nutrition, food and pharmaceuticals. In this section, we will summarize some applications of MS-based lipidomics published in recent 5 years in disease biomarker discovery as well as pathogenic mechanism and therapeutic strategy.
4.1. Disease biomarker discovery
Lipid metabolism in biological systems is strictly regulated, and therefore, the lipid metabolic process has been disturbed or disordered to varying degrees before and during diseases onset. Therefore, lipidomics can provide evidence linking malignant tumors with specific lipid abnormalities. In several recent studies, lipidomic profiles of patients’ matched tumor tissues and adjacent non-tumor tissues were compared directly for the purpose of identifying differential lipid markers that were not affected by individual differences. Eight GPs containing palmitic acyl (C16:0) successfully distinguished hepatocellular carcinoma (HCC) tissues from nontumor tissues [100]. Elevated pre-diagnostic level of PC (30:0) as well as reduced LPC (18:0) was defined to be potential biomarkers for breast, prostate and colorectal cancer [101]. Several sphingomyelins (SMs) and phophatidylinositols (PIs) expressed significantly different levels in malignant and paired nontumor tissues of non-small cell lung cancer (NSCLC) patients [102]. A prospective lipidomics analysis of prostate cancer showed the negative correlation between aggressive cancer progression and several GPLs, cholines and FAs [103]. Our group observed that aberrant accumulation of CEs in patients with prostate cancer based on a lipidomics study of the paired prostate cancer tumor tissues (PCT) and adjacent nontumor tissues (ANT). Independent external verifications further demonstrated that CEs were the most predictive lipids in discriminating between PCT and ANT or prostate hyperplasia (BPH) [104].
Type 2 diabetes (T2D) and cardiovascular disease (CVD) are two most prevalent metabolic diseases globally. A handful of recent studies have indicated that abnormality in lipid metabolism is a risk factor for these diseases. For instance, Cer (d18:0) lipid class was identified and verified as an indicator of metabolic deregulation in the progressing of T2D in view of its impact on impaired glucose tolerance and altered insulin secretion in a population-based prospective cohort study [105]. Similarly, Cer (d18:1) lipid class was quantified as potential risk-related molecules for CVD [106]. Given the diverse and complex factors leading to the disease progressing, a combined biomarker formed by the synergic changes of several lipid species may improve the accuracy of prediction [107]. Lu et al. identified serum lipid predictors for T2D via detecting changes in lipid metabolism before T2D onset using a HPLC-MRM-based high-coverage targeted lipidomics method. Six serum lipids including LPI (16:1), PC (34:3), PE (18:0p/20:4), TAG 50:2(16:2), TAG 51:0(17:0) and TAG 54:7(22:6) were regarded as biomarkers to improve prediction accuracy of diabetes based on a large scale of discovery and validation cohorts from six regions of China [108]. It needs to point out that although the combination of markers will theoretically show better predictive performance than a single marker, from the perspective of economic cost and efficiency, the number of markers should be minimized while ensuring high predictability [109].
Alzheimer’s disease (AD) is a devastating neurodegenerative disease that affects hundreds of millions of people throughout the world. The difficulty in early diagnosis and the lack of effective treatment result in the urgency to determinate AD biomarkers. Cell biological and genetic studies have proved that lipids are extensively involved in the pathogenesis of AD [110,111]. In a UPLC-HRMS-based untargeted lipidomics study, Proitsi et al. identified a panel of 10 plasma lipids as a “fingerprint” to distinguish AD patients from controls, of which 6 were identified as long chain cholesteryl esters (CEs), with approximately 80% accuracy [112]. Another targeted study on brain and blood metabolite signatures of pathology and progression in AD, showed that higher level of sphingolipids (SLs) was positively correlated with AD, indicating SLs could be biologically relevant biomarkers for the early diagnosis of AD [113].
In addition, many lipids were considered to be risk factors for disease aggravation. For instance, the higher concentration of MAG (16:0) combining with the lower concentration of DAG (32:0) and DAG (36:0) would drive kidney disease to end-stage [114]. Prognostic model consisting of Cer (d18:1/24:1), SM (d18:2/16:0) and PC (16:0/16:0) was related to shorter survival of patients with castration-resistant prostate cancer (CRPC) [115]. The combination of Cers with PCs has demonstrated good CVD mortality prediction effect in multiple large-scale cohorts, and the synergistic performance with high-sensitivity troponin-T could be more effective in risk classification [116].
It is well known that lipid metabolism is a systematic interaction process of gene, protein, metabolite, lipid and enzyme. Therefore, integration of multi-omics technology is indispensable for better understanding the body’s systemic response during treatment and better prognosis. In a multi-omics study based on metabolomics, lipidomics and proteomics, Eisfeld et al. analyzed plasma samples from patients with Ebola virus disease (EVD) over the process from diagnosis to recovery or fatality. They reported that 11 biomarkers including 4 lipids, 2 metabolites, 2 cytokines, 2 proteins and 1 clinical index exhibited outstanding predictive ability of survival at the early stage of diagnosis, thereby helping to improve the prognostic treatment of those high-risk patients [117].
Table 1 summarizes recent representative studies of MS-based lipidomics on biomarker discovery for disease prediction, diagnosis and prognosis.
Table 1.
Representative applications of disease biomarkers for prediction, diagnosis and prognosis.
| (1) Prediction | |||||||
|---|---|---|---|---|---|---|---|
| Category | Disease | Sample | Analytical strategy | Analytical platform | Up-regulated lipids | Down-regulated lipids | Ref. |
| Metabolic syndrome | Type 2 diabetes (T2D) | Plasma | Targeted | UHPLC-MS/MS, shotgun | Cer (d18:0) | – | [105] |
| Serum | Targeted | HPLC-MS/MS | LPI 16:1, PC 34:3, TAG 50:2(16:2), TAG 51:0(17:0), TAG 54:7(22:6) |
PE (18:0p/20:4) | [108] | ||
| Serum | Targeted | GC-MS/MS | – | linoleic acid | [128] | ||
| Cardiovascular disease (CVD) | Plasma | Targeted | UHPLC-MS/MS | Cer(d18:1) | – | [106] | |
| Cancer | Prostate | Serum | Untargeted | UHPLC-MS/MS, GC-MS | – | GPs, choline, FFAs | [103] |
| Colorectal | Serum | Targeted | HPLC-MS/MS | PC (30:0) | LPC (18:0) | [101] | |
| Breast |
Serum |
Targeted |
HPLC-MS/MS |
PC (30:0) |
LPC (18:0) |
[101] |
|
| (2) Diagnosis and classification | |||||||
| Metabolic syndrome | T2D | Serum | Pseudo-targeted | UHPLC-MS/MS | TGs, DGs, PEs, LPEs | HexCers, PE-Os, PC-Os, LPC-Os | [97] |
| Nonalcoholic fatty liver disease (NAFLD) | Tissue, plasma, urine | Targeted | LC-MS/MS, GC-MS | SPs | GPs | [129] | |
| Neurological disorder | Alzheimer’s disease (AD) | Plasma | Untargeted | UHPLC-MS/MS | – | CEs | [112] |
| Tissue, serum | Targeted | HPLC-MS/MS | SMs | – | [113] | ||
| Cancer | Breast | Tissue | Untargeted | UHPLC-MS | PCs, PEs, PIs, SM, Cer | TAGs | [130] |
| Kidney | Plasma | Untargeted | UHPLC-MS/MS | Saturated C12–C16 FFAs, TAGs | – | [[131], [132], [133]] | |
| Hepatocellular carcinoma (HCC) | Tissue, cell | Untargeted | HPLC-MS/MS | – | C16:0-containing GPs | [100] | |
| Prostate | Tissue | Untargeted | UHPLC-MS/MS | CEs | – | [104] | |
| Non-small cell lung cancer (NSCLC) | Tissue | Targeted | Shotgun | PI (38:2), PI (38:3), PI (40:3) |
SM (40:1), SM (42:1), SM (36:1) | [102] | |
| Viral disease |
Lassa |
Serum |
Untargeted |
HPLC-MS |
– |
PAFs |
[134] |
| (3) Prognosis evaluation | |||||||
| Metabolic syndrome | CVD | Plasma, serum | Targeted | UHPLC-MS/MS | Cer, PCs | – | [116] |
| Neurological disorder | Stroke | Plasma | Untargeted | LC-MS | – | LPC (20:4) | [135] |
| Cancer | Breast | Tissue | Untargeted | UHPLC-MS/MS | PC(30:0), PC (32:0), PC (32:1), PC (32:2) | – | [130] |
| Kidney | Serum | Untargeted | Shotgun | MAG (16:0) | DAG (32:0), DAG (36:0) | [114] | |
| Prostate | Plasma | Untargeted | LC-MS/MS | Cer(d18:1/24:1), SM(d18:2/16:0), PC(16:0/16:0) | – | [115] | |
| Ovarian | Plasma | Untargeted | UHPLC-MS | – | LPG(20:5) | [136] | |
| Viral disease | Ebola virus disease (EVD) | Plasma | Untargeted | UHPLC-MS/MS | PG(18:1/22:6) | PI(18:0/20:4), Cer(d18:0/24:0), LPC(20:0) | [117] |
4.2. Pathogenic mechanism and therapeutic strategy
As lipids play vital roles in many physio-pathological activities, lipid metabolic disorders are closely connected with pathogenic processes [118]. Consequently, lipidomics can be used to reveal the underlying mechanisms of physio-pathological processes. Integrated multi-omics technique also helps to seek novel potential lipid-related targets for treatments by combining inhibitors of these targets [119].
In a recent study, Thangapandi et al. investigated the pathogenic mechanism of liver fibrosis based on a combined genetic and lipidomic analysis of human liver biopsies across the spectrum of NAFLD and a mouse model with hepatocyte-specific membrane-bound O-acyltransferase domain containing 7 (Mboat7) deletion. The results demonstrated that lipid mediators such as PI and lysoPI may serve as pathogenetic drivers of fibrosis, and indicated that targeting lysoPI and PI signaling may be effective for treatment of NAFLD and liver fibrosis [120]. Compared with non-tumor tissues, the expression of fatty acid synthase (FASN) associated with FA production was upregulated in liver tumors. Silencing of FASN severely impaired AKT-driven HCC progression [121]. Recent studies have shown that ablation of FASN significantly delayed HCC instead of completely inhibiting tumor proliferation. Hepatocytes can take up exogenous FA and promote the absorption through lipoprotein lipase (LPL) to compensate for the blocking of endogenous FA synthesis. A research demonstrated that overexpression of LPL induced poor prognosis of HCC patients. Silencing of LPL enhanced HCC cell growth inhibition and accelerated cell apoptosis in vitro experiments [122]. Modulating cholesterol synthesis was another mechanism. Che et al. reported an increase in CEs, upregulation of the corresponding genes involved in its synthesis and overexpression of the synthetic rate-limiting enzyme HMGCR in FASN knockout mice [123].
Currently, the coronavirus disease 2019 (COVID-19) pandemic, a severe inflammatory disease with no specific drugs or vaccines, is affecting more than 18 million of infected patients and causing widespread mortality. It is of great urgency to find out the pathologic mechanisms behind its great damage to human body. It is well recognized that the eicosanoid class of signaling molecules are implicated in many devastating inflammatory diseases, and therefore may also play functional roles in COVID-19. Schwarz et al. reported that a loss of specific immune regulatory eicosanoids and docosanoids lipid mediators and increased pro-inflammatory species such as AA-derived products of ALOX5 and cytochrome P450 (CYP) contribute the progression from moderate to severe disease based on a combination of LC-MSMS targeted lipidomics, cytokine and chemokine quantitative analysis and single cell RNA sequencing analysis [124]. This study provided not only mechanistic insights into the immune regulation in COVID-19 but also a potentially targetable therapy option for COVID-19.
Bietti’s crystalline dystrophy (BCD) is an autosomal recessive disease characterized by progressive loss of vision due to chorioretinal degeneration. It is caused by mutations in CYP4V2 gene and no effective therapy is available. Inspired by the fact that CYP4V2 gene belongs to a member of the CYP which involved in lipid metabolism, Isobe et al. explored the mechanisms underlying the onset and progression of BCD via LC-MS/MS-based lipidomics, morphological and functional analysis of BCD patient-specific induced pluripotent stem cells with retinal pigment epithelium (iPSC-RPE cells) together with analysis of related enzyme activities. The accumulation of glucosylceramide (GlcCer) and free cholesterol was found to contribute to the RFE damage in BCD. The outcome of the therapeutic experiment on phenotypes in BCD iPSC-RPF cells showed that compounds that enable to reduce the accumulation of free cholesterol could be therapeutic agents for BCD [125].
Colon cancer is one of the leading tumors worldwide with a high mortality rate. Many efforts have been made to explore the pathogenesis and seek the specific lipid regulators for therapeutic intervention of colon cancer. Elevated CYP monooxygenase-generated epoxyoctadecenoic acid (EpOME) was observed both in the plasma and colon tissues of mice model by Wang et al. Treatment with EpOME not only induced inflammation in colon cancer cells, but also exacerbated tumor growth in vivo. Furthermore, ablation of corresponding regulatory genes and treatment of enzyme inhibitors effectively suppressed colon tumorigenesis, which demonstrated the effectiveness of CYP monooxygenase eicosanoid pathway as a therapeutic strategy for colon cancer tumors [126]. The CYP oxidized eicosanoid-associated signal network also modulated the development of metastatic triple negative breast cancer (TNBC). Epoxyeicosatrienoic acid (EET) showed a significantly high level in breast tumors, and its promotion effect on the metastasis of TNBC cells was verified in vitro, which was consistent with the overexpression of upstream CYP epoxygenase. Therefore, modulating the signal network of CYP epoxygenase-EET may provide an effective intervention strategy for TNBC patients.
To sum up, we have reasons to believe that MS-based lipidomics combined with other omics techniques has a bright prospect in studying the pathogenic mechanism and potentially targetable therapy.
5. Conclusion remarks and perspectives
Advances in MS and chromatographic separation facilities have greatly driven the development of lipidomics as a part of metabolomics. Current MS-based analytical techniques allow fast, reliable and sensitive determination of hundreds to thousands of lipids over a broad dynamic range. However, the extreme complexity in lipid structures is an insuperable barrier for the achievement of high-throughput and high-coverage analysis of the complete lipidome. A realistic solution is to choose an appropriate analytical strategy according to the purpose of the study. Untargeted, targeted and pseudotargeted approaches enable effective measurements of the lipidome from various aspects. Untargeted lipidomics is excellent in detection and discovery of novel lipids, and therefore is well suited for the discovery of differential lipids and screening of lipid biomarker candidates. Targeted lipidomics, favorable for accurate quantification of specific or predefined lipid species, is powerful for authenticating the effectiveness of the defined lipid biomarkers and studying lipid molecular mechanisms. Pseudotargeted lipidomics, merging advantages of untargeted and targeted lipidomics, enables to monitor hundreds to thousands of lipids by dynamic MRM. It is appropriate for high-coverage and semi-/accurate-quantitative analysis, therefore, it can be used in the discovery and confirmation stages.
Nevertheless, great efforts are still needed to cope with the complexity of lipids so that the full characterization of the lipidome is approachable. Firstly, innovative MS techniques for comprehensive lipid structural elucidation (e.g., determination of positional isomers of PLs, position of double bonds, position of acyl chain branching and stereo-structure) are required in routine lipidomic analysis. Secondly, HRMS with faster data acquisition speed hyphenated to highly efficient chromatographic separation tools (especially 2D-/3D-LC-based facilities and nano-LC devices) will favor high-throughput, accurate qualitative and quantitative analysis of lipids. Thirdly, the combination of HRMS with other analytical facilities (e.g., IMS) that enables to provide a supplementary separation dimension, will facilitate the discrimination between lipid isomers, thereby greatly improving the lipid coverage. Last but not least, the interlaboratory studies on evaluation of the detected lipids in commercially available SRM 1950 plasma using nonstandardized laboratory-independent workflows, helpful for finding out the metrological questions and/or gaps in current lipidomic measurement, will provide valuable insights into the issues of quantitative accuracy and repeatability of lipidomics analysis [127].
Current state-of-the-art technologies provide wide application prospects for lipidomics. The great potentials of lipidomics in disease biomarker discovery, pathogenesis and therapeutic strategy investigations have been shown over the past years. However, integration of multi-omics analytical techniques is highly essential for better understanding the relationships between individual lipids and lipid classes within a metabolic network and uncovering the role of specific lipid molecules in cellular functions as metabolism process in human body is intertwined with the interactions between genes, proteins, metabolites, lipids, and enzyme. Multi-omics analytical strategies can maximize the power of lipidomics in finding novel lipid biomarkers, understanding disease pathology and monitoring efficacy of drug therapy. To our delight, multi-omics joint research has drawn increasing attention. However, novel tools that can efficiently integrate the lipidomics data with other omics data are still limited. Efforts from statisticians with rich biological background are needed to overcome this issue.
CRediT authorship contribution statement
Tianrun Xu: Writing - original draft, modification. Chunxiu Hu: Conceptualization, Writing - original draft, modification. Qiuhui Xuan: Writing - original draft, Writing - review & editing, Writing, discussion. Guowang Xu: Conceptualization, Funding acquisition, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully acknowledge the financial support from the National Natural Science Foundation of China (No. 21874130, No. 21876169), the Innovation Program (DICP&QIBEBT UN201806) from DICP, CAS and the National Key Research and Development Program of China (2016YFC1303101).
References
- 1.Shevchenko A., Simons K. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010;11(8):593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 2.Han X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016;12(11):668–679. doi: 10.1038/nrendo.2016.98. [DOI] [PubMed] [Google Scholar]
- 3.Fahy E., Subramaniam S., Brown H.A., Glass C.K., Merrill A.H., Murphy R.C., Raetz C.R.H., Russell D.W., Seyama Y., Shaw W., Shimizu T., Spener F., van Meer G., VanNieuwenhze M.S., White S.H., Witztum J.L., Dennis E.A. A comprehensive classification system for lipids. J. Lipid Res. 2005;46(5):839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Murphy R.C., Axelsen P.H. Mass spectrometric analysis OF long-chain lipids. Mass Spectrom. Rev. 2011;30(4):579–599. doi: 10.1002/mas.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M., Wang C., Han R.H., Han X. Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 2016;61:83–108. doi: 10.1016/j.plipres.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harayama T., Riezman H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018;19(5):281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura M.T., Yudell B.E., Loor J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014;53:124–144. doi: 10.1016/j.plipres.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Gross R.W., Han X.L. Lipidomics at the interface of structure and function in systems biology. Chem. Biol. 2011;18(3):284–291. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst R., Ejsing C.S., Antonny B. Homeoviscous adaptation and the regulation of membrane lipids. J. Mol. Biol. 2016;428(24):4776–4791. doi: 10.1016/j.jmb.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Antonny B. Mechanisms of membrane curvature sensing. In: Kornberg R.D., Raetz C.R.H., Rothman J.E., Thorner J.W., editors. vol. 80. Annual Reviews; Palo Alto: 2011. pp. 101–123. (Annual Review of Biochemistry). [DOI] [PubMed] [Google Scholar]
- 11.Baumgart T., Capraro B.R., Zhu C., Das S.L. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. In: Leone S.R., Cremer P.S., Groves J.T., Johnson M.A., editors. vol. 62. Annual Reviews; Palo Alto: 2011. pp. 483–506. (Annual Review of Physical Chemistry). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan E., Reid G.E. Chemical derivatization and ultrahigh resolution and accurate mass spectrometry strategies for “shotgun” lipidome analysis. Accounts Chem. Res. 2016;49(9):1596–1604. doi: 10.1021/acs.accounts.6b00030. [DOI] [PubMed] [Google Scholar]
- 13.Wang M., Hayakawa J., Yang K., Han X.L. Characterization and quantification of diacylglycerol species in biological extracts after one-step derivatization: a shotgun lipidomics approach. Anal. Chem. 2014;86(4):2146–2155. doi: 10.1021/ac403798q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketel K., Krauss M., Nicot A.S., Puchkov D., Wieffer M., Muller R., Subramanian D., Schultz C., Laporte J., Haucke V. A phosphoinositide conversion mechanism for exit from endosomes. Nature. 2016;529(7586):408–412. doi: 10.1038/nature16516. [DOI] [PubMed] [Google Scholar]
- 15.Li L.X., Zhong S.S., Shen X., Li Q.J., Xu W.X., Tao Y.Z., Yin H.Y. Recent development on liquid chromatography-mass spectrometry analysis of oxidized lipids. Free Radic. Biol. Med. 2019;144:16–34. doi: 10.1016/j.freeradbiomed.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Hu C.F., Wang C.Y., He L.J., Han X.L. Novel strategies for enhancing shotgun lipidomics for comprehensive analysis of cellular lipidomes. Trac. Trends Anal. Chem. 2019;120 doi: 10.1016/j.trac.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rustam Y.H., Reid G.E. Analytical challenges and recent advances in mass spectrometry based lipidomics. Anal. Chem. 2018;90(1):374–397. doi: 10.1021/acs.analchem.7b04836. [DOI] [PubMed] [Google Scholar]
- 18.Rampler E., Criscuolo A., Zeller M., El Abiead Y., Schoeny H., Hermann G., Sokol E., Cook K., Peake D.A., Delanghe B., Koellensperger G. A novel lipidomics workflow for improved human plasma identification and quantification using RPLC-MSn methods and isotope dilution strategies. Anal. Chem. 2018;90(11):6494–6501. doi: 10.1021/acs.analchem.7b05382. [DOI] [PubMed] [Google Scholar]
- 19.Wang M., Wang C.Y., Han X.L. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-what, how and why? Mass Spectrom. Rev. 2017;36(6):693–714. doi: 10.1002/mas.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X.L., Gross R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44(6):1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Almeida R., Pauling J.K., Sokol E., Hannibal-Bach H.K., Ejsing C.S. Comprehensive lipidome analysis by shotgun lipidomics on a hybrid quadrupole-orbitrap-linear ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2015;26(1):133–148. doi: 10.1007/s13361-014-1013-x. [DOI] [PubMed] [Google Scholar]
- 22.Annesley T.M. Ion suppression in mass spectrometry. Clin. Chem. 2003;49(7):1041–1044. doi: 10.1373/49.7.1041. [DOI] [PubMed] [Google Scholar]
- 23.Southam A.D., Weber R.J.M., Engel J., Jones M.R., Viant M.R. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nat. Protoc. 2016;12(2):310–328. doi: 10.1038/nprot.2016.156. [DOI] [PubMed] [Google Scholar]
- 24.Yang K., Han X. Accurate quantification of lipid species by electrospray ionization mass spectrometry - meet a key challenge in lipidomics. Metabolites. 2011;1(1):21–40. doi: 10.3390/metabo1010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu C.F., Duan Q., Han X.L. Strategies to improve/eliminate the limitations in shotgun lipidomics. Proteomics. 2020;20(11) doi: 10.1002/pmic.201900070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lintonen T.P.I., Baker P.R.S., Suoniemi M., Ubhi B.K., Koistinen K.M., Duchoslav E., Campbell J.L., Ekroos K. Differential mobility spectrometry-driven shotgun lipidomics. Anal. Chem. 2014;86(19):9662–9669. doi: 10.1021/ac5021744. [DOI] [PubMed] [Google Scholar]
- 27.Levy A.J., Oranzi N.R., Ahmadireskety A., Kemperman R.H.J., Wei M.S., Yost R.A. Recent progress in metabolomics using ion mobility-mass spectrometry. Trac. Trends Anal. Chem. 2019;116:274–281. [Google Scholar]
- 28.Hinz C., Liggi S., Griffin J.L. The potential of Ion Mobility Mass Spectrometry for high-throughput and high-resolution lipidomics. Curr. Opin. Chem. Biol. 2018;42:42–50. doi: 10.1016/j.cbpa.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Yeo H.C., Chen S.W., Ho Y.S., Lee D.Y. An LC-MS-based lipidomics pre-processing framework underpins rapid hypothesis generation towards CHO systems biotechnology. Metabolomics. 2018;14(7) doi: 10.1007/s11306-018-1394-0. [DOI] [PubMed] [Google Scholar]
- 30.Lorenzen W., Bozhuyuk K.A.J., Cortina N.S., Bode H.B. A comprehensive insight into the lipid composition of Myxococcus xanthus by UPLC-ESI-MS. J. Lipid Res. 2014;55(12):2620–2633. doi: 10.1194/jlr.M054593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xuan Q., Zheng F., Yu D., Ouyang Y., Zhao X., Hu C., Xu G. Rapid lipidomic profiling based on ultra-high performance liquid chromatography-mass spectrometry and its application in diabetic retinopathy. Anal. Bioanal. Chem. 2020;412(15):3585–3594. doi: 10.1007/s00216-020-02632-6. [DOI] [PubMed] [Google Scholar]
- 32.Lagerborg K.A., Watrous J.D., Cheng S., Jain M. High-throughput measure of bioactive lipids using non-targeted mass spectrometry. Methods Mol. Biol. 2019;1862:17–35. doi: 10.1007/978-1-4939-8769-6_2. [DOI] [PubMed] [Google Scholar]
- 33.Dugo P., Favoino O., Tranchida P.Q., Dugo G., Mondello L. Off-line coupling of non-aqueous reversed-phase and silver ion high-performance liquid chromatography-mass spectrometry for the characterization of rice oil triacylglycerol positional isomers. J. Chromatogr. A. 2004;1041(1–2):135–142. doi: 10.1016/j.chroma.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 34.Yang Q., Shi X., Gu Q., Zhao S., Shan Y., Xu G. On-line two dimensional liquid chromatography/mass spectrometry for the analysis of triacylglycerides in peanut oil and mouse tissue. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;895–896:48–55. doi: 10.1016/j.jchromb.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 35.Wang S., Li J., Shi X., Qiao L., Lu X., Xu G. A novel stop-flow two-dimensional liquid chromatography-mass spectrometry method for lipid analysis. J. Chromatogr. A. 2013;1321:65–72. doi: 10.1016/j.chroma.2013.10.069. [DOI] [PubMed] [Google Scholar]
- 36.Wang S., Zhou L., Wang Z., Shi X., Xu G. Simultaneous metabolomics and lipidomics analysis based on novel heart-cutting two-dimensional liquid chromatography-mass spectrometry. Anal. Chim. Acta. 2017;966:34–40. doi: 10.1016/j.aca.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Lv W.J., Shi X.Z., Wang S.Y., Xu G.W. Multidimensional liquid chromatography-mass spectrometry for metabolomic and lipidomic analyses. Trac. Trends Anal. Chem. 2019;120 [Google Scholar]
- 38.Zhang X., Kew K., Reisdorph R., Sartain M., Powell R., Armstrong M., Quinn K., Cruickshank-Quinn C., Walmsley S., Bokatzian S., Darland E., Rain M., Imatani K., Reisdorph N. Performance of a high-pressure liquid chromatography-ion mobility-mass spectrometry system for metabolic profiling. Anal. Chem. 2017;89(12):6384–6391. doi: 10.1021/acs.analchem.6b04628. [DOI] [PubMed] [Google Scholar]
- 39.Tu J., Zhou Z.W., Li T.Z., Zhu Z.J. The emerging role of ion mobility-mass spectrometry in lipidomics to facilitate lipid separation and identification. Trac. Trends Anal. Chem. 2019;116:332–339. [Google Scholar]
- 40.Paglia G., Williams J.P., Menikarachchi L., Thompson J.W., Tyldesley-Worster R., Halldórsson S., Rolfsson O., Moseley A., Grant D., Langridge J., Palsson B.O., Astarita G. Ion mobility derived collision cross sections to support metabolomics applications. Anal. Chem. 2014;86(8):3985–3993. doi: 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodds J.N., May J.C., McLean J.A. Correlating resolving power, resolution, and collision cross section: unifying cross-platform assessment of separation efficiency in ion mobility spectrometry. Anal. Chem. 2017;89(22):12176–12184. doi: 10.1021/acs.analchem.7b02827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kylli P., Hankemeier T., Kostiainen R. Feasibility of ultra-performance liquid chromatography-ion mobility-time-of-flight mass spectrometry in analyzing oxysterols. J. Chromatogr. A. 2017;1487:147–152. doi: 10.1016/j.chroma.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 43.Poad B.L.J., Zheng X.Y., Mitchell T.W., Smith R.D., Baker E.S., Blanksby S.J. Online ozonolysis combined with ion mobility-mass spectrometry provides a new platform for lipid isomer analyses. Anal. Chem. 2018;90(2):1292–1300. doi: 10.1021/acs.analchem.7b04091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baglai A., Gargano A.F.G., Jordens J., Mengerink Y., Honing M., van der Wal S., Schoenmakers P.J. Comprehensive lipidomic analysis of human plasma using multidimensional liquid- and gas-phase separations: two-dimensional liquid chromatography-mass spectrometry vs. liquid chromatography-trapped-ion-mobility-mass spectrometry. J. Chromatogr. A. 2017;1530:90–103. doi: 10.1016/j.chroma.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 45.Zhu X., Chen Y., Subramanian R. Comparison of information-dependent acquisition, SWATH, and MS(All) techniques in metabolite identification study employing ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Anal. Chem. 2014;86(2):1202–1209. doi: 10.1021/ac403385y. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., Feng R., He C., Su H., Ma H., Wan J.-B. An integrated strategy to improve data acquisition and metabolite identification by time-staggered ion lists in UHPLC/Q-TOF MS-based metabolomics. J. Pharmaceut. Biomed. Anal. 2018;157:171–179. doi: 10.1016/j.jpba.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y., Feng R.B., Wang R.B., Yang F.Q., Li P., Wan J.B. Enhanced MS/MS coverage for metabolite identification in LC-MSbased untargeted metabolomics by target-directed data dependent acquisition with time-staggered precursor ion list. Anal. Chim. Acta. 2017;992:67–75. doi: 10.1016/j.aca.2017.08.044. [DOI] [PubMed] [Google Scholar]
- 48.Bendall S.C., Hughes C., Campbell J.L., Stewart M.H., Pittock P., Liu S., Bonneil E., Thibault P., Bhatia M., Lajoie G.A. An enhanced mass spectrometry approach reveals human embryonic stem cell growth factors in culture. Mol. Cell. Proteomics. 2009;8(3):421–432. doi: 10.1074/mcp.M800190-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koelmel J.P., Kroeger N.M., Gill E.L., Ulmer C.Z., Bowden J.A., Patterson R.E., Yost R.A., Garrett T.J. Expanding lipidome coverage using LC-MS/MS data-dependent acquisition with automated exclusion list generation. J. Am. Soc. Mass Spectrom. 2017;28(5):908–917. doi: 10.1007/s13361-017-1608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nazari M., Muddiman D.C. Enhanced lipidome coverage in shotgun analyses by using gas-phase fractionation. J. Am. Soc. Mass Spectrom. 2016;27(11):1735–1744. doi: 10.1007/s13361-016-1446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullard G., Allwood J.W., Weber R., Brown M., Begley P., Hollywood K.A., Jones M., Unwin R.D., Bishop P.N., Cooper G.J.S., Dunn W.B. A new strategy for MS/MS data acquisition applying multiple data dependent experiments on Orbitrap mass spectrometers in non-targeted metabolomic applications. Metabolomics. 2015;11(5):1068–1080. [Google Scholar]
- 52.Silva J.C., Denny R., Dorschel C.A., Gorenstein M., Kass I.J., Li G.Z., McKenna T., Nold M.J., Richardson K., Young P., Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77(7):2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 53.Geiger T., Cox J., Mann M. Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell. Proteomics. 2010;9(10):2252–2261. doi: 10.1074/mcp.M110.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillet L.C., Navarro P., Tate S., Rost H., Selevsek N., Reiter L., Bonner R., Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11(6) doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsugawa H., Satoh A., Uchino H., Cajka T., Arita M., Arita M. Mass spectrometry data repository enhances novel metabolite discoveries with advances in computational metabolomics. Metabolites. 2019;9(6) doi: 10.3390/metabo9060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlotterbeck J., Chatterjee M., Gawaz M., Lammerhofer M. Comprehensive MS/MS profiling by UHPLC-ESI-QTOF-MS/MS using SWATH data-independent acquisition for the study of platelet lipidomes in coronary artery disease. Anal. Chim. Acta. 2019;1046:1–15. doi: 10.1016/j.aca.2018.08.060. [DOI] [PubMed] [Google Scholar]
- 57.Drotleff B., Illison J., Schlotterbeck J., Lukowski R., Lammerhofer M. Comprehensive lipidomics of mouse plasma using class-specific surrogate calibrants and SWATH acquisition for large-scale lipid quantification in untargeted analysis. Anal. Chim. Acta. 2019;1086:90–102. doi: 10.1016/j.aca.2019.08.030. [DOI] [PubMed] [Google Scholar]
- 58.Cebo M., Schlotterbeck J., Gawaz M., Chatterjee M., Lammerhofer M. Simultaneous targeted and untargeted UHPLC-ESI-MS/MS method with data-independent acquisition for quantification and profiling of (oxidized) fatty acids released upon platelet activation by thrombin. Anal. Chim. Acta. 2020;1094:57–69. doi: 10.1016/j.aca.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 59.Drotleff B., Hallschmid M., Lammerhofer M. Quantification of steroid hormones in plasma using a surrogate calibrant approach and UHPLC-ESI-QTOF-MS/MS with SWATH-acquisition combined with untargeted profiling. Anal. Chim. Acta. 2018;1022:70–80. doi: 10.1016/j.aca.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 60.Wang R.H., Yin Y.D., Zhu Z.J. Advancing untargeted metabolomics using data-independent acquisition mass spectrometry technology. Anal. Bioanal. Chem. 2019;411(19):4349–4357. doi: 10.1007/s00216-019-01709-1. [DOI] [PubMed] [Google Scholar]
- 61.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., Kanazawa M., VanderGheynst J., Fiehn O., Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12(6):523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin Y., Wang R., Cai Y., Wang Z., Zhu Z.-J. DecoMetDIA: deconvolution of multiplexed MS/MS spectra for metabolite identification in SWATH-MS-based untargeted metabolomics. Anal. Chem. 2019;91(18):11897–11904. doi: 10.1021/acs.analchem.9b02655. [DOI] [PubMed] [Google Scholar]
- 63.Li H., Cai Y., Guo Y., Chen F., Zhu Z.-J. MetDIA: targeted metabolite extraction of multiplexed MS/MS spectra generated by data-independent acquisition. Anal. Chem. 2016;88(17):8757–8764. doi: 10.1021/acs.analchem.6b02122. [DOI] [PubMed] [Google Scholar]
- 64.Chen G., Walmsley S., Cheung G.C.M., Chen L., Cheng C.-Y., Beuerman R.W., Wong T.Y., Zhou L., Choi H. Customized consensus spectral library building for untargeted quantitative metabolomics analysis with data independent acquisition mass spectrometry and MetaboDIA workflow. Anal. Chem. 2017;89(9):4897–4906. doi: 10.1021/acs.analchem.6b05006. [DOI] [PubMed] [Google Scholar]
- 65.Wang M., Wang C., Han X. Selection of internal standards for accurate quantification OF complex lipid species in biological extracts by electrospray ionization mass spectrometry-what, how and why? Mass Spectrom. Rev. 2017;36(6):693–714. doi: 10.1002/mas.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu C., Duan Q., Han X. Strategies to improve/eliminate the limitations in shotgun lipidomics. Proteomics. 2020;20(11) doi: 10.1002/pmic.201900070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gathungu R.M., Larrea P., Sniatynski M.J., Marur V.R., Bowden J.A., Koelmel J.P., Starke-Reed P., Hubbard V.S., Kristal B.S. Optimization of electrospray ionization source parameters for lipidomics to reduce misannotation of in-source fragments as precursor ions. Anal. Chem. 2018;90(22):13523–13532. doi: 10.1021/acs.analchem.8b03436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin M., Wang Z., Wang D., Chen X., Zhang J.-L. Mathematical model-assisted UHPLC-MS/MS method for global profiling and quantification of cholesteryl esters in hyperlipidemic golden hamsters. Anal. Chem. 2019;91(7):4504–4512. doi: 10.1021/acs.analchem.8b05337. [DOI] [PubMed] [Google Scholar]
- 69.Takeda H., Izumi Y., Takahashi M., Paxton T., Tamura S., Koike T., Yu Y., Kato N., Nagase K., Shiomi M., Bamba T. Widely-targeted quantitative lipidomics method by supercritical fluid chromatography triple quadrupole mass spectrometry. J. Lipid Res. 2018;59(7):1283–1293. doi: 10.1194/jlr.D083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Massey K.A., Nicolaou A. Lipidomics of oxidized polyunsaturated fatty acids. Free Radic. Biol. Med. 2013;59:45–55. doi: 10.1016/j.freeradbiomed.2012.08.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., Armando A.M., Quehenberger O., Yan C., Dennis E.A. Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J. Chromatogr. A. 2014;1359:60–69. doi: 10.1016/j.chroma.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang M., Han X. Advanced shotgun lipidomics for characterization of altered lipid patterns in neurodegenerative diseases and brain injury. Methods Mol. Biol. 2016;1303:405–422. doi: 10.1007/978-1-4939-2627-5_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu X.P., Moon S.H., Mancuso D.J., Jenkins C.M., Guan S.P., Sims H.F., Gross R.W. Oxidized fatty acid analysis by charge-switch derivatization, selected reaction monitoring, and accurate mass quantitation. Anal. Biochem. 2013;442(1):40–50. doi: 10.1016/j.ab.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cajka T., Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 2016;88(1):524–545. doi: 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- 75.Giles C., Takechi R., Lam V., Dhaliwal S.S., Mamo J.C.L. Contemporary lipidomic analytics: opportunities and pitfalls. Prog. Lipid Res. 2018;71:86–100. doi: 10.1016/j.plipres.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 76.Strassburg K., Huijbrechts A.M.L., Kortekaas K.A., Lindeman J.H., Pedersen T.L., Dane A., Berger R., Brenkman A., Hankemeier T., van Duynhoven J., Kalkhoven E., Newman J.W., Vreeken R.J. Quantitative profiling of oxylipins through comprehensive LC-MS/MS analysis: application in cardiac surgery. Anal. Bioanal. Chem. 2012;404(5):1413–1426. doi: 10.1007/s00216-012-6226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alshehry Z.H., Mundra P.A., Barlow C.K., Mellett N.A., Wong G., McConville M.J., Simes J., Tonkin A.M., Sullivan D.R., Barnes E.H., Nestel P.J., Kingwell B.A., Marre M., Neal B., Poulter N.R., Rodgers A., Williams B., Zoungas S., Hillis G.S., Chalmers J., Woodward M., Meikle P.J. Plasma lipidomic profiles improve on traditional risk factors for the prediction of cardiovascular events in type 2 diabetes mellitus. Circulation. 2016;134(21):1637–1650. doi: 10.1161/CIRCULATIONAHA.116.023233. [DOI] [PubMed] [Google Scholar]
- 78.Li J., Hu C., Zhao X., Dai W., Chen S., Lu X., Xu G. Large-scaled human serum sphingolipid profiling by using reversed-phase liquid chromatography coupled with dynamic multiple reaction monitoring of mass spectrometry: method development and application in hepatocellular carcinoma. J. Chromatogr. A. 2013;1320:103–110. doi: 10.1016/j.chroma.2013.10.064. [DOI] [PubMed] [Google Scholar]
- 79.Jia Z.-X., Zhang J.-L., Shen C.-P., Ma L. Profile and quantification of human stratum corneum ceramides by normal-phase liquid chromatography coupled with dynamic multiple reaction monitoring of mass spectrometry: development of targeted lipidomic method and application to human stratum corneum of different age groups. Anal. Bioanal. Chem. 2016;408(24):6623–6636. doi: 10.1007/s00216-016-9775-6. [DOI] [PubMed] [Google Scholar]
- 80.Tang H., Fang H., Yin E., Brasier A.R., Sowers L.C., Zhang K. Multiplexed parallel reaction monitoring targeting histone modifications on the QExactive mass spectrometer. Anal. Chem. 2014;86(11):5526–5534. doi: 10.1021/ac500972x. [DOI] [PubMed] [Google Scholar]
- 81.Gallien S., Bourmaud A., Kim S.Y., Domon B. Technical considerations for large-scale parallel reaction monitoring analysis. J Proteomics. 2014;100:147–159. doi: 10.1016/j.jprot.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 82.Gallien S., Kim S.Y., Domon B. Large-scale targeted proteomics using internal standard triggered-parallel reaction monitoring (IS-PRM) Mol. Cell. Proteomics. 2015;14(6):1630–1644. doi: 10.1074/mcp.O114.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bourmaud A., Gallien S., Domon B. Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: principle and applications. Proteomics. 2016;16(15–16):2146–2159. doi: 10.1002/pmic.201500543. [DOI] [PubMed] [Google Scholar]
- 84.Zhou J., Liu C., Si D., Jia B., Zhong L., Yin Y. Workflow development for targeted lipidomic quantification using parallel reaction monitoring on a quadrupole-time of flight mass spectrometry. Anal. Chim. Acta. 2017;972:62–72. doi: 10.1016/j.aca.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 85.Zhang T., Trauger S.A., Vidoudez C., Doane K.P., Pluimer B.R., Peterson R.T. Parallel Reaction Monitoring reveals structure-specific ceramide alterations in the zebrafish. Sci. Rep. 2019;9(1):19939. doi: 10.1038/s41598-019-56466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu D.Y., Rupasinghe T.W.T., Boughton B.A., Natera S.H.A., Hill C.B., Tarazona P., Feussner I., Roessner U. A high-resolution HPLC-QqTOF platform using parallel reaction monitoring for in-depth lipid discovery and rapid profiling. Anal. Chim. Acta. 2018;1026:87–100. doi: 10.1016/j.aca.2018.03.062. [DOI] [PubMed] [Google Scholar]
- 87.Rampler E., Coman C., Hermann G., Sickmann A., Ahrends R., Koellensperger G. LILY-lipidome isotope labeling of yeast: in vivo synthesis of C labeled reference lipids for quantification by mass spectrometry. Analyst. 2017;142(11):1891–1899. doi: 10.1039/c7an00107j. [DOI] [PubMed] [Google Scholar]
- 88.Tague E.D., Woodall B.M., Harp J.R., Farmer A.T., Fozo E.M., Campagna S.R. Expanding lipidomics coverage: effective ultra performance liquid chromatography-high resolution mass spectrometer methods for detection and quantitation of cardiolipin, phosphatidylglycerol, and lysyl-phosphatidylglycerol. Metabolomics : Off. J. Metabol. Soc. 2019;15(4):53. doi: 10.1007/s11306-019-1512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou J., Liu H., Liu Y., Liu J., Zhao X., Yin Y. Development and evaluation of a parallel reaction monitoring strategy for large-scale targeted metabolomics quantification. Anal. Chem. 2016;88(8):4478–4486. doi: 10.1021/acs.analchem.6b00355. [DOI] [PubMed] [Google Scholar]
- 90.Li Y., Ruan Q., Li Y., Ye G., Lu X., Lin X., Xu G. A novel approach to transforming a non-targeted metabolic profiling method to a pseudo-targeted method using the retention time locking gas chromatography/mass spectrometry-selected ions monitoring. J. Chromatogr. A. 2012;1255:228–236. doi: 10.1016/j.chroma.2012.01.076. [DOI] [PubMed] [Google Scholar]
- 91.Chen S., Kong H., Lu X., Li Y., Yin P., Zeng Z., Xu G. Pseudotargeted metabolomics method and its application in serum biomarker discovery for hepatocellular carcinoma based on ultra high-performance liquid chromatography/triple quadrupole mass spectrometry. Anal. Chem. 2013;85(17):8326–8333. doi: 10.1021/ac4016787. [DOI] [PubMed] [Google Scholar]
- 92.Chen W., Gong L., Guo Z., Wang W., Zhang H., Liu X., Yu S., Xiong L., Luo J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant. 2013;6(6):1769–1780. doi: 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
- 93.Luo P., Dai W., Yin P., Zeng Z., Kong H., Zhou L., Wang X., Chen S., Lu X., Xu G. Multiple reaction monitoring-ion pair finder: a systematic approach to Transform nontargeted mode to pseudotargeted mode for metabolomics study based on liquid chromatography-mass spectrometry. Anal. Chem. 2015;87(10):5050–5055. doi: 10.1021/acs.analchem.5b00615. [DOI] [PubMed] [Google Scholar]
- 94.Zhao Y., Hao Z., Zhao C., Zhao J., Zhang J., Li Y., Li L., Huang X., Lin X., Zeng Z., Lu X., Xu G. A novel strategy for large-scale metabolomics study by calibrating gross and systematic errors in gas chromatography-mass spectrometry. Anal. Chem. 2016;88(4):2234–2242. doi: 10.1021/acs.analchem.5b03912. [DOI] [PubMed] [Google Scholar]
- 95.Luo P., Yin P., Zhang W., Zhou L., Lu X., Lin X., Xu G. Optimization of large-scale pseudotargeted metabolomics method based on liquid chromatography-mass spectrometry. J. Chromatogr. A. 2016;1437:127–136. doi: 10.1016/j.chroma.2016.01.078. [DOI] [PubMed] [Google Scholar]
- 96.Wang L., Su B., Zeng Z., Li C., Zhao X., Lv W., Xuan Q., Ouyang Y., Zhou L., Yin P., Peng X., Lu X., Lin X., Xu G. Ion-pair selection method for pseudotargeted metabolomics based on SWATH MS acquisition and its application in differential metabolite discovery of type 2 diabetes. Anal. Chem. 2018;90(19):11401–11408. doi: 10.1021/acs.analchem.8b02377. [DOI] [PubMed] [Google Scholar]
- 97.Xuan Q.H., Hu C.X., Yu D., Wang L.C., Zhou Y., Zhao X.J., Li Q., Hou X.L., Xu G.W. Development of a high coverage pseudotargeted lipidomics method based on ultra-high performance liquid chromatography-mass spectrometry. Anal. Chem. 2018;90(12):7608–7616. doi: 10.1021/acs.analchem.8b01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lv W., Wang L., Xuan Q., Zhao X., Liu X., Shi X., Xu G. Pseudotargeted method based on parallel column two- dimensional liquid chromatography-mass spectrometry for broad coverage of metabolome and lipidome. Anal. Chem. 2020;92(8):6043–6050. doi: 10.1021/acs.analchem.0c00372. [DOI] [PubMed] [Google Scholar]
- 99.Zheng F., Zhao X., Zeng Z., Wang L., Lv W., Wang Q., Xu G. Development of a plasma pseudotargeted metabolomics method based on ultra-high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2020;15(8):2519–2537. doi: 10.1038/s41596-020-0341-5. [DOI] [PubMed] [Google Scholar]
- 100.Lin L., Ding Y., Wang Y., Wang Z.X., Yin X.F., Yan G.Q., Zhang L., Yang P.Y., Shen H.L. Functional lipidomics: palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism. Hepatology. 2017;66(2):432–448. doi: 10.1002/hep.29033. [DOI] [PubMed] [Google Scholar]
- 101.Kuhn T., Floegel A., Sookthai D., Johnson T., Rolle-Kampczyk U., Otto W., von Bergen M., Boeing H., Kaaks R. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016;14 doi: 10.1186/s12916-016-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marien E., Meister M., Muley T., Fieuws S., Bordel S., Derua R., Spraggins J., Van de Plas R., Dehairs J., Wouters J., Bagadi M., Dienemann H., Thomas M., Schnabel P.A., Caprioli R.M., Waelkens E., Swinnen J.V. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int. J. Canc. 2015;137(7):1539–1548. doi: 10.1002/ijc.29517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mondul A.M., Moore S.C., Weinstein S.J., Karoly E.D., Sampson J.N., Albanes D. Metabolomic analysis of prostate cancer risk in a prospective cohort: the alpha-tocolpherol, beta-carotene cancer prevention (ATBC) study. Int. J. Canc. 2015;137(9):2124–2132. doi: 10.1002/ijc.29576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li J., Ren S.C., Piao H.L., Wang F.B., Yin P.Y., Xu C.L., Lu X., Ye G.Z., Shao Y.P., Yan M., Zhao X.J., Sun Y.H., Xu G.W. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Sci. Rep. 2016;6 doi: 10.1038/srep20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wigger L., Cruciani-Guglielmacci C., Nicolas A., Denom J., Fernandez N., Fumeron F., Marques-Vidal P., Ktorza A., Kramer W., Schulte A., Le Stunff H., Liechti R., Xenarios I., Vollenweider P., Waeber G., Uphues I., Roussel R., Magnan C., Ibberson M., Thorens B. Plasma dihydroceramides are diabetes susceptibility biomarker candidates in mice and humans. Cell Rep. 2017;18(9):2269–2279. doi: 10.1016/j.celrep.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 106.Laaksonen R., Ekroos K., Sysi-Aho M., Hilvo M., Vihervaara T., Kauhanen D., Suoniemi M., Hurme R., Marz W., Scharnagl H., Stojakovic T., Vlachopoulou E., Lokki M.L., Nieminen M.S., Klingenberg R., Matter C.M., Hornemann T., Juni P., Rodondi N., Raber L., Windecker S., Gencer B., Pedersen E.R., Tell G.S., Nygard O., Mach F., Sinisalo J., Luscher T.F. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur. Heart J. 2016;37(25):1967–1976. doi: 10.1093/eurheartj/ehw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vvedenskaya O., Wang Y.T., Ackerman J.M., Knittelfelder O., Shevchenko A. Analytical challenges in human plasma lipidomics: a winding path towards the truth. Trac. Trends Anal. Chem. 2019;120 [Google Scholar]
- 108.Lu J.L., Lam S.M., Wan Q., Shi L.X., Huo Y.N., Chen L.L., Tang X.L., Li B.W., Wu X.Y., Peng K., Li M., Wang S.Y., Xu Y., Xu M., Bi Y.F., Ning G., Shui G.H., Wang W.Q. High-coverage targeted lipidomics reveals novel serum lipid predictors and lipid pathway dysregulation antecedent to type 2 diabetes onset in normoglycemic Chinese adults. Diabetes Care. 2019;42(11):2117–2126. doi: 10.2337/dc19-0100. [DOI] [PubMed] [Google Scholar]
- 109.Loke S.Y., Lee A.S.G. The future of blood-based biomarkers for the early detection of breast cancer. Eur. J. Canc. 2018;92:54–68. doi: 10.1016/j.ejca.2017.12.025. [DOI] [PubMed] [Google Scholar]
- 110.Reitz C. Dyslipidemia and the risk of Alzheimer’s disease. Curr. Atherosclerosis Rep. 2013;15(3) doi: 10.1007/s11883-012-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reitz C., Mayeux R. Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem. Pharmacol. 2014;88(4):640–651. doi: 10.1016/j.bcp.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Proitsi P., Kim M., Whiley L., Pritchard M., Leung R., Soininen H., Kloszewska I., Mecocci P., Tsolaki M., Vellas B., Sham P., Lovestone S., Powell J.F., Dobson R.J.B., Legido-Quigley C. Plasma lipidomics analysis finds long chain cholesteryl esters to be associated with Alzheimer’s disease. Transl. Psychiatry. 2015;5(1) doi: 10.1038/tp.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Varma V.R., Oommen A.M., Varma S., Casanova R., An Y., Andrews R.M., O’Brien R., Pletnikova O., Troncoso J.C., Toledo J., Baillie R., Arnold M., Kastenmueller G., Nho K., Doraiswamy P.M., Saykin A.J., Kaddurah-Daouk R., Legido-Quigley C., Thambisetty M. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: a targeted metabolomics study. PLoS Med. 2018;15(1) doi: 10.1371/journal.pmed.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Afshinnia F., Rajendiran T.M., Karnovsky A., Soni T., Wang X., Xie D., Yang W., Shafi T., Weir M.R., He J., Brecklin C.S., Rhee E.P., Schelling J.R., Ojo A., Feldman H., Michailidis G., Pennathur S. Lipidomic signature of progression of chronic kidney disease in the chronic renal insufficiency cohort. Kidney Int Rep. 2016;1(4):256–268. doi: 10.1016/j.ekir.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin H.M., Mahon K.L., Weir J.M., Mundra P.A., Spielman C., Briscoe K., Gurney H., Mallesara G., Marx G., Stockler M.R., Parton R.G., Hoy A.J., Daly R.J., Meikle P.J., Horvath L.G., Consortium P.R. A distinct plasma lipid signature associated with poor prognosis in castration-resistant prostate cancer. Int. J. Canc. 2017;141(10):2112–2120. doi: 10.1002/ijc.30903. [DOI] [PubMed] [Google Scholar]
- 116.Hilvo M., Meikle P.J., Pedersen E.R., Tell G.S., Dhar I., Brenner H., Schöttker B., Lääperi M., Kauhanen D., Koistinen K.M., Jylhä A., Huynh K., Mellett N.A., Tonkin A.M., Sullivan D.R., Simes J., Nestel P., Koenig W., Rothenbacher D., Nygård O., Laaksonen R. Development and validation of a ceramide- and phospholipid-based cardiovascular risk estimation score for coronary artery disease patients. Eur. Heart J. 2020;41(3):371–380. doi: 10.1093/eurheartj/ehz387. [DOI] [PubMed] [Google Scholar]
- 117.Eisfeld A.J., Halfmann P.J., Wendler J.P., Kyle J.E., Burnum-Johnson K.E., Peralta Z., Maemura T., Walters K.B., Watanabe T., Fukuyama S., Yamashita M., Jacobs J.M., Kim Y.M., Casey C.P., Stratton K.G., Webb-Robertson B.J.M., Gritsenko M.A., Monroe M.E., Weitz K.K., Shukla A.K., Tian M.Y., Neumann G., Reed J.L., van Bakel H., Metz T.O., Smith R.D., Waters K.M., N’Jai A., Sahr F., Kawaoka Y. Multi-platform ’omics analysis of human Ebola virus disease pathogenesis. Cell Host Microbe. 2017;22(6):817–829. doi: 10.1016/j.chom.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Butler L., Perone Y., Dehairs J., Lupien L.E., de Laat V., Talebi A., Loda M., Kinlaw W.B., Swinnen J.V. Lipids and cancer: emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020;S0169-409X(20) doi: 10.1016/j.addr.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5(1) doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thangapandi V.R., Knittelfelder O., Brosch M., Patsenker E., Vvedenskaya O., Buch S., Hinz S., Hendricks A., Nati M., Herrmann A., Rekhade D.R., Berg T., Matz-Soja M., Huse K., Klipp E., Pauling J.K., Wodke J.A., Miranda Ackerman J., Bonin M.v., Aigner E., Datz C., von Schonfels W., Nehring S., Zeissig S., Rocken C., Dahl A., Chavakis T., Stickel F., Shevchenko A., Schafmayer C., Hampe J., Subramanian P. Loss of hepatic Mboat7 leads to liver fibrosis. Gut. 2020 doi: 10.1136/gutjnl-2020-320853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li L., Pilo G.M., Li X., Cigliano A., Latte G., Che L., Joseph C., Mela M., Wang C., Jiang L., Ribback S., Simile M.M., Pascale R.M., Dombrowski F., Evert M., Semenkovich C.F., Chen X., Calvisi D.F. Inactivation of fatty acid synthase impairs hepatocarcinogenesis driven by AKT in mice and humans. J. Hepatol. 2016;64(2):333–341. doi: 10.1016/j.jhep.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cao D., Song X., Che L., Li X., Pilo M.G., Vidili G., Porcu A., Solinas A., Cigliano A., Pes G.M., Ribback S., Dombrowski F., Chen X., Li L., Calvisi D.F. Both de novo synthetized and exogenous fatty acids support the growth of hepatocellular carcinoma cells. Liver Int. 2017;37(1):80–89. doi: 10.1111/liv.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Che L., Chi W., Qiao Y., Zhang J., Song X., Liu Y., Li L., Jia J., Pilo M.G., Wang J., Cigliano A., Ma Z., Kuang W., Tang Z., Zhang Z., Shui G., Ribback S., Dombrowski F., Evert M., Pascale R.M., Cossu C., Pes G.M., Osborne T.F., Calvisi D.F., Chen X., Chen L. Cholesterol biosynthesis supports the growth of hepatocarcinoma lesions depleted of fatty acid synthase in mice and humans. Gut. 2020;69(1):177–186. doi: 10.1136/gutjnl-2018-317581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schwarz B., Sharma L., Roberts L., Peng X., Bermejo S., Leighton I., Massana A.C., Farhadian S., Ko A., DelaCruz C., Bosio C.M. 2020. Severe SARS-CoV-2 Infection in Humans Is Defined by a Shift in the Serum Lipidome Resulting in Dysregulation of Eicosanoid Immune Mediators, medRxiv : the Preprint Server for Health Sciences. [Google Scholar]
- 125.Hata M., Ikeda H.O., Iwai S., Iida Y., Gotoh N., Asaka I., Ikeda K., Isobe Y., Hori A., Nakagawa S., Yamato S., Arita M., Yoshimura N., Tsujikawa A. Reduction of lipid accumulation rescues Bietti’s crystalline dystrophy phenotypes. Proc. Natl. Acad. Sci. U.S.A. 2018;115(15):3936–3941. doi: 10.1073/pnas.1717338115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang W., Yang J., Edin M.L., Wang Y., Luo Y., Wan D., Yang H., Song C.-Q., Xue W., Sanidad K.Z., Song M., Bisbee H.A., Bradbury J.A., Nan G., Zhang J., Shih P.-A.B., Lee K.S.S., Minter L.M., Kim D., Xiao H., Liu J.-Y., Hammock B.D., Zeldin D.C., Zhang G. Targeted metabolomics identifies the cytochrome P450 monooxygenase eicosanoid pathway as a novel therapeutic target of colon tumorigenesis. Canc. Res. 2019;79(8):1822–1830. doi: 10.1158/0008-5472.CAN-18-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bowden J.A., Heckert A., Ulmer C.Z., Jones C.M., Koelmel J.P., Abdullah L., Ahonen L., Alnouti Y., Armando A.M., Asara J.M., Bamba T., Barr J.R., Bergquist J., Borchers C.H., Brandsma J., Breitkopf S.B., Cajka T., Cazenave-Gassiot A., Checa A., Cinel M.A., Colas R.A., Cremers S., Dennis E.A., Evans J.E., Fauland A., Fiehn O., Gardner M.S., Garrett T.J., Gotlinger K.H., Han J., Huang Y., Neo A.H., Hyotylainen T., Izumi Y., Jiang H., Jiang H., Jiang J., Kachman M., Kiyonami R., Klavins K., Klose C., Kofeler H.C., Kolmert J., Koal T., Koster G., Kuklenyik Z., Kurland I.J., Leadley M., Lin K., Maddipati K.R., McDougall D., Meikle P.J., Mellett N.A., Monnin C., Moseley M.A., Nandakumar R., Oresic M., Patterson R., Peake D., Pierce J.S., Post M., Postle A.D., Pugh R., Qiu Y., Quehenberger O., Ramrup P., Rees J., Rembiesa B., Reynaud D., Roth M.R., Sales S., Schuhmann K., Schwartzman M.L., Serhan C.N., Shevchenko A., Somerville S.E., John-Williams L.S., Surma M.A., Takeda H., Thakare R., Thompson J.W., Torta F., Triebl A., Troetzmueller M., Ubhayasekera S.J.K., Vuckovic D., Weir J.M., Welti R., Wenk M.R., Wheelock C.E., Yao L., Yuan M., Zhao X.H., Zhou S. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in Frozen Human Plasma. J. Lipid Res. 2017;58(12):2275–2288. doi: 10.1194/jlr.M079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu J.H.Y., Marklund M., Imamura F., Tintle N., Korat A.V.A., de Goede J., Zhou X., Yang W.S., Otto M.C.D., Kroger J., Qureshi W., Virtanen J.K., Bassett J.K., Frazier-Wood A.C., Lankinen M., Murphy R.A., Rajaobelina K., Del Gobbo L.C., Forouhi N.G., Luben R., Khaw K.T., Wareham N., Kalsbeek A., Veenstra J., Luo J.H., Hu F.B., Lin H.J., Siscovick D.S., Boeing H., Chen T.A., Steffen B., Steffen L.M., Hodge A., Eriksdottir G., Smith A.V., Gudnason V., Harris T.B., Brouwer I.A., Berr C., Helmer C., Samieri C., Laakso M., Tsai M.Y., Giles G.G., Nurmi T., Wagenknecht L., Schulze M.B., Lemaitre R.N., Chien K.L., Soedamah-Muthu S.S., Geleijnse J.M., Sun Q., Harris W.S., Lind L., Arnlov J., Riserus U., Micha R., Mozaffarian D., Cohorts H., Aging Res G. Omega-6 fatty acid biomarkers and incident type 2 diabetes: pooled analysis of individual-level data for 39 740 adults from 20 prospective cohort studies. Lancet Diabetes Endocrinol. 2017;5(12):965–974. doi: 10.1016/S2213-8587(17)30307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gorden D.L., Myers D.S., Ivanova P.T., Fahy E., Maurya M.R., Gupta S., Min J., Spann N.J., McDonald J.G., Kelly S.L., Duan J.J., Sullards M.C., Leiker T.J., Barkley R.M., Quehenberger O., Armando A.M., Milne S.B., Mathews T.P., Armstrong M.D., Li C.J., Melvin W.V., Clements R.H., Washington M.K., Mendonsa A.M., Witztum J.L., Guan Z.Q., Glass C.K., Murphy R.C., Dennis E.A., Merrill A.H., Russell D.W., Subramaniam S., Brown H.A. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 2015;56(3):722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hilvo M., Denkert C., Lehtinen L., Muller B., Brockmoller S., Seppanen-Laakso T., Budczies J., Bucher E., Yetukuri L., Castillo S., Berg E., Nygren H., Sysi-Aho M., Griffin J.L., Fiehn O., Loibl S., Richter-Ehrenstein C., Radke C., Hyotylainen T., Kallioniemi O., Iljin K., Oresic M. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Canc. Res. 2011;71(9):3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 131.Afshinnia F., Rajendiran T.M., Soni T., Byun J., Wernisch S., Sas K.M., Hawkins J., Bellovich K., Gipson D., Michailidis G., Pennathur S. Impaired -oxidation and altered complex lipid fatty acid partitioning with advancing CKD. J. Am. Soc. Nephrol. 2018;29(1):295–306. doi: 10.1681/ASN.2017030350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen H., Chen L., Liu D., Chen D.-Q., Vaziri N.D., Yu X.-Y., Zhang L., Su W., Bai X., Zhao Y.-Y. Combined clinical phenotype and lipidomic analysis reveals the impact of chronic kidney disease on lipid metabolism. J. Proteome Res. 2017;16(4):1566–1578. doi: 10.1021/acs.jproteome.6b00956. [DOI] [PubMed] [Google Scholar]
- 133.Rhee E.P., Souza A., Farrell L., Pollak M.R., Lewis G.D., Steele D.J.R., Thadhani R., Clish C.B., Greka A., Gerszten R.E. Metabolite profiling identifies markers of uremia. J. Am. Soc. Nephrol. 2010;21(6):1041–1051. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gale T.V., Horton T.M., Grant D.S., Garry R.F. Metabolomics analyses identify platelet activating factors and heme breakdown products as Lassa fever biomarkers. PLoS Neglected Trop. Dis. 2017;11(9) doi: 10.1371/journal.pntd.0005943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jove M., Mauri-Capdevila G., Suarez I., Cambray S., Sanahuja J., Quilez A., Farre J., Benabdelhak I., Pamplona R., Portero-Otin M., Purroy F. Metabolomics predicts stroke recurrence after transient ischemic attack. Neurology. 2015;84(1):36–45. doi: 10.1212/WNL.0000000000001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li J.N., Xie H.Y., Li A., Cheng J.L., Yang K., Wang J.T., Wang W.J., Zhang F., Li Z.Z., Dhillon H.S., Openkova M.S., Zhou X.H., Li K., Hou Y. Distinct plasma lipids profiles of recurrent ovarian cancer by liquid chromatography-mass spectrometry. Oncotarget. 2017;8(29):46834–46845. doi: 10.18632/oncotarget.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]