Abstract
Cell shape is regulated by cell adhesion and cytoskeletal and membrane dynamics. Cell shape, adhesion, and motility have a complex relationship and understanding them is important in understanding developmental patterning and embryogenesis. Here we show that the lipid kinase phosphatidylinositol 4-kinase III beta (PI4KIIIβ) regulates cell shape, migration, and focal adhesion (FA) number. PI4KIIIβ generates phosphatidylinositol 4-phosphate (PI4P) from phosphatidylinositol and is highly expressed in a subset of human breast cancers. PI4KIIIβ and the PI4P it generates regulate a variety of cellular functions, ranging from control of Golgi structure, fly fertility, and Akt signaling. Here, we show that loss of PI4KIIIβ expression decreases cell migration and alters cell shape in NIH3T3 fibroblasts. The changes are accompanied by an increase in the number of FA in cells lacking PI4KIIIβ. Furthermore, we find that PI4P-containing vesicles move to the migratory leading edge during migration and that some of these vesicles tether to and fuse with FA. Fusion is associated with FA disassembly. This suggests a novel regulatory role for PI4KIIIβ and PI4P in cell adhesion and cell shape maintenance.
INTRODUCTION
A culture of genetically identical NIH3T3 fibroblasts displays a striking visual diversity. A single microscopic field of view is populated with elongated cells, round cells, and yet others with extraordinarily complex geometries. Furthermore, some NIH3T3 cells are solitary while others cluster themselves into multicellular groups. Last, some fibroblasts are stationary while others are motile. These moving cells display marked differences in speed and directionality. This fascinating architectural and behavioral diversity is at the root of processes such as organismal development and patterning.
Cell shape has important implications in cell function (Bellas and Chen, 2014; Gilbert and Weaver, 2017). For example, the spreading of a cell in two-dimensional culture regulates both sensitivity to apoptosis and proliferative capacity (Chen et al., 1997). The spreading and shape of a cell results from a complex and dynamic interaction among its cytoskeleton, the plasma membrane, and the extracellular matrix (ECM) (Mogilner and Keren, 2009). In fish keratinocytes, which assume a hemispherical appearance in two-dimensional culture, cell shape is determined by a balance between the protrusive force of actin polymerization and tension in the plasma membrane (Keren et al., 2008). In other cells, cell shape is regulated by lipid constituents of the plasma membrane (Wen et al., 2018) and by membrane-binding proteins that induce curvature (Nishimura et al., 2018). In migratory fibroblasts, which assume a wedge-shaped form, cell shape is additionally regulated by the presence and strength of adhesive structures at the cell periphery (Satulovsky et al., 2008). These adhesive structures provide sites for attachment to the ECM and the sensing of its mechanical properties. Cellular adhesions also serve as sites for the generation of contractile forces (Bosch-Fortea and Martin-Belmonte, 2018).
Perhaps the most important of mammalian adhesive structures are focal adhesions (FAs) (Burridge, 2017). FA are oval-shaped, multi-protein complexes, generally 2 μm wide and 3–10 μm long, coupling the actin cytoskeleton to the ECM via transmembrane integrin heterodimeric receptors (Parsons et al., 2010). During cell migration, FAs are created from smaller integrin-containing structures described as either focal complexes or nascent adhesions (Parsons et al., 2010). In a mature FA, the cytoplasmic tails of the integrin molecules are linked to actin filaments via adapter proteins such as talin, paxillin, zyxin, and vinculin (Kanchanawong et al., 2010). FA are sites of mechano-transduction and activate a diverse array of signal transduction pathways (Geiger et al., 2009). FA number and size are critical in the maintenance of migratory capacity (Kim and Wirtz, 2013a), cell spreading (Kim and Wirtz, 2013b), and cell shape (Chen et al., 2003; Mogilner and Keren, 2009).
In this report, we find that the lipid kinase phosphatidylinositol 4-kinase III beta (PI4KIIIβ) has an important role in regulating cell shape, migration, and FA number. A Golgi-resident enzyme, PI4KIIIβ is one of four mammalian proteins that generate phosphatidylinositol 4-phosphate (PI4P) from PI (Balla, 1998, 2013). PI4 kinases have been implicated in several human cancers (Waugh, 2012). PI4KIIIβ is a likely human oncogene based on its high expression in a subset of human breast tumors and its ability to co-operate with the Rab11a GTPase to activate signaling through Akt (Morrow et al., 2014). PI4KIIIβ and its homologues have multiple physiological roles, ranging from controlling Golgi structure in yeast to regulating male fertility and development in Drosophila (Godi et al., 1999; Polevoy et al., 2009). PI4KIIIβ also regulates in vitro morphogenesis of human breast cells (Pinke and Lee, 2011).
RESULTS
PI4P vesicles move to the migratory leading edge
To explore a role for PI4P in cell migration, we used two PI4P biosensors, GFP-FAPP1 and GFP-P4M, to study intracellular PI4P localization during NIH3T3 migration. The GFP-FAPP1 biosensor consists of GFP conjugated to the Pleckstrin Homology domain of FAPP1 (four-phosphate-adaptor protein 1) (Balla, 2007). The GFP-P4M biosensor consists of GFP conjugated to a single P4M domain containing residues 546–647 of the Legionella pneumophila SidM protein (Hammond et al., 2014). Both biosensors bind to PI4P directly. However, FAPP1 binds to PI4P only when complexed to the Arf-1 GTPase (Balla et al., 2005).
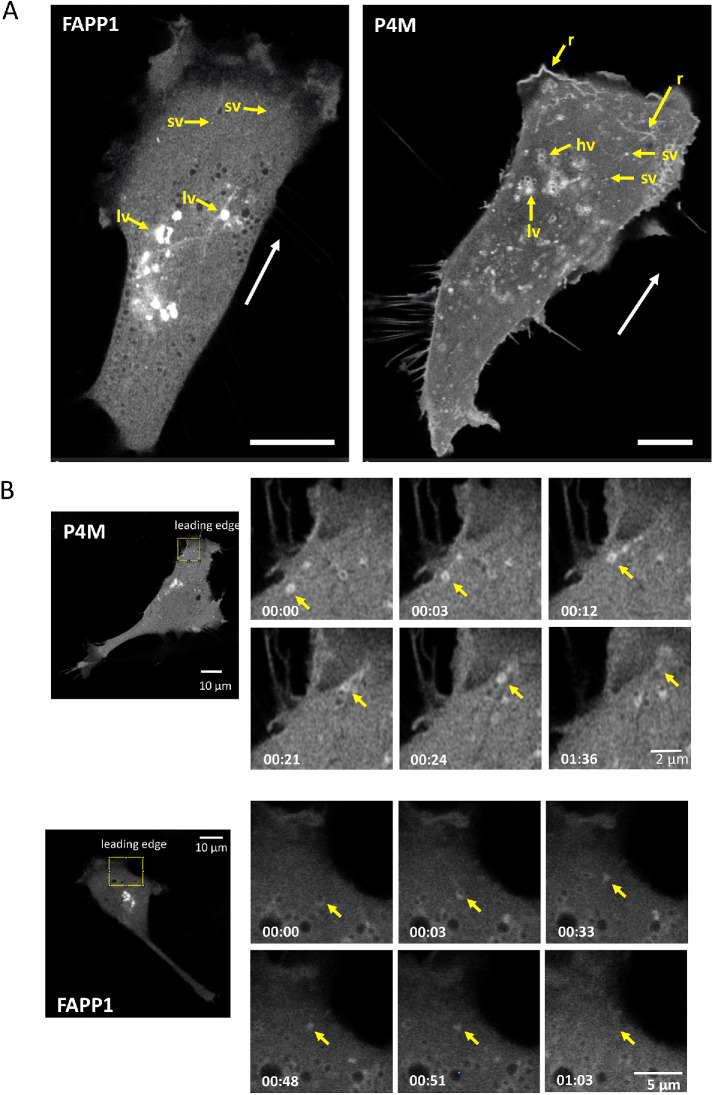
In migrating NIH3T3 cells, FAPP1 shows a broadly perinuclear distribution consistent with Golgi localization (Figure 1A; Supplemental Video S1). Large (0.8–1.6 µm diameter) and small vesicles (sv) (0.2–0.45 µm diameter) are visible. The large vesicles (lv) are predominantly perinuclear while the sv localize to the cell periphery. P4M shows a somewhat similar localization pattern with many lv in the perinuclear region. Different from FAPP1, many of these perinuclear vesicles appear hollow (hv) (Figure 1A; Supplemental Video S2). As is the case with FAPP1, there are sv in the cell periphery but these seem to be much more numerous in P4M-transfected cells. In direct contrast to FAPP1 and as observed previously in COS7 cells (Hammond et al., 2014), the P4M reporter localizes to plasma membrane structures. Some plasma membrane features at the migratory leading edge features are reminiscent of ruffles (r), actin-rich structures that move away from the leading edge and derive from poor lamellar adhesion to the growth substrate (Borm et al., 2005). The difference between FAPP1 and P4M staining presumably reflects the requirement that FAPP1 interact with Arf–1-associated PI4P pools.
FIGURE 1:
PI4P-containing vesicles move to and fuse with the migratory leading edge. (A) PI4P-contining vesicles are visualized in NIH3T3 fibroblasts undergoing migration (direction indicated by white arrow) using FAPP1 and P4M reporters. In FAPP1 cells, large and small vesicles, labeled lv and sv, respectively, are seen. In P4M cells, hv and linear structures reminiscent of ruffles can also be seen. Scale bar is 10 μm. (B) PI4P-contining vesicles in motile cells move to and fuse with the leading edge. An individual vesicle is identified by the yellow arrow. Elapsed time and scale bars are indicated.
Video S1.
Migrating NIH3T3 fibroblast transfected with FAPP1 to visualize PI4P vesicles.
Video S2.
Migrating NIH3T3 fibroblast transfected with P4M to visualize PI4P vesicles.
During migration, the lv of both P4M and FAPP1 show only limited intracellular movement and remain generally perinuclear (Supplemental Videos S1 and S2). In contrast, the peripheral sv show a high degree of mobility (Supplemental Videos S1 and S2). In addition, we observe multiple P4M and FAPPI sv moving to the migratory leading edge and then disappearing (Figure 1B; Supplemental Videos S3 and S4). This is consistent with vesicular delivery of PI4P-containing vesicles, both Arf-1 associated and Arf-1 free forms, to the plasma membrane. The absence of FAPP1 staining at the plasma membrane suggests that once the vesicle delivers the PI4P/Arf-1 cargo, either Arf-1 dissociates or PI4P is converted by PI4P kinases to another PI. In the case of P4M, the persistence of PI4P in some parts of the leading edge indicate that some of the membrane delivered PI4P remains as PI4P. In addition to their movement to the plasma membrane, many small P4M and FAPP1 vesicles show considerable directional freedom. Both retrograde and anterograde movements are observed (Supplemental Videos S3 and S4). However, the movement of sv is generally toward the migratory leading edge.
Video S3.
Migrating NIH3T3 fibroblast transfected with P4M to visualize delivery and fusion of PI4P vesicles with the plasma membrane during migration.
Video S4.
Migrating NIH3T3 fibroblast transfected with FAPP1 to visualize delivery and fusion of PI4P vesicles with the plasma membrane during migration.
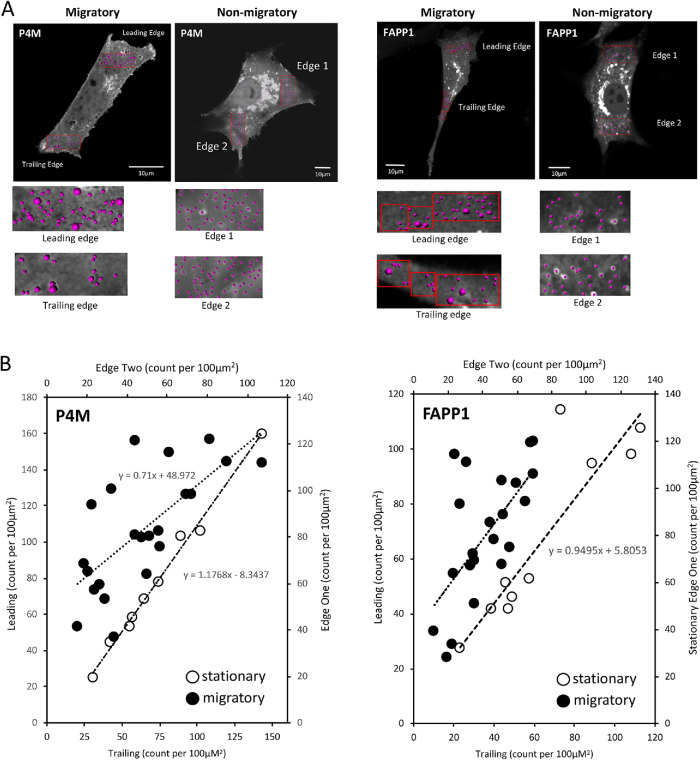
The movement of vesicles toward the migratory leading edge is mirrored in the visibly unequal intracellular distribution of both FAPP1 and P4M vesicles in the migratory leading edge compared with the trailing edge. To further explore this, we used automated image analysis to quantify the distribution of PI4P vesicles in migratory and nonmigratory cells (Supplemental Videos S5–S8 show representative cells). In migratory cells, we counted vesicles in both the leading and trailing edges. In stationary cells, we counted vesicles present in two opposing edges of the cell. In stationary cells there are a generally equal number of P4M and FAPP1 marked PI4P vesicles at two opposite edges (Figure 2B, open circles; Supplemental Videos S6 and S8). On the other hand, migratory cells show a greater number of vesicles at their leading edge compared with the trailing edge (Figure 2B, closed circles; Supplemental Videos S5 and S7). This is consistent with the idea that the transport of PI4P to the migratory leading edge is a part of the directional cell migration machinery.
FIGURE 2:
PI4P containing vesicles have polarized distribution in migrating cells. (A) PI4P-containing vesicles are visualized in NIH3T3 fibroblasts that are either migratory (leading and trailing edges indicated) or nonmigratory. Vesicles are counted by volume rendering in Imaris. (B) The number of PI4P vesicles, either P4M or FAPP1 labeled, at the leading edge is plotted as a function of the vesicles at the trailing edge in migratory cells (•). For nonmigratory cells, the number of vesicles is plotted for two opposite edges (o). The line of best fit is shown.
Video S5.
Migrating NIH3T3 fibroblast transfected with P4M to visualize the distribution of PI4P vesicles in the cytoplasm.
Video S6.
Non-migrating NIH3T3 fibroblast transfected with P4M to visualize the distribution of PI4P vesicles in the cytoplasm.
Video S7.
Migrating NIH3T3 fibroblast transfected with FAPP1 to visualize the distribution of PI4P vesicles in the cytoplasm.
Video S8.
Non-migrating NIH3T3 fibroblast transfected with FAPP1 to visualize the distribution of PI4P vesicles in the cytoplasm.
PI4KIIIβ deletion decreases cell migration
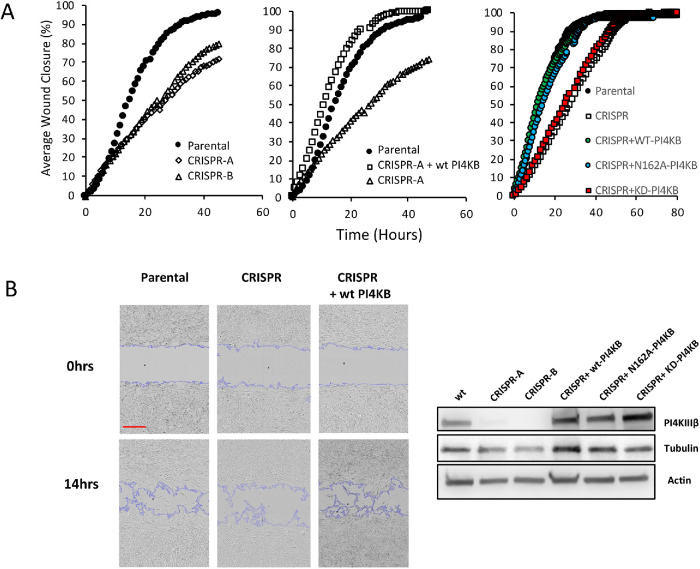
To explore a role for PI4P in cell migration, we used CRISPR to delete the PI4KIIIβ gene in mouse NIH3T3 fibroblasts. PI4KIIIβ generates PI4P in the Golgi (Balla and Balla, 2006; Balla, 2013). We then tested two independent PI4KIIIβ null lines for migratory capacity in wound healing assays. As shown in Figure 3A (left panel), the two CRISPR lines close a wound much slower that parental cells. Wild-type (WT) NIH3T3 cells close half the wound in ∼18 h, while the null lines take ∼25 h. This defect in wound closure is rescued by re-expression of WT PI4KIIIβ (Figure 3A, middle panel).
FIGURE 3:
Loss of PI4KIIIβ impairs migration. (A) (Left panel) The ability to close an in vitro wound is shown in two independent lines of PI4KIIIβ-deleted cells compared with parental cells. (Center panel) Attenuated wound healing is rescued in the CRISPR line by re-expression of WT PI4KIIIβ (wtPI4KB). (Right panel) Attenuated wound healing is rescued in the CRISPR line by re-expression of wild-type PI4KIIIβ (WT-PI4KB) and KD (KD-PI4KB) but not a Rab11-binding mutant (N162A-PI4KB). Experiments are representative of triplicates. (B) (Left panels) Inset shows a representative image from the wound closure experiment in the center panel of A. Scale bar in red is 300 μm. (Right panel). Protein expression of PI4KIIIβ and tubulin and actin control in the cell lines.
The best characterized function of PI4KIIIβ is the generation of PI4P from PI (Balla, 1998). However, PI4KIIIβ also interacts with the Rab11a GTPase and is part of the Rab11a-dependent pathway controlling endosome function (de Graaf et al., 2004; Polevoy et al., 2009; Burke et al., 2014). To determine which of these functions were involved in migration regulation, we expressed wt-PI4KIIIβ, kinase dead PI4KIIIβ (KD-PI4KIIB) (Zhao et al., 2000) or a PI4KIIIβ mutant that does not interact with Rab11a (N162A) (Burke et al., 2014) in a CRISPR line. These lines were then tested for migratory capacity. As shown in Figure 3A (right panel), a CRIPSR line expressing wt-PI4KIIIβ or the Rab11a-binding mutant N162A had similar closure times to those of parental fibroblasts. On the other hand, KD-PI4KIIIβ was unable to rescue would closure kinetics. This indicates that the ability to generate PI4P, rather than Rab11a interaction, is required PI4KIIIβ-mediated control of migration. Figure 3B shows protein expression of the PI4KIIIβ-rescued cell lines.
PI4KIIIβ regulates stress fiber appearance
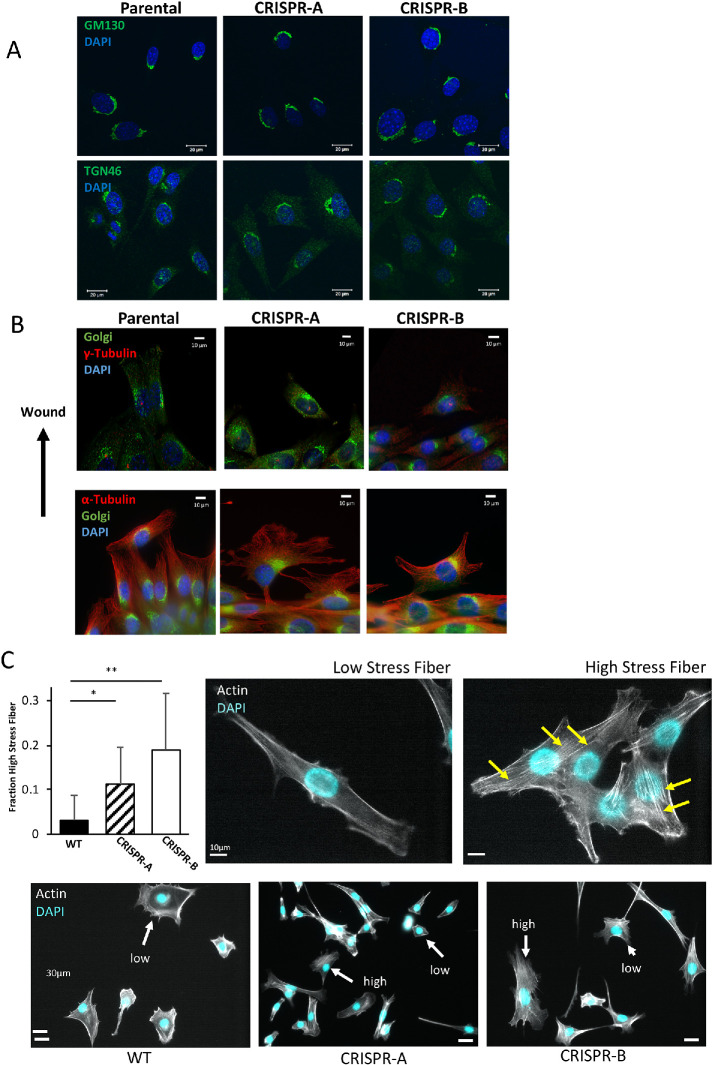
In yeast, the PI4KIIIβ homologue Pik1 is required for maintaining Golgi structure (Walch-Solimena and Novick, 1999; de Graaf et al., 2004). Because cell migration involves repositioning of the Golgi between the nucleus and the leading edge (Kupfer et al., 1982) and functional alterations of the Golgi affect migration (Xing et al., 2016; Ahat et al., 2019), we next investigated whether or not PI4KIIIβ depletion affected Golgi appearance. When stained for markers of either the cis- or the trans-Golgi (GM130 and TGN46, respectively), NIH3T3 cells lacking PI4KIIIβ have Golgi that are visibly indistinguishable from WT cells (Figure 4A). Moreover, in scratch wound assays, PI4KIIIβ depletion does not visibly affect Golgi positioning to face the wound (Figure 4B). Microtubule structure also appears normal as does centrosome alignment away from the wound (Figure 4B). One the other hand, PI4KIIIβ depletion causes an increase in the number of cells with numerous stress fibers (Figure 4C). In a population of normal NIH3T3 cells, most cells show few stress fibers with only 3.1 ± 5% of the population having more than six fibers per cells. In contrast, the PI4KIIIβ-depleted lines have a much larger fraction (11.2 ± 8% and 18.9 ± 13%) in their population with high stress fiber content.
FIGURE 4:
Loss of PI4KIIIβ alters stress fiber appearance. (A) Cis- and trans-Golgi structure as visualized by GM130 and TGN46 staining, respectively, in WT NIH3T3 cells and two independent lines of PI4KIIIβ-deleted cells. (B) Golgi, centrosome (γ-tubulin) and microtubule (α-tubulin) structure visualized in cells entering a scratch wound. (C) CRISPR cell lines both show significantly (t test, p < 0.0001) more cells with high numbers of stress fibers compared with WT cells. Representative cells with respectively low or high numbers of stress fibers (yellow arrows) are shown in the right panels. Bottom panels show representative fields of WT and CRISPR lines showing cells with high or low stress fiber content.
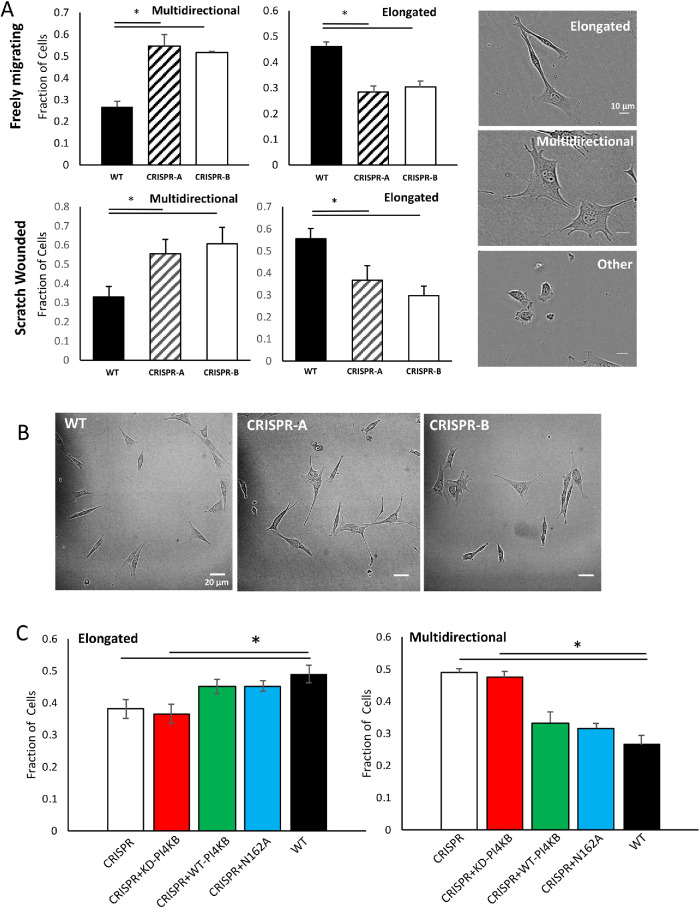
PI4KIIIβ regulates cell shape
In our studies of the cell cytoskeleton and cell migration, we noticed that cultures of the CRIPSR-deleted NIH3T3 cells had a very different morphological appearance under phase than either parental or rescued cells. In our experience, most cultured NIH3T3 cells assume one of three broad shapes. The first is an elongated form (Figure 5A) and approximately half of WT NIH3T3 cells assume this shape (Figure 5A, right panel). The second most common shape is what we term “multidirectional.” These cells are roughly rectangular in shape with multiple pseudopodial protrusions. Approximately 25% of WT cells are of this type (Figure 5A, right panel). The remaining cells, which have a smaller, generally spherical appearance, we classified as “other.” The loss of PI4KIIIβ leads to a redistribution of cell shapes in both of the CRISPR lines. In freely migrating conditions, the number of elongated cells in the PI4KIIIβ-deleted cells decreases by almost 50% and the number of multidirectional cells more than doubles (Figure 5A, top panel). Similarly, cells present in the wound of a scratch migration assay show an increase in the number of multidirectional cells and a decrease in elongated ones (Figure 5A, bottom panel). Representative fields are shown in Figure 5B. As is the case with the wound healing assay, wt-PI4KIIB and the Rab11-binding mutant (N162A) were able to restore WT shape distribution to the CRISPR lines, while the KD-PI4KIIIβ did not (Figure 5C). Parental cells and CRISPR lines rescued with either WT PI4KIIB or N162A had 45–50% of cells as elongated, while the KD-rescued cells had nearly 35% as multidirectional, similar to the original CRSPR line. Similarly, parental cells and CRISPR lines rescued with either WT PI4KIIIβ or N162A had 25–35% of cells as multidirectional, while the CRISPR- and KD-rescued cells had nearly 50% as multidirectional. Thus, cell shape control by PI4KIIIβ, like wound healing, is dependent on PI4P generation rather than Rab11a interaction.
FIGURE 5:
Loss of PI4KIIIβ alters cell shape distribution. (A) Two independent lines of PI4KIIIβ-deleted cells have different population shape distributions (depicted in the right panels) in both freely migrating conditions and in the wound of a scratch. The fraction of elongated cells in either line of CRISPR cells is significantly (p < 0.001, t test) lower than in a population of WT cells. Similarly, the fraction of elongated cells in either line of CRISPR cells is significantly (p < 0.001, t test) higher than in a population of WT cells. Results are the mean and SD of triplicate independent measurements of at least 200 cells each from a minimum of 20 randomly selected fields (B) Representative fields of each cell type. Scale bar is 20 μm. (C) Re-expression of either WT- or N162A-PI4KIIIβ restores elongated and multidirectional shape distribution to the CRISPR cells. Expression of KD-PI4KIIIβ does not rescue cell shape changes and their elongated of multidirectional composition is similar to the CRSPR control and significantly different from WT cells (p < 0.05, t test). Results are the mean and SD of quadruplicate independent measurements of at least 200 cells from at least 20 randomly selected fields (D) Distance traveled over time was monitored for 50 WT NIH3T3 cells with either elongated or multidirectional morphologies. Tracks for 10 cells of each type are shown. Multidirectional cells have a significantly reduced average angular rotation that elongated cells (p < 0.05, t test). (E) Quantification of migration patterns in WT cells with either multidirectional or elongated morphologies. Mean angular rotation or velocity is presented as the mean and SD of 50 cells collected over at least 30 min. Angular rotation is significantly lower in the multidirectional population compared with the elongated one (p < 0.05, t test). Comparison between Angular Rotation and Velocity between WT and CRISPR lines. Mean angular rotation or velocity is presented as the mean and SD of 50 cells collected over at least 30 min.
We hypothesized that this change in population cell shape was related to migratory defects in the CRISPR knockout. As shown in Figure 5D, elongated WT NIH3T3 cells exhibit different migratory behavior that the multidirectional ones. Multidirectional cells tend to make sharp and frequent turns, while elongated cells oscillate back and forth only a more or less straight line (Figure 5D). This means that although their migratory velocity is similar (Figure 5D), elongated cells travel a further distance from their origin than their multidirectional counterparts (Figure 5D). The behavior of individual CRIPSR cells, in either elongated or multidirectional shape classes, is similar to their WT counterparts in that they have similar turning behaviors and recorded velocities (Figure 5E). This suggests that the loss of PI4KIIIβ is not affecting the migratory machinery per se. Rather, it is affecting cell shape, which changes the migratory behavior of the cell population.
PI4KIIIβ regulates focal adhesions
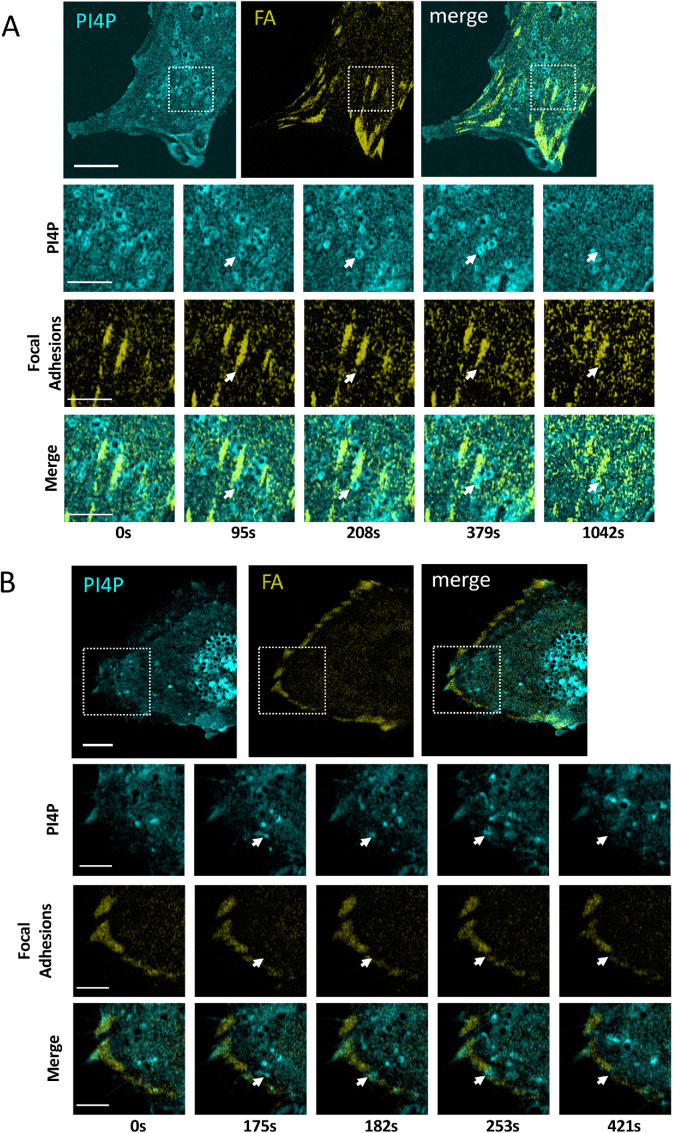
To further explore the regulation of cell shape and migration by PI4KIIIβ, we investigated what cellular structures might be interacting with PI4P-containing vesicles. We reasoned that FA might be one such structure since FA-dependent adhesion is likely to be involved in cell shape, migration control, and stress fiber formation (Chen et al., 2003; Kim and Wirtz, 2013a). To investigate possible FA and PI4P interaction during cell migration, we simultaneously imaged them in migrating cells using fluorescent Talin and P4M reporters. At the migratory leading edge (Figure 6A; Supplemental Video S9), PI4P vesicles can be observed moving to FA. Vesicles dock with FA and then disappear, presumably delivering their cargo to FA and the PI4P becoming metabolized. Similar behavior occurs at FA of the trailing edge (Figure 6B; Supplemental Video S10).
FIGURE 6:
PI4P Vesicles move to and fuse with FAs. (A) Visualization of PI4P and FAs in the leading edge of a migrating NIH3T3 cells. The time series indicates a single vesicle observed from the boxed inset. The fusing vesicle (white arrow) appears at 95 s. Scale bar in the large image is 10 μm and is 3 μm in the enlarged images. (B) Visualization of PI4P and FA in the trailing edge of a migrating NIH3T3 cells. The time series indicates a single vesicle seen from the boxed inset. The fusing vesicle (white arrow) appears at 175s. Scale bar in the large image is 10 μm and is 3 μm in the enlarged images. (C) BLLSM imaging of PI4P vesicles (green P4M reporter) fusing with FA (red Vinculin reporter) in a migrating cell. Histogram shows that the majority of fusions events (n = 50) cause FA destruction within 3–6 s. The bottom panels show enlarged fields of the top panel where two fusion events, one causing FA destruction and the other not, can be seen.
Video S9.
Migrating NIH3T3 fibroblast transfected with GFP-P4M and mApple-Talin to simultaneously visualize FAand PI4P vesicles. Video shows the migratory leading edge.
Video S10.
Migrating NIH3T3 fibroblast transfected with GFP-P4M and mApple-Talin to simultaneously visualize FA and PI4P vesicles. Video shows the migratory trailing edge.
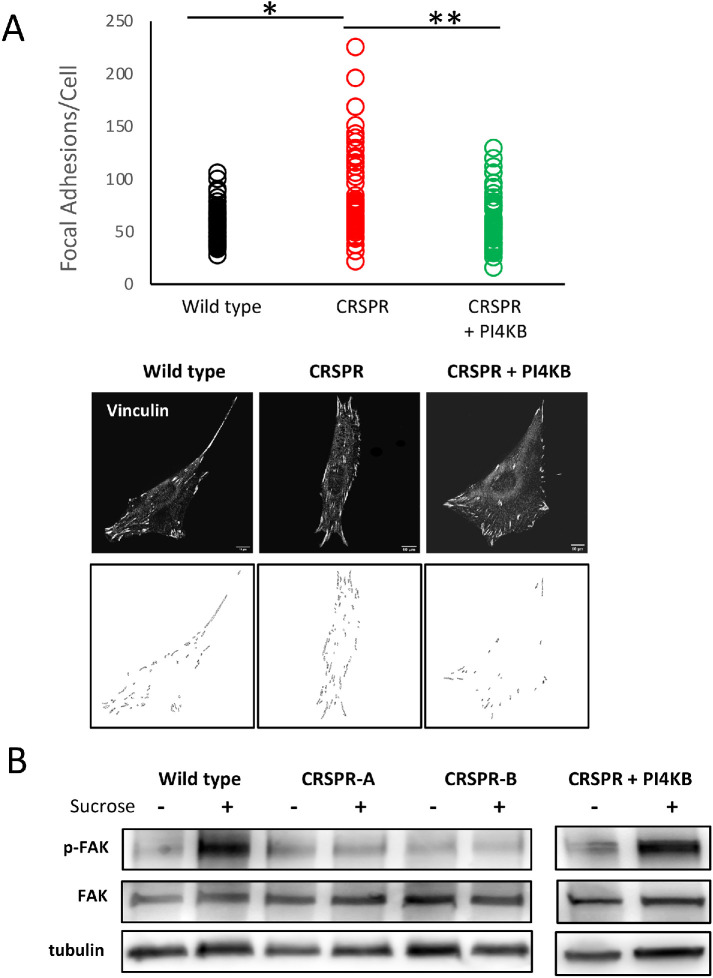
To further explore delivery of PI4P vesicles to FA, we used a Bessel Lattice Light Sheet microscope (BLLSM) to image migrating NIH3T3 cells. The BLLSM illuminates a sample using a thin structured illumination beam and allows for long-term imaging with high axial and temporal resolution and with minimal photobleaching (Planchon et al., 2011). Using the BLLSM, we visualized 50 PI4P vesicle-FA fusion events in five independent migrating WT NIH3T3 cells at 3 s/vol. Cells were observed for ∼25 min each. As shown in Figure 6C, approximate two-thirds of fusion events were associated with disappearance of the FA within 3–6 s. This suggests that PI4P or cargo in PI4P vesicles is involved in FA disassembly. To test this idea, we used an automated image analysis program to count FA in WT and CRISPR-deleted NIH3T3 cells (Horzum et al., 2014). As shown in Figure 7A, WT NIH3T3 cells (n = 60) had an average of 56.7 ± 17 FA per cell. CRISPR-deleted cells (n = 59), on the other hand, had significantly (t test, p < 1 × 10–5) more FA, averaging 84.6 ± 42 per cell. Rescue of the CRISPR line by PI4KIIIβ re-expression returned the FA number to WT levels (58.4 ± 24). Thus, the number of FA/cell is dependent, at least in part, on PI4KIIIβ.
FIGURE 7:
Loss of PI4KIIIβ Increased the number of focal adhesions per cell. (A) The number of FA per cell is shown for WT cells (n = 60), a CRSPR-deleted line (n = 59), and a line rescued with WT-PI4KIIIβ (n = 60). The number of FA in the CRPSR line is significantly greater than in either the WT line of the one expressing WT PI4KIIIβ (respectively, p < 1 × 10–5, p < 2 × 10–7, t test). Bottom images show representative images of FA staining and image segmentation used to count FA for each line. (B) Blot showing that CRISPR cell lines have a defect in p-Y397 FAK phosphorylation in response to 400 mM sucrose. This defect is rescued by wt-PI4KB expression.
To assay FA signaling capacity in the CRISPR lines, we subjected cells to hyperosmotic stress by culturing them in 400 mM sucrose. Hyperosmotic stress causes cell shrinkage and activates FA kinase (FAK) autophosphorylation at tyrosine 397 (Lunn and Rozengurt, 2004) and Src-mediated phosphorylation of multiple FAK residues (Calalb et al., 1996; Westhoff et al., 2004). FAK hyperphosphorylation is part of Src-mediated FA disassembly (Calalb et al., 1996; Westhoff et al., 2004). As shown in Figure 7B, parental cells have strong FAK phosphorylation (Y-861) in response to sucrose treatment but PI4KIIIβ-deleted cells do not. This reduced phosphorylation is rescued by expression of wt-PI4KIIIβ. Thus, loss of PI4KIIIβ allows for more FA per cell but these have impaired signaling consistent with attenuated disassembly.
DISCUSSION
The central findings of this paper are that the PI4KIIIβ is an important regulator of cell shape, cell migration, and FA disassembly. The best known function of PI4KIIIβ is the generation of PI4P from PI (Balla and Balla, 2006; Balla, 2013). In addition, PI4KIIIβ binds to the Rab11a GTPase in a kinase-independent manner (de Graaf et al., 2004; Polevoy et al., 2009; Burke et al., 2014) and is involved in Rab11a-dependent control of Akt signaling (Jeganathan et al., 2008; Morrow et al., 2014) and endosome function (de Graaf et al., 2004; Polevoy et al., 2009). In our case, a catalytically inactive version of PI4KIIIβ rescues neither the reduced migration nor the cell shape defects in PI4KIIIβ-deleted cells. On the other hand, a PI4KIIIβ mutant that has kinase activity but does not bind the Rab11a GTPase (Burke et al., 2014) does rescue motility and cell shape distribution to a level equivalent to WT. This indicates that control of cell migration and shape is independent of Rab11a interaction but requires PI4P generation.
Consistent with an importance for PI4P in migration, our live-cell imaging shows directed movement and fusion of PI4P-containing vesicles to the migratory leading edge and to FA. There are also more PI4P vesicles between the nucleus and the leading edge than between the nucleus and the trailing edge. These vesicles are likely to derive from the Golgi and therefore could be produced by both PI4KIIIβ and PI4KIIβ (Balla, 1998) in the Golgi. PI4P is a precursor for two phosphoinositides, PI(4,5)P2 and PI(3,4,5)P3, that have a well-documented role in cell migration control (Schink et al., 2016). Some of the delivered PI4P could be converted to PI(4,5)P2 and PI(3,4,5)P3 at the leading edge. While PI4P is in large intracellular molar excess over PI(4,5)P2 and PI(3,4,5)P3, localized pools of PI(4,5)P2 in the plasma membrane that regulate ion transport are dependent on an influx of Golgi-derived PI4P (Dickson et al., 2014). We propose that pools of PI(4,5)P2 and PI(3,4,5)P3 regulating cell migration could similarly be dependent on PI4P transport from the Golgi.
Our observation that PI4KIIIβ deletion alone produces defects in cell migration suggests that the PI4P produced by PI4KIIIβ is an important part of cell migration that is not duplicated by PI4KIIβ or other PI4P-generating enzymes. Silencing PI4KIIIβ in a breast tumor cell line has previously been shown to attenuate cell migration (Tokuda et al., 2014). Here we describe a similar role for PI4KIIIβ in the migration of a noncancer line. We believe that migratory deficiency in PI4KIIIβ-deleted cells is directly related to the changes of cell shape we observe. The majority of WT NIH3T3 cells display an extended and elongated morphology in culture. On the other hand, a population of PI4KIIIβ-deleted cells show fewer elongated cells and more with multiple pseudopodia. We term the multi-pseudopod phenotype “multidirectional” because the cells attempt to move in several directions at once. Elongated cells, on the other hand, generally move along a single axis. The net consequence of the two different types of motion in the different cells is that the multidirectional cells cover smaller distances than elongated ones because they are constantly making small angle turns. Thus, in a wound healing assay, a population of elongated cells will close the wound faster than multidirectional ones. It is important to note that, as a group, both elongated and multidirectional PI4KIIIβ-deleted cells have the same migratory parameters as their respective elongated and multidirectional counterparts in WT cells. They travel at the same velocity and have the same turning behaviors. We propose that deletion of PI4KIIIβ is not impairing the intrinsic migration machinery per se but rather is altering the relative balance of fast-moving and slow-moving cells in the overall population.
The increase in FA number we observe in PI4KIIIβ-deleted cells indicates that specific PI4KIIIβ cargo is necessary for normal FA disassembly. FA link the actin cytoskeleton to the ECM. During cell migration, FA are created immediately behind the migratory leading edge and are disassembled at later stages of migration (Parsons et al., 2010; Burridge, 2017). Our live-cell imaging indicates that PI4P-containing vesicles move to and fuse with FA at both the leading and the trailing edges. The majority of these fusion events are associated with rapid FA disassembly and we favor a model where PI4KIIIβ regulates FA number by directing the delivery of cargo that initiates FA disassembly. Further consistent with the idea that FA disassembly involves a PI4P-dependent process is the impaired FAK tyrosine phosphorylation in PI4KIIIβ-deleted cells. Hyperphosphorylation of FAK by Src is associated with movement of FAK away from FA and a decrease in FA number (Westhoff et al., 2004; Hamadi et al., 2010). Possibly, PI4P-containing vesicles could facilitate the movement of Src to FAK on FA to potentiate adhesion disassembly.
Another well-characterized process of FA disassembly is the endocytic destruction of a FA via microtubule-mediated delivery of dynamin (Ezratty et al., 2005, 2009). This activates FA destruction through clathrin-mediated endocytosis. We suggest that delivery of endocytic machinery to sites of FA may be dependent on PI4P-mediated transport. Alternatively, the transport of proteases such as calpain or MMP1 that destroy FA components (Franco et al., 2004; Takino et al., 2006) could be transported in or on PI4P vesicles.
Another possibility is that PI4P itself, rather than PI4P-dependent cargo, is responsible for FA destruction. Consistent with this idea, depletion of either PI4KIIIβ or IQSec1 results in similar motility impairment and FA disassembly attenuation (D’Souza et al., 2020). D’Souza et al. (2020) propose a model where IQSec1, on activation by the Ca2+ channel Orai1, induces FA disassembly by activating the Arf5 GTPase through the ORP3 lipid exchanger. ORP3 exchanges plasma membrane PI4P for phosphatidyl choline in the ER. Thus, the delivery of PI4P itself could mediate FA destruction through the ORP3/IQSec1 complex.
We favor the idea that the increase in FA number by the loss of PI4KIIIβ is directly responsible for changes in cell shape and migration. The change in FA number is likely responsible for the increase in stress fiber appearance (Figure 4C). Stress fibers are long bundles of actin polymers that are typically linked to FA at one or both ends (Burridge and Guilluy, 2016). FA regulate migration because they mediate connection to the ECM and intracellular contractile forces (Parsons et al., 2010; Kim and Wirtz, 2013a). While some adhesion is necessary for cell migration, too many FA could impair migration because of excessive adhesion. FA have a reciprocal relationship with cell shape in that the positioning of adhesions determines cell shape but altering the cell shape itself modifies FA location in a cell (Chen et al., 2003; Lehnert et al., 2004). However, it is a possibility that the changes in cell shape, migration, and FA number we observe are regulated by independent PI4KIIIβ and PI4P-dependent pathways.
In summary, we have identified an import role for PI4KIIIβ in cell shape, migration, and adhesion. PI4KIIIΒ is likely to be a human oncogene (Waugh, 2012; Morrow et al., 2014) and we propose that its role in cancer is related to its regulation of these processes. In the future it will be important to identify the cargo involved.
MATERIALS AND METHODS
Cell lines and culture
The NIH3T3 fibroblast cell line was obtained from ATCC (Manassas, VA). NIH3T3 cells were cultured in DMEM (Sigma Aldrich) supplemented with 10% fetal bovine serum (FBS) (Thermo Scientific), 1 mM sodium pyruvate (Thermo Scientific), and 1 mM penicillin and streptomycin (Thermo Scientific). Cells are passaged following treatment with 0.25% trypsin protease (GE Healthcare Life Sciences) and counted using TC20 cell counter (Bio-Rad). The PI4KIIIβ-targeted RNA sequences for CRISPR deletion of PI4KIIIβ exons 4-5 were 5′-CAGACCGTGTACTCCGAATT-3′, 5′-GGCTCCCTACCTGATCTACG-3′, 5′-ATAAGCTCCCTGCCCGAGTC-3′ (Santa Cruz sc-430739).
Wound healing and cell tracking
For wound healing, cells were seeded at a density of 1 × 105 cells/ml in a 24-well ImageLock (EssenBio) plate containing a culture 2-insert well (ibidi) and incubated at 37°C at 5% CO2; 24 h later, the insert was removed. Images were acquired at a magnification of 10× every 30 min using the IncuCyte ZOOM Scratch Wound tool (EssenBio). For cell tracking, cells were seeded at 5000 cells/ml in a 24-well ImageLock plate (EssenBio) and images were acquired at a magnification of 10 min using the IncuCyte ZOOM (EssenBio). Cells were individually tracked and analyzed in ImageJ.
Western blot
Cells were lysed in radioimmunoprecipitation assay buffer (Tris-HCl, pH 7.4, 50 mM; NaCl, 150 mM; NP-40 1%; sodium deoxycholate, 0.5%; sodium dodecyl sulfate, 0.1%; EDTA, 2 mM; sodium fluoride, 50 mM) supplemented with protease and phosphatase inhibitor cocktails (Roche, Mississauga, Canada). Protein concentrations were determined by Bradford protein assay (Bio-Rad, Mississauga, Canada). Loading buffer was added to 30 μg of protein lysate and resolved by SDS–PAGE. The protein was then transferred onto a polyvinylidene difluoride membrane (Millipore, Toronto, Canada) and probed using antibodies for PI4KIIIβ (BD Biosciences 611817; Mississauga, Canada), pan-actin (Cell Signaling Technology 4968), tubulin (1:1000, Cell Signaling Technology 3873; Whitby, Canada), mouse α-FAK (1:200 Thermo Fisher #396500), rabbit α-Phospho-FAK (1:200 ThermoFisher #44-626), as well as anti-rabbit HRP-linked (Cell Signaling Technology catalogue no. 7074). Bands were detected with a MicroChemi chemiluminescent system (DNR Bio-Imaging Systems, Toronto, Canada) and intensities were quantified by densitometry using GelQuant (DNR Bio-Imaging Systems).
Plasmids and transfections
P4M-GFP and mApple-Talin-N-10 were obtained from Addgene (51469 and 54951, respectively). FAPP-GFP was a gift from Tamas Balla. Cells were grown and transfected on µ-Dish 35-mm, high-wall dishes (ibidi). For individual P4M and FAPP1 experiments, cells were transfected with 1 µg of DNA mixed with Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocol. For P4M and FA imaging, cells were transfected with 1.5 μg of P4M, 1.5 μg of Talin, and 12 μl of Lipofectamine 2000 reagent in 250 μl.
Sucrose stimulation of FAK phosphorylation
Proliferating NIH3T3 cells and derivatives thereof were serum starved for 2 h and then incubated for 10 min in serum-free DMEM media (Sigma) or stimulated for 10 min in serum-free media containing 400 mM sucrose (Sigma). Cells were collected and pelleted in ice-cold phosphate-buffered saline (PBS). Cell pellets were lysed in ice-cold RIPA buffer containing protease inhibitors (Complete, Millipore Sigma #11873580001) and phosphatase inhibitors (PhosSTOP, Millipore Sigma #4906845001) and frozen in 5× LSB.
Microscopy
For fixed-cell microscopy, NIH3T3 cells were seeded on High Precision 1.5H cover glass (Deckglaser) in 12-well cluster plates (Corning). Cells were fixed with 3.7% paraformaldehyde/PBS for 10 min, permeabilized for 10 min in 0.1% Triton X-100/PBS, and incubated for 1 h in 3% FBS/0.1% Triton X-100/PBS at room temperature. Rabbit α-GM130 (ThermoFisher PA5-95727) and mouse α-tubulin (Cell Signaling 3873S) or γ-tublin (Abcam 27074) were diluted 1:300 in blocking buffer and incubated overnight at 4°C. Mouse α-Vinculin (Millipore Sigma V9131) was diluted 1:300 in blocking buffer and incubated on the cells for 2 h (RT). After triplicate PBS washes, Goat anti-Mouse IgG (H+L) Antibody, Alexa Fluor 647 (Thermo Fisher A21235) was diluted 1:500 in PBS and incubated on cells for 1 h (RT). Cells were mounted on microscope slides (Fisher Scientific 12-544-7) with EverBrite Mounting Medium with DAPI (Biotium 23002) and sealed with CoverGrip Coverslip Sealant (Biotium 23005). Epifluorescent images were acquired with a Zeiss AxioObserver Z1 or a Zeiss AxioObserver M2 microscope with a 63× Plan-Apochromat 1.4 NA oil objective and Zen Blue 2.3 software. Image processing was carried out using ImageJ. The identification of FA immunostained for vinculin were processed as described by Horzum et al. (2014).
Live-cell microscopy
Time-lapse series of cells in phenol-free DMEM were recorded at 37°C on a Zeiss LSM880-AxioObserver Z1 microscope equipped with 63× Plan Aprochromat (NA 1.4) oil objective. We used an AiryScan detector in FAST mode and the bandpass emission filter 495–620 nm. Live-cell time lapse series were also captured using a Bessel Beam Lattice Light Sheet microscope equipped with detection objective (Semrock; NA of 0.54). A 5-mm coverslip was loaded to a piezo stage and submerged in a 37°C water bath. Time-lapse series of 175 Z-planes and 350 vol were captured at 63× magnification with S Piezo Offset 50 and an interval of 0.4 µm. Raw data were assembled into .tif hyperstacks using ImageJ. Three-dimensional renders were then generated for analysis using Imaris.
Supplementary Material
Acknowledgments
The authors thank Skye McBride and Chloe van Oostende for training and assistance with microscopy and Vera Tang for help with the flow cytometry of the CRSPR lines. We thank the Advanced Imaging Center at Janelia Farms for the use of the BLLSM. We thank Teng-Leong Chew, Satya Khuon, John Hedlleston, Blair Rossetti, and Eric Wait for help with the BLLSM and subsequent image analysis. The PI4KIIIβ-N162A plasmid was a generous gift from John Burke. FAPP1-GFP was a gift from Tamas Balla. We thank Spencer MacDonald for technical assistance and Redaet Daniel and John Copeland for helpful discussion and critical reading of this manuscript. This work was supported by an operating grant from the Natural Sciences and Engineering Research Council of Canada (J.M.L.).
Abbreviations used:
- BLLSM
Bessel Lattice Light Sheet microscope
- ECM
extracellular matrix
- FA
focal adhesion
- FAK
focal adhesion kinase
- FAPP1
four-phosphate-adaptor protein 1
- FBS
fetal bovine serum
- hv
hollow vesicle
- KD
kinase dead
- lv
large vesicle
- PBS
phosphate-buffered saline
- PI4KIIIβ
phosphatidylinositol 4-kinase III beta
- PI4P
phosphatidylinositol 4-phosphate
- r
ruffle
- sv
small vesicle
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-11-0600) on June 17, 2020.
REFERENCES
- Ahat E, Xiang Y, Zhang X, Bekier ME, 2nd, Wang Y. (2019). GRASP depletion-mediated Golgi destruction decreases cell adhesion and migration via the reduction of alpha5beta1 integrin. Mol Biol Cell , 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. (1998). Phosphatidylinositol 4-kinases. Biochim Biophys Acta , 69–85. [DOI] [PubMed] [Google Scholar]
- Balla T. (2007). Imaging and manipulating phosphoinositides in living cells. J Physiol , 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev , 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Balla T. (2006). Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol , 351–361. [DOI] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. (2005). A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell , 1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellas E, Chen CS. (2014). Forms, forces, and stem cell fate. Curr Opin Cell Biol , 92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borm B, Requardt RP, Herzog V, Kirfel G. (2005). Membrane ruffles in cell migration: indicators of inefficient lamellipodia adhesion and compartments of actin filament reorganization. Exp Cell Res , 83–95. [DOI] [PubMed] [Google Scholar]
- Bosch-Fortea M, Martin-Belmonte F. (2018). Mechanosensitive adhesion complexes in epithelial architecture and cancer onset. Curr Opin Cell Biol , 42–49. [DOI] [PubMed] [Google Scholar]
- Burke JE, Inglis AJ, Perisic O, Masson GR, McLaughlin SH, Rutaganira F, Shokat KM, Williams RL. (2014). Structures of PI4KIIIbeta complexes show simultaneous recruitment of Rab11 and its effectors. Science , 1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K. (2017). Focal adhesions: a personal perspective on a half century of progress. FEBS J , 3355–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Guilluy C. (2016). Focal adhesions, stress fibers and mechanical tension. Exp Cell Res , 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calalb MB, Zhang X, Polte TR, Hanks SK. (1996). Focal adhesion kinase tyrosine-861 is a major site of phosphorylation by Src. Biochem Biophys Res Commun , 662–668. [DOI] [PubMed] [Google Scholar]
- Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. (2003). Cell shape provides global control of focal adhesion assembly. Biochem Biophys Res Commun , 355–361. [DOI] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. (1997). Geometric control of cell life and death. Science , 1425–1428. [DOI] [PubMed] [Google Scholar]
- D’Souza RS, Lim JY, Turgut A, Servage K, Zhang J, Orth K, Sosale NG, Lazzara MJ, Allegood J, Casanova JE. (2020). Calcium-stimulated disassembly of focal adhesions mediated by an ORP3/IQSec1 complex. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf P, Zwart WT, van Dijken RA, Deneka M, Schulz TK, Geijsen N, Coffer PJ, Gadella BM, Verkleij AJ, van der Sluijs P, van Bergen en Henegouwen PM. (2004). Phosphatidylinositol 4-kinasebeta is critical for functional association of rab11 with the Golgi complex. Mol Biol Cell , 2038–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson EJ, Jensen JB, Hille B. (2014). Golgi and plasma membrane pools of PI(4)P contribute to plasma membrane PI(4,5)P2 and maintenance of KCNQ2/3 ion channel current. Proc Natl Acad Sci USA , E2281–E2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG. (2009). Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J Cell Biol , 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Partridge MA, Gundersen GG. (2005). Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol , 581–590. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Rodgers MA, Perrin BJ, Han J, Bennin DA, Critchley DR, Huttenlocher A. (2004). Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat Cell Biol , 977–983. [DOI] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. (2009). Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol , 21–33. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Weaver VM. (2017). Cellular adaptation to biomechanical stress across length scales in tissue homeostasis and disease. Semin Cell Dev Biol , 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. (1999). ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol , 280–287. [DOI] [PubMed] [Google Scholar]
- Hamadi A, Deramaudt TB, Takeda K, Ronde P. (2010). Hyperphosphorylated FAK delocalizes from focal adhesions to membrane ruffles. J Oncol . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GR, Machner MP, Balla T. (2014). A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol , 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzum U, Ozdil B, Pesen-Okvur D. (2014). Step-by-step quantitative analysis of focal adhesions. MethodsX , 56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan S, Morrow A, Amiri A, Lee JM. (2008). Eukaryotic elongation factor 1A2 cooperates with phosphatidylinositol-4 kinase III beta to stimulate production of filopodia through increased phosphatidylinositol-4,5 bisphosphate generation. Mol Cell Biol , 4549–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. (2010). Nanoscale architecture of integrin-based cell adhesions. Nature , 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, Theriot JA. (2008). Mechanism of shape determination in motile cells. Nature , 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Wirtz D. (2013a). Focal adhesion size uniquely predicts cell migration. FASEB J , 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Wirtz D. (2013b). Predicting how cells spread and migrate: focal adhesion size does matter. Cell Adh Migr , 293–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer A, Louvard D, Singer SJ. (1982). Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci USA , 2603–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert D, Wehrle-Haller B, David C, Weiland U, Ballestrem C, Imhof BA, Bastmeyer M. (2004). Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J Cell Sci , 41–52. [DOI] [PubMed] [Google Scholar]
- Lunn JA, Rozengurt E. (2004). Hyperosmotic stress induces rapid focal adhesion kinase phosphorylation at tyrosines 397 and 577. Role of Src family kinases and Rho family GTPases. J Biol Chem , 45266–45278. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Keren K. (2009). The shape of motile cells. Curr Biol , R762–R771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow AA, Amir Alipour M, Bridges D, Yao Z, Saltiel AR, Lee JM. (2014). The lipid kinase PI4KIIIbeta is highly expressed in breast tumors and activates Akt in cooperation with Rab11a. Mol Cancer Res DOI: 10.1158/1541-7786.MCR-13-0604. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Morone N, Suetsugu S. (2018). Membrane re-modelling by BAR domain superfamily proteins via molecular and non-molecular factors. Biochem Soc Trans , 379–389. [DOI] [PubMed] [Google Scholar]
- Parsons JT, Horwitz AR, Schwartz MA. (2010). Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol , 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinke DE, Lee JM. (2011). The lipid kinase PI4KIIIbeta and the eEF1A2 oncogene co-operate to disrupt three-dimensional in vitro acinar morphogenesis. Exp Cell Res , 2503–2511. [DOI] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. (2011). Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods , 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoy G, Wei HC, Wong R, Szentpetery Z, Kim YJ, Goldbach P, Steinbach SK, Balla T, Brill JA. (2009). Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J Cell Biol , 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satulovsky J, Lui R, Wang YL. (2008). Exploring the control circuit of cell migration by mathematical modeling. Biophys J , 3671–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink KO, Tan KW, Stenmark H. (2016). Phosphoinositides in Control of Membrane Dynamics. Annu Rev Cell Dev Biol , 143–171. [DOI] [PubMed] [Google Scholar]
- Takino T, Watanabe Y, Matsui M, Miyamori H, Kudo T, Seiki M, Sato H. (2006). Membrane-type 1 matrix metalloproteinase modulates focal adhesion stability and cell migration. Exp Cell Res , 1381–1389. [DOI] [PubMed] [Google Scholar]
- Tokuda E, Itoh T, Hasegawa J, Ijuin T, Takeuchi Y, Irino Y, Fukumoto M, Takenawa T. (2014). Phosphatidylinositol 4-phosphate in the Golgi apparatus regulates cell-cell adhesion and invasive cell migration in human breast cancer. Cancer Res , 3054–3066. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. (1999). The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol , 523–525. [DOI] [PubMed] [Google Scholar]
- Waugh MG. (2012). Phosphatidylinositol 4-kinases, phosphatidylinositol 4-phosphate and cancer. Cancer Lett , 125–131. [DOI] [PubMed] [Google Scholar]
- Wen PC, Mahinthichaichan P, Trebesch N, Jiang T, Zhao Z, Shinn E, Wang Y, Shekhar M, Kapoor K, Chan CK, Tajkhorshid E. (2018). Microscopic view of lipids and their diverse biological functions. Curr Opin Struct Biol , 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff MA, Serrels B, Fincham VJ, Frame MC, Carragher NO. (2004). SRC-mediated phosphorylation of focal adhesion kinase couples actin and adhesion dynamics to survival signaling. Mol Cell Biol , 8113–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Peterman MC, Davis RL, Oegema K, Shiau AK, Field SJ. (2016). GOLPH3 drives cell migration by promoting Golgi reorientation and directional trafficking to the leading edge. Mol Biol Cell , 3828–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XH, Bondeva T, Balla T. (2000). Characterization of recombinant phosphatidylinositol 4-kinase beta reveals auto- and heterophosphorylation of the enzyme. J Biol Chem , 14642–14648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.