Abstract
Reactive oxygen species (ROS)–induced oxidative stress has been associated with diseases such as amyotrophic lateral sclerosis, stroke, and cancer. While the effect of ROS on mitochondria and endoplasmic reticulum (ER) has been well documented, its consequence on the Golgi apparatus is less well understood. In this study, we characterized the Golgi structure and function in HeLa cells after exposure to hydrogen peroxide (H2O2), a reagent commonly used to introduce ROS to cells. Treatment of cells with 1 mM H2O2 for 10 min resulted in the degradation of Arl1 and dissociation of GRIP domain–containing proteins Golgin-97 and Golgin-245 from the trans-Golgi. This effect could be rescued by treatment of cells with a ROS scavenger N-acetyl cysteine or protease inhibitors. Structurally, H2O2 treatment reduced the number of cisternal membranes per Golgi stack, suggesting a loss of trans-Golgi cisternae. Functionally, H2O2 treatment inhibited both anterograde and retrograde protein transport, consistent with the loss of membrane tethers on the trans-Golgi cisternae. This study revealed membrane tethers at the trans-Golgi as novel specific targets of ROS in cells.
INTRODUCTION
In eukaryotic cells the reduction of O2 to H2O during ATP generation in the mitochondria necessarily leads to the production of reactive oxygen species (ROS). ROS can damage various cellular components such as lipids, proteins, and nucleic acids. Oxidative stress of cells occurs when damage to cellular components is high enough to lead to a constellation of system failures. For example, treatment of hemoglobin and rat globular basement membrane with hydrogen peroxide (H2O2) makes them more susceptible to proteolytic cleavage (Fligiel et al., 1984; Davies, 2016). In addition, oxygen free radicals such as superoxide anion, singlet oxygen, hydroxyl, and perhydroxyl, either generated by the cell or from an external source, can cause the death of multiple cell types in vitro and are thought to contribute to many inflammatory conditions (Ahsan et al., 2003; Singh et al., 2007). High levels of oxidative stress are often associated with diseases such as cancer, diabetes, atherosclerosis, stroke, and neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS), in which there is a failure in the cells to reduce ROS (Nindl et al., 2004; Manoharan et al., 2016).
ROS can damage membrane organelles. For example, oxidation and thereby inactivation of protein tyrosine phosphatases can increase steady-state protein phosphorylation of eIF2α and endoplasmic reticulum (ER) stress through redox inhibition of protein phosphatase 1 (Santos et al., 2016). H2O2 treatment inactivates catalase and disrupts mitochondria in Hs27 human fibroblasts, and increases the secretion of matrix metalloproteases (Koepke et al., 2008). H2O2 can also damage the fluidity and function of the plasma membrane in vascular endothelial cells (Block, 1991). So far, how ROS affects the structure and function of the Golgi apparatus has not been well documented.
The Golgi is a membrane organelle with a stacked structure that functions as a trafficking hub in the cell for the delivery of proteins and lipids to their final destinations. In anterograde trafficking, the cis-Golgi cisternae receive proteins synthesized by the ER in COPII vesicles and sends them to the endosomal/lysosomal system, the plasma membrane, or outside of the cell (Glick and Luini, 2011). In retrograde trafficking, the trans face of the Golgi receives vesicles from the plasma membrane and endosomes for subsequent delivery to the ER (Huang and Wang, 2017). These functions rely on a group of long coiled-coil proteins called golgins, which act as membrane tethers to capture vesicles and facilitate their fusion with the Golgi membranes (Lupashin and Sztul, 2005; Xiang and Wang, 2011; Witkos and Lowe, 2015; Muschalik and Munro, 2018). These include GM130 in the cis-Golgi that functions in ER-to-Golgi trafficking, and a group of four GRIP (“Golgin-97, ranBP2alpha, imh1p, and p230/golgin-245”) domain–containing proteins, Golgin-97, Golgin-245, GCC88, and GCC185, in the trans-Golgi (Jackson, 2003; Lu and Hong, 2003; Luke et al., 2003).
The GRIP domain–containing golgins are important for both anterograde and retrograde trafficking at the trans-Golgi (Munro, 2011). GCC88 (GRIP and coiled-coil domain containing 88 kDa) and GCC185 (GRIP and coiled-coil domain containing 185 kDa [also called GCC2]) have been shown to differ in their membrane binding properties and localize to distinct subdomains of the trans-Golgi, and facilitate the sorting and delivery of distinct cargo sets to and from the trans-Golgi network (TGN; Derby et al., 2004). Golgin-97 and Golgin-245 localize to overlapping but nonidentical subdomains of the TGN and mediate endosome-to-Golgi trafficking (Lu et al., 2004; Yoshino et al., 2005; Wong and Munro, 2014), but only Golgin-97 has been implicated in the anterograde trafficking of E-cadherin to the cell surface (Lock et al., 2005). Therefore, the GRIP domain tethers regulate the transport of diverse cargo molecules. The GRIP domain of Golgin-97 and Golgin-245 anchors these golgins to the trans-Golgi by the interaction with Arl1 (ADP-ribosylation factor-like protein 1), a member of the ARF/Arl family of small GTPases that is localized on trans-Golgi membranes. Depletion of Arl1 by RNAi causes dissociation of Golgin-97 and Golgin-245 from the trans-Golgi membranes and impairs both anterograde and retrograde trafficking at the TGN (Nishimoto-Morita et al., 2009).
In this study we determined the effect of ROS on Golgi structure and function by acute treatment of cells with H2O2. We found that H2O2 treatment resulted in a reduction of the number of Golgi cisternae in the stacks, a rapid loss of Arl1, and dissociation of Golgin-97 and Golgin-245 from the trans-Golgi, which impaired both anterograde and retrograde trafficking at the Golgi. This study revealed the trans-Golgi and trafficking at the trans-Golgi as novel targets of ROS in cells, which may help understand the toxicity of ROS in human diseases.
RESULTS
H 2O 2 treatment causes specific degradation of Arl1 and its binding partners Golgin-97 and Golgin-245
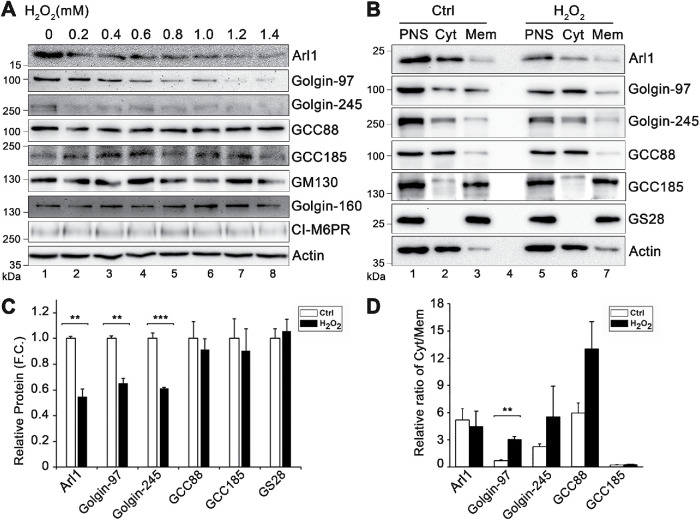
Oxidative stress is most frequently studied using hydrogen peroxide (H2O2), a membrane-permeable source of the peroxide ion (O22−) that is relatively stable in aqueous solutions (Stone and Yang, 2006). To investigate the effect of ROS on the Golgi, we applied H2O2 to HeLa cells in culture for 10 min with increasing concentrations. Because it has been previously shown that ROS triggers protein degradation (Pajares et al., 2015), we first examined the effect of H2O2 treatment on the level of Golgi structural proteins by Western blotting. As shown in Figure 1A and Supplemental Figure S1, H2O2 treatment at 1.4 mM, the highest concentration we tested, had no significant effects on most Golgi structural proteins, including GCC88, GCC185, GM130, GRASP65, GRASP55, Golgin-160, cation-independent mannose-6-phosphate receptor (CI-M6PR), and syntaxin 6. However, Arl1, Golgin-97, and Golgin-245 were significantly reduced. In this experiment we also tested α-tubulin, a microtubule protein that has previously been shown to be degraded upon 200 µM H2O2 treatment for 1 or 4 h (Hu and Lu, 2014). However, total α-tubulin level was unaffected in our experiments, possibly because we used a shorter time for the treatment, indicating that our experimental condition is milder than that of previous studies. Syntaxin 6 is an important SNARE protein in TGN trafficking (Bock et al., 1997), but the level of syntaxin 6 remained unchanged after H2O2 treatment at all concentrations (Supplemental Figure S1). Based on these results, we chose to use 1 mM H2O2 and 10 min for the treatment in most of our following experiments; at this condition Arl1, Golgin-97, and Golgin-245 are degraded, whereas GM130 and other Golgi structural proteins did not change.
FIGURE 1:
H2O2 treatment causes a loss of Arl1, Golgin-97, and Golgin-245 in cells. (A) Effect of H2O2 treatment on indicated proteins. HeLa cells were treated with the indicated concentrations of H2O2 for 10 min and blotted for the indicated proteins. (B) Distribution of indicated proteins in membrane and cytosolic fractionations. Control (Ctrl) cells and cells treated with 1 mM H2O2 for 10 min were homogenized with a ball-bearing homogenizer followed by a low-speed centrifugation to prepare postnuclear supernatant (PNS). The PNS was then subjected to ultracentrifugation to separate membranes (Mem) from the cytosol (Cyt). Equal proportions of each fraction were loaded onto the gel. GS28 and actin were used to denote membrane and cytosol fractions, respectively. (C) Relative protein level in the PNS in H2O2-treated cells vs. that in control cells based on the results shown in B. F.C., fold change. (D) Relative ratio of the protein level in the cytosolic fraction vs. that in the membrane fraction. Results are shown as mean ± SEM; statistical analyses were performed using two-tailed Student’s t tests (**, p ≤ 0.01; ***, p ≤ 0.001).
As an alternative approach, we performed high-speed centrifugation to separate cytosolic proteins from cellular membranes. This approach allowed us to examine not only the protein level, but also the membrane association of Arl1 and other proteins. As controls we used GS28 and actin as markers for membranes and cytosol, respectively. As shown in Figure 1B, Arl1, Golgin-97, GCC88, and GCC185 were all partially found in the membrane fraction of control cells. After H2O2 treatment, the protein levels of Arl1 and Golgin-97 in the postnuclear supernatant (PNS) were significantly reduced compared with the control cells, while GCC88 and GCC185 were less affected (Figure 1C). We also analyzed the ratio of each protein in the cytosol versus that in the membrane fraction. H2O2 treatment significantly reduced Golgin-97 in the membrane fractions; other proteins exhibited a similar trend, though statistically less significant (Figure 1D). Taken together, these results demonstrate that H2O2 treatment causes specific degradation of Arl1 and its associated golgins.
H 2O 2 treatment reduces membrane association of GRIP domain–containing golgins in trans-Golgi
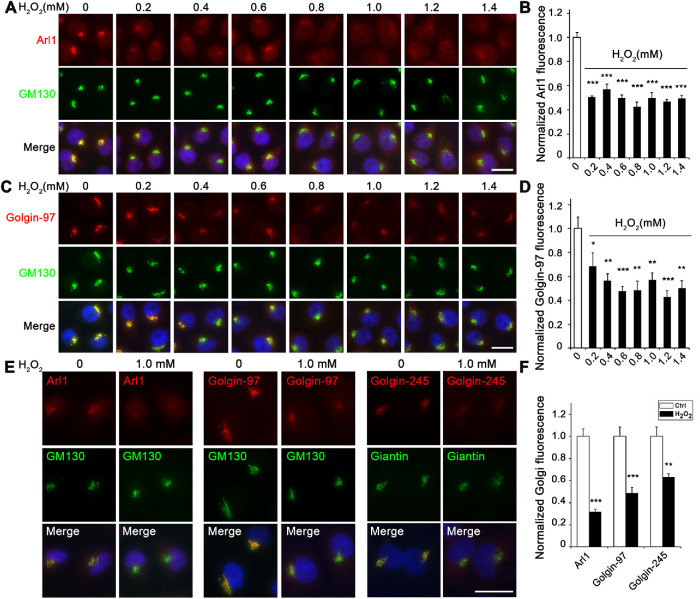
Our results showed that H2O2 treatment reduced the protein level of Arl1, Golgin-97, and Golgin-245 but not the other two GRIP domain–containing golgins, GCC88 and GCC185 (Figure 1). To further confirm that H2O2 treatment reduces the level of GRIP domain–containing golgins in the Golgi by an alternative method, we analyzed their subcellular localization by immunofluorescence microscopy. As shown in Figure 2, A–D, H2O2 treatment reduced the level of Arl1 and Golgin-97 in the Golgi region indicated by GM130 in a dose-dependent manner. Similarly, Golgin-245 also reduced its level in the Golgi region (Figure 2, E and F). Unlike Golgin-97 and Golgin-245, GCC88 and GCC185 signals reduced in the Golgi region but increased in the cytosol after H2O2 treatment (Supplemental Figure S2, A and B). These results indicate that GCC88 and GCC185 were partially dissociated from the Golgi, although they were not degraded, consistent with the subcellular fractionation results (Figure 1, B–D). Taken together, our results indicate that H2O2 treatment reduces the level of membrane tethers in the trans-Golgi membranes.
FIGURE 2:
H2O2 treatment reduces the Golgi localization of Arl1 and its associated golgins. (A) H2O2 treatment reduces the Golgi localization of Arl1. HeLa cells were treated with the indicated concentrations of H2O2 for 10 min, fixed, and stained for Arl1 (red), GM130 (green),and DNA (blue). (B) Quantitation of A for the relative fluorescence level of Arl1 signal in the Golgi area outlined by the GM130 signal, with the control normalized to 1. (C) H2O2 treatment reduces the Golgi localization of Golgin-97. HeLa cells were treated as above and stained for Golgin-97, GM130, and DNA. (D) Quantitation of C for the relative fluorescence level of Golgin-97 signal in the Golgi area defined by the GM130 signal, with the control normalized to 1. (E) H2O2 treatment reduces the Golgi localization of Golgin-245. HeLa cells were treated with 1 mM H2O2 for 10 min and stained for indicated proteins. Note that Arl1, Golgin-97, and Golgin-245 all have reduced Golgi-localized signal after H2O2 treatment. (F) Quantitation of E for the relative fluorescence level of Golgi protein signal in the Golgi area defined by the GM130 or giantin signal, with the controls normalized to 1. Scale bars in all panels = 20 µm. All quantitation results are shown as mean ± SEM; statistical analyses were performed using two-tailed Student’s t tests (*, p ≤ 0.05;**, p ≤ 0.01; ***, p ≤ 0.001).
H 2O 2 treatment reduces the number of cisternae per Golgi stack
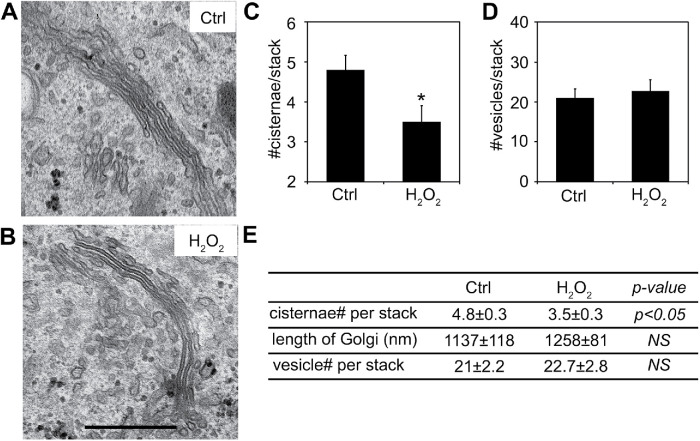
Despite the degradation of Arl1, Golgin-97, and Golgin-245, and dissociation of other GRIP domain–containing golgins, no significant change was observed in the Golgi morphology by immunofluorescence microscopy using GM130 and TGN46 as markers (Figure 2 and Supplemental Figure S2, C and D). Given that an intact Golgi ribbon structure requires an organized microtubule cytoskeleton, this observation is consistent with the result that H2O2 treatment does not cause α-tubulin degradation in our experimental system (Figure 1A). Because Arl1 and its associated golgins play important roles in Golgi structure formation and function within the stacks, the functional units of the Golgi, we performed electron microscopy (EM) to analyze the ultrastructure of the Golgi stacks in more detail. As shown in Figure 3, the average number of Golgi cisternae per stack was reduced by H2O2 treatment (3.5 ± 0.3) compared with that of control cells (4.8 ± 0.3), indicating a loss of Golgi cisternae. Previously, we have observed that depletion of GRASP65 and/or GRASP55 reduces the number of cisternae in the Golgi stack and the length of the Golgi cisternae, resulting in vesiculation of Golgi membranes (Xiang and Wang, 2010; Bekier et al., 2017). However, unlike GRASP depletion, the number of vesicles surrounding each Golgi stack did not significantly change after H2O2 treatment, nor did the average length of the stacks (Figure 3 and Supplemental Figure S3). The shape of the Golgi cisternae appeared to be normal, with flat, well-aligned cisternae within the stacks, and with narrow and uniform gaps between them. The reduction in cisterna number in the stacks and the loss of trans-Golgi membrane tethers suggest a possibility that trans-Golgi cisternae were removed or reduced after H2O2 treatment.
FIGURE 3:
H2O2 treatment reduces the number of cisternae per Golgi stack. (A, B) Representative electron micrographs of Golgi profiles in HeLa cells either untreated (Ctrl; A) or treated with 1 mM H2O2 for 10 min (H2O2; B). Note that the Golgi stacks contain fewer cisternae in the H2O2 treatment compared with Ctrl cells. Scale bar = 0.5 µm. (C, D) Quantitation of Golgi stack morphological features of cells in A and B. (E) Summary of quantitation of the morphological features of Golgi stacks on the EM images represented in A and B. Results are shown as mean ± SEM; statistical analyses were performed using two-tailed Student’s t tests (*, p ≤ 0.05; NS, not significant).
H 2O 2-induced Arl1 and Golgin-97 degradation is ROS dependent
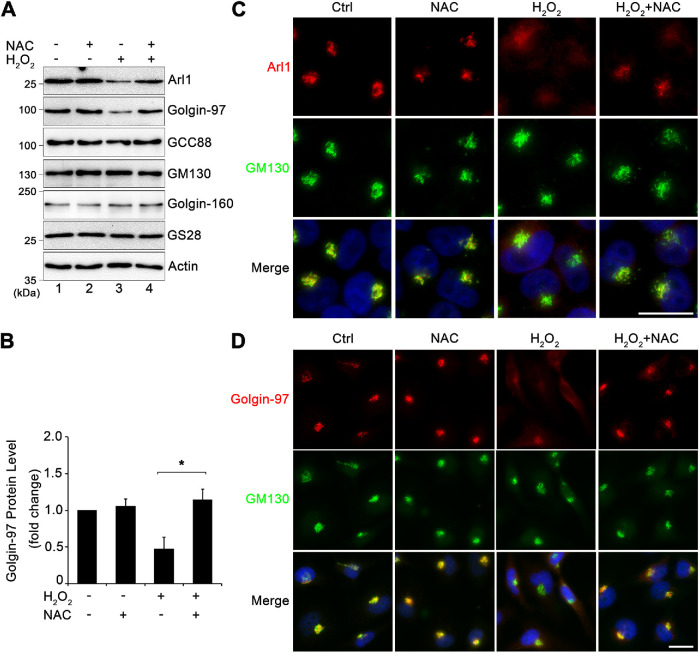
While H2O2 is widely used to induce ROS and cell stress, H2O2 itself has been observed to have a signaling role in cells (Veal et al., 2007). For example, it was recently discovered that H2O2 plays a significant role in T-cell signaling (Kim et al., 2017). To determine whether H2O2 induces Arl1 and Golgin-97 degradation through ROS, we used N-acetyl cysteine (NAC), a commonly used membrane-permeable antioxidant, to reduce ROS after H2O2-treatment. We cotreated cells with 5 mM NAC and 1 mM H2O2 for 10 min and determined the level of Arl1 and golgins by Western blotting. While NAC treatment alone had no effect on the protein levels of Arl1 and Golgin-97, cotreatment of cells with NAC inhibited H2O2-induced Arl1 and Golgin-97 degradation (Figure 4, A, lane 4 vs. 3, and B). In this and all following experiments we focused on Arl1 and Golgin-97 as they were the most affected proteins by H2O2 treatment, as described above. Further immunofluorescence microscopy confirmed that the addition of NAC not only significantly inhibited H2O2-induced Golgin-97 reduction, but also rescued the Golgi pool of Arl1 and Golgin-97 (Figure 4, C and D). These results confirmed that H2O2-induced Arl1 and Golgin-97 degradation is ROS dependent.
FIGURE 4:
H2O2-induced Arl1 and Golgin-97 degradation is ROS dependent. (A) HeLa cells cotreated with or without 5.0 mM NAC and 1 mM H2O2 for 10 min were analyzed by Western blotting. (B) Quantitation of Golgin-97 Western blot in A with densitometric analysis, with the control normalized to 1. Results are shown as mean ± SEM from three independent experiments. Statistical analyses were performed using two-tailed Student’s t tests (*, p ≤ 0.05). (C) HeLa cells treated as in A were stained for Arl1 (red), GM130 (green), and Hoechst (blue). Note the diffused pattern of Arl1. (D) Cells treated as in A were stained for Golgin-97 (red), GM130 (green), and DNA (blue). Note the diffuse pattern of Golgin-97 but not GM130 in H2O2-treated cells. Scale bars = 20 µm.
H 2O 2 treatment causes Arl1 and Golgin-97 degradation by cytoplasmic proteases
As demonstrated above, Arl1, Golgin-97, and Golgin-245 are degraded upon H2O2 treatment; therefore, we determined whether this degradation was mediated by proteasomes or lysosomes, two major protein degradation pathways in the cell. Because it has been previously shown that oxidative stress triggers autophagy, a cellular process that delivers cargo molecules to lysosomes for degradation, we first inhibited autophagy and lysosomal degradation with bafilomycin A1 (BafA1) and determined the effect on H2O2-induced Golgi protein degradation. BafA1 treatment increased LC3-II and p62 signals (Figure 3A, lane 3), indicating that it effectively blocked autophagic flux. In contrast, BafA1 treatment had no effect on H2O2-induced Arl1 and Golgin-97 degradation (Supplemental Figure S4, B, lane 4 vs. 3, and C), indicating that H2O2-induced Arl1 and Golgin-97 degradation is not through autophagy or lysosomes. This result was confirmed by immunofluorescence microscopy; addition of BafA1 had no effect on H2O2-induced Golgin-97 reduction in the Golgi region (Supplemental Figure S4D).
Proteasomal degradation is a process in which cells target ubiquitinated proteins for degradation by the proteasome (Eisenberg-Lerner et al., 2020). By examining our Western blots, we did not detect ubiquitinated forms of Arl1 and Golgin-97. In addition, inhibition of proteasomal degradation by MG132, which caused accumulation of ubiquitinated proteins (Supplemental Figure S4A, lane 2), had no effect on H2O2-induced Arl1 and Golgin-97 degradation as examined by Western blotting or fluorescence microscopy (Supplemental Figure S4, E–G). Taken together, these results demonstrated that ROS-induced Arl1 and Golgin-97 degradation is not via lysosomes or proteasomes.
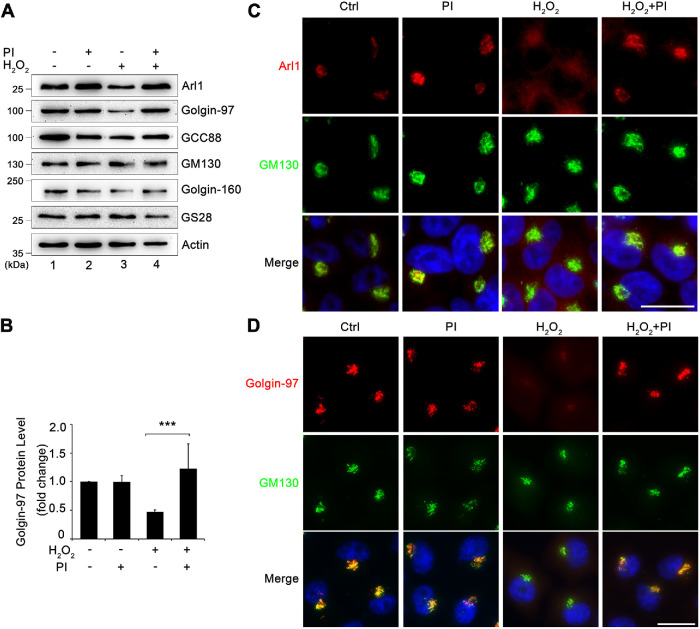
In addition to proteasomal and lysosomal degradation pathways, cytosolic proteases are known to degrade Golgi proteins such as Golgin-160 (Mancini et al., 2000). Therefore, we added a protease inhibitor tablet (Roche) into the treatment to determine whether it could rescue H2O2-induced Arl1 degradation. Such a tablet contains a proprietary set of chemicals that can effectively inhibit a broad spectrum of serine and cysteine proteases. As shown in Figure 5, A and B, the addition of the protease inhibitors blocked H2O2-induced Arl1 and Golgin-97 degradation. Furthermore, immunofluorescence microscopy confirmed that adding protease inhibitors effectively inhibited H2O2-induced Arl1 and Golgin-97 reduction in the Golgi region (Figure 5, C and D). Taken together, these results indicate that the degradation of Arl1 and Golgin-97 upon H2O2 treatment is mediated by cytoplasmic proteases.
FIGURE 5:
H2O2 treatment causes Arl1 and Golgin-97 degradation by cytoplasmic proteases. (A) HeLa cells cotreated with or without a protease inhibitor cocktail and 1 mM H2O2 for 10 min were analyzed by Western blotting. (B) Quantitation of Golgin-97 Western blot in A with densitometric analysis, with the control normalized to 1. Results are shown as mean ± SEM from three independent experiments. Statistical analyses were performed using two-tailed Student’s t tests (***, p ≤ 0.001). (C) Cells treated as in A were stained for Arl1 (red), GM130 (green), and Hoechst (blue). (D) Cells treated as in A were stained for Golgin-97 (red), GM130 (green), and DNA (blue). Scale bars = 20 µm.
H 2O 2 treatment reduces Golgi-mediated membrane trafficking
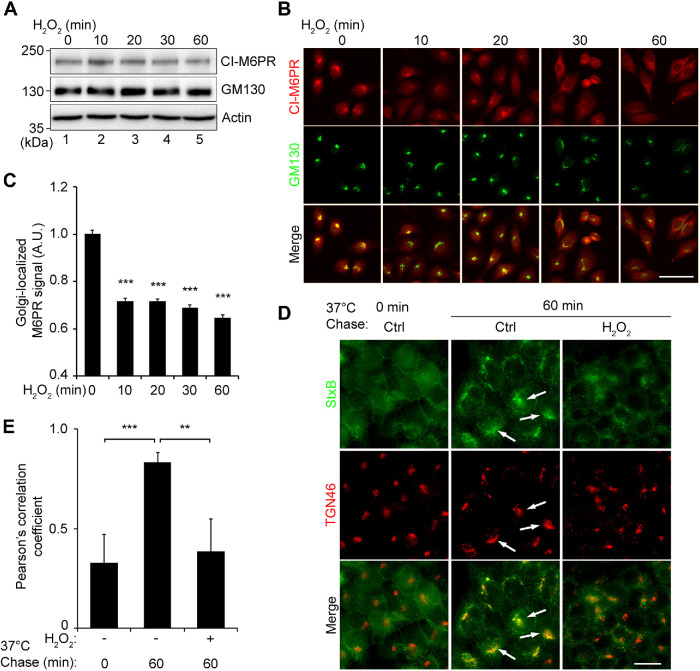
Arl1 and the GRIP domain–containing golgins are best known for their roles in tethering vesicles from the endosomes to facilitate their fusion with Golgi membranes. To determine the effect of H2O2 treatment on retrograde trafficking, we first looked at the level and subcellular localization of CI-M6PR, which is constantly recycled between the TGN and late endosomes. Treatment of cells with 1 mM H2O2 for up to 60 min did not significantly affect the CI-M6PR protein level (Figure 6A), but reduced its signal in the Golgi region over time (Figure 6, B and C). The reduction of the CI-M6PR signal in the Golgi region indicates an inhibition of CI-M6PR trafficking from the endosomes to the Golgi in H2O2-treated cells. As a complementary approach, we used Shiga toxin 1 B subunit (StxB) as a tool to measure retrograde trafficking (Selyunin and Mukhopadhyay, 2015). Cells were first incubated with StxB at 4°C to allow it to bind to the cell surface, and then warmed up to 37°C to allow StxB trafficking toward the Golgi (chase). After 60 min chase, StxB was accumulated in the Golgi region in control cells (Figure 6, D, white arrows, and E). In contrast, StxB existed mostly as cytoplasmic puncta in H2O2-treated cells. Consistent with the loss of Arl1 tethering complexes, our results demonstrate that H2O2 treatment impairs the function of the trans-Golgi in retrograde trafficking.
FIGURE 6:
H2O2 treatment impairs retrograde trafficking. (A) HeLa cells treated with 1 mM H2O2 for indicated times were analyzed by Western blotting. (B) HeLa cells treated as in A were stained for CI-M6PR (red) and GM130 (green). Scale bar = 50 µm. (B) Quantitation of B for the CI-M6PR intensity in the Golgi area defined by the GM130 signal, with the control normalized to 1. (D) Live cells were incubated with Shiga toxin (StxB) on ice followed by a 60-min incubation at 37°C to allow StxB trafficking to the Golgi. Cells were fixed and stained for StxB (green) and TGN46 (red). Scale bar = 20 µm. Note the dispersed StxB signals in H2O2-treated cells, which is more concentrated in the Golgi region in control cells indicated by arrows. (E) Pearson’s correlation coefficients of StxB and TGN46 signals in D. All quantitation results are shown as mean ± SEM from three independent experiments. Statistical analyses were performed using two-tailed Student’s t tests in comparison with the control (**, p ≤ 0.01; ***, p ≤ 0.001).
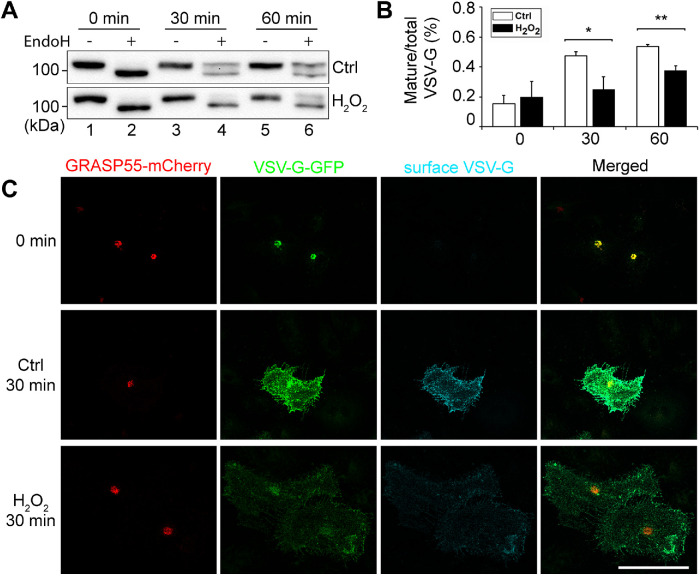
Because TGN also play essential roles in anterograde trafficking, we determined the effect of H2O2 treatment on vesicular stomatitis virus–glycoprotein (VSV-G) trafficking using the RUSH system (Boncompain et al., 2012). Cells were transfected with a plasmid that encodes both the invariant chain of the major histocompatibility complex (Ii, an ER protein) fused to core streptavidin and VSV-G fused to streptavidin-binding peptide (SBP). Under normal conditions the interaction between streptavidin and SBP retains VSV-G in the ER. Upon the addition of biotin, this interaction is displaced by the strong binding of biotin with streptavidin, resulting in the release of the VSV-G reporter from the ER for trafficking to the plasma membrane. Because VSV-G is a glycoprotein, we used endoglycosidase H (EndoH) to distinguish its core (ER and cis-Golgi) and complex (post-Golgi) glycosylation forms as an indicator of trafficking. As shown in Figure 7, A and B, H2O2 treatment decreased the EndoH-resistant post-Golgi forms of VSV-G at 30 and 60 min time points after release compared with that in the control, demonstrating that H2O2 treatment decreased VSV-G trafficking through the Golgi.
FIGURE 7:
H2O2 treatment impairs anterograde trafficking. (A) VSV-G trafficking assay using the RUSH system. Cells transfected with the Str-Ii_VSVG-SBP-EGFP plasmid were treated with or without 1 mM H2O2 for 10 min followed by VSV-G release from the ER for indicated times. Cell lysate was treated with or without EndoH and blotted for GFP. (B) Bands in A were quantified using densitometry analysis. The intensity of the upper EndoH-resistant band was divided by that of the total of the upper and lower bands to calculate the mature/total ratio. Results are shown as mean ± SEM from three independent experiments. Statistical analyses were performed using two-tailed Student’s t tests in comparison with the control (*, p ≤ 0.05; **, p ≤ 0.01). (C) HeLa cells were cotransfected with Str-Ii_VSVG-SBP-EGFP and GRASP55-mCherry for 24 h, and treated with 40 µM biotin at 20°C for 2 h to accumulate VSV-G in the Golgi, which is shown as the 0 min. Cells are then treated with complete medium at 37°C, first with 1 mM H2O2 for 10 min and then without H2O2 for 20 min. Control (Ctrl) cells were treated with complete medium at 37°C without H2O2 for 30 min. Cells were fixed without permeabilization and stained with an antibody to the lumenal domain of VSVG-G, and imaged for GRASP55 (mCherry), VSV-G (GFP), and surface VSV-G. Scale bar = 20 µm.
Given that H2O2 treatment results in the loss of the trans-Golgi, we also tested the effect of H2O2 treatment on TGN–to–plasma membrane trafficking by combining the RUSH system with a temperature block and release. It has been shown that anterograde cargo molecules are blocked at the TGN when cells are incubated at 20°C (Matlin and Simons, 1983), which can be released upon shifting the temperature back to 37°C. In this experiment, we cotransfected HeLa cells with the Str-Ii_VSVG-SBP-EGFP as described above, and N1-GRASP55-mCherry to indicate the Golgi localization. We first treated the cells with 40 µM biotin at 20°C for 2 h to accumulate VSV-G in the TGN. We then treated cells with complete medium with 1 mM H2O2 at 37°C H2O2 for 10 min, washed it out, and further incubated the cells in growth medium without H2O2 for 20 min (total 30 min chase). After fixation without permeabilization, we stained cell surface VSV-G with an antibody to its lumenal domain, and analyzed total VSV-G (GFP signal), GRASP55 (mCherry), and cell surface VSV-G by fluorescence microscopy. As shown in Figure 7C, there was more cell surface VSV-G signal in the control group compared with the H2O2-treated group at 30 min release, suggesting that anterograde trafficking from Golgi to plasma membrane is impaired during H2O2 treatment. Taken together, H2O2 treatment reduces both anterograde and retrograde trafficking at the TGN.
DISCUSSION
In this study, we revealed that H2O2 treatment causes a rapid degradation of Golgi tethering proteins on the trans-Golgi, including Arl1, Golgin-97, and Golgin-245, which could be rescued by the ROS-scavenging drug NAC or protease inhibitors, indicating that H2O2-induced ROS activates a cytoplasmic protease to selectively degrade Arl1, Golgin-97, and Golgin-245 on the trans-Golgi. This results in a reduction of the cisternal number per Golgi stack and impairs both anterograde and retrograde trafficking. Our results revealed a novel mechanism by which oxidative stress induces rapid and selective degradation of Arl1 and its associated golgins on the trans-Golgi, which impairs both anterograde and retrograde trafficking. Not all proteins in the trans-Golgi were tested in this study, and there may be others involved in the trafficking phenotype we observed. For example, ARFRP1 functions upstream of Arl1 and Arl5, and has been shown to regulate the recruitment of membrane tethers to the TGN (Nishimoto-Morita et al., 2009; Ishida and Bonifacino, 2019). However, our study has identified the Golgi as a novel site of ROS to exert its toxicity in cells.
It is interesting to see that the degradation of Arl1 and its associated golgins is not by proteasomes or lysosomes, but rather by cytoplasmic proteases. How proteins in the Golgi are degraded is so far not well understood. Theoretically, Golgi proteins can be degraded by the following pathways: 1) by lysosome-mediated protein degradation, which requires delivery of Golgi proteins to the lysosomes either by direct trafficking or by autophagy; 2) by ER-associated degradation, in which particular Golgi proteins traffic back to the ER for degradation by proteasomes; 3) by direct ubiquitination at the Golgi and degradation by the proteasomes on site; and 4) by cytoplasmic proteases. We originally speculated lysosomal degradation as the most likely mechanism for Arl1 degradation, as it has previously been shown that oxidative stress triggers autophagy (Zhang et al., 2016) and that there is a link between Golgi function and autophagy (Zhang et al., 2018, 2019). However, this possibility was ruled out by BafA1 treatment, which inhibits lysosomal degradation but not Arl1 and Golgin-97 degradation in H2O2-treated cells. Similarly, inhibition of proteasomes by MG132 also had no effect on Arl1 degradation. It is possible that a cytoplasmic protease may be more readily accessible than proteasomes or lysosomes for Arl1, Golgin-97, and Golgin-245. The identity of this cytoplasmic protease is as yet unknown and remains a subject for future investigation. One possible candidate is calpain, which is known to be activated by ROS in intermittent hypoxia (Bailey et al., 2015). Calpain cleavage specificity is likely mediated by substrate tertiary structure and not primary protein sequence, which increases the difficulty to predict the cleavage sites on its target proteins.
In contrast to some other Golgi stresses, such as thapsigargin treatment that triggers Golgi fragmentation (Ireland et al., 2020), H2O2 does not cause obvious Golgi fragmentation, or apoptosis at the concentrations and times that we tested here. The loss of Arl1 and its associated golgins, together with the lack of Golgi fragmentation and the impairment of anterograde and retrograde trafficking in H2O2-treated cells, indicates that Arl1 and its associated golgins play a role in membrane trafficking, but may not be essential for Golgi structure formation. Alternatively, Golgi fragmentation may take longer than 10 min at which our assays were performed. Indeed, H2O2 treatment reduced the number of cisternae in the Golgi stack and Golgi-mediated trafficking, indicating that trafficking defects occur before Golgi fragmentation. The identity of the lost cisternae remains unknown; but given that Arl1 and its associated golgins reside in the trans-Golgi, it is more likely that some trans cisternae are lost upon H2O2 treatment. An interesting question remains as to where trans-Golgi resident proteins go when the trans-Golgi cisternae are missing. One possibility is that the original trans-Golgi proteins are reequilibrated into the remaining cisternae; another is that they are now associated with vesicles or tubular membrane structures. The exact answer requires further investigation.
Our study revealed that the trans-Golgi is uniquely sensitive to oxidative stress in cells, which may help understand the toxicity of ROS in human diseases. ROS has been found to occur in several human diseases such as ALS, cancer, and diabetes (Nindl et al., 2004; Manoharan et al., 2016), and Golgi structural and functional defects have been observed in Parkinson’s (Mizuno et al., 2001), Huntington’s (Hilditch-Maguire et al., 2000), and Alzheimer’s (Joshi et al., 2014, 2015; Joshi and Wang, 2015) diseases and ALS (Mourelatos et al., 1996; Gonatas et al., 1998; Fujita and Okamoto, 2005). It would be interesting to test whether Arl1 and its associated golgins are degraded in these diseases in future studies.
MATERIALS AND METHODS
Reagents and plasmids
All reagents used in this study were purchased from Sigma-Aldrich (St. Louis, MO), Roche (Basel, Switzerland), EMD Millipore (Burlington, MA), or ThermoFisher (Waltham, MA), unless otherwise stated. H2O2 (30% in water) was from Fisher Chemical. Trypan blue was from Life Technologies (Dublin, Ireland). NAC was from Sigma. The cOmplete, EDTA-free protease inhibitor (PI) tablets were from Roche. StxB was from BEI Resources (Manassas, VA). D-Biotin was from VWR Life Science (Radnor, PA). BafA1 was from Fisher Scientific. MG132 was from EMD Millipore. The Str-Ii_VSVG-SBP-EGFP plasmid was provided by Franck Perez (Institut Curie). An EGFP-GCC185 plasmid from Paul Gleeson (University of Melbourne) was ectopically expressed in Supplemental Figure S2A.
Antibodies
The following primary antibodies were used. Monoclonal antibodies against β-actin and GFP (Sigma-Aldrich); Shiga toxin B and ubiquitin (ThermoFisher); GM130, Gos28, Golgin-245, and syntaxin 6 (BD Biosciences, Franklin Lanes, NJ); α-tubulin (Developmental Studies Hybridoma Bank, University of Iowa); Arl1 (Abcam); VSV-G extracellular domain (David Sheff, University of Iowa Carver College of Medicine). Polyclonal antibodies against GCC88, Golgin-160, CI-M6PR, GRASP55, and GRASP65 (Proteintech); GCC185 (Abcam); GM130 (“N73,” from J. Seemann, University of Texas Southwestern Medical Center); TGN46 (Bio-Rad); LC3B (Cell signaling). Secondary antibodies were purchased from Jackson Laboratory (Bar Harbor, ME). Secondary antibodies used for fluorescence microscopy include fluorescence-labeled goat anti-mouse, goat anti-rabbit, and goat anti-sheep antibodies. Secondary antibodies used for Western blot include HRP-conjugated goat anti-mouse and goat anti-rabbit antibodies.
Cell culture and drug treatments
HeLa were obtained from ATCC (Manassas, VA), cultured in DMEM (ThermoFisher) supplemented with 10% fetal bovine serum (Gemini Bio-Products, Sacramento, CA), and 100 units/ml penicillin–streptomycin at 37°C with 5% CO2, and routinely screened for mycoplasma contamination. Cells were grown on glass coverslips according to standard tissue culture methods (Tang et al., 2011). Coverslips were precoated with polylysine (Life Technologies) to aid in cell attachment. H2O2, NAC, and PI solutions were made fresh in Milli-Q water. All other drugs were dissolved in dimethyl sulfoxide (DMSO) and stock solution aliquoted and stored at −20°C until use. Stock solutions were diluted into working solutions of DMEM or water at the time of the experiment. Depending on the chemical, H2O or DMSO was used as control in the experiments.
Protein biochemistry
For immunoblotting, cells from a 6-cm dish were washed three times with ice-cold phosphate-buffered saline (PBS) and collected with a cell scraper. Cells were pelleted and lysed with 100 µl lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100 [Bio-Rad, Hercules, CA], 20 mM glycerolphosphate and protease inhibitor cocktail). Samples, excluding GCC185 samples (see below), were mixed with 6X SDS–PAGE buffer with fresh dithiothreitol (DTT), denatured at 95°C for 4 min, and then analyzed by SDS–PAGE. Protein was transferred to nitrocellulose membranes using semidry transfer at a constant 16 V. Membranes were blocked for 10 min with 3% milk in 0.2% Tween-20 in phosphate-buffered saline (PBST) and immunoblotted. GCC185 and Golgin-245 samples were denatured at 50°C for 5 min before loading onto a SDS–PAGE and transferred to nitrocellulose membranes by a wet transfer at 50 V for 2 h. Western blots were captured with enhanced chemiluminescence (ECL) dye reagent (ThermoFisher) in the FluorChem M chemiluminescent imager (ProteinSimple).
Subcellular fractionation
Fractionation of membranes was performed as described (Xiang et al., 2013). Briefly, control (Ctrl) HeLa cells and cells treated with 1 mM H2O2 for 10 min (H2O2) were washed with PBS, collected in HBS buffer (0.25 M sucrose, 10 mM HEPES, pH 7.2, 1 mM Mg(OAc)2, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) with a cell scraper, and homogenized using a ball-bearing (Ø = 8.008) homogenizer (HGM Lab Equipment) to ∼80% cell breakage (Tang et al., 2010, 2011). Plasma membrane disruption efficiency was examined by a trypan blue exclusion method. Homogenate was centrifuged at low speed for 10 min at 1000 × g at 4°C. PNS was then subjected to ultracentrifugation for 1 h in a TLA55 rotor at 40,300 rpm (100,000 × g) at 4°C to separate membranes from cytosol. Equal proportions of the membranes and cytosol fractions were loaded onto SDS–PAGE and analyzed by Western blot using the indicated antibodies (Xiang et al., 2013).
Immunofluorescence microscopy
For fluorescence microscopy, cells were rinsed three times in ice-cold PBS, fixed with 4% (wt/vol) paraformaldehyde (PFA), quenched with 50 mM NH4Cl in PBS, permeabilized in 0.2% (vol/vol) Triton X-100 in PBS, and blocked for 1 h with PBSB (PBS supplemented with 1% [wt/vol] bovine serum albumin (BSA) fraction V; ThermoFisher; Tang et al., 2016). Cells were incubated with a primary antibody diluted in PBSB overnight, gently rocking at 4°C, thoroughly washed with PBS, and incubated with an FITC-, TRITC-, or CY5-labeled secondary antibody (1:200 dilution in PBSB) at room temperature for 30 min. Cells were then washed three times with PBS and stained with 1:10,000 Hoechst dye for 3 min, and then mounted on glass slides with Moviol plus fluorescence brightener (DABCO; Acros Organics). Images were captured with a Zeiss Observer fluorescent microscope with a 63× oil objective lens and Axiocam Mrm camera. TIF files were exported with AxioVision software (Zeiss). For the data in Figure 6, B and C, images were collected at random locations on the coverslip with the autofocus function of ZEN software at 20× magnification.
Parameters used to quantify Golgi fragmentation on fluorescence images
Briefly, cells were evaluated by eye under a microscope according to predefined fragmentation criteria, with at least 300 cells counted in each reaction (Ireland et al., 2020). A Golgi was considered intact or fragmented based on the following: 1) If the Golgi exists as a single piece of connected membrane, it is intact. 2) If a Golgi exhibits several items that are connected by visible membrane bridges, even though these bridges might be faint, the Golgi is considered intact. 3) If a Golgi exhibits ≥3 disconnected parts (no visible bridges connecting them), then the Golgi is considered fragmented. 4) Mitotic cells, defined by the DNA pattern, and overlapping cells in which the Golgi pattern is difficult to define, are not counted. Hoechst was used to identify individual mitotic and overlapping cells.
EM
All EM-related reagents were from Electron Microscopy Sciences (EMS; Hatfield, PA). Cells were fixed in prewarmed serum-free DMEM, 20 mM HEPES, pH 7.4, 2% glutaraldehyde at 4°C overnight as previously described (Wang et al., 2005; Tang et al., 2010). Cells were washed twice with 0.1 M sodium cacodylate, and postfixed on ice in 1% (vol/vol) reduced osmium tetroxide, 0.1 M sodium cacodylate (wt/vol), and 1.5% cyanoferrate (wt/vol) in water. Cells were rinsed three times with 50 mM maleate buffer, pH 5.2, three times with water, scraped, and pelleted in microcentrifuge tubes for embedding. The EMBED 812 protocol was used to embed cells and resin blocks were sectioned to 60 nm with a diamond knife and mounted on Formvar-coated copper grids (Tang et al., 2010). Samples were double contrasted with 2% uranyl acetate, then with lead citrate and rinsed with copious amounts of water. Grids were imaged using a Philips (Amsterdam, Netherlands) transmission electron microscope. Golgi images were captured at 11,000× magnification. Golgi stacks were identified using morphological criteria and quantified using standard stereological techniques. A Golgi cisterna was identified as a perinuclear membrane within a Golgi stack ≥4 times longer than its width. Stack length was measured for the longest cisterna within a Golgi stack using the ruler tool in Photoshop Elements 13. For the number of cisternae per stack, the number of cisternae was counted. For the number of vesicles per stack, round objects no greater than 80 μm in diameter within 0.5 μm distance to a Golgi stack were counted. Golgi morphology was quantified from at least 20 cells in each experiment.
StxB transport assay
Shiga toxin assay was performed as previously described (Selyunin and Mukhopadhyay, 2015). In short, cells were plated onto polylysine-coated coverslips and cultured overnight, incubated with 4 µg/ml purified StxB DMEM at 4°C for 30 min, and then washed thoroughly with cold PBS. Cells were then incubated with H2O2 in growth medium for 10 min at 37°C, washed with growth medium, and further incubated in growth medium without H2O2 for an additional 50 min at 37°C. After treatment, cells were fixed and permeabilized as described above, stained with indicated primary and secondary antibodies for 1.5 h each at room temperature, and analyzed by fluorescence microscopy.
VSV-G protein trafficking and exofacial labeling
For EndoH treatment and Western blotting, cells in a dish were transfected with the Str-Ii_VSVG-SBP-EGFP plasmid for 16 h followed by a 10-min treatment with 1 mM H2O2. Cells were then incubated with fresh growth medium containing 40 µM biotin (chase) for the indicated times. Cells were lysed with denaturing buffer (0.5% SDS, 40 mM DTT), boiled at 90°C for 10 min, and treated with (+) or without (−) EndoH in G5 buffer (50 mM sodium citrate, pH 5.5) at 37°C for 1 h. Reaction was mixed with SDS buffer and analyzed by Western blot for GFP. Bands on Western blots were quantified using densitometry analysis. The intensity of the upper EndoH-resistant band was divided by that of the total of the upper and lower bands to calculate the mature/total ratio.
To specifically analyze TGN-to-plasma membrane trafficking, HeLa cells grown on polylysine-coated coverslips were cotransfected with the Str-Ii_VSVG-SBP-EGFP and N1-GRASP55-mCherry plasmids for 24 h and treated with 40 µM biotin in growth medium at 20°C for 2 h to accumulate VSV-G in the TGN. Cells were then treated with complete medium with 1 mM H2O2 at 37°C for 10 min, washed to remove H2O2, and further incubated in growth medium at 37°C for 20 min. For control, cells from the 20°C block were directly changed to growth medium prewarmed to 37°C and further incubated at 37°C for 30 min. Cells were then fixed in 1% PFA without permeabilization, blocked with 1% BSA in PBS, and incubated with an anti–VSV-G antibody that recognizes its lumenal domain overnight at 4°C. Cells are then stained with a secondary antibody and processed for immunofluorescence microscopy. Images were taken using a Nikon C2+ laser scanning confocal microscope and processed using ImageJ software.
Quantitation and statistics
All data represent the mean ± SEM of at least three independent experiments unless noted. A statistical analysis was conducted with two-tailed Student’s t test in the Excel program (Microsoft, Redmond, WA). Differences in means were considered statistically significant if p ≤ 0.05. Significance levels are as follows: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. Figures were assembled with Photoshop (Adobe, San Jose, CA). For Figure 6C, automatic thresholding was performed in ImageJ, and the Golgi-localized CI-M6PR signal was plotted. More than 300 cells were evaluated for each timepoint. Pearson’s colocalization coefficient values were computed using the “Coloc 2” function in ImageJ software.
Supplementary Material
Acknowledgments
We thank Paul Gleeson (University of Melbourne), Carolyn Machamer (Johns Hopkins University), Franck Perez (Institut Curie), Joachim Seemann (UT Southwestern), David Sheff (University of Iowa), and Haoxing Xu (University of Michigan) for reagents. We thank Gregg Sobocinski for microscope and equipment assistance, and Dana Holcomb and Zeiss for microscopy help and use of the Observer 7 demo unit. This work was supported by the National Institutes of Health (Grants no. GM112786 and no. GM130331), MCubed, and the Fastforward Protein Folding Disease Initiative of the University of Michigan (Y.W.). S.I. is a graduate student who was partially supported by the Mary Sue and Kenneth Coleman Endowed Fellowship Fund and the Edward’s Fellowship from the University of Michigan. H.H. was an undergraduate who received the Underwood-Alger award from The University of Michigan, MCDB/LS&A. Many thanks to past and current members of the Wang lab, in particular Sarah Bui and Shun Enomoto, for ideas, reagents, and technical support.
Abbreviations used:
- ALS
amyotrophic lateral sclerosis
- Arl1
ADP-ribosylation factor-like protein 1 (also called ARL1)
- EM
electron microscopy
- EndoH
endoglycosidase H
- ER
endoplasmic reticulum
- GCC185
GRIP and coiled-coil domain containing 185 kDa (also called GCC2)
- GCC88
GRIP and coiled-coil domain containing 88 kDa
- GM130
Golgi matrix protein of 130 kDa (also called golgin subfamily A member 2, Golga2)
- Golgin-245/p230
golgin subfamily A member 4 (GOLGA4)
- Golgin-97
golgin subfamily A member 1 (GOLGA1)
- GRASP55
Golgi reassembly-stacking protein of 55 kDa (also called GORASP2)
- GRASP65
Golgi reassembly-stacking protein of 65 kDa (also called GORASP1)
- GRIP domain
“Golgin-97, ranBP2alpha, imh1p and p230/golgin-245” domain
- H 2O 2
hydrogen peroxide
- CI-M6PR
cation-independent mannose-6-phosphate receptor
- NAC
N-acetyl cysteine
- PBS
phosphate-buffered saline
- PI
protease inhibitor cocktail
- ROS
reactive oxygen species
- StxB
Shiga toxin subunit 1B
- TGN
trans-Golgi network
- TGN46
trans-Golgi network integral membrane protein 2 (TGOLN2)
- VSV-G
vesicular stomatitis virus G glycoprotein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-01-0063) on June 17, 2020.
REFERENCES
- Ahsan H, Ali A, Ali R. (2003). Oxygen free radicals and systemic autoimmunity. Clin Exp Immunol , 398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AP, Koster G, Guillermier C, Hirst EM, MacRae JI, Lechene CP, Postle AD, Gould AP. (2015). Antioxidant role for lipid droplets in a stem cell niche of Drosophila. Cell , 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekier ME, 2nd, Wang L, Li J, Huang H, Tang D, Zhang X, Wang Y. (2017). Knockout of the Golgi stacking proteins GRASP55 and GRASP65 impairs Golgi structure and function. Mol Biol Cell , 2833–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block ER. (1991). Hydrogen peroxide alters the physical state and function of the plasma membrane of pulmonary artery endothelial cells. J Cell Physiol , 362–369. [DOI] [PubMed] [Google Scholar]
- Bock JB, Klumperman J, Davanger S, Scheller RH. (1997). Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell , 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, Mercanti V, Jollivet F, Raposo G, Perez F. (2012). Synchronization of secretory protein traffic in populations of cells. Nat Methods , 493–498. [DOI] [PubMed] [Google Scholar]
- Davies MJ. (2016). Protein oxidation and peroxidation. Biochem J , 805–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby MC, van Vliet C, Brown D, Luke MR, Lu L, Hong W, Stow JL, Gleeson PA. (2004). Mammalian GRIP domain proteins differ in their membrane binding properties and are recruited to distinct domains of the TGN. J Cell Sci , 5865–5874. [DOI] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Benyair R, Hizkiahou N, Maor R, Kramer M, Shmueli M, Zigdon N, Nudel N, Lev MC, Ulman A, et al. (2020). Fine-tuning regulation of Golgi organization is mediated by proteasomal degradation. Nat Commun (inpress). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fligiel SE, Lee EC, McCoy JP, Johnson KJ, Varani J. (1984). Protein degradation following treatment with hydrogen peroxide. Am J Pathol , 418–425. [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Okamoto K. (2005). Golgi apparatus of the motor neurons in patients with amyotrophic lateral sclerosis and in mice models of amyotrophic lateral sclerosis. Neuropathology , 388–394. [DOI] [PubMed] [Google Scholar]
- Glick BS, Luini A. (2011). Models for Golgi traffic: a critical assessment. Cold Spring Harb Perspect Biol , a005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatas NK, Gonatas JO, Stieber A. (1998). The involvement of the Golgi apparatus in the pathogenesis of amyotrophic lateral sclerosis, Alzheimer’s disease, and ricin intoxication. Histochem Cell Biol , 591–600. [DOI] [PubMed] [Google Scholar]
- Hilditch-Maguire P, Trettel F, Passani LA, Auerbach A, Persichetti F, MacDonald ME. (2000). Huntingtin: an iron-regulated protein essential for normal nuclear and perinuclear organelles. Hum Mol Genet , 2789–2797. [DOI] [PubMed] [Google Scholar]
- Hu WG, Lu QP. (2014). Impact of oxidative stress on the cytoskeleton of pancreatic epithelial cells. Exp Ther Med , 1438–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang Y. (2017). Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000Res , 2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland S, Ramnarayanan S, Fu M, Zhang X, Zhang J, Li J, Emebo D, Wang Y. (2020). Cytosolic Ca2+ modulates Golgi structure through PKCα-mediated GRASP55 phosphorylation. iScience , 100952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M, Bonifacino JS. (2019). ARFRP1 functions upstream of ARL1 and ARL5 to coordinate recruitment of distinct tethering factors to the trans-Golgi network. J Cell Biol , 3681–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL. (2003). Membrane traffic: Arl GTPases get a GRIP on the Golgi. Curr Biol , R174–R176. [DOI] [PubMed] [Google Scholar]
- Joshi G, Bekier ME, 2nd, Wang Y. (2015). Golgi fragmentation in Alzheimer’s disease. Front Neurosci , 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Chi Y, Huang Z, Wang Y. (2014). Aβ-induced Golgi fragmentation in Alzheimer’s disease enhances Aβ production. Proc Natl Acad Sci USA , E1230–E1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G, Wang Y. (2015). Golgi defects enhance APP amyloidogenic processing in Alzheimer’s disease. Bioessays , 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lim J, Jang YS, Shin EC, Kim HR, Seoh JY, Lee JS, Lee SN, Kang JL, Choi YH. (2017). Exogenous hydrogen peroxide induces lipid raft-mediated STAT-6 activation in T cells. Cell Physiol Biochem , 2467–2480. [DOI] [PubMed] [Google Scholar]
- Koepke JI, Wood CS, Terlecky LJ, Walton PA, Terlecky SR. (2008). Progeric effects of catalase inactivation in human cells. Toxicol Appl Pharmacol , 99–108. [DOI] [PubMed] [Google Scholar]
- Lock JG, Hammond LA, Houghton F, Gleeson PA, Stow JL. (2005). E-cadherin transport from the trans-Golgi network in tubulovesicular carriers is selectively regulated by golgin-97. Traffic , 1142–1156. [DOI] [PubMed] [Google Scholar]
- Lu L, Hong W. (2003). Interaction of Arl1-GTP with GRIP domains recruits autoantigens Golgin-97 and Golgin-245/p230 onto the Golgi. Mol Biol Cell , 3767–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Tai G, Hong W. (2004). Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-Golgi network. Mol Biol Cell , 4426–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke MR, Kjer-Nielsen L, Brown DL, Stow JL, Gleeson PA. (2003). GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J Biol Chem , 4216–4226. [DOI] [PubMed] [Google Scholar]
- Lupashin V, Sztul E. (2005). Golgi tethering factors. Biochim Biophys Acta , 325–339. [DOI] [PubMed] [Google Scholar]
- Mancini M, Machamer CE, Roy S, Nicholson DW, Thornberry NA, Casciola-Rosen LA, Rosen A. (2000). Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J Cell Biol , 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan S, Guillemin GJ, Abiramasundari RS, Essa MM, Akbar M, Akbar MD. (2016). The role of reactive oxygen species in the pathogenesis of Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease: a mini review. Oxid Med Cell Longev , 8590578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin KS, Simons K. (1983). Reduced temperature prevents transfer of a membrane glycoprotein to the cell surface but does not prevent terminal glycosylation. Cell , 233–243. [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Hattori N, Kitada T, Matsumine H, Mori H, Shimura H, Kubo S, Kobayashi H, Asakawa S, Minoshima S, Shimizu N. (2001). Familial Parkinson’s disease. Alpha-synuclein and parkin. Adv Neurol , 13–21. [PubMed] [Google Scholar]
- Mourelatos Z, Gonatas NK, Stieber A, Gurney ME, Dal Canto MC. (1996). The Golgi apparatus of spinal cord motor neurons in transgenic mice expressing mutant Cu,Zn superoxide dismutase becomes fragmented in early, preclinical stages of the disease. Proc Natl Acad Sci USA , 5472–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. (2011). The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol , 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschalik N, Munro S. (2018). Golgins. Curr Biol , R374–R376. [DOI] [PubMed] [Google Scholar]
- Nindl G, Peterson NR, Hughes EF, Waite LR, Johnson MT. (2004). Effect of hydrogen peroxide on proliferation, apoptosis and interleukin-2 production of Jurkat T cells. Biomed Sci Instrum , 123–128. [PubMed] [Google Scholar]
- Nishimoto-Morita K, Shin HW, Mitsuhashi H, Kitamura M, Zhang Q, Johannes L, Nakayama K. (2009). Differential effects of depletion of ARL1 and ARFRP1 on membrane trafficking between the trans-Golgi network and endosomes. J Biol Chem , 10583–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares M, Jimenez-Moreno N, Dias IHK, Debelec B, Vucetic M, Fladmark KE, Basaga H, Ribaric S, Milisav I, Cuadrado A. (2015). Redox control of protein degradation. Redox Biol , 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CX, Hafstad AD, Beretta M, Zhang M, Molenaar C, Kopec J, Fotinou D, Murray TV, Cobb AM, Martin D, et al. (2016). Targeted redox inhibition of protein phosphatase 1 by Nox4 regulates eIF2α-mediated stress signaling. EMBO J , 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selyunin AS, Mukhopadhyay S. (2015). A conserved structural motif mediates retrograde trafficking of Shiga toxin types 1 and 2. Traffic , 1270–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Sharma H, Singh N. (2007). Hydrogen peroxide induces apoptosis in HeLa cells through mitochondrial pathway. Mitochondrion , 367–373. [DOI] [PubMed] [Google Scholar]
- Stone JR, Yang S. (2006). Hydrogen peroxide: a signaling messenger. Antioxid Redox Signal , 243–270. [DOI] [PubMed] [Google Scholar]
- Tang D, Xiang Y, De Renzis S, Rink J, Zheng G, Zerial M, Wang Y. (2011). The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nat Commun , 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, Wang Y. (2010). Reconstitution of the cell cycle-regulated Golgi disassembly and reassembly in a cell-free system. Nat Protoc , 758–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Zhang X, Huang S, Yuan H, Li J, Wang Y. (2016). Mena-GRASP65 interaction couples actin polymerization to Golgi ribbon linking. Mol Biol Cell , 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal EA, Day AM, Morgan BA. (2007). Hydrogen peroxide sensing and signaling. Mol Cell , 1–14. [DOI] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G. (2005). Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem , 4921–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkos TM, Lowe M. (2015). The Golgin family of coiled-coil tethering proteins. Front Cell Dev Biol , 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Munro S. (2014). Membrane trafficking. The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science , 1256898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. (2010). GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol , 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. (2011). New components of the Golgi matrix. Cell Tissue Res , 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang X, Nix DB, Katoh T, Aoki K, Tiemeyer M, Wang Y. (2013). Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat Commun , 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino A, Setty SR, Poynton C, Whiteman EL, Saint-Pol A, Burd CG, Johannes L, Holzbaur EL, Koval M, McCaffery JM, Marks MS. (2005). tGolgin-1 (p230, golgin-245) modulates Shiga-toxin transport to the Golgi and Golgi motility towards the microtubule-organizing centre. J Cell Sci , 2279–2293. [DOI] [PubMed] [Google Scholar]
- Zhang X, Cheng X, Yu L, Yang J, Calvo R, Patnaik S, Hu X, Gao Q, Yang M, Lawas M, et al. (2016). MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat Commun , 12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang L, Ireland SC, Ahat E, Li J, Bekier ME, 2nd, Zhang Z, Wang Y. (2019). GORASP2/GRASP55 collaborates with the PtdIns3K UVRAG complex to facilitate autophagosome-lysosome fusion. Autophagy , 1787–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang L, Lak B, Li J, Jokitalo E, Wang Y. (2018). GRASP55 senses glucose deprivation through O-GlcNAcylation to promote aAutophagosome-lysosome fusion. Dev Cell , 245–261.e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.