SUMMARY
Rotavirus (RV) encounters intestinal epithelial cells amidst diverse microbiota, opening possibilities of microbes influencing RV infection. While RV clearance typically requires adaptive immunity, we unintentionally generated RV-resistant immune-deficient mice, which we hypothesized reflected select microbes protecting against RV. Accordingly, such RV-resistance was transferred by co-housing and fecal transplant. RV-protecting microbiotas were interrogated by heat, filtration, and antimicrobial agents, followed by limiting dilution transplant to germ-free mice and microbiome analysis. This approach revealed that segmented filamentous bacteria (SFB) was sufficient to protect mice against RV infection and associated diarrhea. Such protection was independent of previously-defined RV-impeding factors including interferon, IL-17, and IL-22. Colonization of ileum by SFB induced changes in host gene expression and accelerated epithelial cell turnover. Incubation of RV with SFB-containing feces reduced infectivity in vitro, suggesting direct neutralization of RV. Thus, independent of immune cells, SFB confers protection against certain enteric viral infections and associated diarrheal disease.
Keywords: Microbiota-viral interactions, germ-free mice, infectious diarrhea, fecal transplant, segmented filamentous bacteria, rotavirus
Graphical Abstract
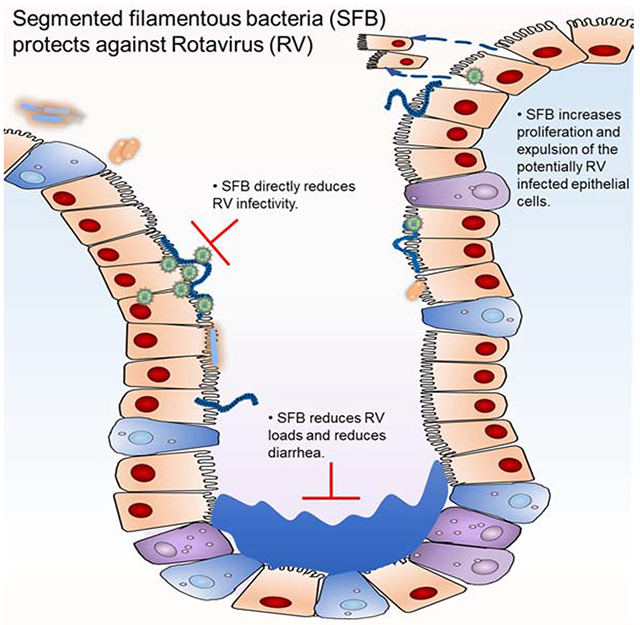
Abstract figure. GSU Rag1-KO mice microbiota protect against acute RV infection and cure chronic RV infection in adult mice and protect neonatal mice from RV-induced diarrhea. The antiviral activity is not confined to RV but is effective against other viruses, including reovirus in vivo and IAV and VSV in vitro. 16S sequencing yielded a new strain of SFB, named SFB-G, and SFB-G mono-associated fecal samples are comparable to GSU Rag1-KO mice microbiota in conferring RV-resistance to GF recipient mice. SFB-G isolated from GSU Rag1-KO mice microbiota, induces intestinal epithelium proliferation and confers anti-RV activity to both immunocompetent and immunocompromised mice.
Introduction
Rotavirus (RV), a nonenveloped dsRNA virus, remains a major world-wide scourge, causing life-threatening diarrhea in hundreds of thousands of children annually (Tate et al., 2016). RV enters its host orally and primarily infects ileal villous epithelial cells, which exist amidst a large diverse ecosystem of microorganisms collectively called gut microbiota (Boshuizen et al., 2003). Gut microbiota protect against pathogenic bacteria. For example, antibiotic-treated hosts are highly prone to colonization and disease by bacterial pathogens (Ribet and Cossart, 2015). Such colonization resistance is thought to reflect that specific families of bacteria prevent colonization by specific classes of pathogens (Kamada et al., 2013). In contrast, the extent to which microbiota impacts enteric viral infection is less clear.
Ablation of microbiota in mice, via germfree (GF) or antibiotic approaches, impedes infection of mouse mammary tumor virus, poliovirus, reovirus, and RV (Kane et al., 2011; Kuss et al., 2011; Uchiyama et al., 2014). However, administration of bacterial products, including LPS, CpG DNA, and flagellin can protect against viral infection (Jorgensen et al., 2003; Zhang et al., 2014). In particular, we discovered that flagellin induced TLR5-mediated innate lymphoid cell (ILC) IL-22 production and NLRC4-mediated IL-18 release that prevented and cured RV infection (Zhang et al., 2014). Such protection was independent of adaptive immunity in that it was maintained in Rag1-KO mice, which, like humans with severe immunodeficiencies, develop chronic RV infections (Bogaert et al., 2016; Oishi et al., 1991; Rosenfeld et al., 2017; Yoshikawa et al., 2019).
In breeding Rag1-KO mice for our studies, we unintentionally developed a breeding colony of Rag1-KO mice that was highly resistant to RV infection. We hypothesized that these mice had acquired a microbiota that resulted in ILC activation that conferred resistance to RV. In fact, we found that their RV-resistance was independent of ILCs. Nor did it require interferon (IFN), which is critical for control of most viruses. Rather, their RV-resistance was mediated by segmented filamentous bacteria (SFB), which altered both RV and the ileum in manner that prevented or cured RV infection, suggesting a new mechanism by which gut microbiota can improve health.
RESULTS
Transmissible Microorganisms Protect Immune-Deficient Mice Against Rotavirus Infection
RV primarily infects villus epithelial cells, causing severe diarrhea in young hosts and moderate distress in adults (Ramig, 2004). While RV is typically cleared within 10 days, immune deficient humans and mice, including C57BL/6 Rag1-KO mice, develop chronic RV infections (Riepenhoff-Talty et al., 1987). Accordingly, Rag1-KO mice, purchased from The Jackson Laboratory (JAX) perpetually shed RV antigens into their feces following oral inoculation with RV, indicating chronic infection (Figure 1A). In contrast, Rag1-KO mice bred at Georgia State University (GSU) after being purchased from JAX did not shed RV antigens. We speculated that these mice had acquired a microbiota that made them resistant to RV infection. A second colony of Rag1-KO mice, for which husbandry was performed by the authors of this study, using autoclaved food and water, maintained Rag1-KO mice that became chronically infected with RV when inoculated with this virus. We refer to such RV-susceptible Rag1-KO mice as “JAX-RAG” and the aforementioned RV-resistant Rag1-KO mice as “GSU-RAG.”
Figure 1. Transmissible Microorganisms Protect Immune-Deficient Mice Against Rotavirus Infection.
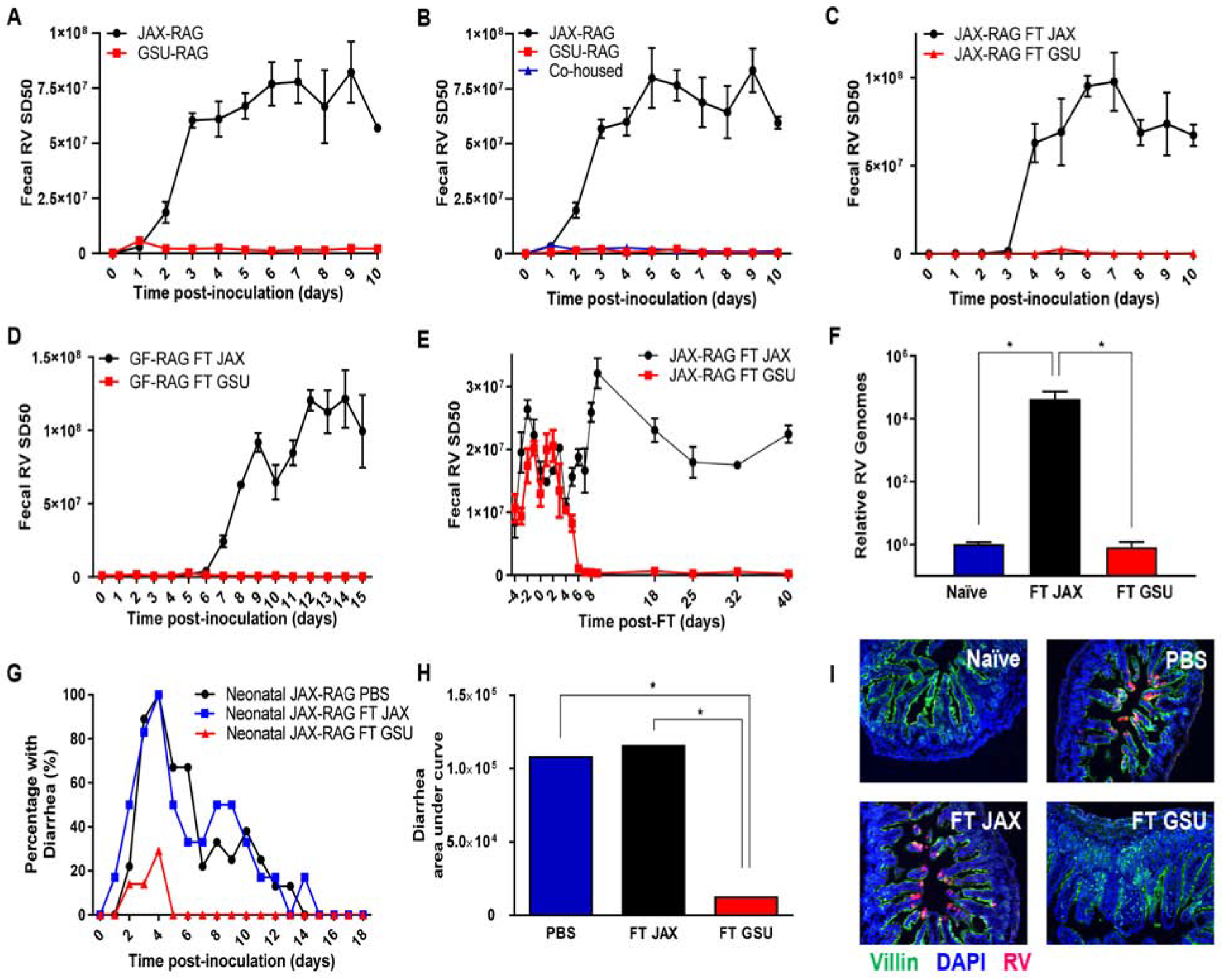
Rag1-KO mice were orally inoculated with RV, EC strain. Feces were collected daily, and fecal RV antigen quantified by ELISA.
(A) Comparison of mice from two colonies of Rag1-KO mice (GSU and JAX).
(B) JAX, GSU, and JAX co-housed with GSU, Rag1-KO mice.
(C) JAX Rag1-KO mice received fecal transplants (FT) from JAX or GSU Rag1-KO mice one week prior to RV inoculation.
(D) Germ-free (GF) Rag1-KO mice received FT from JAX or GSU Rag1-KO mice one week prior to RV inoculation.
(E–F) JAX Rag1-KO with established chronic RV infections received FT from JAX or GSU Rag1-KO mice. Panel E shows fecal RV antigens, while panel F shows levels of intestinal RV genomes quantified by qRT-PCR at day 62 post-transplantation.
(G–I) Seven-day-old Rag1-KO mice pups received PBS or FT from JAX or GSU Rag1-KO mice 6 d and 1 d prior to RV inoculation. Panels G and H show the incidence of diarrhea, while I shows RV antigen by fluorescence microscopy 5 weeks post-inoculation.
Data shown are means ± SEM of individual experiments and were significant by two-way analysis of variance (ANOVA); A; n = 3, P < 0.0001. B; n = 3–4, P < 0.0001. C-D; n = 4, P < 0.0001. E-F; n = 6, P < 0.0001. G, FT GSU differed significantly from both FT JAX and the control (FT WT) groups by one-way ANOVA, n = 12, *, P < 0.0001; H, FT GSU differed significantly from both FT JAX and the control (PBS) groups by χ2 test, n=12, *, P < 0.01. I, representative images (n= 4 mice per condition). Each experiment was performed 2–3 times and yielded an identical pattern of results.
Co-housing of JAX-RAG and GSU-RAG mice, or orally administering GSU-RAG feces to JAX-RAG mice, resulted in JAX-RAG mice acquiring RV-resistance, implicating gut microbiota (Figures 1B and 1C). To determine whether transmissible microbes were sufficient to yield RV resistance, as opposed to requiring both resident and transferred species, we transplanted GSU-RAG fecal suspensions into germfree (GF) Rag1-KO mice. GF Rag1-KO mice receiving JAX-RAG feces developed chronic RV infection, while GF Rag-1KO mice receiving GSU-RAG feces were fully resistant to RV (Figure 1D). Furthermore, administration of GSU-RAG feces, but not JAX-RAG feces, to JAX-RAG mice with established chronic RV infection resulted in cessation of RV antigen shedding, indicating cure of chronic RV infection (Figure 1E). Quantification of RV genomes in the ileum by PCR confirmed absence of RV infection (Figure 1F). Infection of suckling RAG pups with RV results in watery diarrhea (Kuklin et al., 2000). RV-induced Such diarrhea was almost eliminated by GSU-RAG, but not JAX-RAG feces (Figures 1G and 1H). Moreover, both control groups (i.e., PBS- and JAX-RAG feces-treated), but not the GSU-RAG feces-treated group, displayed RV antigen by immunostaining (Figure 1I), providing additional evidence that GSU-RAG feces prevented RV infection in a highly susceptible host. Thus, transmissible agents present feces of GSU-RAG feces protect against RV infection and its major associated disease.
GSU-RAG Feces Transiently Protect Immune Competent Mice from RV
Regardless of husbandry procedures, wild-type (WT) C57 BL/6 mice bred at GSU remained prone to RV infection, suggesting the protective agent(s) in GSU-RAG mice was held in-check by adaptive immunity. To test this hypothesis, we determined whether administration of GSU-RAG feces protected WT mice against RV infection. Administration of GSU-RAG feces to adult (8-week-old) WT mice one week before RV inoculation resulted in a marked reduction, albeit not complete absence, of RV infection relative to mice receiving JAX-RAG feces (Figure 2A). Analogous partial protection against RV infection and associated diarrhea was observed in neonate WT mice (Figures 2B and S1B). In accord with the notion that the transient nature of this protection reflected clearance of the protective agent, shortening the time from fecal transplant to RV administration from 7 days to 2 or 4 days markedly enhanced the level of protection (Figure 2C). Another factor influencing the extent of protection was age in that 3-week-old WT mice appeared fully protected against RV administered 2–7 days post-FT (Figure 2D). Assay of serum anti-RV antibodies 14 days post-inoculation observed that, in WT FT recipients, lack of RV shedding correlated with lack of RV antibodies providing further evidence that mice not shedding RV antigens had not ever been infected by this virus (Figures S1 C, D). Analogous to studies in Rag-KO, administering GSU-RAG but not JAX-RAG feces protected adult GF-WT mice from RV infection in both C57BL/6 and Swiss-Webster genetic backgrounds, confirming that fecally transmissible microbes are sufficient to protect against RV (Figures 2E and S1A).
Figure 2. Bacterial Constituents of GSU RAG Feces Partially Protect Immune-Competent Mice from RV.
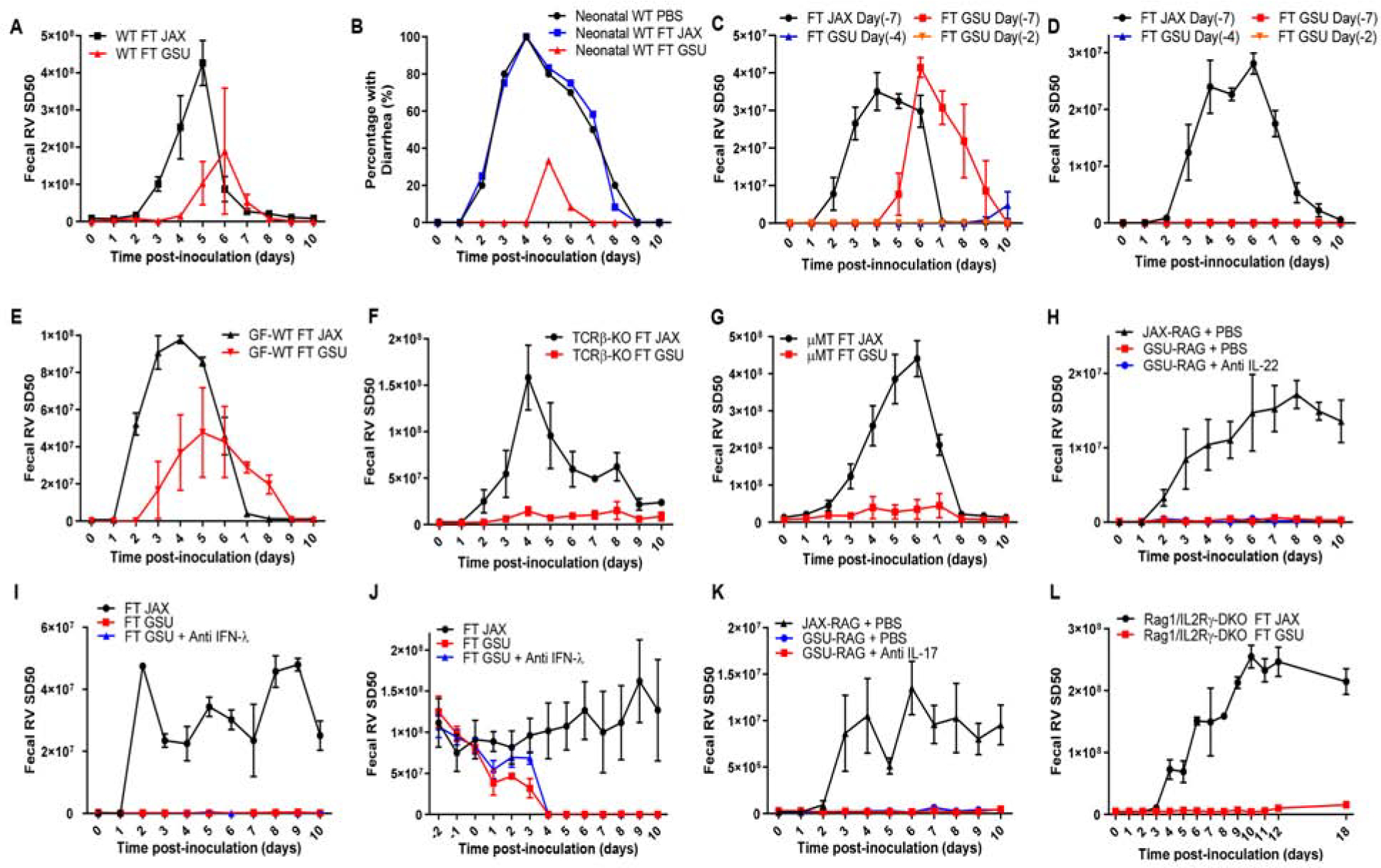
Mice were orally inoculated with RV, EC strain. Feces were collected daily, and fecal RV antigen was quantified by ELISA.
(A) WT C57BL/6J mice received FT from JAX or GSU Rag1-KO mice one week prior to RV inoculation.
(B) Seven-day-old WT C57BL/6J mice pups received PBS or FT from JAX or GSU Rag1-KO mice twice 6 d and 1 d prior to RV inoculation. Panels B shows the incidence of diarrhea.
(C–D) (C) Eight-week-old or (D) Three-week-old WT mice received FT from JAX or GSU Rag1-KO mice. Mice were then inoculated 2, 4, or 7 days later with RV (7-days for recipients of JAX FT).
(E) GF WT C57BL/6J mice received FT from JAX or GSU Rag1-KO mice one week prior to RV inoculation.
(F) TCRβ-KO mice received FT from JAX or GSU Rag1-KO mice one week prior to RV inoculation. (G) μMT (Ighm-KO) mice received FT from JAX or GSU Rag1-KO mice one week prior to RV inoculation.
(H) JAX or GSU colony of Rag1-KO mice were inoculated intraperitoneally with PBS or IL-22 neutralizing antibody (150 μg daily for 2 days) neutralizing antibody shortly before RV inoculation.
(I) JAX colony of Rag1-KO mice were inoculated intraperitoneally daily with PBS or IFN-λ neutralizing antibody (1 μg per gram of body weight/day) from day −1 to day 10 following RV inoculation.
(J) RV chronically infected JAX Rag1-KO mice received FT with GSU mice microbiota or GSU microbiota with intraperitoneal inoculation of IFN-λ neutralizing antibody (1μg per gram of body weight/day) from day −1 to day 10 post-FT.
(K) JAX or GSU colony of Rag1-KO mice were inoculated intraperitoneally with PBS or IL-17 neutralizing antibody (100μg daily for 2days) neutralizing antibody shortly before RV inoculation.
(L) ILC-deficient (Rag1/IL2Rγ-DKO) received FT from JAX or GSU Rag1-KO mice one week prior to RV inoculation.
Data shown are means ± SEM (two-way ANOVA). A; n = 3, P < 0.01. C; n=3, P < 0.0001. D; n=5, P < 0.0001. E; n = 4, P < 0.05. F; n = 3, P < 0.0001. G; n = 3, P < 0.0001. H-I; n = 4, P < 0.0001. J; n=3, P < 0.0001. K; n=3, P < 0.01. L; n = 4, P < 0.0001. B, FT GSU differed significantly from both FT JAX and the control (FT WT) groups by one-way ANOVA, n = 12, *, P < 0.01. Each experiment was performed 2–3 times and yielded an identical pattern of results.
We next investigated whether the capacity of GSU-RAG microbes to protect Rag1-KO mice against RV reflected absence of T and/or B-lymphocytes. Mice deficient in T-cell receptor β (TCRβKO), which lack conventional T-cells, and μMT mice, which lack functional B-cells, were administered JAX-RAG (control) or GSU-RAG feces and, one week later, orally inoculated with RV. Either humoral or cell-mediated immunity affords RV clearance (Corthesy et al., 2006; Jaimes et al., 2005). Accordingly, both TCRβKO and μMT mice administered control JAX-RAG feces exhibited a week-long course of RV infection (Figures 2F and 2G). In contrast, both strains of mice transplanted with GSU-RAG feces remained protected against RV infection. This suggests that microbes in GSU RAG feces that might otherwise protect against RV infection are normally constrained by a mechanism involving humoral and cell-mediated immunity.
GSU-RAG Microbiota May Directly Interact with Viruses to Reduce Infectivity
We next considered the possibility that JAX-RAG and GSU-RAG mice might display differences in intestinal leukocytes that mediated resistance to RV. Flow cytometric analysis of lamina propria (LP) revealed elevated neutrophils in GSU-RAG mice (Figures S2A and S2B). However, neutrophil numbers did not change following fecal transplant arguing against this parameter mediating RV-resistance. Administering mice bacterial flagellin activates innate lymphoid cell (ILC) production of IL-22, which impedes RV infection (Zhang et al., 2014). Therefore, we reasoned that organisms present in GSU-RAG microbiota might also activate IL-22 production that would protect against RV infection. However, transplant of GSU-RAG feces into JAX-RAG mice did not increase IL-22 expression (Figure S2C). Nor did such transfer result in elevations in IL-18, which acts in concert with IL-22 to prevent or cure RV infection in response to flagellin. Moreover, antibody-mediated neutralization of IL-22, which blocks flagellin’s protection against RV infection (Zhang et al., 2014), did not reduce RV-resistance in GSU-RAG mice (Figure 2H). RV is relatively resistant to type I IFN (Barro and Patton, 2007) but is constrained by type III IFN, also known as IFN-λ RV (Pott et al., 2011), which can provide sterilizing antiviral immunity in the absence of adaptive immunity (Nice et al., 2015). IFN-λ and IL-22 have closely related receptors and cooperate in host defense against RV (Hernandez et al., 2015). Thus, even though transplant of GSU-RAG feces did not elicit IFN-λ expression (Figure S2C), we determined whether GSU feces could protect against or clear RV via a mechanism requiring IFN-λ. Antibody-mediated neutralization of IFN-λ, via an established protocol (Ferguson et al., 2019), did not impair the capacity of GSU-RAG feces to prevent or treat RV infection (Figures 2I and 2J). As an alternate means of investigating a potential role for IFN-λ, we used mice with genetic ablations of both Rag1 and IFN-λ-receptor (R)1 that were established and maintained at Washington University (Wash-U). Interestingly, such Rag1/IFN-λ-R1-DKO mice (Baldridge et al., 2017), as well as the control Rag1-KO mice bred in this vivarium, phenocopied GSU-RAG mice in that they were resistant to RV infection (Figure S2D). These results suggest that the tendency of Rag1-KO mice to acquire microbes that confer resistance to RV infection is not specific to GSU and, moreover, such RV resistance is not mediated by IFN-λ. We also considered the possibility that GSU-RAG fecal transplant might elevate type II IFN expression but our results did not support this possibility (Figure S2C). While flagellin also increases ILC production of IL-17 (Van Maele et al., 2010), neutralization of IL-17 did not reduce resistance of GSU-RAG mice to RV infection (Figure 2K). Moreover, Rag1/IL2Rγ-DKO mice, which lack innate and adaptive lymphocytes, remained RV-resistant following administration of GSU-RAG feces, indicating lack of ILC involvement (Figure 2L). Lastly, we examined the capacity of LP cells from GSU-RAG to dampen RV infection in JAX-RAG mice chronically infected with RV. However, we found that transfer of these cells had no effect; only transfer of WT cells with a functional Rag-1 gene cleared RV infection (Figure S2E).
We next examined if GSU-RAG feces had the capacity to directly disable RV. Aliquots of cell-culture adapted RV, rhesus RV (RRV), were incubated at 37°C with 5 μm- or 0.2 μm-filtered feces from JAX-RAG or GSU-RAG mice (as discussed below, only the 5 μm-filtered feces can protect mice against RV). Four hours later, the RV-microbiota suspensions were passed through 0.2 μm filters, and the RRV-containing filtrate adsorbed to HT-29 epithelial cells, which support RV replication. Twenty hours later, cells inoculated with RRV that had been pre-exposed to JAX-RAG feces expressed RRV antigen, as assessed by immunoblotting (Figure 3A), and cell death (Figure 3B), indicating productive infection. Pre-exposure to 5 μm-filtered, but not 0.2 μm-filtered, GSU-RAG feces markedly reduced RV infection with markedly lower levels of RV genomes (Figure 3C). In contrast, JAX-RAG microbiota did not alter RRV infection in vitro (Figure S3A). These results suggest that microorganism(s) present in GSU-RAG microbiota reduced RRV infectivity. Such reduction correlated with reduced levels of RV attachment to epithelial cells, as determined by qRT-PCR of epithelial cells inoculated with RV at 4°C (Figure S3B). To investigate the nature of microbiota-mediated RV neutralization, we examined relative effects of GSU-RAG and JAX-RAG feces on RV levels by qRT-PCR over time following incubation in feces, but we found that fecal samples from both types of mice moderately, and similarly, degraded RV genomes. Moreover, GSU-RAG and JAX-RAG feces did not differ significantly in the capacity to bind RV (Figures S3C and S3D). These results are in accord with the possibility that microbes present in GSU feces alter a surface component of RV in a manner that impedes initial interactions with epithelial cells.
Figure 3. GSU-RAG Microbiota May Directly Interact with Viruses to Reduce Infectivity.
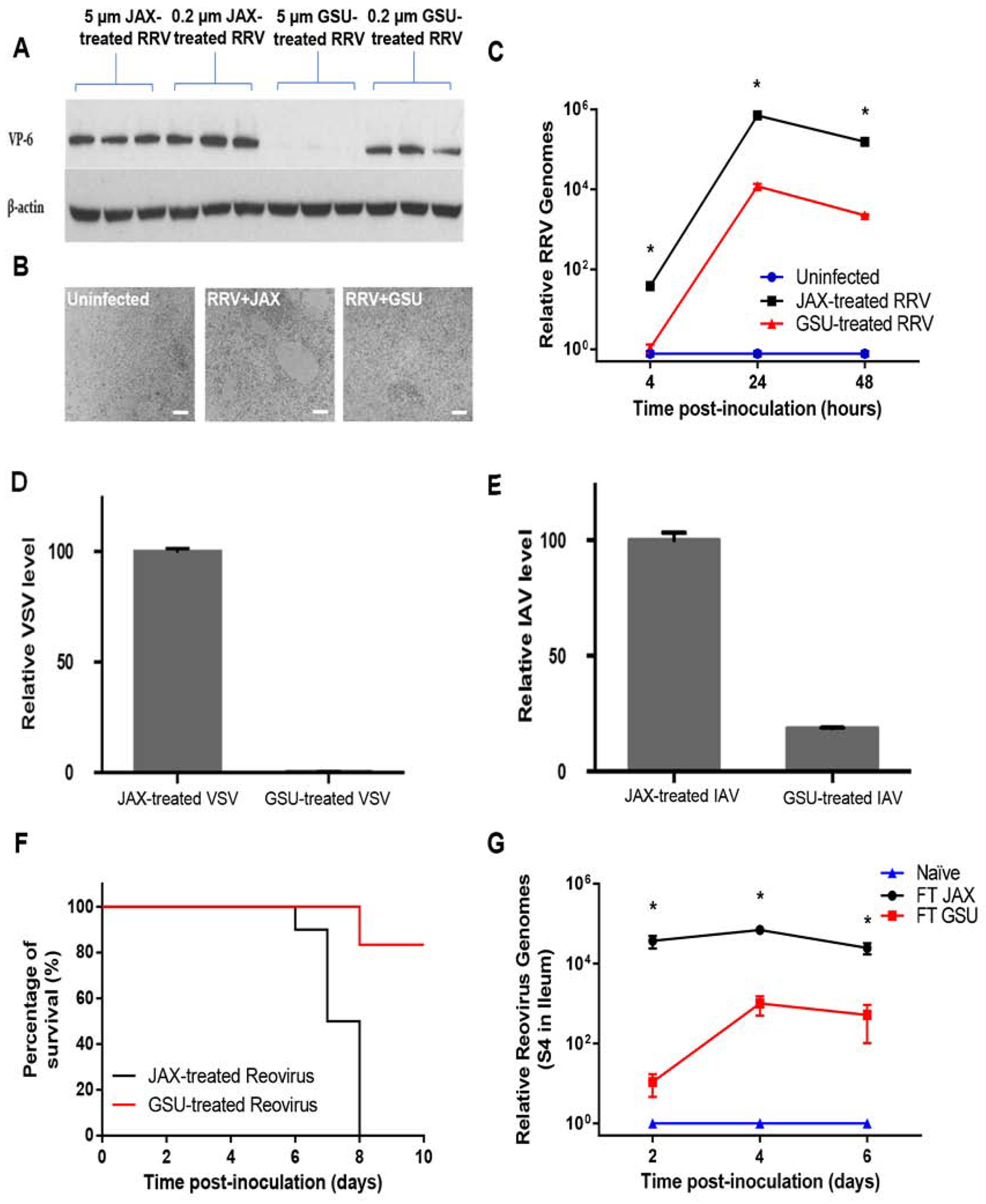
The viruses tested were incubated with 5 μm filtered JAX or GSU Rag1-KO mice fecal samples at 37°C. The mixtures were then filtered through 0.2 μm filters to remove the bacteria cells and used for in vitro challenges.
(A-C) RRV was pre-incubated with 5 μm or 0.2 μm filtered JAX or GSU Rag1-KO mice fecal samples at 37°C for 4 hours prior to inoculating HT-29 cells in vitro. (A) Immunoblotting for RV VP6 at 20 hours after infection with RRV. (B) Microscopic analysis of HT-29 cells at 20 hours after infection with treated RRV (5 μm filtered JAX or GSU Rag1-KO mice fecal samples), scale bar: 100 μm. (C) qRT-PCR results at the time points shown after infection with treated RRV (5 μm filtered JAX or GSU Rag1-KO mice fecal samples).
(D-E) Effect of fecal microbiota (5 μm filtered JAX or GSU Rag1-KO mice fecal samples) on VSV
(D) and IAV WSN (E) infection of BEAS-B cells 24 hours post-infection in vitro.
(F) Survival of IFN-αR-KO mice following inoculation with reovirus treated with either 5 μm-filtered JAX or GSU Rag1-KO mice fecal samples (37°C, 4 hours).
(G) qRT-PCR assay of reovirus RNA in the small intestine of Rag1-KO mice following transplant of JAX or GSU Rag1-KO mice fecal samples 1 week prior to oral inoculation.
Results are shown as mean ± SEM (two-way ANOVA). A-B, representative images (n= 3 per condition). C; GSU treated RRV differed significantly from control (JAX treated RRV) group by two-way ANOVA, n = 4, *, P < 0.0001. D; n = 4, P < 0.0001. E; n = 4, P < 0.0001. F; χ2 test, n = 10, P < 0.0001. G; FT GSU group differed significantly from control (FT JAX) group by two-way ANOVA, n = 3, *, P < 0.0001. Each experiment was performed 2–3 times and yielded an identical pattern of results.
To determine whether exposure to GSU-RAG feces diminishes infection of other viruses, we quantified viral loads in cultured cells inoculated with recombinant vesicular stomatitis virus (VSV) or influenza A virus (A/WSN/1933 [H1N1]) reporter virus strains (Yan et al., 2015; Yoon et al., 2018) that had been pre-exposed to feces from JAX-RAG or GSU-RAG mice. The capacity of GSU-RAG feces to suppress viral infection in vitro was not specific for RV but also extended to VSV and IAV (Figures 3D and 3E). We also tested whether GSU-RAG microbiota alters infection with reovirus, which, like RV, replicates in the murine intestine. Reovirus (T1L strain) was incubated with 5 μm-filtered feces from JAX-RAG or GSU-RAG mice and orally inoculated into type I IFN receptor-deficient mice, which are highly susceptible to this virus. 100% of mice inoculated with JAX-RAG feces-treated reovirus died within 8 days of inoculation, while most mice inoculated with GSU-RAG feces-treated reovirus survived this period (Figure 3F). Moreover, transplant of GSU-RAG feces substantially reduced reovirus loads relative to mice transplanted with JAX-RAG feces, especially at early times after inoculation (Figure 3G). These results suggest that microbes in GSU-RAG feces may have broad antiviral activity.
RV-Resistance Correlates with Transfer of Segmented Filamentous Bacteria (SFB)
We next sought to define the protective component of GSU-RAG microbiota. We prepared frozen and thawed aliquots of pooled GSU-RAG feces that maintained capacity to confer protection against RV infection (Figure 4A). Such aliquots were subjected to filtration, heating, and antimicrobial compounds. The protective agent passed through a 5 μm but not a 0.22 μm filter, withstood heating at 60°C, but not 75°C (Figure 4A, B), and was inactivated ex vivo by a cocktail of antibacterial but not anti-fungal agents (Figure 4C). Individually, ampicillin, neomycin, and streptomycin, but not kanamycin, eliminated the capacity of GSU-RAG feces to confer RV-resistance. (Figure 4D). Yet, prolonged consumption of antibiotics by GSU-RAG mice did not render them susceptible to RV infection (Figure S4A). Together, these results suggest that GSU-RAG mice were protected from RV infection by heat-stable, possibly spore-forming, bacteria that are readily transmitted to immune-deficient mice from which it may be difficult to clear once established.
Figure 4. RV-Resistance Correlates with Transfer of Segmented Filamentous Bacteria (SFB).
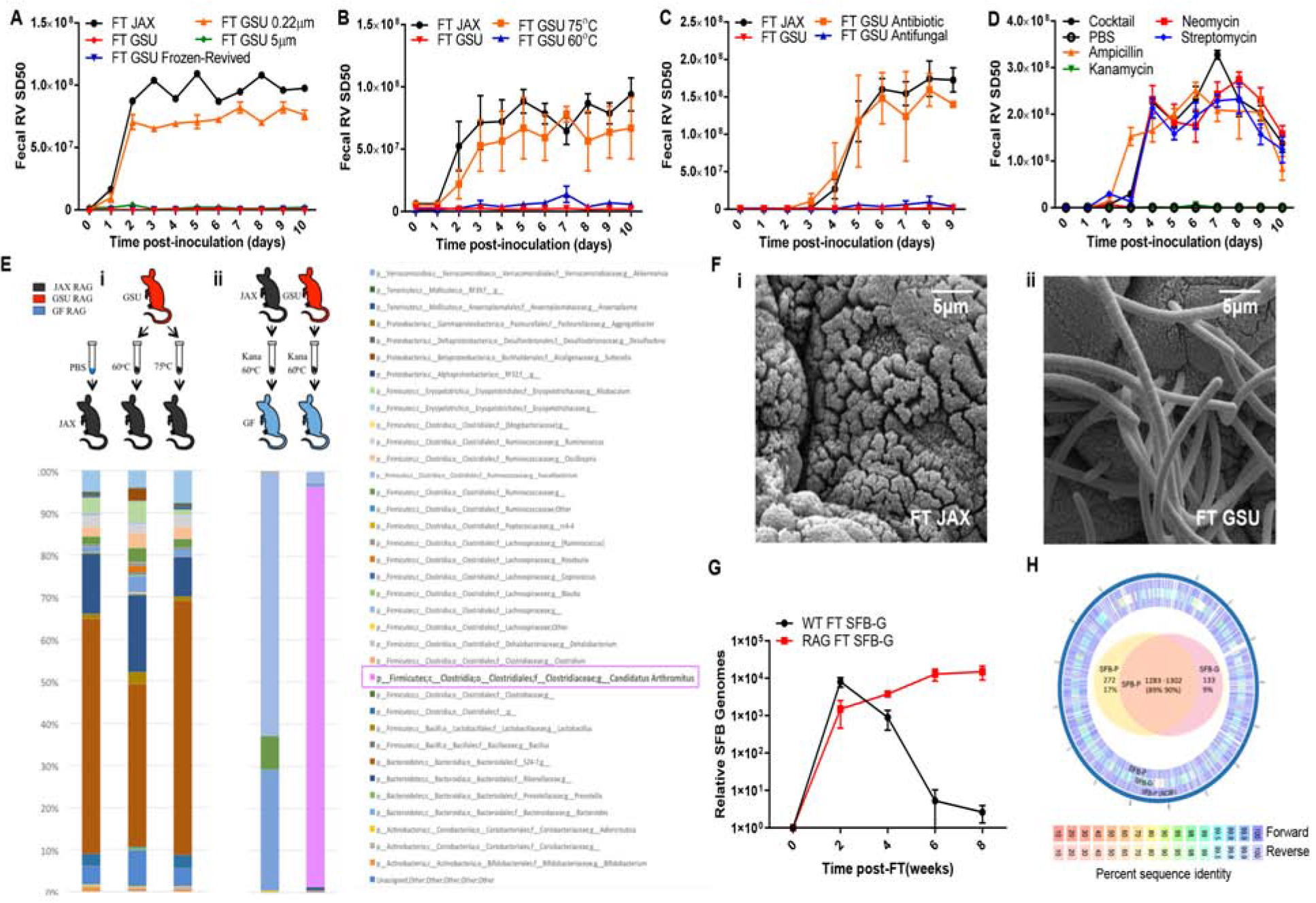
JAX Rag1-KO mice were orally inoculated with RV, EC strain. Feces were collected daily, and fecal RV antigen was quantified by ELISA.
(A) JAX Rag1-KO mice received FT from JAX or GSU Rag1-KO mice with fresh, −80°C revived, 0.22 μm filtered, or 5 μm filtered fecal samples for 1 week prior to RV inoculation.
(B) JAX Rag1-KO mice received FT from JAX or GSU Rag1-KO mice with untreated or heat-treated (60°C or 75°C for 10 minutes) fecal samples for 1 week prior to RV inoculation.
(C) JAX Rag1-KO mice received FT with JAX or GSU Rag1-KO mice fecal samples that were treated with an antibiotic cocktail or anti-fungal cocktail (37°C for 4 hours) 1 week prior to RV inoculation.
(D) JAX Rag1-KO mice received FT with PBS, ampicillin, kanamycin, neomycin, streptomycin, or all four antibiotics combined (37°C for 4 hours) GSU Rag1-KO mice fecal samples 1 week prior to RV inoculation.
(E) (i) 16s rRNA sequencing results of the conventional JAX Rag1-KO mice that received PBS or 60°C-, or 75°C-treated GSU Rag1-KO mice fecal samples. (ii) Fecal 16s rRNA sequencing results of GF Rag1-KO mice that were transplanted with diluted kanamycin-treated, heat-treated (60°C for 10 minutes) JAX or GSU Rag1-KO mice fecal samples.
(F) Scanning electron micrographs of ileal sections from mice in E-ii. Those receiving GSU Rag1-KO were abundantly colonized by SFB.
(G) Fecal SFB levels by qPCR in WT or Rag-1KO mice following FT with GSU-SFB.
(H) Genomes of SFB strain in E-ii (SFB-G) and reference strain provided by Pasteur Institute (SFB-P) were sequenced and compared to SFB-P genome sequence data in the NCBI. Outer ring: Pan-genome analysis of forward and reverse assembly summary. Inner ring: Venn diagram representing the number of sequences unique or shared between the two assembled strains (90%–95% identity between the two strains at the protein level).
Results are shown as mean ± SEM (two-way ANOVA). A-B; n = 4, P < 0.0001. C-D; n = 3, P < 0.0001. H; n=4, P < 0.01. Experiments A-D were performed 2–3 times and yielded an identical pattern of results.
Since Rag1-KO mice are frequently infected with protozoa, for example Tritrichomonas muris (Escalante et al., 2016), which is also susceptible to some antibiotics, we also investigated the possibility that protozoa contributed to the RV-resistance of GSU-RAG mice. Microscopic analysis of intestinal contents of GSU-RAG mice revealed protozoa, which albeit not T. muris (two verified sets of T. muris PCR primers failed to yield amplicons). However, such protozoa were not transferred by our fecal transplant procedure (Figure S4B), perhaps because of failure to pass through a 5.0 μm filter, thus suggesting that protozoa did not mediate RV-resistance.
We next sought to identify bacterial candidates mediating RV-resistance via microbiome analysis. We examined the microbiomes of conventional JAX-RAG mice receiving PBS or GSU-RAG feces that had been heated to 60°C, which protected against RV, or feces heated to 75°C, which did not. This limited transfer protocol yielded about 50 species whose abundance was markedly increased by transfer of 60°C heated feces (Figure 4E–i). However, candidates selected from this list failed to significantly alter RV infection. Therefore, to more specifically isolate bacteria that protected GSU-RAG mice from RV infection, JAX-RAG and GSU-RAG feces were filtered, diluted, and heated to 60°C for 10 minutes, treated with kanamycin, and then administered to GF Rag1-KO mice. One week later, these mice were inoculated with RV. Like conventional JAX-RAG mice, GF Rag1-KO mice receiving such transplants from GSU-RAG, but not JAX-RAG, mice were fully protected against RV infection (Figure S4C). Analysis of the microbiome in these transplanted mice revealed that a striking percentage (95%) of 16S sequences corresponded to a single species of bacteria, namely Candidatus Arthromitus, more commonly referred to as segmented filamentous bacteria (SFB). In contrast, SFB was not detected in mice receiving similarly processed JAX-RAG feces (Figure 4E–ii). Assay of SFB by qPCR indicated that SFB was uniformly present in feces of GSU-RAG, but not JAX-RAG, mice (Figure S4D). Furthermore, examination of the ileum by electron microscopy revealed prominent SFB in recipients of GSU-RAG but not JAX-RAG feces (Figure 4F). Moreover, SFB was uniformly present in RV-resistant Wash-U Rag1-KO and Rag1/IFN-λ-R1-DKO mice (Figure S4E). These results indicate that SFB is present in RV-resistant immune-deficient mice tand can be transferred via fecal transplants that confer RV-resistance suggesting that SFB might contribute to RV-resistance. That SFB can readily colonize WT mice, which remained susceptible to RV infection, seemed inconsistent with this conclusion. However, we observed that while both WT and Rag1-KO mice acquired from Jackson Labs were similarly prone to initial colonization by SFB, WT, but not Rag1-KO, mice constrained SFB loads by 1000-fold from peak levels of colonization (Figure 4G), thus potentially explaining why GSU-RAG feces provided lasting protection of JAX-RAG mice but only temporary protection of WT mice.
To interrogate the SFB strain associated with RV-resistance, its genome was sequenced and analyzed in parallel with a previously-sequenced reference strain provided to us by Cerf-Bensussan and colleagues (Flannigan et al., 2017). While assembling genomes of bacteria maintained in monoassociated mice is more difficult than doing so from bacteria cultured in vitro, we were able to assemble genomes of both strains at over 94% coverage. Relative to the reference strain (Schnupf et al., 2013; Schnupf et al., 2015), referred to here as “Pasteur-SFB” (SFB-P), the “GSU-SFB” (SFB-G) strain displayed a more compact genome, 12.2% smaller than SFB-P (Figure 4H). SFB-G lacked 272 genes that were present in SFB-P, most of which were present in five other SFB strains with sequences in NCBI. Nonetheless, SFB-G contained 133 genes not present (at a similarity > 90%) in SFB-P, of which 28 were absent in any previously sequenced SFB isolate. While genes unique to SFB-G were of broad function (Table S1), several were classified as being phage-related. Genes absent in SFB-G (relative to SFB-P) also were broadly categorized but included many genes related to cell wall biosynthesis and modification suggesting possible differences in surface structures.
We also analyzed the non-SFB sequencing reads present in fecal samples of mice monoassociated with SFB-G and SFB-P as a potential means of identifying additional microbes that might contribute to RV-resistance. Since both strains had been amplified in monoassociated mice fed autoclaved chow, non-SFB sequences common to both strains likely reflected dead microbes present their diet. As expected, an array of viral, bacterial (non-SFB), and fungal sequences were detected. However, such sequences did not differ significantly in mice monoassociated with these two SFB strains (Figure S5A). Analogously, while sequences of eukaryotic parasites, such as protozoa, are less easily filtered from sequencing reads, we did not identify any differences in sequences analogous to protozoa such as T. muris in SFB-G and SFB-P. Overall, these data argue against a role for non-SFB microbes in potential phenotypic differences induced by these two SFB isolates.
GSU-RAG Microbiota and SFB-G Alter Gut Epithelium
SFB is very challenging to culture in vitro (Schnupf et al., 2015). Therefore, as an alternate means of generating potentially cleaner stocks of GSU-SFB, we passaged it in immune-competent GF mice, thus removing any agents that cannot persist in WT mice. Next, we sought to determine the extent to which such SFB protected against RV. In vitro, SFB-G recapitulated the capacity of GSU-RAG feces to reduce RV infection of epithelial cells (Figure S5B). That SFB, which is a strict anaerobe, would likely not be viable in this assay, in which oxygen was not excluded, suggests that a surface component of SFB may alter RV to diminish its infectivity. On the other hand, since SFB is thought to intimately interact with epithelial cells, we considered the possibility that host responses to SFB might contribute to RV resistance. The impact of SFB on gene expression in gut epithelium was examined via RNA-Seq. JAX-RAG mice were orally administered PBS, JAX-RAG feces, GSU-RAG feces, or SFB-G. Two weeks later, mice were euthanized, and gene expression assayed by RNA-seq (25×106 mRNA reads per mouse per ileum). Analysis of relative average expression of all detected genes revealed few differences in mice transplanted with PBS or JAX-RAG feces, indicating that the fecal transplant procedure itself did not have a significant lasting effect on ileal gene expression (Figure 5A). In contrast, relative to control mice, administration of GSU-RAG feces induced an array of changes in gene expression, which was partially mimicked by addition of SFB-G (Figure 5A). Analysis of these data by principle coordinate analysis (PCA), wherein PC1 accounted for 91.5% of the difference among the four groups, indicated that samples from mice treated with PBS and JAX-RAG feces were similar to each other and distinct from those mice treated with GSU-RAG feces and SFB-G, which were similar to each other (Figure 6B and Table S2). To further assess the differences between these groups, we constructed a heat map of all genes that were significantly changed (P < 0.05) by more than 2-fold on average in any of the three treatment groups relative to PBS-treated mice. This analysis further supported the conclusion that, qualitatively, GSU-RAG feces and SFB-G induced similar changes in gene expression, although the extent of such changes were greater in mice administered GSU-RAG feces (Figure 6C). We also quantified levels of SFB genomes in the same ileal samples used to assay host gene expression. There was a clear correlation in levels of SFB and the extent of changes in host gene expression, both within, and especially between, conditions wherein administration of GSU-RAG feces resulted in about 10-fold higher level of SFB-G than did administration of SFB-G itself (Figure 6D). Transcripts in greater abundance in mice treated with GSU-RAG feces and SFB-G comprised several functional categories (Figure S6A), including those related to cellular turnover (Figure 6A), which supported our observation of increased ileal enterocyte proliferation (Figures 5A and 5B). Assay of gene expression via qRT-PCR confirmed RNA seq results (Figure S6B). Collectively, these results suggest that other microbes present in GSU-RAG mice aid colonization of SFB and subsequently enhance SFB-induced changes in host gene expression that might contribute to the RV-resistant phenotype.
Figure 5. GSU-RAG Microbiota and SFB-G Alter Host Gene Expression.
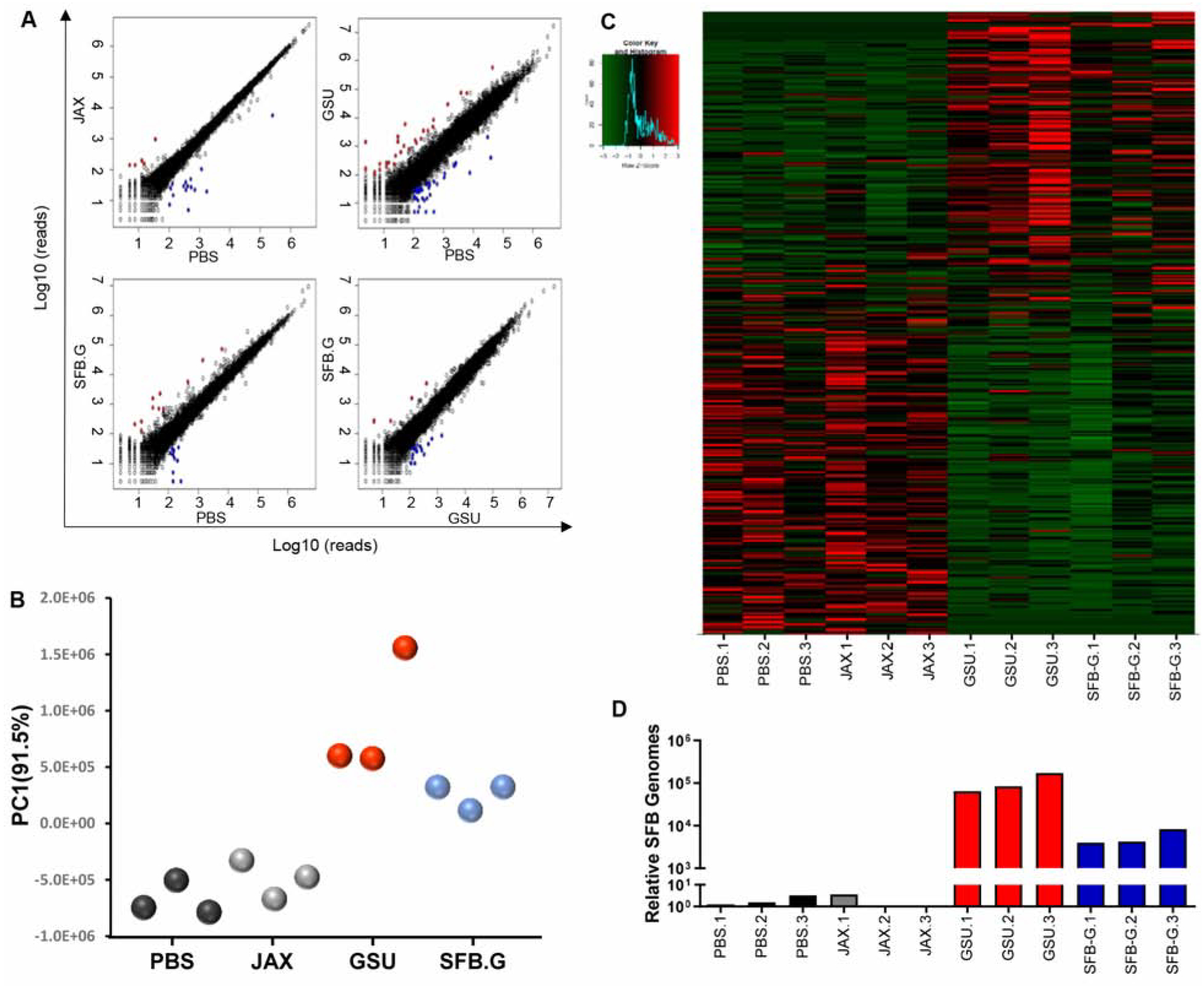
Rag1-KO mice were orally administered PBS or feces from JAX-RAG, GSU-RAG, or SFB-G-monoassociated mice. Ileum was harvested at 2 weeks post-FT for RNAseq analysis.
(A) Linear gene expression comparison.
(B) PCA plot of PC1.
(C) Heatmap of genes significantly altered (P < 0.05) by two-fold in any group relative to the PBS control group.
(D) Levels of SFB genomes in the same ileal samples determined by real-time PCR. N=3 mice per condition.
Figure 6. GSU-RAG Microbiota and SFB-G Increase Gut Epithelial Proliferation and Shedding.
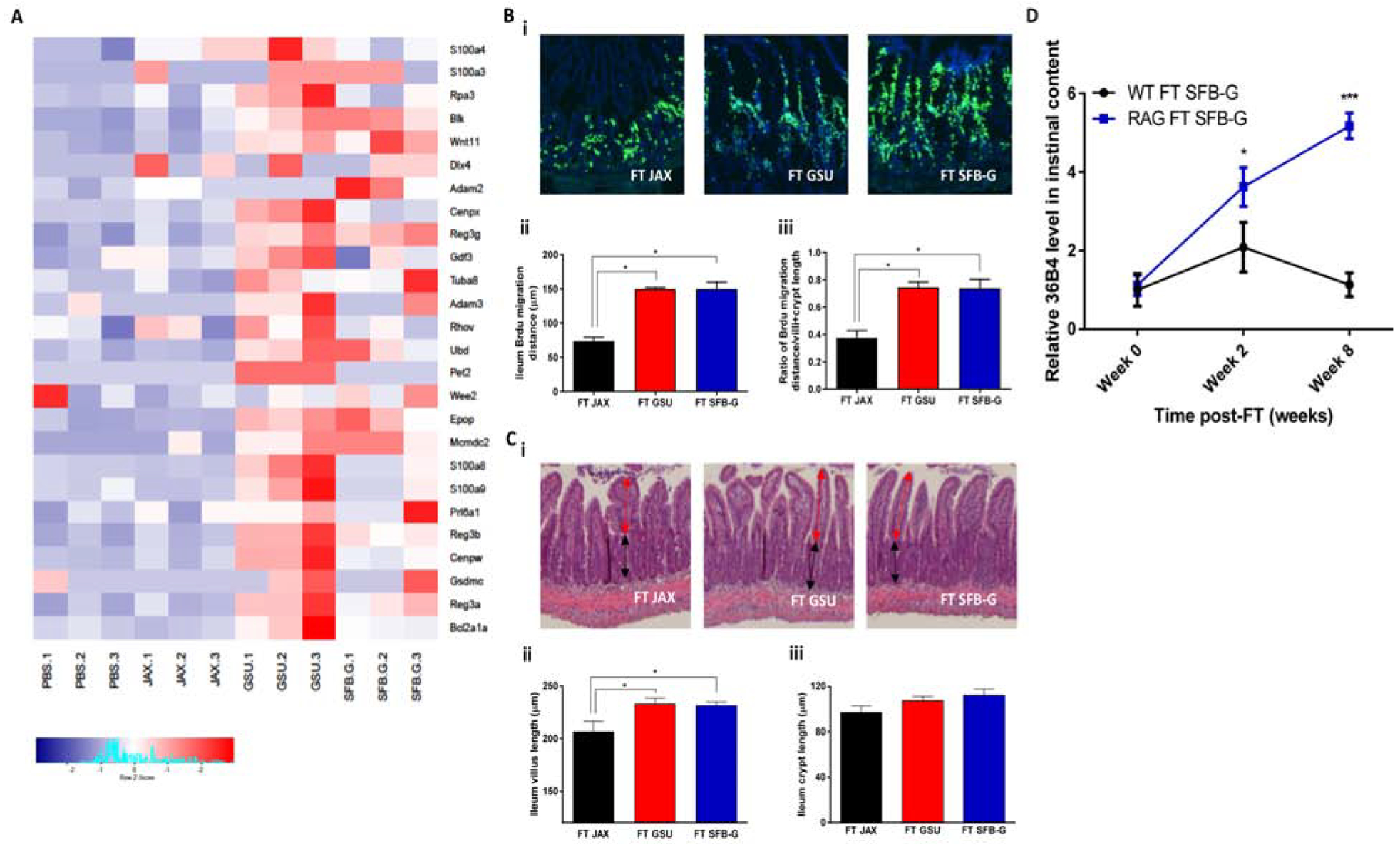
Rag1-KO mice or WT mice were orally transplanted with feces from JAX-RAG, GSU-RAG, or SFBG-monoassociated mice. Ileum was harvested at 1 week.
(A) Heatmap of genes, which are related with cell cycle, cell phase, cell proliferation, and cell death that are significantly altered (P < 0.05) by two-fold in any group relative to the PBS control group. N=3 mice per condition.
(B) BrdU was inoculated intraperitoneally into transplanted Rag1-KO mice 24 hours before harvesting the tissues.
(C) (i) H&E staining of Rag1-KO mice ileum. (ii) Villus length, indicated by the red arrow, was quantified. (iii) Crypt length, indicated by the black arrow, was quantified.
(D) Ileum contents were harvested at 0, 2, and 4 weeks post-transplantation, and cells were quantified by qPCR.
Results are shown as mean ± SEM (Student’s t test). A; n = 6, *, P < 0.001. B; n = 6, *, P < 0.05. C; n = 4, *, P < 0.05. D; n = 4, *, P < 0.05; ***, P < 0.001. Each experiment was performed two times and yielded an identical pattern of results.
Our observation that SFB enriched expression of genes related to cell proliferation/turnover (Figure 5E) is consistent with reports that SFB promotes enterocyte proliferation and migration along the crypt-villus axis (Leppkes et al., 2014; Okumura and Takeda, 2017; Park et al., 2016). We have recently come to appreciate that bacterial flagellin and IL-22 protect against RV infection, in part, by stimulating enterocyte proliferation, thus increasing luminal shedding of villus tip epithelial cells, which are the primary target of RV infection (Zhang et al., 2019). While RV-resistance of GSU-RAG mice does not involve IL-22 (Fig 2h), SFB might increase epithelial proliferation by other means, which we hypothesized might contribute to RV resistance. Hence, we assayed whether administration of GSU-RAG feces affects enterocyte migration and shedding as well as the extent to which such effects would be recapitulated by administration of SFB-G. JAX-RAG mice were orally inoculated with GSU-RAG feces or SFB-G and, two weeks later, were injected with BrdU, which allows enterocyte proliferation and migration to be imaged. Both treatments increased relative and absolute enterocyte migration and resulted in modest villus elongation (Figures 6B, C). Such enhanced enterocyte migration increased shedding of villus epithelial cells, referred to as anoikis, which might protect against RV infection. Therefore, we determined whether SFB-G increased shedding of host cells into the ileal lumen. Administration of SFB-G and GSU-RAG feces lastingly increased cell shedding in JAX-RAG mice (Figure 6D). In contrast, SFB-G only transiently increased this parameter in WT mice, which correlates with SFB levels at these time points (Figure 4G). These results suggest that SFB promotion of enterocyte proliferation, migration, and subsequent luminal shedding contributed to its protection against RV.
SFB-G Confers Resistance to RV Infection
Having characterized the effect of SFB-G on RV-infectivity and gut epithelium, we next investigated the extent to which it was sufficient to promote RV-resistance. First, we administered SFB-G to GF Rag1-KO mice, which were inoculated with RV one week later. GF Rag1-KO that had received SFB-G or GSU-RAG feces were fully protected from RV infection (Figure 7A). As a further means to decrease the possibility that SFB-G, which is passaged in vivo, contained other microorganisms, we treated it with chloroform, which does not inactivate SFB spores. Chloroform did not reduce the capacity of SFB-G to protect GF mice from RV infection, providing further evidence that protection was indeed mediated by SFB-G itself. Next, we examined whether administration of SFB-G to conventional JAX-RAG mice, which have an endogenous microbiota, protected against subsequent RV infection. In accord with our initial observations, administration of GSU-RAG feces fully prevented fecal RV shedding. In contrast, SFB-G reduced but did not entirely prevent RV infection (Figure 7B). Analysis of RV genomes in the ileum confirmed lack of detectable infection in mice administered GSU-RAG feces and indicated a 50-fold reduction in RV levels in mice administered GSU-SFB (Figure 7C), in concordance with mice administered GSU-RAG feces displaying higher SFB levels (Figure 6D). Relative to SFB-G, SFB-P only partially protected against RV infection in GF mice and modestly prevented RV infection in conventional Rag1-KO mice, suggesting that differences in these SFB strains contribute to RV-resistance (Figure 7A–C). In contrast to SFB, administration of another bacterial species that associates closely with the intestinal mucosa, Akkermansia muciniphilia, did not impact RV infection (Figure S7A). Nor were RV levels altered by Lactobacillus rhamnosis GG, which ameliorates RV-induced diarrhea (Figure S7B) (Zhang et al., 2013).
Figure 7. SFB Confers Strain-Specific Resistance to RV Infection.
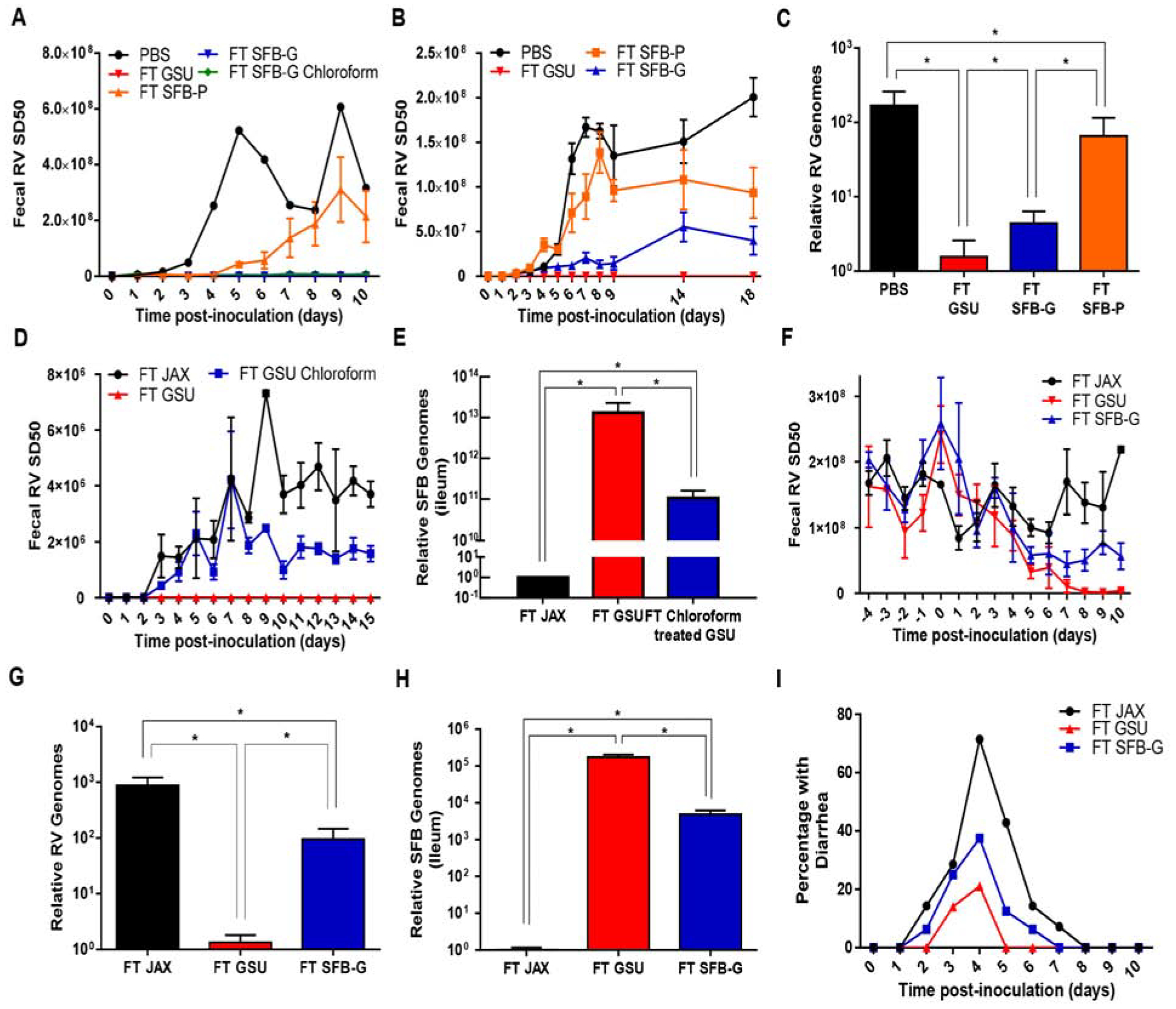
Rag1-KO mice were orally inoculated with RV, EC strain. Feces were collected daily, and fecal RV antigen was quantified by ELISA. Ileum or fecal samples were collected for assaying SFB or RV levels by qPCR.
(A) GF Rag1-KO mice received FT from PBS, GSU Rag1-KO mice, Pasteur-SFB (SFB-P), SFB isolated from GSU Rag1-KO mice (SFB-G), or chloroform-treated (3% at RT for 1 hour) SFB-G for 1 week prior to RV inoculation.
(B-C) (B) Conventional JAX Rag1-KO mice received FT from PBS, GSU Rag1-KO mice, SFB-P, or SFB-G for 1 week prior to RV inoculation. (C) Relative RV genome levels in the recipient mice.
(D-E) (D) Conventional JAX Rag1-KO mice received FT from PBS, GSU, or chloroform treated (3% at RT for 1 hour) GSU Rag1-KO mice for 1 week prior to RV inoculation.
(E) Ileum SFB level by q-PCR.
(F-H) SFB-G significantly reduced RV shedding in chronically infected recipients. (F) Rag1-KO mice chronically infected with RV received FT from JAX, GSU Rag1-KO mice, or SFB-G. (G) qRT-PCR results of ileum RV genome levels at day 10 post-transfer. (H) Ileum SFB levels 1-week post FT.
(I) Seven-day-old WT C57BL/6 mice pups received FT from JAX, GSU, or SFB-G mono-associated Rag1-KO mice 6 d and 1 d prior to RV inoculation.
Results are shown as mean ± SEM (two-way ANOVA). A; n = 3, P < 0.0001. B; n = 3, P < 0.0001. C; Student’s t test, n = 3, *, P < 0.01. D; n = 3, P < 0.01. E; Student’s t test, n = 3, *, P < 0.05. F; n = 3, P < 0.001. G-H; Student’s t test, n = 3, *, P < 0.01. I; Both FT GSU and FT SFB-G differed significantly from control (FT JAX) group by one-way ANOVA, n = 8 mice, P < 0.05. Each experiment was performed 2–3 times and yielded an identical pattern of results.
To determine whether other microbes present in GSU-RAG feces enhance SFB colonization, GSU-RAG feces were treated with chloroform, which destroys many bacterial species but not SFB spores. This treatment reduced but did not eliminate the capacity of GSU-RAG feces to prevent RV infection and resulted in a 100-fold reduction in the levels of SFB adhered to the ileum 3 weeks post-administration (Figures 7D and 7E). An analogous pattern of results was observed in the context of established chronic RV infection in that 10 days following administration of GSU-RAG feces to conventional JAX-RAG mice that had been chronically infected with RV resulted in elimination of RV, as assessed by both fecal RV antigen and RV RNA in ileal lysates, while administration of SFB-G reduced but did not completely eliminate RV infection (Figures 7F and 7G). Again, effects of these treatments on RV levels correlated with levels of SFB colonization (Figure 7H). These results support the conclusion that levels of SFB in the intestine are correlated with the extent of the RV-resistant phenotype and that other microbes present in GSU-RAG mice promote SFB colonization. Lastly, we determined whether SFB-G colonization of the ileum was sufficient to ameliorate RV-induced disease in immune-competent hosts. We found that SFB-G largely recapitulated the capacity of GSU-RAG feces to suppress RV-induced diarrhea in newborn WT mice (Figure 7I), highlighting the potential utility of select gut microbiota members to prevent and treat viral infections.
DISCUSSION
Enteric viral infections remain a major public health challenge. Persons with immune dysfunction are susceptible to developing chronic infections by normally transiently-infecting diarrheagenic viruses such as RV and norovirus (Haessler and Granowitz, 2013; Hennewig et al., 2007). Moreover, despite the advent and dissemination of vaccines, RV continues to cause millions of hospitalizations and over 200,000 pediatric deaths annually in children not known to be immune-compromised (Crawford et al., 2017). While most humans are exposed to RV multiple times, there is substantial heterogeneity in disease penetrance and severity both geographically and within a given population. Some RV infections result in only mild disease, while others cause severe life-threatening diarrhea. Environmental determinants of RV disease heterogeneity are not well defined. Since enteric viruses first encounter host cells amidst microbiota, we hypothesized that microbiota composition might influence susceptibility to RV infection. Studies seeking to broadly define the role of the microbiota in enteric viral infections revealed that mice lacking a microbiota are protected against RV and other enteric viruses, suggesting that such viral pathogens have developed a means to exploit the gut microbiota to aid their infection (Karst, 2016; Pfeiffer and Virgin, 2016; Shi and Gewirtz, 2018). However, we found that the extent to which the microbiota aids or impedes viral infection is dependent on the specific microbiota composition of the host with one bacterial species in particular, SFB, being a strong contributor to resistance to RV infection and disease. Thus, microbiota composition appears to be a key environmental variable in dictating the severity of enteric viral infection.
The notion that gut microbiota composition can determine resistance to RV infection stemmed from Rag1-KO mice, which serve as a model of chronic RV infection, unexpectedly developing profound resistance to RV infection. Development of a tractable means of transferring RV resistance by fecal transplant enabled us to discover that such RV resistance was mediated by transfer of gut bacteria, particularly SFB, which protected against RV infection. Such unintended acquisition of SFB colonization and RV-resistance was first appreciated at GSU but also occurred in a colony of immunodeficient mice at Wash-U., suggesting that lateral microbial transfers occur frequently but may have a particularly significant effect in immune-compromised hosts.
Administration of two distinct SFB strains, SFB-G and SFB-P, which were isolated from distinct mouse colonies and vivaria, to GF Rag1-KO mice reduced RV infectivity, suggesting that SFB alone confers RV resistance. However, the protection conferred by SFB-G was much stronger than that conferred by SFB-P, highlighting a potential role for bacterial strain-specific differences in this phenotype. When administered to mice that have a microbiota, specifically JAX-RAG mice, SFB-G only partially recapitulated the complete protection against RV infection conferred by GSU-RAG feces, indicating a role for additional microbes in mediating RV-resistance. We cannot exclude a function for unidentified microbes in protecting against RV infection regardless of the presence of SFB. However, since removal of non-SFB microbes, which was achieved by use of either feces from SFB-G monoassociated mice or chloroform treatment, markedly reduced levels of SFB in the ileum, the contribution of non-SFB microbes to RV resistance can, at least theoretically, be explained by such microbes supporting RV colonization. While specific mechanisms by which one bacterial species can promote growth of another are not well defined, it is possible that select bacteria provide key nutrients to SFB, alter the host in a manner that aids SFB growth or adherence, or a combination of these properties.
SFB administration reduced RV shedding and decreased the incidence of RV-induced diarrhea in conventionally raised immune-competent neonatal mice. This observation provides a foundation for the development of new approaches to prevent and treat RV infection. However, since SFB promotes development of Th17 cells, which is associated with, and may exacerbate, chronic inflammatory diseases, such approaches should be considered with caution. Nonetheless, the lack of overt pathogenicity by SFB, even in severely immunocompromised hosts, and its capacity in WT mice to confer immune-mediated protection against the bacterial pathogen, Citrobacter rodentium, suggest that SFB should be considered a commensal if not a beneficial organism (Schnupf et al., 2017). Accordingly, we observed that SFB was uniformly present in “pet-store mice” (Figure S7C), which display immune responses more reminiscent of humans than laboratory mice (Beura et al., 2016). Moreover, we observed that among mice purchased from Taconic Biosciences, which offers C57BL/6 mice with or without SFB, the SFB-containing mice cleared RV more efficiently, thus supporting an antiviral benefit of this bacterial species even in immune-competent hosts (Figure S7D). In any case, since the capacity of either GSU-RAG mice microbiota or SFB to protect against RV does not depend on Th17 cells, it may be possible to stimulate RV-resistance without inducing potentially disease-promoting adaptive immunity. Limited studies suggest that SFB is a common constituent of the microbiota of infants and young children in China, a country that seems to lack the severe RV-induced disease burden seen in some countries despite limited use of RV vaccines (Chen et al., 2018).
Mechanisms by which SFB protect against RV infection remain unclear. Our results support non-mutually exclusive possibilities that SFB alters RV directly and/or acts on the intestinal epithelial cells that RV infects. In support of the former, we observed that incubation of RV with SFB results in reduced capacity of RV to bind and infect epithelial cells in vitro. Such disabling of RV, which is a highly stable virus, was not accompanied by enhanced degradation of RV genomes or proteins. Nor did our results suggest that RV was directly bound by SFB or GSU-RAG feces, which, in any case, seems at least as likely to stabilize virions and, consequently, promote, rather than impede infection (Aguilera et al., 2019; Erickson et al., 2018). Based on this in vitro evidence, we speculate that SFB might impede a surface component of RV used in binding to epithelial cells, which might contribute to the capacity of SFB to impede RV infection.
Conversely, the effects of SFB on gut epithelium, which were observed in vivo, highly likely contribute to, and may well be the predominant mediator of, the RV-resistance phenotype. This idea is in accord with our initial hypothesis whereupon realizing that GSU-RAG mice were resistant to RV, we thought that they had acquired a microbiota enriched in motile bacteria that resulted in TLR5-mediated production of IL-22 and NLCR4-mediated maturation of IL-18, which confer RV-resistance in response to bacterial flagellin. Investigation of this possibility yielded results that excluded roles for these cytokines. However, our recent investigation of the mechanism by which these cytokines, particularly IL-22, impede RV infection suggests that SFB may mediate RV-resistance by a similar process. IL-22 accelerates the transport of enterocytes from the crypt to the villus tip, where these cells are normally shed while dying, such that infected cells might be sloughed before they release infectious virus. Despite not inducing or requiring IL-22, SFB-mediated induction of RV-resistance was associated with enhanced enterocyte proliferation and shedding suggesting that SFB confers RV resistance using a mechanism analogous to IL-22. SFB induces the expression of several genes governing cell proliferation. SFB did not appear to induce expression of genes with known antiviral functions, but some genes, e.g., IL-22, have indirect antiviral activities that are not fully appreciated. Thus, it will be important to identify the key genes activated by SFB that lead to RV resistance.
Improved understanding of mechanisms by which SFB promotes RV-resistance may ultimately lead to new approaches to prevent and treat viral infections. Investigation of the effect of SFB on the host may reveal previously unappreciated innate antiviral signaling pathways, while the microbiota itself might be a possible source of antiviral agents. Consistent with the latter concept, searching a marine microbial ecosystem for antiviral compounds yielded cyanovirin-N, a protein produced by a Cyanobacteria strain isolated from algae, which displays direct antiviral activity against HIV (Boyd et al., 1997). Yet, while environmental sources of microbiota produce compounds with enormous biochemical diversity, microbiota that have co-evolved with relevant hosts and viruses are more likely to produce molecules that impede infection by pathogenic viruses, directly or via effects on the host. While viruses may have adapted to some of these products to promote infection, e.g., some enteric viruses bind LPS to facilitate entry, we speculate that other microbiota products yet to be discovered diminish viral infection and could be harnessed to develop new, potentially broad-spectrum antiviral strategies.
LEAD CONTACT AND MATERIALS AVAILABILITY
The mouse line, Rag1/IL2Rγ-DKO was generated and remains housed at GSU. These mice, feces from GSU-RAG and SFB-G monoassociated mice are available subject to a standard academic MTA. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Andrew T. Gewirtz (agewirtz@gsu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice used in this study were 6 to 8-weeks-old adult mice of C57BL/6 background, unless otherwise specified (diarrhea studies used 7–8 days old mice, early FT studies used 3-weeks-old mice). All individual experiments used gender-matched mice with overall study using approximately similar amounts of male and female mice. Breeding colonies of WT C57BL/6 and Rag1-KO (RAG) and IFNαR1-KO, and Il2rγ-KO were established from mice purchased from Jackson Laboratories (Bar Harbor, ME) and maintained at GSU by vivarium staff following American Association for Laboratory Animal Care procedures. Mice referred to as “JAX-RAG” were bred at GSU using autoclaved cages, chow (LabDiet 5010/5021) and water by the authors of this manuscript. C57 BL/6 Tcrβ-KO, Ighm-KO mice (commonly referred to as μMT) were not bred at GSU but purchased directly from Jackson Labs and used for experiments within two weeks of arrival at GSU. Rag1/IFN-λ-R1-DKO, along with control Rag1-KO mice, were bred at Wash-U. GF mice (C57BL/6 and Swiss-Webster) were purchased from Taconic Biosciences (Rensselaer, NY). GF C57BL/6 Rag1-KO mice were generously provided by Dr. Lora Hooper (UT Southwestern). GF mice were maintained in glovebox isolators (Park Bioservices, Groveland MA) and IsoCages (Techniplast Inc. Buguggiate (VA Italy). Genetic authenticity of all KO mice breeding colonies was confirmed by a commercial provider (Transnetyx, Memphis TN). All experiments using mice were conducted under the surveillance of the Georgia State University Institutional Animal Care and Use Committee. As part of such surveillance, sentinel mice that were exposed to used bedding from GSU-RAG mice were subjected to serologic testing by IDEXX Analytics (Columbia MO), specifically their “Edx Comprehensive Profile”, and found to be negative for all microbes except mouse norovirus, which is commonly present in many mouse vivaria.
Viruses
Murine RV EC strain, which was used for all mouse studies, and cell-culture adapted RRV, which was used for in vitro infections, were propagated and titered as previously described. (Burns et al., 1995; Zhang et al., 2014). Mouse RV strain EC was kindly provided by Dr. Mary Estes (Baylor College of Medicine). Reovirus strain type 1 Lang (T1L) was propagated as described (Brown et al., 2018). Recombinant vesicular stomatitis virus harboring a nano luciferase ORF as an additional transcription unit and influenza virus A/WSN/1933 (H1N1) encoding nano luciferase in the NS1 segment were engineered and propagated as described (Yan et al., 2015; Yoon et al., 2018). Viral titers were determined by plaque assays using Vero-E6 and MDCK cells, respectively.
Cell lines
HT29 (HTB38™) and MA-104 (CRL2378.1™) cell lines were obtained from ATCC® and maintained with 10% heat-inactivated FBS (cellgro 35–010-CV) in DMEM (cellgro 10–013-CV).
METHOD DETAILS
Virus Infection
Acute RV infection: Age and sex-matched mice were orally administered 100 μl 1.33% sodium bicarbonate (sigma S5761) followed by oral inoculation with 104-105 SD50 of murine rotavirus EC strain in 100 μl PBS. Chronic RV infection: 3-week-old Rag1-KO mice were orally administered 100 μl 1.33% sodium bicarbonate (sigma S5761) followed by oral inoculation with 105 in 100 μl PBS. RV fecal antigens were measured by specific ELISA 3-weeks post RV challenge (4) (Jiang et al., 2008). Diarrhea model: 8-day-old pups were inoculated with 500 SD50 in 50 μl PBS. Acute reovirus infection: mice were orally inoculated with 1–4 × 108 PFU of reovirus strain T1L in 200 μl PBS (5) (Johansson et al., 2007). In vitro RRV infection: HT-29 cells were washed with serum-free RPMI-1640 medium 3 times and incubated serum-free RPMI-1640 medium for 1 hour before viral challenge. RRV was trypsin-activated (10 μg/ml) for 1 hour in 37°C water bath. Then, virus media of a MOI=1 was prepared in serum-free RPMI-1640 (cellgro 10–040) and used to inoculate the HT-29 cells. 1-hour post-inoculation, the viral media was removed, and fresh serum-free RPMI-1640 was applied to the infected cells (Frias et al., 2010). For virus treatment with microbiota, trypsin-treated RRV or reovirus were incubated for 4 hours at 37°C with 5 μm- or 0.2 μm-filtered mice feces; such RV-microbiota suspensions were passed through 0.2 μm filters to remove bacterial cells and then used for in vitro and ex vivo experiments. VSV and IAV WSN were co-incubated with 5 μm-prefiltered mouse feces for 4 hours at 37°C, followed by 0.2 μm filtration and transfer of the filtrates to human respiratory BEAS-2B cells in a 96-well plate format. After 24-hour incubation at 37°C, luciferase activities were determined in a Synergy H1 microplate reader (BioTek) using Nano-Glo luciferase substrate (Promega). The substrate was directly added to each well and bioluminescence quantified after a 3-minute incubation for signal stabilization.
RV-Antigen-Specific ELISA
A sandwich enzyme-linked immunosorbent assay (ELISA) was performed to detect rotavirus antigen in mouse feces as previously described (Zhang et al., 2014). Briefly, mouse Fecal Rotavirus shedding was analyzed via Sandwich Elisa. Ninety-six well EIA/RIA plates (Costar, 3590) were coated with Rabbit Anti-Rotavirus Group-A (Biorad, AHP1360) capture antibody at 1:1000 dilution in PBS overnight RT, and subsequently blocked by 1% BSA PBS. Mouse fecal homogenates were prepared at 100 mg/mL concentration and centrifuged at 10,000g to remove debris. Supernatants of the homogenates were then incubated in test tubes with serial dilutions of stock mouse Rotavirus utilized as control. Hyperimmune guinea pig anti-RRV serum (Prfal) at 1:1000 dilution in 1% BSA PBS was incubated as detection antibody followed by the incubation of HRP Donkey Anti-Guinea Pig Antibody (Jackson ImmunoResearch, 706-035-148) secondary antibody at 1:10000 dilution in 1% BSA PBS. All incubation steps following capture antibody were at one hour at room temperature. TMB ELISA Substrate Solution (Invitrogen, 00420156) was utilized to develop the signal, followed by addition of TMB Stop Solution (KPL, 50-85-04) after ten minutes of substrate incubation. OD readings were taken at 450nm with 540nm OD subtracted as a correction (Zhang et al., 2014).
Microbiota Transplantation
Donor fecal samples were freshly collected and then suspended in 20% glycerol PBS (21–040-CV) solution at a concentration of 20 mg/ml, passed through a 5 μm filter, aliquoted, and stored in −80°C. Such frozen fecal suspensions were administered to recipient mice by oral gavage using 400 μl per adult mouse and 50 μl per suckling mice. Adult recipients received a single transplant at 6–8 weeks of age. Suckling mice received transplants 2 and 7 days after birth at which times their dams were transplanted via oral gavage and spread of fecal suspension (200 μl) on their belly fur, as a potential means of reducing incidence of maternal rejection of their pups. For antibiotic treatment, before FT, fecal microbiota was firstly prepared as described above, and then incubated with Ampicillin Sigma A6140 50 mg/ml, Neomycin Sigma N6386 50 mg/ml, Streptomycin Sigma S9137 50 mg/ml, Kanamycin Sigma K1377 50mg/ml, or the 4 antibiotics combined at 37°C for 4 hours. For anti-fungal treatment, before FT, fecal microbiota was firstly prepared as described above, and then incubated with anti-fungal cocktail (amphotericin Sigma A9528 2 mg/ml, 5-fluorocytosine Sigma F7129 2 mg/ml, fluconazole Sigma F8929 2 mg/ml) at 37°C for 4 hours.
Antibiotic and Antifungal Drinking Water Treatment
Antibiotic cocktail drinking water (ampicillin, Sigma A6140, 1 g/L, neomycin, Sigma N6386, 1 g/L, streptomycin, Sigma S9137, 1g/L, and kanamycin, Sigma K1377, 1g/L) and/or anti-fungal cocktail drinking water (amphotericin, Sigma A9528, 0.2 g/L, fluconazole, Sigma F8929, 0.5 g/L, and 5-fluorocytosine, Sigma F7129, 0.5 g/L) were prepared and provided for consumption for 3 weeks before RV inoculation.
Neutralization of IL-17, IL-22, and IFN-λ
IL-17 (100 μg), IL-22 (150 μg) neutralizing mAb (Genentech, Inc) were administered by intraperitoneal inoculation shortly prior to RV inoculation and thereafter every other day until day 10 post-infection. Neutralizing mAb against IFN-λ (R&D MAB17892, 1μg/gram of body weight) was administered by intraperitoneal inoculation to mice one day prior to RV inoculation or fecal transplantation and continued for the entire experiment. The dosing regimen of these antibodies neutralizes bioactivity of these cytokines (Basu et al., 2012; Crotta et al., 2013; Kezic et al., 2012).
Fluorescence Immunohistochemistry
Intestine tissue was immediately isolated after euthanasia and imbedded into O.C.T. compound on dry ice. The samples were then sliced into 6μm thickness onto glass slides, which were then incubated in 4% paraformaldehyde for 15 minutes, followed by 5 minutes washing of PBS twice. The slides were then incubated in 100% methanol for 5 minutes, followed by 5 minutes PBS washing 3 times. Then, 3% BSA-PBS was used to block the samples for 1 hour at RT. The slides were then washed with PBS for 5 minutes, followed by incubation of primary antibodies cocktail (Villin-1 Antibody, Cell Signal 2369 1:100, Hyperimmune guinea pig anti-RRV serum, Prfal, 1:100), in 1% BSA-PBS overnight at 4°C. The slides were then washed 3 times with PBS, 5 minutes each time. Then, secondary antibodies cocktail (Jacksonimmuno 706-586-148 Amax: 591 nm Emax: 614 nm 1:500; 711-095-152 Amax: 492 nm Emax: 520 nm) 1%BSA-PBS was applied for 1–2 hours at RT. The samples were then 3 X washed with PBS. Images were generated using a UV microscope under the relative excitation wavelengths as given by the manufacturer.
H&E and BrdU Staining
Mouse ileum was fixed in 10% buffered formalin and then embedded in paraffin. The tissues were sectioned at 5 μm thickness and stained with hematoxylin and eosin (H&E). For BrdU staining, the mice were I.P. injected with BrdU (50 μg of BrdU/g). One day post-injection, ileum was harvested and embedded in OCT. The tissues were divided into 5 μm sections and fixed with 4% formaldehyde for 30 min at room temperature. Villi and crypts, as well as epithelial cell migration and villi length, were determined using Image J.
Bacterial and Viral Binding/Degradation Assay
Fecal samples from mice were collected and suspended in HEPES (20mM)-Glycerol (20%)-PBS into a concentration of 10mg/ml. The fecal suspensions were then filtered through 5 μm filter to remove large food debris. Purified RRV was then incubated with the fecal filtrate for various times, and total RNAs were extracted for RV genome detection.
qRT-PCR
Intestine tissue was homogenized in TRIzol™ Reagent, and mRNA was isolated according to the manufacturer’s instruction (Thermofisher 15596026). Intestine DNA was also isolated using DNeasy Blood & Tissue Kits (Qiagen 69504). Stool bacterial DNA was isolated with QIAamp DNA Stool Mini Kit (Qiagen 51504). Stool total RNA was isolated using RNeasy PowerMicrobiome Kit (Mobio 26000–50), and qRT-PCR was performed using the Biorad iScriptTM One-Step RT-PCR Kit with SYBR green (Bio-Rad 170–8893), and qPCR was performed with QuantiFast SYBR Green PCR Kit (Qiagen 204054) in Bio-Rad CFX96 apparatus (Bio-Rad 170–8892). For quantitation of luminal enterocytes (i.e., cell shedding), the ileum content was collected. Total RNA was then extracted, and qRT-PCR was performed on the 36B4 gene to determine relative shedding for different groups. The sense and antisense oligonucleotides primers used were:
EC.C 5′-GTTCGTTGTGCCTCATTCG-3′ and 5′-TCGGAACGTACTTCTGGAC-3′
36B4 5′-TCCAGGCTTTGGGCATCA-3′ and 5′-CTTTATTCAGCTGCACATCACTCAGA-3′
IFN-λ 5′-AGCTGCAGGCCTTCAAAAAG-3′ and 5′-TGGGAGTGAATGTGGCTCAG-3′
IL-22 5′-GTGCTCAACTTCACCCTGGA-3′ and 5′-TGGATGTTCTGGTCGTCACC-3′
IL-17 5′-TGAGCTTCCCAGATCACAGA 3′ and 5′ TCCAGAAGGCCCTCAGACTA 3′
16S rRNA: 5′-AGAGTTTGATCCTGGCTCAG-3′ and 5′-CTGCTGCCTCCCGTAGGAGT-3′
16S rRNA sequencing: 5′-AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCAGCMGCCGCGGTAA −3′ and 5′-CAAGCAGAAGACGGCATACGAGATxrefxrefAGTCAGTCAGCCGGACTACHVG GGTWTCTAAT-3′
IFN-γ 5′- GTCTCTTCTTGGATATCTGGAGGAACT-3’ and 5’ GTAGTAATCAGGTGTGATTCAATGACGC −3′
SFB: 5′-GACGCTGAGGCATGAGAGCAT-3’ and 5’-GACGGCACGGATTGTTATTCA-3′
Reovirus 5′-CGCTTTTGAAGGTCGTGTATCA-3′ and 5′-CTGGCTGTGCTGAGATTGTTTT-3′
The quantification results were analyzed to relative housing keeping genes (36B4) or feces weight, as previously described (Zhang et al., 2014).
Lamina Propria Transplantation and Flow Cytometry Analysis
Mouse intestinal lamina propria cells were isolated using a Lamina Propria Dissociation Kit (130-097-410) purchased from Miltenyi biotec (Bergisch, Gladbach, Germany) according to the manufacturer’s protocol. The prepared cells were then injected into recipient mice intravenously. For flow cytometry, the prepared cells were then blocked with 1 μg/million cells 2.4G2 (anti-CD16/anti-CD32 ATCC) in 100μl FACS buffer for 5 minutes at 4°C, followed by 1-time wash of FACS buffer to remove residue 2.4G2 and then incubated in conjugated mAbs Alexa Fluor 700-CD45.2, FITC- CD103, PE- F4/80, PerCP-cy5.5- CD11b, APC-MHC class II, PE-Cy7- CD11c, and Pacific blue-CD103 (BD Biosciences). The labeled cells were then washed 3 times in FACS buffer and then analyzed using a Becton-Dickinson LSR II flow cytometer. Data were analyzed using FlowJo (TreeStar, Ashland, OR) (Zhang et al., 2014).
SDS-PAGE and Immunoblotting
Cells were harvested in RIPA buffer. Rabbit anti-rotavirus group-A (Biorad, AHP1360) at 1/1000 dilution in blocking buffer was used as a primary antibody. 2nd antibody (GE healthcare NA934V) was used at 1/5000 dilution to detect the primary antibody. Western blot membrane was developed using detection reagents (GE healthcare RPN2106V1) (Zhang et al., 2014).
16S rRNA Sequencing
Bacterial 16S rRNA sequencing was performed as previously described (Chassaing et al., 2015a). Briefly, fecal microbiome DNA was extracted with QIAamp DNA Stool Mini Kit (Qiagen 51504). Then, the stool DNA was amplified by 4 rounds of PCR targeting the V4 region located within 16S rRNA gene. The intensified examples were then consolidated and purified with Ampure attractive XP beads (Beckman/Agencourt). The samples were then evaluated using gel electrophoresis for examining sample quality and quantified by utilizing a Quant-iT PicoGreen dsDNA assay (BIOTEK Fluorescence Spectrophotometer). The generated DNA pool was then sequenced on Illumina MiSeq sequencer with paired-end reads (Cornell University, Ithaca). The sequencing data was analyzed using Quantitative Insights into Microbial Ecology software (QIIME, version 1.8.0) (Chassaing et al., 2015).
SFB Genome Sequencing and Analysis
SFB genome sequencing was performed by The CHOP Microbiome Center. Briefly, DNA was extracted from cecal and fecal material using the DNeasy PowerSoil kit (Qiagen, Germantown, MD). Sequencing libraries were generated using the Nextera XT DNA Library Preparation Kit and were sequenced on the HiSeq 2500 using 2×125 bp chemistry (Illumina, San Diego, CA). Raw sequencing data were processed using the Sunbeam pipeline (Clarke et al., 2019), including quality control, removing host sequences, and mask low-complexity regions. Due to the high host mouse DNA percentage, we merged the processed reads from 2 cecum samples and 1 fecal sample for better coverage of the SFB genomes. We then aligned reads to all 13 SFB genomes in NCBI and collected all the mapped reads, using BWA (Li and Durbin, 2009), as “SFB reads”, and did de novo assembly using Spades (Bankevich et al., 2012). To evaluate the assembled contigs, we analyzed all the available SFB genomes on NBCI and as well as the assembled draft genomes for GSU and Pasteur-SFB strains using CheckM (Parks et al., 2015), and the estimated completeness for our assembled Pasteur and GSU-SFB strains are 99.01% and 93.89% (five reference genomes were removed from the phylogenetic analysis due to poor genome quality and completeness < 90%). Comparative genome analysis: Pairwise proteome comparison was carried out using Patric (Wattam et al., 2014). The pangenome analysis was carried out for two assembled genomes and eight SFB genomes using Roary (Page et al., 2015). The phylogenetic tree was built on the core genomes using FastTree (Price et al., 2009).
Ileum RNAseq
Total RNA from ileum was extracted using TRIzol™ Reagent (Thermofisher 15596026) and was further purified using Qiagen RNeasy MinElute Cleanup Kit (Cat No./ID: 74204). The prepared RNA samples were then sent to Molecular Evolution Core of Georgia Institute of Technology for library preparation and sequencing on the NextSeq instrument utilizing a high output 2×75 bp run. FASTQC was used for sequencing reads quality screening. The RNAseq data were then analyzed using the Galaxy server. Briefly, the alignment of RNAseq data was performed using HISAT2, and the aligned data were then assigned to the host genome (Ensembl mouse GTF) using HTSeq. The processed data were then analyzed on software R for preparations of linear gene expression comparison, PCA, and heatmap figures.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were plotted in GraphPad Prism (La Jolla, CA). Statistical significance was assessed by 2-way ANOVA, 1-way ANOVA, Student’s t-test, and Chi-square analysis.
DATA AND CODE AVAILABILITY
Unprocessed 16S sequencing data are deposited in the European Nucleotide Archive under accession numbers PRJEB34108. Unprocessed raw SFB genome sequencing data are deposited in the European Nucleotide Archive under accession numbers PRJEB34149. Unprocessed raw Ileum RNAseq sequencing data are deposited in the European Nucleotide Archive under accession numbers PRJEB34133.
Supplementary Material
HIGHLIGHTS.
Some mouse colonies developed spontaneous resistance to rotavirus (RV) infection.
Fecal microbial transplantation transfers RV-resistance.
Protection against RV was mediated by segmented filamentous bacteria (SFB).
SFB impeded RV by increasing epithelial cell turnover.
In brief - A bacterium can protect against rotavirus infection in mice independent of the immune system
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grants DK099071 (ATG), DK083890 (ATG), and AI038296 (TSD). BC is supported by Crohn’a and Colitis and Kenneth Rainin foundation. JZ is supported American Diabetes Association. H. Ingle is supported by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital Postdoctoral Research grant (MI-F-2018-712). M. Baldridge was supported by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (MI-II-2019-790). We thank Solomiia Khomandiak for technical support and Yang Wu for helpful advice and figure preparation.
Abbreviations:
- RV
Rotavirus
- SFB
Segmented filamentous bacteria
- FT
Fecal transplantation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
REFERENCES
- Aguilera ER, Nguyen Y, Sasaki J, and Pfeiffer JK (2019). Bacterial Stabilization of a Panel of Picornaviruses. mSphere 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge MT, Lee S, Brown JJ, McAllister N, Urbanek K, Dermody TS, Nice TJ, and Virgin HW (2017). Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro M, and Patton JT (2007). Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J Virol 81, 4473–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, Hatton RD, and Weaver CT (2012). Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37, 1061–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, et al. (2016). Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert D, Van Schil K, Taghon T, Bordon V, Bonroy C, Dullaers M, De Baere E, and Haerynck F (2016). Persistent rotavirus diarrhea post-transplant in a novel JAK3-SCID patient after vaccination. Pediatr Allergy Immunol 27, 93–96. [DOI] [PubMed] [Google Scholar]
- Boshuizen JA, Reimerink JH, Korteland-van Male AM, van Ham VJ, Koopmans MP, Buller HA, Dekker J, and Einerhand AW (2003). Changes in small intestinal homeostasis, morphology, and gene expression during rotavirus infection of infant mice. J Virol 77, 13005–13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O’Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM, et al. (1997). Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother 41, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JJ, Short SP, Stencel-Baerenwald J, Urbanek K, Pruijssers AJ, McAllister N, Ikizler M, Taylor G, Aravamudhan P, Khomandiak S, et al. (2018). Reovirus-Induced Apoptosis in the Intestine Limits Establishment of Enteric Infection. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JW, Krishnaney AA, Vo PT, Rouse RV, Anderson LJ, and Greenberg HB (1995). Analyses of homologous rotavirus infection in the mouse model. Virology 207, 143–153. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, and Gewirtz AT (2015). Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Chen H, Shu X, Yin Y, Li J, Qin J, Chen L, Peng K, Xu F, Gu W, et al. (2018). Presence of Segmented Filamentous Bacteria in Human Children and Its Potential Role in the Modulation of Human Gut Immunity. Front Microbiol 9, 1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke EL, Taylor LJ, Zhao C, Connell A, Lee JJ, Fett B, Bushman FD, and Bittinger K (2019). Sunbeam: an extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome 7, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthesy B, Benureau Y, Perrier C, Fourgeux C, Parez N, Greenberg H, and Schwartz-Cornil I (2006). Rotavirus anti-VP6 secretory immunoglobulin A contributes to protection via intracellular neutralization but not via immune exclusion. J Virol 80, 10692–10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford SE, Ramani S, Tate JE, Parashar UD, Svensson L, Hagbom M, Franco MA, Greenberg HB, O’Ryan M, Kang G, et al. (2017). Rotavirus infection. Nat Rev Dis Primers 3, 17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, and Wack A (2013). Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog 9, e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, and Pfeiffer JK (2018). Bacteria Facilitate Enteric Virus Co-infection of Mammalian Cells and Promote Genetic Recombination. Cell Host Microbe 23, 77–88 e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante NK, Lemire P, Cruz Tleugabulova M, Prescott D, Mortha A, Streutker CJ, Girardin SE, Philpott DJ, and Mallevaey T (2016). The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J Exp Med 213, 2841–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SH, Foster DM, Sherry B, Magness ST, Nielsen DM, and Gookin JL (2019). Interferon-lambda3 Promotes Epithelial Defense and Barrier Function Against Cryptosporidium parvum Infection. Cell Mol Gastroenterol Hepatol 8, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannigan KL, Ngo VL, Geem D, Harusato A, Hirota SA, Parkos CA, Lukacs NW, Nusrat A, Gaboriau-Routhiau V, Cerf-Bensussan N, et al. (2017). IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol 10, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias AH, Vijay-Kumar M, Gentsch JR, Crawford SE, Carvalho FA, Estes MK, and Gewirtz AT (2010). Intestinal epithelia activate anti-viral signaling via intracellular sensing of rotavirus structural components. Mucosal Immunol 3, 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haessler S, and Granowitz EV (2013). Norovirus gastroenteritis in immunocompromised patients. N Engl J Med 368, 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennewig U, Schulz A, Adams O, Friedrich W, Gobel U, and Niehues T (2007). Severe combined immunodeficiency signalized by eosinophilia and lymphopenia in rotavirus infected infants. Klin Padiatr 219, 343–347. [DOI] [PubMed] [Google Scholar]
- Hernandez PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, Gronke K, Ryffel B, Hoelscher C, Dumoutier L, et al. (2015). Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol 16, 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes MC, Feng N, and Greenberg HB (2005). Characterization of homologous and heterologous rotavirus-specific T-cell responses in infant and adult mice. J Virol 79, 4568–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JQ, He XS, Feng N, and Greenberg HB (2008). Qualitative and quantitative characteristics of rotavirus-specific CD8 T cells vary depending on the route of infection. J Virol 82, 6812–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C, Wetzel JD, He J, Mikacenic C, Dermody TS, and Kelsall BL (2007). Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J Exp Med 204, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JB, Johansen LH, Steiro K, and Johansen A (2003). CpG DNA induces protective antiviral immune responses in Atlantic salmon (Salmo salar L.). J Virol 77, 11471–11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, and Nunez G (2013). Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, and Golovkina TV (2011). Successful transmission of a retrovirus depends on the commensal microbiota. Science 334, 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM (2016). The influence of commensal bacteria on infection with enteric viruses. Nat Rev Microbiol 14, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kezic JM, Glant TT, Rosenbaum JT, and Rosenzweig HL (2012). Neutralization of IL-17 ameliorates uveitis but damages photoreceptors in a murine model of spondyloarthritis. Arthritis Res Ther 14, R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuklin NA, Rott L, Darling J, Campbell JJ, Franco M, Feng N, Muller W, Wagner N, Altman J, Butcher EC, et al. (2000). alpha(4)beta(7) independent pathway for CD8(+) T cell-mediated intestinal immunity to rotavirus. J Clin Invest 106, 1541–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, and Pfeiffer JK (2011). Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334, 249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes M, Roulis M, Neurath MF, Kollias G, and Becker C (2014). Pleiotropic functions of TNF-alpha in the regulation of the intestinal epithelial response to inflammation. Int Immunol 26, 509–515. [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, and Virgin HW (2015). Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I, Kimura T, Murakami T, Haruki K, Yamazaki K, Seto Y, Minekawa Y, and Funamoto H (1991). Serial observations of chronic rotavirus infection in an immunodeficient child. Microbiol Immunol 35, 953–961. [DOI] [PubMed] [Google Scholar]
- Okumura R, and Takeda K (2017). Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med 49, e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, and Parkhill J (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kotani T, Konno T, Setiawan J, Kitamura Y, Imada S, Usui Y, Hatano N, Shinohara M, Saito Y, et al. (2016). Promotion of Intestinal Epithelial Cell Turnover by Commensal Bacteria: Role of Short-Chain Fatty Acids. PLoS One 11, e0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, and Tyson GW (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JK, and Virgin HW (2016). Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, and Hornef MW (2011). IFN-λambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A 108, 7944–7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig RF (2004). Pathogenesis of intestinal and systemic rotavirus infection. J Virol 78, 10213–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, and Cossart P (2015). How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect 17, 173–183. [DOI] [PubMed] [Google Scholar]
- Riepenhoff-Talty M, Dharakul T, Kowalski E, Michalak S, and Ogra PL (1987). Persistent rotavirus infection in mice with severe combined immunodeficiency. J Virol 61, 3345–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld L, Mas Marques A, Niendorf S, Hofmann J, Gratopp A, Kuhl JS, Schulte JH, von Bernuth H, and Voigt S (2017). Life-threatening systemic rotavirus infection after vaccination in severe combined immunodeficiency (SCID). Pediatr Allergy Immunol 28, 841–843. [DOI] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, and Cerf-Bensussan N (2013). Host interactions with Segmented Filamentous Bacteria: an unusual trade-off that drives the post-natal maturation of the gut immune system. Semin Immunol 25, 342–351. [DOI] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, and Sansonetti PJ (2015). Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, Sansonetti PJ, and Cerf-Bensussan N (2017). Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr Opin Microbiol 35, 100–109. [DOI] [PubMed] [Google Scholar]
- Shi Z, and Gewirtz AT (2018). Together Forever: Bacterial-Viral Interactions in Infection and Immunity. Viruses 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, and World Health Organization-Coordinated Global Rotavirus Surveillance, N. (2016). Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clin Infect Dis 62 Suppl 2, S96–S105. [DOI] [PubMed] [Google Scholar]
- Uchiyama R, Chassaing B, Zhang B, and Gewirtz AT (2014). Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J Infect Dis 210, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Maele L, Carnoy C, Cayet D, Songhet P, Dumoutier L, Ferrero I, Janot L, Erard F, Bertout J, Leger H, et al. (2010). TLR5 signaling stimulates the innate production of IL-17 and IL-22 by CD3(neg)CD127+ immune cells in spleen and mucosa. J Immunol 185, 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, et al. (2014). PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42, D581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Weisshaar M, Lamb K, Chung HK, Lin MZ, and Plemper RK (2015). Replication-Competent Influenza Virus and Respiratory Syncytial Virus Luciferase Reporter Strains Engineered for Co-Infections Identify Antiviral Compounds in Combination Screens. Biochemistry 54, 5589–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JJ, Toots M, Lee S, Lee ME, Ludeke B, Luczo JM, Ganti K, Cox RM, Sticher ZM, Edpuganti V, et al. (2018). Orally Efficacious Broad-Spectrum Ribonucleoside Analog Inhibitor of Influenza and Respiratory Syncytial Viruses. Antimicrob Agents Chemother 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Ihira M, Higashimoto Y, Hattori F, Miura H, Sugata K, Komoto S, Taniguchi K, Iguchi A, Yamada M, et al. (2019). Persistent systemic rotavirus vaccine infection in a child with X-linked severe combined immunodeficiency. J Med Virol 91, 1008–1013. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chassaing B, Shi Z, Uchiyama R, Zhang Z, Denning TL, Crawford SE, Pruijssers AJ, Iskarpatyoti JA, Estes MK, et al. (2014). Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science 346, 861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xiang Y, Li N, Wang B, Ai H, Wang X, Huang L, and Zheng Y (2013). Protective effects of Lactobacillus rhamnosus GG against human rotavirus-induced diarrhoea in a neonatal mouse model. Pathog Dis 67, 184–191. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zou J, Shi1 Z, Zhang B, Etienne-Mesmin L, Chassaing B, and Gewirtz AT (2019). IL-22 induced cell extrusion and IL-18-induced pyroptosis prevent and cure rotavirus infection. BioRix doi: 10.1101/774984 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unprocessed 16S sequencing data are deposited in the European Nucleotide Archive under accession numbers PRJEB34108. Unprocessed raw SFB genome sequencing data are deposited in the European Nucleotide Archive under accession numbers PRJEB34149. Unprocessed raw Ileum RNAseq sequencing data are deposited in the European Nucleotide Archive under accession numbers PRJEB34133.


