Abstract
The cardiac homeobox transcription factor Nkx2-5 is a major determinant of cardiac identity and cardiac morphogenesis. Nkx2-5 operates as part of a complex and mutually reinforcing network of early transcription factors of the homeobox, GATA zinc finger and MADS domain families to initiate the program of cardiac development and differentiation, particularly in outflow tract precursor cells in the second heart field (SHF). We have now found evidence for another aspect of cardiac transcription factor cooperativity between Nkx2-5 and the cardiac enriched MADS domain transcription factor Srf. Specifically, Srf interaction with an evolutionarily conserved binding site in the Nkx2-5 CpG island-like proximal promoter is required for cardiac specific expression mediated by an SHF enhancer, and for combinatorial activation of these elements by cardiac transcription factors. These results provide further insight into cooperative gene regulation during cardiogenesis at the level of promoter-enhancer interactions.
Keywords: Outflow tract, Srf, Nkx2–5, promoter, transcription
Introduction
Initially identified as an early marker of cardiac mesoderm induction [1, 2], the mammalian homolog of the Drosophila Tinman homeobox gene, Nkx2–5, is a key element of the early cardiogenic program. Nkx2–5 is one of the earliest genes to be activated by cardiogenic signals in mesodermal precursors [3], and its function and regulation are particularly important for normal development of the cardiac outflow tract (OFT) and right ventricle (RV) [4–6]. In mice, experimental manipulation of Nkx2–5 expression levels results in dose-specific OFT and RV defects that range from near complete aplasia of the OFT and RV to double outlet right ventricle (DORV) stemming from defective looping of the heart tube. These phenotypes are attributed to altered proliferation vs. differentiation of cells of the second heart field (SHF), a specific population of cardiac progenitor cells that contribute to heart tube formation and looping beginning at mid-gestation (E8.5–9.5) of murine heart development. [5, 7–9]. In humans, hypomorphic point mutations of Nkx2–5 coding regions are associated with various types of familial and sporadic CHD affecting the OFT, likely due to an inability to regulate downstream target genes during SHF development [7, 8].
An important facet of Nkx2–5 function during SHF development is its transcriptional regulation in response to cardiac inducing factors like Bmp, Wnt and Fgf, [10–14] and its interaction with the overall network of transcription factors regulating cardiac fate and differentiation [15]. We previously defined enhancers of the mouse and chick Nkx2–5 genes that are activated by Bmp, and that regulate reporter transgene expression first in the SHF, and subsequently in SHF-derived OFT and RV [10, 16]. These enhancers are strongly conserved in non-overlapping fashion in placental mammals vs. non-placental avian, amphibian and reptile vertebrate species in the 5’ or 3’ flanking regions, respectively, of their cognate Nkx2–5 genes. While the mammalian and non-placental vertebrate conserved regions have limited homology, they both contain elements we identified as essential for SHF-expression and Bmp response in the chick. These include transcription factor (TF) binding sites for cardiac Gata proteins, for the Tgfβ/Bmp signal-transducing Smad4 protein, and for the zinc finger TF YY1. In addition to being activated by Bmp, these enhancers are subject to greatly amplified activation in the presence of multiple co-expressed cardiac transcription factors like the cardiogenic Gatas, Mef2c, Srf, Myocardin and the p300 transcriptional co-activator, as well as Nkx2–5 itself [10, 15].
We also noted that heart field-specific expression was contingent upon interactions with proximal promoter elements: while both 3’ chick and 5’ mouse conserved enhancers regulated SHF-specific expression in transgenic mice when coupled to an Nkx2–5 minimal promoter, they supported more promiscuous expression in both FHF and SHF when combined with the heterologous Hsp68 proximal promoter [10, 17]. The identity of such proximal promoter site-specific DNA binding factors governing this transcriptional activation was unclear, as plausible TATA, initiator or other sequences that could provide an interaction site for basal transcription factors were absent in this region [18]. Instead, mapping of the transcriptional initiation site by 5’ RACE and S1 nuclease protection found evidence that the Nkx2–5 promoter behaved like a dispersed, rather than a focused promoter [18], with major and minor transcriptional initiation sites scattered over an approximately 50 bp span in a GC-rich, CpG island-like region [10]. While dispersed promoters have classically been associated with “housekeeping genes,” whose rate of transcription is regulated mainly by cellular metabolic conditions, some tissue-specific dispersed promoters have been found to rely upon promoter proximal interaction with one or more sequence-specific DNA binding proteins in lieu of strong recruitment of basal transcription factors like the TATA binding proteins to canonical initiation sites [18].
We now present evidence that SHF-specific cardiac expression of Nkx2–5 is dependent upon binding of the cardiac lineage-enriched MADS domain transcription factor, Serum Response Factor (Srf) to an evolutionarily conserved binding site in the proximal promoter.
Materials and Methods
Vectors and Plasmids
Chicken Nkx2.5 reporters. Cloning and characterization of wildtype chicken Nkx2–5 reporter constructs including: CNkx2.5-lux-SHF and CNkx2.5-lacZ-SHF, have been previously described [10, 15]. CNkx2.5-lux-SHF-SRFmut and CNkx2.5-lacZ-SHF-CArGmut were created by PCR mutagenesis substituting bps −104–5 (relative to the principle chick Nkx2–5 start site) using the oligonucleotide and its complement: 5’-ATG TAT TAC TAT GCT AGG TGG CAT GGA A-3’ (IDTDNA, Coralville, IA) and the QuikChange site-specific mutagenesis kit (Stratagene, La Jolla, CA). Mutagenized BMPRE and CAR3 cassettes were ligated 3’ of the Nkx2–5 promoter-reporter constructs expressing lacZ, or luciferase coding region derived from the pGL3 basic plasmid (Promega, Madison, WI) or NLS cre recombinase (a kind gift of F. Alt) fused in-frame with the first 10 amino acids of the chicken Nkx2–5 coding region. The CNkx2.5-SHF-lacZnm reporter additionally contained a 5’ HS4 insulator sequence from the chicken β-globin locus [19] (a kind gift of G. Felsenfeld).
Transient transgenic mouse assays
CNkx2.5-SHF wild-type and CArGm Srf site mutant reporter constructs were purified from their pBluescript backbones by restriction digestion, gel electrophoresis and extraction using QIAEX bead affinity purification (Qiagen, Valencia, CA). Linear DNA was introduced by pronuclear injection into a one-cell staged FVB mouse embryos according to standard methods. F0 embryos were collected at 9.5–10.5 days post-injection following maternal sacrifice, fixed and stained for β-galactosidase activity according to previously described methods [20]. Transgenic status of individual embryos was determined by PCR from DNA derived from yolk sacs and embryo fragments using oligonucleotides for lacZ: 5’-CGG CCA GGA CAG TCG TTT GCC GTC TG-3’ and 5’-CCT GAC CAT GCA GAG GAT GAT GCT CG-3’; and, for the constitutive gene Prx1: 5’- CCT GAG TTA CCT GCA CTC TG’3’ and 5’-AGG ACT GAG GAG GAT TCT TG-3’. Results presented are representative of five transient transgenic embryos (n = 2 at E9.5, n = 3 at E10.5).
In vivo cellular reporter assays
Bmp and overexpression reporter gene assays were performed in P19CL6 cells as previously described [10]. All assay results are presented as fold induction of duplicate unstimulated versus Bmp stimulated activities; all transfections are representative of 3 independent experiments. pCS2Nkx2.5 was a kind gift from S.Izumo and H. Kasahara. pCS2 MT-Smad1, pCS2 MT/Flag Smad4 and were a kind gift from M. Whitman. pcDNA rat GATA-4 was derived from pCG-GATA-4, a kind gift of M. Nemer. pcDNA mouse GATA-6 was a kind gift of T. Collins. pcDNA SRF was a kind gift of E. Olson. pCMV-SPORT6 Mef2c (based on cDNA accession number BC026841) was purchased from Thermo Scientific (OpenBiosystems, Huntsville, AL). Recombinant Bmp4 and Noggin was purchased from R&D Systems (Minneapolis, MN). Statistical significance of reporter gene activation was calculated using Student’s t-test with significance threshold of p≤ 0.01.
Gel shift assays
Gel shift assays were performed as previously described [10] using nuclear extracts prepared from control HEK293 cells transfected with empty pCS2 vector, or programmed to express Srf, using 0.5–1.0ug dI/dC (Pharmacia), resolved on 5% acrylamide/1X TBE (Invitrogen, Carlsbad, CA) at 4°C, dried and autoradiographed on Kodak XAR X-ray film for digitization. Gel shifts were performed using the following end-P32 labeled double stranded oligonucleotides and their complements: wild-type CArG SRE: 5’-TAC-TAT-ATT-AGG-TGA-3’; mutant CArG/SRE: 5’-TAC TAT GCT AGG TGA-3’.
Chromatin Immunoprecipitation Assays (ChIP)
Chromatin extracted from wild-type FVB E10.5 mouse SHF-containing pharyngeal arch regions and hearts were assayed by ChIP according to previously described methods [21] using anti-Srf antibody (Sigma-Aldrich, #HPA005533) and primers 5’ CTC TGC TGT GTG GCC TTG TA-3’ and 5’-CGA CAG GAA ACT CGG AGC TA-3’ (rev) (mNkx2.5 SHF region); 5’-CTC CTG CAA GGA GAT TGC TC-3’ (for) and 5’-CCT CAC CAG CCC ATT TAG TG-3’ (rev) (mNkx2.5 promoter region); and, 5’-CTA CAA GTG CAA GCG ACA GC-3’ (for) and 5’-GCG TTG TAG CCA TAG GCA TT-3’ (rev) (mNkx2.5 exon 2). Results are expressed as fold enrichment based upon ΔC(t) calculations relative to control nonspecific Ig and are representative of three independent chromatin isolations and immunoprecipitations.
Results and Discussion
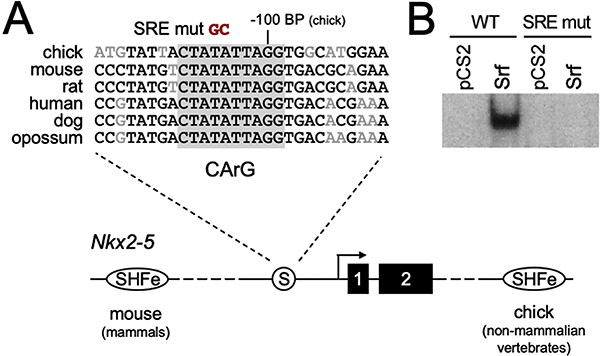
Sequence alignment of the Nkx2–5 promoter proximal region from multiple vertebrate species detected the presence of several areas of significant conservation within the −200 bp from the transcriptional start site we previously mapped for the chick Nkx2–5 gene [10] (Fig. 1A). In particular, we noted the presence of an AT-rich sequence, 5’-CTATATTAGG-3’, at approximately −109 to −100 bp from the start site that appeared to be a strong consensus binding sequence for the MADS-domain transcription factor Serum Response Factor (Srf). Gel shift mobility shift assays confirmed that this sequence is capable of binding to Srf expressed in cell extracts (Fig. 1B, left lanes). This interaction could be abrogated by replacement of the central TA residues in this consensus site with GC (Fig 1A, B: SRE mut, right lanes).
Figure 1. Nkx2–5 proximal promoter CArG box Srf binding site.
A. Multiple species alignment of the promoter proximal region surrounding a conserved CArG consensus (boxed in gray) approximately -100bp upstream of the chick Nkx2–5 transcriptional start site. The alignment is shown over a schematic of the Nkx2–5 locus with the CArG site/Srf binding site (circled S) and relative positions of the two Nkx2–5 coding exons and the 5’ and 3’ SHF-specific enhancers found in mammals and non-mammalian vertebrates [15]. B. Gel shifts obtained using HEK 293 cell whole cell extracts transfected with control empty vector (pCS2) or Srf expression vector, and radiolabeled double stranded oligonucleotides representing the -100bp CArG/Srf binding site (WT), or a mutated sequence bearing a GC substitution of the central AT base pairs shown in A.
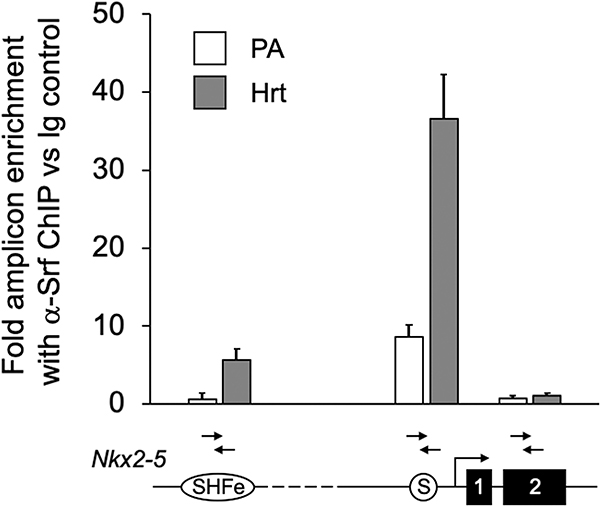
To test whether Srf bound the putative promoter-proximal consensus CArG site in vivo, we performed chromatin immunoprecipitation (ChIP) using chromatin from embryonic day E9.5 SHF-containing pharyngeal arch (PA) tissue and 9.5 mouse hearts (Hrt). As shown in Fig. 2, ChIP with anti-Srf antibody greatly enriched for amplicons surrounding the putative Nkx2–5 proximal promoter Srf consensus site in both PA and heart. Interestingly, more modest but significant in vivo binding was also detected in heart to an SHF-specific enhancer (SHFe) we previously characterized as being activated by a multiple transcription factor complex (multi-TF), despite the absence within that region of a canonical CArG box binding site [15]. By contrast, amplicons within the nearby Nkx2–5 Exon 2 genomic region were not enriched by anti-Srf ChIP in either heart or PA regions.
Figure 2. Srf is bound to the conserved Nkx2–5 promoter proximal CArG site region in vivo.
qPCR assay of chromatin recovered from E9.5 SHF-containing pharyngeal arch (PA) and heart (Hrt) using anti-Srf ChIP relative to non-specific IgG reveals highly significant enrichment of amplicons representing the promoter proximal region near the conserved CArG box/Srf binding consensus (PA: 8.66 ±1.51 fold; Hrt: 36.63 ± 5.66 fold) as compared to control Exon 2 regions (PA: 0.71 ± 0.4 fold; Hrt: 1.02 ± 0.46 fold). Enrichment was also detected for amplicons within the conserved 5’ Nkx2–5 SHF enhancer in embryonic heart (PA: 0.64 ± 0.77 fold; Hrt: 5.65 ± 1.45 fold). Results are shown above a schematic of the interrogated regions of the Nkx2–5 gene.
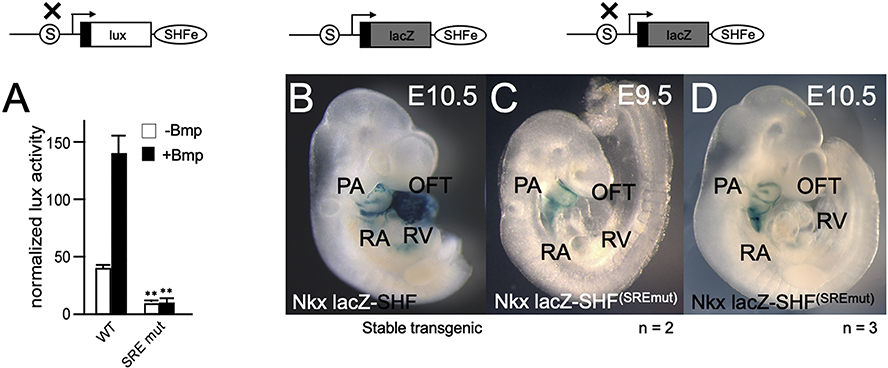
We then tested the requirement for the Srf binding site for Nkx2–5 activation in vitro and in vivo. We previously showed that a reporter gene driven by an SHF-specific enhancer (CNkx2–5–SHF-lux) could be activated by both Bmp and by a combination of cardiac-specific transcription factors (TF) [15]. Alteration of the promoter-proximal putative Srf binding site to the non-binding sequence (see Fig. 1A) in a parallel reporter gene, CNkx2–5–SHF-lux–SREmut, resulted in almost complete loss of the activation of an Nkx2–5 SHF reporter gene by either Bmp or by a combination of cardiac TFs (Fig. 3A). Similarly, alteration of the promoter proximal Srf binding site to the non-binding mutation had a dramatic effect on lacZ expression mediated by a Nkx2–5-lacZ-SHFSREmut reporter gene as compared to the wild-type reporter (Fig. 3B): while residual lacZ expression was observed in PA arch regions, reporter gene expression in OFT and RV was completely lost at both E9.5 (Fig. 3C) and E10.5 (Fig. 3D) confirming the requirement for the promoter proximal Srf binding consensus site to maintain cardiac expression. These findings further supported the requirement for Srf binding to the Nkx2–5 proximal promoter for cardiac-specific expression.
Figure 3. The conserved proximal promoter CArG consensus site is necessary for in vitro and in vivo activation of an Nkx2–5 SHF enhancer reporter.
A. Nkx2–5 SHF minigene reporter assay results obtained in P19CL6 cells using either a chick wildtype Nkx2–5-lux-SHF reporter gene (WT) [15], or a parallel reporter bearing a site-specific mutation of the promoter proximal CArG box/Srf binding element (SRE mut) (schematic above). Results shown reflect activation by Bmp4 (black vs. white bars) in the presence of co-expression of multiple cardiac transcription factors (multi-TF) previously shown to greatly amplify Nkx2–5-lux-SHF activation in vitro [15] (WT: 38.8±3.5; WT+Bmp: 139.3±15.75; mut: 8.0+3.25; mut: 8.8±5.0; ** p<0.02). Transient transgenic embryos are shown displaying expression patterns obtained using an Nkx2–5-lacZ-SHFSREmut reporter gene, as compared to expression from a stable transgenic line bearing a wild-type Nkx2–5-lacZ-SHF reporter (B) at E10.5. While weak expression is retained in pharyngeal arch regions, expression is eliminated in developing heart populations at E9.5 (n = 2 transient TG embryos) (C) and E10.5 (n = 3 transient TG embryos) (D). Transgene expression results are shown above a schematic of the Nkx2–5-SHF reporters assayed.
It has previously been shown that heart field-specific knock-out of Srf using an Nkx2–5 Cre driver (Srfcko) results in a primarily SHF-specific phenotype: severe developmental defects were noted at the onset of heart looping, including misplaced anterior portions of the developing outflow tract, cranially retained RV/OFT and reduced expression of SHF markers [22]. The observed failure of looping and hypoplastic development of RV and OFT segments was strongly reminiscent of the defects observed in Nkx2–5−/− constitutive null embryos due to lack of proliferation and differentiation of SHF progenitors [5, 6, 23]. While Nkx2–5 mRNA expression was retained in remaining first heart field-derived cardiac myocytes of Srfcko, Nkx2–5 mRNA expression was significantly diminished in cardiac-differentiated Srf knockout ES cells, perhaps due to specific loss of expression in SHF lineages [22]. Interestingly, the deficit of Nkx2–5 expression in differentiated ES cells could not be rescued by viral re-expression of Srf mutants specifically compromised for protein-protein association with other cardiac transcription factors like Nkx2–5, Gata4 and Myocardin, but competent to bind DNA [22].
This last finding underscored a significant facet of Srf transcriptional activity; namely, the modulation of Srf-mediated transcriptional activation via its interactions with other transcription factors and/or transcriptional regulators. Interaction of Srf with the myocardin-related transcription factors (MRTFs) and ternary complex factors (TCFs) modulates major aspects of serum-stimulated growth response, development and contractile behavior of fibroblasts and smooth muscle lineages [24]. In developing cardiac myocytes, Srf additionally participates in protein-protein interactions with one or more early cardiac transcription factors, especially Nkx2–5 itself and the cardiac Gata proteins, to regulate cardiac muscle gene expression [25–27]. As an example, in our prior studies, we found that Bmp-activation of SHF-specific Nkx2–5 expression was greatly amplified by co-expression of a complex of transcriptional activators including Nkx2–5, Gata4 and 6, Myocardin, Srf and p300, particularly in differentiating myocytes arising from SHF progenitors [15].
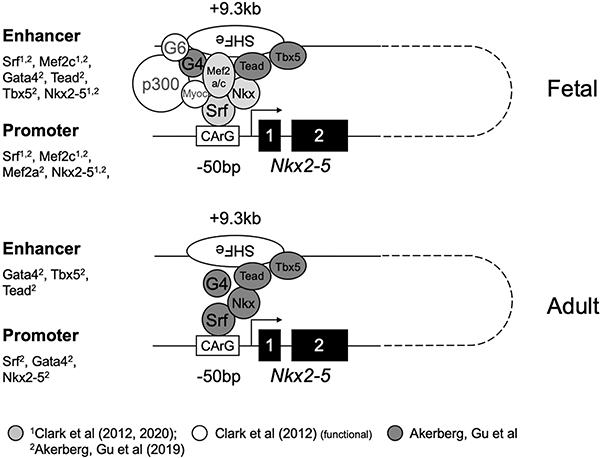
The cooperative action of cardiac-specifying and/or differentiating factors in activating the cardiac program was recently underscored by genome-wide analysis of cardiac TF occupancy in embryonic and adult hearts in mice [28]. Data from this study independently corroborate both our findings of Srf occupancy of the Nkx2–5 proximal promoter region, as well as our previously reported findings of Nkx2–5 and Mef2c binding to a distal 5’ enhancer region [15] (Fig. 4). As well, they detect continued Srf occupancy of the proximal promoter in adult as well as fetal heart, consistent with our finding that abrogation of Srf interaction with the proximal promoter resulted in loss of reporter signal in differentiated myocytes (Fig. 3).
Figure 4. Model for Nkx2–5 promoter-enhancer interaction in SHF-derived heart.
Shown is a conceptual model for promoter-enhancer interaction via multiple cardiac transcription factors, anchored on the SHF enhancer by DNA-bound Gata4/6 and Nkx2–5, and on the proximal promoter by DNA-bound Srf [15]. Assembly may culminate in the recruitment of Srf-associated myocardin and p300 acetylase, and may be further influenced by additional yet uncharacterized enhancer and/or promoter binding elements. Abbreviations: G4: Gata4; G6: Gata6; Nkx: Nkx2–5; Myoc: Myocardin; Srf: Srf. The figure includes combined data from this study, a previous study of conserved Nkx2–5 SHF regulation [15], and manually curated data from a genome-wide TF occupancy analysis [28] (gray, white and dark gray symbols), as noted at the bottom of the figure. Summary of ChIP data from these studies detecting TFs associated with SHF or promoter proximal amplicons is lifted at left for both fetal and adult stages. Not shown are YY1 and associated Bmp Smad1/4 complexes previously shown to mediate Bmp activation of SHF enhancers. Putative associations of Gata6, Myocardin and p300 with fetal SHF are inferred from prior functional activation data [10, 15].
Taken as a whole, our results support a role for Srf in the SHF-specific regulation of Nkx2–5 via interactions with its proximal promoter. While a principal role for Srf in transcriptional initiation from a diffuse, CpG island-like promoter has not previously been described, Srf has frequently been found associated with TATA-initiated promoters and start site-proximal basal promoters regulating the expression of growth signal activated-genes. As well, prior promoter studies have noted that Srf can interact directly with basal transcription factors like TFII-F [29] and TFII-I [30, 31] in promoter complexes, and that Srf can act at basal promoter elements to facilitate alternative transcriptional initiation from competing TATA and Initiator elements [32]. These findings collectively illustrate Srf crossover functions as both a general and tissue-specific transcription factor. These results resonate with our findings of concomitant Srf interaction with both Nkx2–5 SHF enhancer (Fig. 2;[15]) and proximal promoter regions, and implicating Srf as a key bridging factor for enhancer-promoter interactions (Fig. 4). The extent to which Srf acts similarly in other specific instances of cardiac or other gene regulation to mediate enhancer-promoter interaction and gene activation remains an ongoing objective of future studies.
HIGHLIGHTS.
Cardiac development requires the cooperative action of multiple early lineage-specific transcriptional regulators to reinforce cardiac fate and differentiation.
Nkx2–5 homeobox transcription factor is essential for early right heart development from second heart field progenitors.
Serum Response Factor (Srf), itself a key regulator of early cardiac differentiation, binds to an evolutionarily conserved promoter proximal consensus binding site, as well as to a distal conserved 5’ second heart field enhancer of Nkx2–5.
Loss of Srf binding to the Nkx2–5 promoter results in failure to maintain expression in differentiated right heart lineages, indicating its essential role, along with other cardiac TFs, in reinforcing cell fate in the developing right heart through regulation of Nkx2–5.
Acknowledgements
We wish to thank E. Olson, M. Whitman, S. Izumo, W. Pu, H. Kasahara, T. Collins, M. Nemer and G. Felsenfeld for expression and insulator plasmids. B. Lee, K. Doherty and K. Floras provided technical assistance. We thank A. Foley for critical reading of the manuscript. Work was supported by the American Heart Association (12BGIA12060120), the SC COBRE in Developmentally-based Cardiovascular Disease, (P20-RR016434-06), and facilities and services at the Darby Children’s Research Institute at MUSC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Bodmer R, Jan LY, and Jan YN, A new homeobox-containing gene, msh-2, is transiently expressed early during mesoderm formation in Drosophila. Development, 1990. 110: p. 661–669. [DOI] [PubMed] [Google Scholar]
- 2.Schultheiss TM, Xydas S, and Lassar AB, Induction of avian cardiac myogenesis by anterior endoderm. Development (Cambridge, England), 1995. 121(12): p. 4203–14. [DOI] [PubMed] [Google Scholar]
- 3.Schultheiss TM, Burch JBE, and Lassar AB, A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes and Development, 1997. 11: p. 451–462. [DOI] [PubMed] [Google Scholar]
- 4.Lyons I, et al. , Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev, 1995. 9(13): p. 1654–66. [DOI] [PubMed] [Google Scholar]
- 5.Prall OW, et al. , An Nkx2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell, 2007. 128(5): p. 947–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka M, et al. , The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development, 1999. 126(6): p. 1269–80. [DOI] [PubMed] [Google Scholar]
- 7.McElhinney DB, et al. , NKX2.5 mutations in patients with congenital heart disease. Journal Of The American College Of Cardiology, 2003. 42(9): p. 1650–5. [DOI] [PubMed] [Google Scholar]
- 8.Goldmuntz E, Geiger E, and Benson DW, NKX2.5 mutations in patients with tetralogy of fallot. Circulation, 2001. 104(21): p. 2565–8. [DOI] [PubMed] [Google Scholar]
- 9.Benson DW, et al. , Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. The Journal Of Clinical Investigation, 1999. 104(11): p. 1567–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee KH, et al. , SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development, 2004. 131(19): p. 4709–23. [DOI] [PubMed] [Google Scholar]
- 11.Schultheiss TM, Burch JB, and Lassar AB, A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev, 1997. 11(4): p. 451–62. [DOI] [PubMed] [Google Scholar]
- 12.Waldo KL, et al. , Conotruncal myocardium arises from a secondary heart field. Development (Cambridge, England), 2001. 128(16): p. 3179–88. [DOI] [PubMed] [Google Scholar]
- 13.Park EJ, et al. , Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development (Cambridge, England), 2006. 133(12): p. 2419–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutson MR, et al. , Arterial pole progenitors interpret opposing FGF/BMP signals to proliferate or differentiate. Development, 2010. 137(18): p. 3001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark CD, et al. , Evolutionary Conservation of Nkx2.5 Autoregulation in the Second Heart Field. Developmental Biology, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly RG, Brown NA, and Buckingham ME, The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Developmental Cell, 2001. 1(3): p. 435–440. [DOI] [PubMed] [Google Scholar]
- 17.Lien CL, et al. , Control of early cardiac-specific transcription of Nkx2–5 by a GATA-dependent enhancer. Development (Cambridge, England), 1999. 126(1): p. 75–84. [DOI] [PubMed] [Google Scholar]
- 18.Kadonaga JT, Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol, 2012. 1(1): p. 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung J, Whiteley M, and Felsenfeld G, A 5’ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 1993. 74(3): p. 505–14. [DOI] [PubMed] [Google Scholar]
- 20.Zimmerman L, et al. , Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron, 1994. 12(1): p. 11–24. [DOI] [PubMed] [Google Scholar]
- 21.Barth JL, et al. , Jarid2 is among a set of genes differentially regulated by Nkx2.5 during outflow tract morphogenesis. Developmental Dynamics: An Official Publication Of The American Association Of Anatomists, 2010. 239(7): p. 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu Z, et al. , Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc Natl Acad Sci U S A, 2008. 105(46): p. 17824–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lints TJ, et al. , Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development (Cambridge, England), 1993. 119(3): p. 969. [DOI] [PubMed] [Google Scholar]
- 24.Gualdrini F, et al. , SRF Co-factors Control the Balance between Cell Proliferation and Contractility. Mol Cell, 2016. 64(6): p. 1048–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CY and Schwartz RJ, Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Molecular And Cellular Biology, 1996. 16(11): p. 6372–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belaguli NS, et al. , Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Molecular and Cellular Biology, 2000. 20(20): p. 7550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, et al. , Identification of a novel serum response factor cofactor in cardiac gene regulation. The Journal Of Biological Chemistry, 2004. 279(53): p. 55626–32. [DOI] [PubMed] [Google Scholar]
- 28.Akerberg BN, et al. , A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat Commun, 2019. 10(1): p. 4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joliot V, Demma M, and Prywes R, Interaction with RAP74 subunit of TFIIF is required for transcriptional activation by serum response factor. Nature, 1995. 373(6515): p. 632–5. [DOI] [PubMed] [Google Scholar]
- 30.Grueneberg DA, et al. , A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev, 1997. 11(19): p. 2482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer zu Reckendorf C, et al. , Proteomic analysis of SRF associated transcription complexes identified TFII-I as modulator of SRF function in neurons. Eur J Cell Biol, 2016. 95(1): p. 42–56. [DOI] [PubMed] [Google Scholar]
- 32.Xu M, Gonzalez-Hurtado E, and Martinez E, Core promoter-specific gene regulation: TATA box selectivity and Initiator-dependent bi-directionality of serum response factor-activated transcription. Biochim Biophys Acta, 2016. 1859(4): p. 553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]