Abstract
Fusobacterium nucleatum (Fn) is frequently found in colorectal cancers (CRCs). High loads of Fn DNA are detected in CRC tissues with microsatellite instability-high (MSI-H), or with the CpG island hypermethylation phenotype (CIMP). Fn infection is also associated with the inflammatory tumor microenvironment of CRC. A subtype of CRC exhibits inflammation-associated microsatellite alterations (IAMA), which are characterized by microsatellite instability-low (MSI-L) and/or an elevated level of microsatellite alterations at selected tetra-nucleotide repeats (EMAST). Here we describe two independent CRC cohorts in which heavy or moderate loads of Fn DNA are associated with MSI-H and L/E CRC respectively. We also show evidence that Fn produces factors that induce γ-H2AX, a hallmark of DNA double strand breaks (DSBs), in the infected cells.
Keywords: Fusobacterium nucleatum, Colorectal cancer, Microsatellite instability, CpG island methylator phenotype, MSI-L, EMAST, MSH3, Mismatch repair, Inflammation
Main text
Fn is a common resident in the human gut mucosa and is an anaerobic bacterium that colonizes CRC tumors more frequently than adjacent normal mucosa. To date, most epidemiological studies using 16s rRNA sequencing or metagenomic sequencing methods have detected an increased level of Fn DNA and/or RNA in colorectal adenoma/carcinoma tissues or stools from tumor bearing patients, as compared with normal controls [1]. Furthermore, Fn infection is associated with specific subtypes of CRC that exhibits CIMP or MSI-H [2, 3]. These observations might suggest that Fn infection may contribute to a serrated pathway of CRC development [4]. On the other hand, tumor tissue infected with Fn exhibits an inflamed tumor microenvironment, rich in inflammatory factors such as IL6 or reactive oxygen species [5], leading to the assumption that Fn infection might also contribute to the generation of IAMA or L/E positive CRC [6–8]. Despite a strong association between Fn infection and colorectal cancer, there has been no evidence of Fn infection damaging the DNA of colon tissues. In this study, we show evidence that a degree of Fn infection may determine molecular characteristics of CRC, and that Fn infection may be carcinogenic.
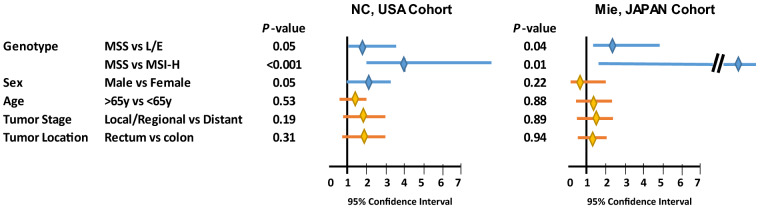
A total of 304 cases of unselected sporadic CRC from North Carolina [9, 10] were analyzed for MSI-H, MSI-L and EMAST [11, 12]. The amount of Fn DNA per nanogram of tumor tissue DNA was also determined by qPCR (see Additional file 1: Additional Materials and Methods). Thirty-eight cases (12.5%), 129 cases (42.4%) and 137 cases (45.1%) exhibited MSI-H, L/E and MSS, respectively. Fn DNA was detected in 116 of 304 (38%) CRC tumor tissues, ranging from 0.002 to 880 pg/ng of tissue DNA. When the quantity of Fn DNA was compared among MSI-H, L/E and MSS CRC, the Fn DNA load in MSI-H was the highest (MSI-H > L/E, p = 0.028; MSI-H > MSS, p = 0.000085) and the Fn DNA load in L/E was higher than in MSS (L/E > MSS, p = 0.028) (Fig. 1a). We then determined whether Fn infection was associated with MSI-H and/or L/E compared to MSS using a logistic regression model. In univariate analysis, Fn infection was associated with MSI-H at an odds ratio (OR) of 4.21 (p < 0.001) and was also associated with L/E at an OR of 1.74 (p = 0.03). When adjusted for sex, age, tumor location and tumor stage, MSI-H (OR = 3.99, 95%CI 1.85–8.9, p < 0.001) and L/E (OR = 1.68, 95% CI 1.00–2.84, p = 0.05) were independently associated with Fn infection (Fig. 2). To validate the above results, we analyzed 174 cases of CRC from Mie, Japan. Thirteen (7.4%), 69, (39.7%) and 92 cases (52.9%) exhibited MSI-H, L/E and MSS, respectively. Fn DNA was detected in 131 of 174 (75%) tumors, ranging from 0.0003 to 200 pg/ug tissue DNA. The quantity of Fn DNA was highest in MSH-H compared to L/E (p = 0.02) or MSS (p = 0.0005), and the Fn load was higher in L/E than MSS (p = 0.015) (Fig. 1b). Fn infection was associated with MSI-H at OR = 13.83 (p = 0.007) and with L/E at OR = 2.35 (p = 0.02) in univariate logistic regression analysis. Multivariate analysis adjusted by sex, age. tumor location and stage showed that MSI-H (OR = 13.67, 95% CI 1.63–1789.13, p = 0.01) and L/E (OR = 2.23, 95% CI 1.05–4.95, p = 0.04) were significantly associated with Fn infection compared to MSS (Fig. 2).
Fig. 1.
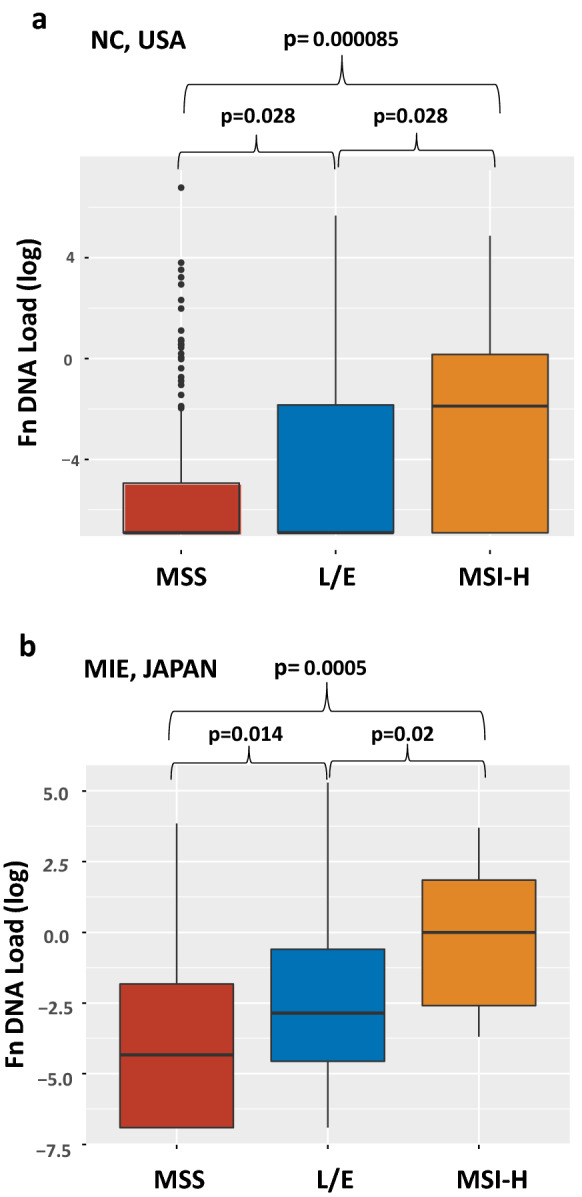
Fn DNA load in MSS, L/E and MSI-H CRCs. (a) Discovery cohort from North Carolina, USA. (b) Validation cohort from Mie, Japan. Absolute Fn DNA weight was normalized by the absolute tumor DNA weight present in the same DNA sample. Normalized values were converted to logarithmic scales. The data is depicted in the boxplot. Brown, blue and orange boxes represent the distribution of Fn DNA in MSS, L/E and MSI-H CRC, respectively. The thick horizontal line within in each box represents the median Fn DNA load. Dots in MSS column in the USA cohort represents outliers. The statistical difference in Fn DNA loads among MSS, L/E and MSI-H was tested using the Wilcoxon rank sum test. A p-value that is less than 0.05 is considered significant
Fig. 2.
Logistic regression model for association between Fn infection and CRC molecular subtypes. Discovery cohort from North Carolina, USA (left) and validation cohort from Mie, Japan (right). Association was modeled by comparing MSS versus L/E or MSI-H. Positive for Fn infection was designated when the sample gave detectable Fn-specific PCR products by qPCR. The association was adjusted by sex, age, tumor location and tumor stage. The X-axis represents a range of 95% confidence interval (CI). Each horizontal bar indicates 95% CI for that variable. A blue or orange bar represents the significance or insignificance, respectively, between each variable and Fn infection. The middle diamond shape represents the value for the odds ratio. The P-values are shown after each variable
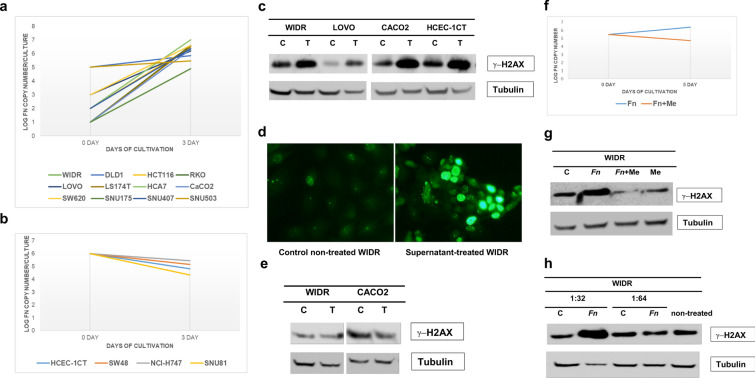
To explore whether Fn infection causes cellular DNA damage (see Additional file 1: Additional Materials and Methods), we first determined the ability of human colon cancer cells to support infection. When each of 16 human colon cancer cell lines was co-cultured with Fn in 5% CO2/21% O2 conditions, Fn grew aerobically in 12 of 16 cell lines (Fig. 3a), but not in 4 cell lines (Fig. 3b). Furthermore, there was a difference in the ability to support aerobic growth of Fn among the 12 cell lines. Some cell lines such as WIDR required less Fn (MOI of 0.001) to initiate successive Fn growth whereas SNU503 required Fn at MOI of 10 (Fig. 3a). The supernatants from co-cultures between WIDR and Fn, where Fn grew, induced γ-H2AX in various colon cancer cell lines (Fig. 3c, d), a hallmark of DNA double strand breaks (DSBs) and suggesting that Fn infection may be carcinogenic to infected tissues, whereas the supernatants from co-cultures between HCEC-1CT and Fn, where Fn growth was not permissive, did not induce γ-H2AX in the exposed colon cancer cell lines (Fig. 3e). Inclusion of the antibiotic metronidazole [13] in co-cultures between WIDR and Fn inhibited Fn growth (Fig. 3f) and abolished the supernatants’ ability to induce γ-H2AX in WIDR cells (Fig. 3g). Finally, the bacterial culture medium where Fn was anaerobically grown induced γ-H2AX in WIDR cells (Fig. 3h), indicating that Fn produces a factor that may cause DNA DSBs in mammalian cells.
Fig. 3.
Aerobic growth of Fn and induction of γ-H2AX by Fn. (a) Twelve colon cancer cell lines (WIDR, DLD1, HCT116, RKO, LOVO, LS174T, HCA7, CaCO2, SW620, SNU175, SNU407, and SNU503) were infected with Fn strain EAVG_002. During a 3-day incubation, an increase in Fn copy number was observed in co-cultures with these cell lines. (b) No increase in Fn copy number was observed in co-cultures with 4 different colon cancer cell lines (HCEC-1CT, SW48, NCI-H747 and SNU81). X-axis: days of cultivation; Y-axis: log of Fn copy number per culture. (c) WIDR cells were infected with Fn EAVG_002 strain at a multiplicity of infection of 1 under 5%CO2/21%O2 for 1 week. Supernatants were collected, centrifuged, and filtered through a 0.2 μm porous membrane. 2 × 105 cells (WIDR, LOVO, CaCO2 and HCEC-1CT) were exposed to the supernatants for 9 hr. Cell lysates were analyzed for induction of γ-H2AX by Western blotting using anti-γ-H2AX mouse monoclonal antibodies. C: control non-treated; T: supernatant treated. Treated cells expressed more γ-H2AX than control non-treated cells. (d) Immunofluorescent staining of Fn supernatant-treated WIDR cells with anti-γ-H2AX mouse monoclonal antibodies. Brighter nuclear γ-H2AX signal is evident in treated cells. (e) Supernatant from HCEC-1CT cell culture infected with Fn EAVG_002 strain at an MOI of 1 under 5%CO2/21%O2 conditions failed to induce γ-H2AX in supernatant-treated WIDR and CaCO2 cells. C: control non-treated; T: supernatant treated. There was no difference in the amount of γ-H2AX between control non-treated and supernatant treated cells. (f) Treatment of co-culture between WIDR and Fn with metronidazole inhibited Fn aerobic growth (red line). Blue line represents growth of Fn without metronidazole. (g) Metronidazole abolished γ-H2AX induction by Fn. Supernatants were collected from 4 cultures (C: control non-treated cells; Fn: Fn infected cells; Fn + Me: Fn infected cells treated with metronidazole; Me: non-infected cells treated with metronidazole alone) and tested for induction of γ-H2AX in WIDR cells. (h) Bacterial medium in which Fn grew anaerobically was tested for γ-H2AX induction. The Fn grown medium and control fresh bacterial medium was diluted by Dulbecco's modified Eagle medium with 10% fetal bovine serum at 1:32 and 1:64 ratio and then exposed to WIDR cells. C: WIDR cells were treated with a diluted fresh bacterial medium; Fn: WIDR cells were treated with diluted Fn grown medium. Induction of γ-H2AX was detected at 1:32 but not at 1:64 dilutions of Fn grown medium
Here we provide the initial report showing that heavy or moderate loads of Fn DNA are associated with MSI-H and L/E CRC, respectively. We have also identified evidence that Fn infection may cause DNA damage in infected colon tissues. It remains to be determined whether different degrees of Fn infection directly or indirectly impair DNA mismatch repair differently, resulting in MSI-H or L/E, or whether Fn opportunistically heavily infects MSI-H compared to L/E or MSS CRC. The host cell-dependent aerobic growth of Fn observed in this study may give further clues to answer these questions. However, our observation that Fn infection may trigger cellular DNA damage strongly suggests that Fn infection causes genetic and/or epigenetic alterations, initiating and/or promoting colorectal carcinogenesis. The identity and origin of the DNA damaging factor generated by Fn infection will need to be investigated.
Supplementary information
Additional file 1. Additional materials and methods
Acknowledgements
Not applicable for this study.
Abbreviations
- Fn
Fusobacterium nucleatum
- CRCs
Colorectal cancers
- MSI-H
Microsatellite instability-high
- CIMP
CpG island hyper-methylation phenotype
- IAMA
Inflammation associated microsatellite alterations
- MSI-L
Microsatellite instability-low
- EMAST
Elevated level of microsatellite alterations at selected tetra-nucleotide repeats
- L/E
MSI-L/EMAST
- OR
Odd ratio
- DSBs
Double strand breaks
- MMR
Mismatch repair
- CI
Confidential interval
- MOI
Multiplicity of infection
- NC
North Carolina
Authors’ contributions
YO, MK, and JMC conceived and designed experiments. YO, MK, and RR performed experiments. BM, EK, EMS, JAG, ANM, TOK, YO, TK, YT, and EM contributed reagents/materials/analysis tools. YO, MK, and JMC wrote and edited the manuscript. All authors read and approved the final manuscript.
Funding
This study is supported by the United States Public Health Service (NIH grant CA206010) and the A. Alfred Taubman Medical Research Institute of the University of Michigan (to J.M.C.).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author by reasonable request.
Ethics approval and consent to participate
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective analysis study was conducted according to the World Medical Association Declaration of Helsinki and was approved by the Internal Review Board of the University of North Carolina and Mie University. Since the collection of archival tissue was done through an un-identifiable approach, no consent form was needed for this study. None of the cell lines used in the present study required ethics approval for their use.
Consent for publication
Not applicable for this study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yoshiki Okita and Minoru Koi contributed equally to this work
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13099-020-00384-3.
References
- 1.Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158(2):322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, et al. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139(6):1318–1326. doi: 10.1002/ijc.30168. [DOI] [PubMed] [Google Scholar]
- 5.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koi M, Tseng-Rogenski S, Carethers JM. Inflammation-associated microsatellite alterations: mechanisms and significance in the prognosis of patients with colorectal cancer. World J Gastrointest Oncol. 2018;10(1):1–14. doi: 10.4251/wjgo.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng-Rogenski SS, Hamaya Y, Choi DY, Carethers JM. Interleukin 6 alters localization of hMSH3, leading to DNA mismatch repair defects in colorectal cancer cells. Gastroenterology. 2015;148(3):579–589. doi: 10.1053/j.gastro.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tseng-Rogenski SS, Munakata K, Choi DY, Martin PK, Mehta S, Koi M, Zheng W, Zhang Y, Carethers JM. The human DNA MMR protein MSH3 contains nuclear localization and export signals that enable nuclear-cytosolic shuttling in response to inflammation. Mol Cell Biol. 2020;40:e00029–e120. doi: 10.1128/MCB.00029-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gethings-Behncke C, Coleman HG, Jordao HW, Longley DB, Crawford N, Murray LJ, et al. Fusobacterium nucleatum in the colorectum, and its association with cancer risk and survival: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2020;29(3):539–548. doi: 10.1158/1055-9965.EPI-18-1295. [DOI] [PubMed] [Google Scholar]
- 10.Devaraj B, Lee A, Cabrera BL, Miyai K, Luo L, Ramamoorthy S, et al. Relationship of EMAST and microsatellite instability among patients with rectal cancer. J Gastrointest Surg. 2010;14(10):1521–1528. doi: 10.1007/s11605-010-1340-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munakata K, Koi M, Kitajima T, Tseng-Rogenski S, Uemura M, Matsuno H, et al. Inflammation-associated microsatellite alterations caused by MSH3 dysfunction are prevalent in ulcerative colitis and increase with neoplastic advancement. Clin Transl Gastroenterol. 2019;10(12):e00105. doi: 10.14309/ctg.0000000000000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raeker MO, Pierre-Charles J, Carethers JM. Tetranucleotide microsatellite mutational behavior assess in real time: implications for future microsatellite panels. Cell Mol Gastroenterol Hepatol. 2020;9:689–704. doi: 10.1016/j.jcmgh.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional materials and methods
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author by reasonable request.