Abstract
Premise
Motivated to make sensible interpretations of the massive volume of data from the Australian Microbiome Initiative (AusMic), we characterize the soil mycota of Australia. We establish operational taxonomic units (OTUs) from the data and compare these to GenBank and a data set from the National Herbarium of Victoria (MEL), Melbourne, Australia. We also provide visualizations of Agaricomycete diversity, drawn from our analyses of the AusMic sequences and taxonomy.
Methods
The AusMic internal transcribed spacer (ITS) data were filtered to create OTUs, which were searched against the National Center for Biotechnology Information Nucleotide database and the MEL database. We further characterized a portion of our OTUs by graphing the counts of the families and orders of Agaricomycetes. We also graphed AusMic species determinations for Australian Agaricomycetes against latitude.
Results
Our filtering process generated 192,325 OTUs; for Agaricomycetes, there were 27,730 OTUs. Based on the existing AusMic taxonomy at species level, we inferred the diversity of Australian Agaricomycetes against latitude to be lowest between −20 and −25 decimal degrees.
Discussion
BLAST comparisons provided reciprocal insights between the three data sets, including the detection of unusual root‐associated species in the AusMic data, insights into mushroom morphology from the MEL data, and points of comparison for the taxonomic determinations between AusMic, GenBank, and MEL. This study provides a tabulation of Australian fungi, different visual snapshots of a subset of those taxa, and a springboard for future studies.
Keywords: Australasia, bioinformatics, metagenomics, mushrooms, sequestrate fungi, truffle‐like fungi
Fungi constitute a hyperdiverse kingdom representing an array of ecological lifestyles, including human pathogens, ectomycorrhizae, lichens, and many more (Burgess et al., 2006; Blackwell, 2011; Li et al., 2016; Medeiros et al., 2017; Crossay et al., 2018; Chang et al., 2019; Mujic et al., 2019). Due to their fundamentally microscopic nature and their usually ephemeral reproductive structures (e.g., mushrooms, apothecia, etc.), the identification of fungi has historically been exceptionally difficult, relying on often artificial groupings based on limited morphological features. The advent and maturation of molecular approaches has revolutionized mycology; in the past two decades, a significant number of new orders, classes, and even phyla have been described (Schüβler et al., 2001; Zalar et al., 2005; Hosaka et al., 2006; Schoch et al., 2009; Rosling et al., 2011; Hodkinson et al., 2014). Correspondingly, a massive number of fungal families, genera, and species have been described during this period (Smith et al., 2006; Halling et al., 2012; Wu et al., 2016; Torres‐Cruz et al., 2017; Willis, 2018). Although sequence‐based approaches are not without pitfalls and controversy (Hofstetter et al., 2019), they remain major catalysts for the description and identification of fungal diversity.
Although the study of fungal diversity has been biased toward Northern Hemisphere temperate ecosystems, there have been major efforts to reconcile the gap in knowledge regarding tropical and Southern Hemisphere fungi, with major foci of activity in the eastern paleotropics (Luo et al., 2016; Vadthanarat et al., 2018, 2019; Sukarno et al., 2019), Central and South America (Kuhar et al., 2017; Accioly et al., 2018; Kaishian and Weir, 2018; Ovrebo et al., 2019), Africa (Castellano et al., 2016; Buyck et al., 2018, 2019; Jami et al., 2018), and Australia (Midgley et al., 2018; Davoodian et al., 2019; Ji et al., 2019; Khmelnitsky et al., 2019).
In line with many of the works cited above, systematic mycology studies generally rely on herbaria as sources of samples and repositories for new specimens, allowing the tethering of names and concepts to physical vouchers. Herbaria serve a critical role in the preservation and curation of biological resources and heritage, and ongoing efforts to digitize these resources will continue to positively impact biodiversity sciences (Willis et al., 2017; Thiers and Halling, 2018). Indeed, the acceleration of mycology via molecular approaches has reciprocally enhanced herbarium‐based approaches, with herbaria providing curated collections and molecular techniques providing new insights into these resources.
The Australian Microbiome Initiative (AusMic; https://www.australianmicrobiome.com/) is a broadscale collaboration elucidating the microbial diversity of Australia, a nation‐continent that is geographically large, highly biodiverse, and ecologically heterogeneous. The AusMic project is a merger of two previous Australian microbiome characterization efforts: the Marine Microbes project (https://data.bioplatforms.com/organization/pages/bpa‐marine‐microbes) and the Biomes of Australian Soil Environments (BASE) project (https://bioplatforms.com/projects/soil‐biodiversity/). Data from these projects are publicly available, and have been utilized in a wide range of studies (Delgado‐Baquerizo et al., 2017; Midgley et al., 2017; Bissett and Brown, 2018; Raes et al., 2018).
To explore the mycota of Australia, we downloaded an AusMic data set of internal transcribed spacer (ITS1) DNA sequences from fungi found in Australian soils (Bissett et al., 2016). We examined the taxonomic results reported by AusMic, which are derived from the UNITE database taxonomy (Nilsson et al., 2019), then applied filtration steps to the AusMic sequences to allow for the sensible biological interpretation of these data. Next, we compared the filtered sequences against available sequences on GenBank using BLAST (Johnson et al., 2008; Benson et al., 2018). The AusMic data were then compared with a data set of 591 partial ITS sequences from specimens housed in the National Herbarium of Victoria (MEL) at the Royal Botanic Gardens Victoria, Melbourne, Australia; given the research interests of the authors, nearly all of the MEL specimens utilized were epigeous and hypogeous macrobasidiomycetes of the class Agaricomycetes. Furthermore, we generated two different visualizations of diversity for the Australian Agaricomycetes: one provides an overview using filtered operational taxonomic units (OTUs) and the AusMic taxonomy (Fig. 1), while the other is a plot of species diversity based on the AusMic taxonomy against latitude (Fig. 2). Each of our different approaches provided complementary insights, the highlights of which are discussed below.
Figure 1.
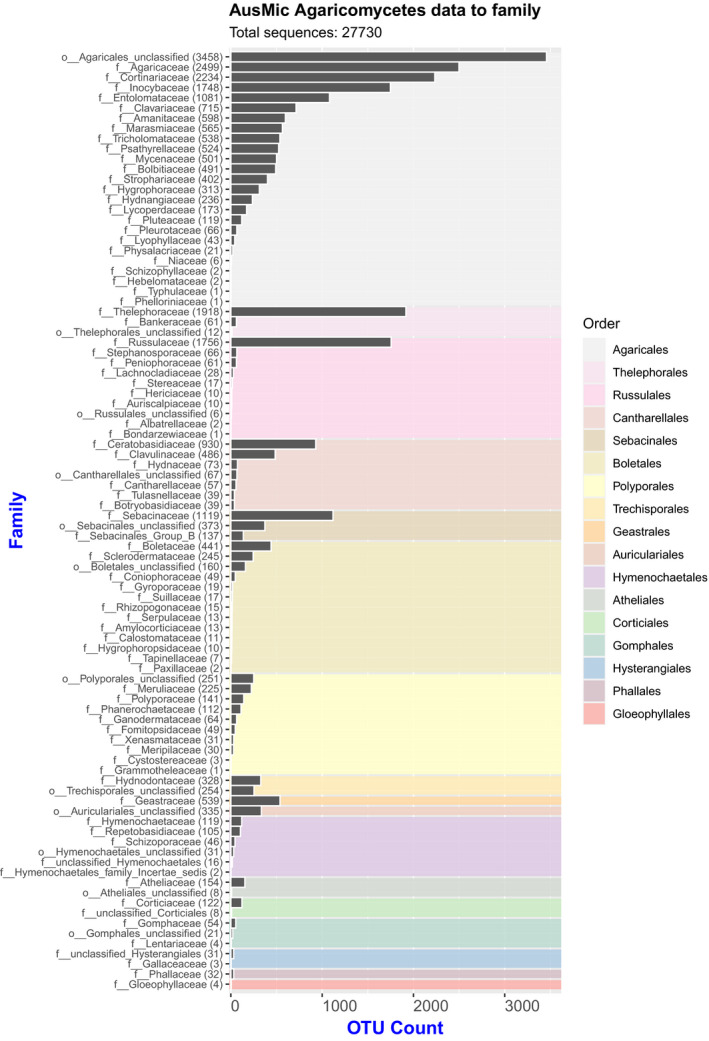
Histogram of Australian Microbiome Initiative (AusMic) Agaricomycete operational taxonomic units (OTUs) compiled to family and organized by order (background color‐coded based on legend) (total sequences: 27,730).
Figure 2.
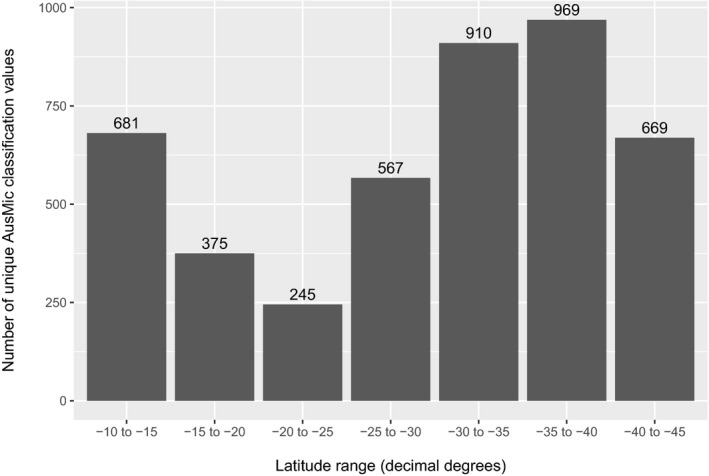
Histogram of unique taxonomic classifications (species) derived from the AusMic taxonomy for Agaricomycetes per bins of five decimal degrees latitude, estimating the relative diversity of Agaricomycetes across Australia. A total of 1263 unique taxonomic determinations were available within Agaricomycetes; each geographic bin was populated from this pool (counts for each bin are shown above each bar on the graph).
METHODS
ITS amplicon data and associated metadata generated by AusMic were downloaded from the BioPlatforms Australia data portal (https://data.bioplatforms.com/bpa/otu/ [accessed 14 March 2019]) using the ITS1FITS4_fungi amplicon filter. Consequently, 1,170,628 sequences were recovered. Due to sequencing issues during the construction of the AusMic fungal ITS data set (Bissett et al., 2016), these sequences correspond to forward Illumina reads only (maximum sequence length 301 bp, N50 182 bp). Sequences from some Antarctic samples included in the AusMic project were identified using the sample/latitude values in the metadata file, and were subsequently removed along with the duplicated sequences, leaving 195,177 amplicons. These data were clustered using usearch version 11.0.667_i86linuSx32 (Edgar, 2010) and the results were filtered to remove the nested sequences, resulting in 192,325 remaining sequences (OTUs) with a maximum length of 301 bp and an N50 of 189 bp. Thus, our OTUs represent a subset of the initial AusMic OTUs. By clustering the nested sequences from the initial data set, our OTUs reduce the potential overestimation of diversity that might occur if using the unfiltered AusMic data (which contains nested but otherwise identical sequences).
The sequences were searched against both the National Center for Biotechnology Information (NCBI) Nucleotide (nt) database and an in‐house (MEL) database of fungal ITS sequences using BLASTN version 2.9.0+ with the settings max_target_seqs = 1 and evalue = 1e‐5. In each case, the top BLAST hit was retained if the BLAST alignment covered more than 95% of the query length and the BLAST high‐scoring segment pair identity was greater than 97%. These results were output to a .csv file (Appendix S1). The in‐house database is derived from gDNA extracts of macromycete collections housed at MEL, with the ITS sequences acquired by PCR amplification using the ITS1 (or ITS1‐F) and ITS4 primers (White et al., 1990; Gardes and Bruns, 1993) under the following thermocycling protocol: 95°C for 5 min, 38 cycles of 94°C for 35 s, 50°C for 60 s, and 72°C for 60 s, with a final extension at 72°C for 60 s. After manually inspecting all of the MEL chromatograms to ensure quality and retaining only sequences assembled from multiple reads (i.e., forward and reverse primers and/or multiple sequencing attempts) with unambiguous base calls (a small number of ambiguous base calls were marked as N), a total of 591 ITS sequences were found to be of sufficiently high quality to include in this study. Because AusMic retained only data corresponding to ITS1 (derived from forward reads) for public release (Bissett et al., 2016), we manually trimmed our sequences to correspond to ITS1 as well (some portions of adjacent regions, such as 5.8S, were also retained during the trimming process).
Agaricomycete OTUs present in the filtered AusMic data (192,325 sequences) were visualized with ggplot2 (Wickham, 2016), using R version 3.6.1 (R Core Team, 2019) within RStudio version 1.2.1335 (RStudio Team, 2019). Briefly, the .csv file (Appendix S1) was used as input, and OTUs for which the AusMic classification included ‘phylum = k__fungi_unclassified’ (that is, the sequence could be assigned to the kingdom Fungi, but not to a specific phylum) were removed. This data set was further filtered to include only sequences where the AusMic classification included ‘class = c__Agaricomycetes’, leaving 35,905 sequences. For visualization purposes, OTUs with the classification ‘c__Agaricomycetes_unclassified’, ‘f__Agaricomycetes_family_Incertae_sedis’, or ‘f__unclassified_Agaricomycetes’ were removed. Moreover, in several cases where classification to family level was uncertain, counts of OTUs were merged under the corresponding order name (see Appendix S2). The resultant 27,730 Agaricomycete OTUs were visualized (Fig. 1, Appendix S3).
The following steps were carried out to create a plot of species determinations (based on the AusMic taxonomy) against latitude. Sequences from Antarctica were removed from the AusMic data set as described above. AusMic OTUs with an AusMic classification of ‘phylum = k__fungi_unclassified’ were removed, and the data set was filtered to include only sequences where the AusMic classification included ‘class = c__Agaricomycetes', leaving 216,295 sequences. Latitude information for each sequence was extracted from the metadata file downloaded from BioPlatforms Australia, and for each latitude ‘bin’ of five decimal degrees a list of unique AusMic classifications was created based on the Species column of the table presented in Appendix S4. Counts of unique classifications per bin were visualized using ggplot2 and R version 3.6.1, as described above (Fig. 2).
RESULTS
The results of the BLAST analysis are included in Appendix S1. Of the 192,325 OTUs BLASTed against the NCBI nt database, 46,099 (~24%) retrieved hits. Against the MEL data (591 ITS sequences), 935 (<1% of the 192,325 OTUs) hits were retrieved. A graph of the Agaricomycete OTUs grouped to family and order is shown in Fig. 1. Of the Agaricomycetes present in Australia’s soil mycota (27,730 OTUs), Agaricales was inferred to be the most diverse order (16,337 OTUs), followed by Thelephorales (1991 OTUs), Russulales (1957 OTUs), Cantharellales (1691 OTUs), Sebacinales (1629 OTUs), Boletales (1002 OTUs), and Polyporales (907 OTUs). Overall, 17 orders of Agaricomycetes were inferred. Counts of the Agaricomycete OTUs at familial and ordinal levels are included in Appendix S3. A graph of Australian Agaricomycete species (derived from a pool of 1263 unique taxonomic determinations from AusMic) plotted against binned latitude values is presented in Fig. 2.
DISCUSSION
Agaricales, which includes many familiar gilled mushrooms (e.g., species of Agaricus L., Amanita Pers., and numerous “weedy” groups that occur on wood matter, leaf litter, and soil such as Mycena (Pers.) Roussel, Marasmius Fr., and Psathyrella (Fr.) Quél.), is inferred to be the most diverse order of Agaricomycetes in Australia’s soil mycota, with 16,337 OTUs (Fig. 1). This is significantly higher than the other 16 orders of Agaricomycetes inferred in our study. It should be noted that the AusMic classifications, which are reflected in Fig. 1, should be interpreted with some caution, as the taxonomic assignment was performed using a bootstrap‐based confidence cut‐off value of at least 60% (see Bissett et al., 2016 for details on OTU taxonomic assignment for AusMic).
Using the AusMic taxonomic determinations, we observed a general decrease in Australian Agaricomycete species diversity toward the −20.0 to −25.0 latitudes (decimal degrees), which harbor the lowest diversity of all the latitude bins (Fig. 2). This is likely due to these areas comprising substantial portions of Australia’s arid interior. Using the AusMic taxonomy alone underestimates diversity, as each taxonomic determination can correspond to many OTUs.
The BLAST comparisons of the AusMic data to GenBank and the MEL data set yielded ecological, morphological, and taxonomic insights. One example of an ecological insight arose from the 43 AusMic OTUs that matched a Mycena sequence from GenBank (AY627835.1) that was generated from fungal material associated with roots of Epacris pulchella Cav. (Ericaceae). Although the genus Mycena is generally known to be non‐mycorrhizal, this result aligns with other instances of Mycena species reported to be in mycorrhizal or mycorrhizal‐like relationships with Ericaceae and other groups (Zhang et al., 2012; Grelet et al., 2017). We are confident in the generic determination of Mycena in this case because (a) the AusMic classification placed the 43 OTUs in Mycena (except for two OTUs that fell into “unclassified Agaricales”), (b) a post‐hoc BLAST search of GenBank using AY627835.1 retrieved additional Mycena sequences, and (c) four of the 43 AusMic OTUs hit a Mycena specimen in the MEL data.
For morphological insights, the MEL data set was especially useful. MEL houses many specimens of sequestrate fungi (enclosed or truffle‐like, and often buried in soil), which are diverse and abundant throughout Australia. Our BLAST analysis of the AusMic OTUs against the MEL data retrieved numerous hits for the sequestrate genus Zelleromyces Singer & A. H. Sm. (Russulaceae). While corresponding GenBank hits and AusMic determinations occasionally reported sequestrate genera, the majority of GenBank and AusMic determinations corresponding to MEL specimens of sequestrate Russulaceae were for the epigeous mushroom genera Russula Pers. and Lactarius Pers. As such, the MEL data set elucidates the morphological form of some of our study organisms. Although the majority of sequestrate Russulaceae genera are polyphyletic within the family (Vidal et al., 2019), indicating the need for systematic revision, the retention of “field identification” names on herbarium specimens adds another layer of information to this study.
Our analysis provides insights into the differences in taxonomic classification between the AusMic, GenBank, and MEL taxon determinations. Many MEL specimens have been identified by taxonomic specialists, therefore, in some cases MEL hits provided informative names where AusMic and GenBank did not. For example, the AusMic sequence 459338 (Appendix S1) is determined as “Agaricales unclassified” and retrieved no GenBank hits; however, a MEL sequence for Richoniella Costantin & L. M. Dufour (a sequestrate genus of the Entolomataceae) was retrieved. A post‐hoc BLAST search of the AusMic sequence 459338 against GenBank to include identities as low as 82% retrieved taxonomic determinations at various levels, such as Entoloma (Fr.) P. Kumm. and “Uncultured Agaricales.” In other cases, the determinations of MEL specimens at high taxonomic ranks were better achieved using AusMic or GenBank. For example, the AusMic sequence 407378 retrieved a specimen submitted to MEL with only the ordinal identification of Agaricales; however, AusMic determined it as a Crepidotus (Fr.) Staude, and while no GenBank hits above a 97% identity were found, a post‐hoc search below 97% retrieved numerous Crepidotus hits.
The use of ITS sequences for fungal metagenomics is powerful, but not without problems. The existence of extensive intragenomic variation in fungal ITS sequences due to multiple polymorphic copies within a species has been reported for various orders of Ascomycota and Basidiomycota (e.g., Vydryakova et al., 2012; Stadler et al., 2020). The causes of this are likely heterogeneous, and may include the release of concerted evolution and/or the multinucleate (dikaryotic) condition of the Ascomycota and Basidiomycota (Roper et al., 2011; Roberts and Gladfelter, 2015). This variability in fungal ITS sequences can lead to the overestimation of species diversity (Lindner and Banik, 2011), which may be a caveat for the interpretation of the AusMic data. The utilization of alternative loci for fungal metagenomics, such as rpb2 (Větrovský et al., 2016) or 28S (Kivlin et al., 2011), may provide a fruitful basis of comparison and calibration for ITS‐based studies in the future.
In summary, our work outlines protocols to (a) establish OTUs from large metagenomic data sets while avoiding a potential overestimation of diversity, (b) cross‐reference these OTUs against existing taxonomies and sequence data, (c) use geographic metadata and taxonomic determinations from metagenomic studies to analyze diversity against geographic variables, and (d) provide useful outputs and visualizations of these findings. We draw insights into the Australian fungi, especially Agaricomycetes. Our methods can be applied and expanded with the data used in this study or similar data.
AUTHOR CONTRIBUTIONS
N.D. contributed to the conception, planning, and writing of this article and generated a portion of the MEL data. C.J.J. contributed to the planning of this article and conducted the bioinformatic analyses, including the data visualization. G.D.H. generated most of the MEL data and contributed to conversations reflected in the paper. T.L. contributed to the planning of this study, generated a portion of the MEL data, and contributed to the information in the paper.
Supporting information
APPENDIX S1. Output file (.csv) for the BLAST analyses of the filtered Australian Microbiome Initiative (AusMic) sequences versus the National Center for Biotechnology Information (NCBI) and the National Herbarium of Victoria (MEL) data sets, showing the filtered operational taxonomic units (OTUs) (the AusMic identifiers in the first column and the sequences in the second column), the AusMic classification for each OTU (third column), the NCBI hits (fourth column) with percent identity (fifth column), and the MEL hits (sixth column) with percent identity (seventh column).
APPENDIX S2. Merged and omitted operational taxonomic unit (OTU) categories.
APPENDIX S3. Counts of Agaricomycete operational taxonomic units (OTUs) at familial and ordinal levels. The “freq” column shows the numbers of OTUs for the corresponding taxonomic classification in the “family” column, which also shows the same OTU count in parentheses. The “group_sum” column shows the total OTU counts for the corresponding orders.
APPENDIX S4. Latitudes (last column) with corresponding occurrences of Agaricomycetes indicated by Australian Microbiome Initiative (AusMic) taxonomic classifications, along with associated environmental sample IDs and operational taxonomic unit (OTU) counts. The “species” column contains 1263 unique classifications (this includes 219 unique determinations at ranks above species).
APPENDIX S5. FASTA file of the National Herbarium of Victoria (MEL) data.
Acknowledgments
The authors acknowledge support from Bioplatforms Australia through funding from the Australian Government National Collaborative Research Infrastructure Strategy, through use of the Biomes of Australian Soil Environments (BASE) data, and through contributions toward sequencing efforts for the National Herbarium of Victoria (MEL) database. This study was also supported in part by an Australian Biological Resources Study grant (RF217‐63).
Davoodian, N. , Jackson C. J., Holmes G. D., and Lebel T.. 2020. Continental‐scale metagenomics, BLAST searches, and herbarium specimens: The Australian Microbiome Initiative and the National Herbarium of Victoria. Applications in Plant Sciences 8(9): e11392.
Data Availability
The AusMic data utilized in this study were downloaded from the BioPlatforms Australia data portal (https://data.bioplatforms.com/bpa/otu/ [accessed 14 March 2019]), using the ITS1FITS4_fungi amplicon filter (see the Methods for details on further processing). Our subsequent analyses using the AusMic data are available as Appendices S1–S4. The FASTA file containing the MEL data utilized in our study is included here as Appendix S5; the sequences were also deposited in GenBank (MT703045–MT703635).
LITERATURE CITED
- Accioly, T. , Cruz R. H., Assis N. M., Ishikawa N. K., Hosaka K., Martín M. P., and Baseia I. G.. 2018. Amazonian bird’s nest fungi (Basidiomycota): Current knowledge and novelties on Cyathus species. Mycoscience 59: 331–342. [Google Scholar]
- Benson, D. A. , Cavanaugh M., Clark K., Karsch‐Mizrachi I., Ostell J., Pruitt K. D., and Sayers E. W.. 2018. GenBank. Nucleic Acids Research 46(D1): D41–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissett, A. , Fitzgerald A., Court L., Meintjes T., Mele P. M., Reith F., Dennis P. G., et al. 2016. Introducing BASE: The Biomes of Australian Soil Environments soil microbial diversity database. GigaScience 5: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissett, A. , and Brown M. V.. 2018. Alpha‐diversity is strongly influenced by the composition of other samples when using multiplexed sequencing approaches. Soil Biology and Biochemistry 127: 79–81. [Google Scholar]
- Blackwell, M. 2011. The Fungi: 1, 2, 3… 5.1 million species? American Journal of Botany 98(3): 426–438. [DOI] [PubMed] [Google Scholar]
- Burgess, J. W. , Schwan W. R., and Volk T. J.. 2006. PCR‐based detection of DNA from the human pathogen Blastomyces dermatitidis from natural soil samples. Medical Mycology 44: 741–748. [DOI] [PubMed] [Google Scholar]
- Buyck, B. , Henkel T. W., and Hofstetter V.. 2018. The Cantharellus isabellinus species complex (Cantharellales, Hydnaceae) in tropical Africa. Mycosphere 9: 1209–1221. [Google Scholar]
- Buyck, B. , Henkel T. W., and Hofstetter V.. 2019. Epitypification of the Central African Cantharellus densifolius and C. luteopunctatus allows for the recognition of two additional species. MycoKeys 49: 49–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano, M. A. , Elliott T. F., Truong C., Séné O., Dentinger B. T., and Henkel T. W.. 2016. Kombocles bakaiana gen. sp. nov. (Boletaceae), a new sequestrate fungus from Cameroon. IMA Fungus 7: 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. , Desirò A., Na H., Sandor L., Lipzen A., Clum A., Barry K., et al. 2019. Phylogenomics of Endogonaceae and evolution of mycorrhizas within Mucoromycota. New Phytologist 222: 511–525. [DOI] [PubMed] [Google Scholar]
- Crossay, T. , Cilia A., Cavaloc Y., Amir H., and Redecker D.. 2018. Four new species of arbuscular mycorrhizal fungi (Glomeromycota) associated with endemic plants from ultramafic soils of New Caledonia. Mycological Progress 17: 729–744. [Google Scholar]
- Davoodian, N. , Bougher N. L., Fechner N. A., Bergemann S. E., and Halling R. E.. 2019. Three new species of Gyroporus (Boletales, Basidiomycota) from Australia. Muelleria 37: 101–107. [Google Scholar]
- Delgado‐Baquerizo, M. , Bissett A., Eldridge D. J., Maestre F. T., He J. Z., Wang J. T., Hamonts K., et al. 2017. Palaeoclimate explains a unique proportion of the global variation in soil bacterial communities. Nature Ecology and Evolution 1: 1339–1347. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Gardes, M. , and Bruns T. D.. 1993. ITS primers with enhanced specificity for basidiomycetes: Application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Grelet, G. A. , Ba R., Goeke D. F., Houliston G. J., Taylor A. F. S., and Durall D. M.. 2017. A plant growth‐promoting symbiosis between Mycena galopus and Vaccinium corymbosum seedlings. Mycorrhiza 27: 831–839. [DOI] [PubMed] [Google Scholar]
- Halling, R. E. , Nuhn M., Fechner N. A., Osmundson T. W., Soytong K., Arora D., Hibbett D. S., et al. 2012. Sutorius: A new genus for Boletus eximius . Mycologia 104: 951–961. [DOI] [PubMed] [Google Scholar]
- Hodkinson, B. P. , Moncada B., and Lücking R.. 2014. Lepidostromatales, a new order of lichenized fungi (Basidiomycota, Agaricomycetes), with two new genera, Ertzia and Sulzbacheromyces, and one new species, Lepidostroma winklerianum . Fungal Diversity 64: 165–179. [Google Scholar]
- Hofstetter, V. , Buyck B., Eyssartier G., Schnee S., and Gindro K.. 2019. The unbearable lightness of sequenced‐based identification. Fungal Diversity 96: 243–284. [Google Scholar]
- Hosaka, K. , Bates S. T., Beever R. E., Castellano M. A., Colgan W. III, Domínguez L. S., Nouhra E. R., et al. 2006. Molecular phylogenetics of the gomphoid‐phalloid fungi with an establishment of the new subclass Phallomycetidae and two new orders. Mycologia 98: 949–959. [DOI] [PubMed] [Google Scholar]
- Jami, F. , Marincowitz S., Crous P. W., Jacobsohn A., and Wingfield M. J.. 2018. A new Cytospora species pathogenic on Carpobrotus edulis in its native habitat. Fungal Systematics and Evolution 2: 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X. , Wu D. M., Song C. G., Liu S., Si J., and Cui B. K.. 2019. Two new Neofomitella species (Polyporaceae, Basidiomycota) based on morphological and molecular evidence. Mycological Progress 18: 593–602. [Google Scholar]
- Johnson, M. , Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., and Madden T. L.. 2008. NCBI BLAST: A better web interface. Nucleic Acids Research 36(Web Server issue): W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaishian, P. , and Weir A.. 2018. New species of Prolixandromyces (Laboulbeniales) from South America. Mycologia 110: 222–229. [DOI] [PubMed] [Google Scholar]
- Khmelnitsky, O. , Davoodian N., Singh P., Raspé O., Lee S. M. L., Fechner N., Bonito G., et al. 2019. Ionosporus: A new genus for Boletus longipes (Boletaceae), with a new species, I. australis, from Australia. Mycological Progress 18: 439–451. [Google Scholar]
- Kivlin, S. N. , Hawkes C. V., and Treseder K. K.. 2011. Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biology and Biochemistry 43: 2294–2303. [Google Scholar]
- Kuhar, F. , Smith M. E., Mujic A., Truong C., and Nouhra E.. 2017. A systematic overview of Descolea (Agaricales) in the Nothofagaceae forests of Patagonia. Fungal Biology 121: 876–889. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Gu X., Kasson M. T., Bateman C. C., Guo J., Huang Y., Li Q., et al. 2016. Distribution, host records, and symbiotic fungi of Euwallacea fornicatus (Coleoptera: Curculionidae: Scolytinae) in China. Florida Entomologist 99: 801–804. [Google Scholar]
- Lindner, D. L. , and Banik M. T.. 2011. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus . Mycologia 103: 731–740. [DOI] [PubMed] [Google Scholar]
- Luo, Z. L. , Bahkali A. H., Liu X. Y., Phookamsak R., Zhao Y. C., Zhou D. Q., Su H. Y., et al. 2016. Poaceascoma aquaticum sp. nov. (Lentitheciaceae), a new species from submerged bamboo in freshwater. Phytotaxa 253: 71–80. [Google Scholar]
- Medeiros, I. D. , Kraichak E., Lücking R., Mangold A., and Lumbsch H. T.. 2017. Assembling a taxonomic monograph of tribe Wirthiotremateae (lichenized Ascomycota: Ostropales: Graphidaceae). Fieldiana Life and Earth Sciences 9: 1–32. [Google Scholar]
- Midgley, D. J. , Greenfield P., Bissett A., and Tran‐Dinh N.. 2017. First evidence of Pezoloma ericae in Australia: Using the Biomes of Australia Soil Environments (BASE) to explore the Australian phylogeography of known ericoid mycorrhizal and root‐associated fungi. Mycorrhiza 27: 587–594. [DOI] [PubMed] [Google Scholar]
- Midgley, D. J. , Sutcliffe B., Greenfield P., and Tran‐Dinh N.. 2018. Gamarada debralockiae gen. nov. sp. nov.—The genome of the most widespread Australian ericoid mycorrhizal fungus. Mycorrhiza 28: 379–389. [DOI] [PubMed] [Google Scholar]
- Mujic, A. B. , Zheng N., Kim K., Spatafora J. W., Castellano M. A., and Smith M. E.. 2019. The Cedrus‐associated truffle Trappeindia himalayensis is a morphologically unique and phylogenetically divergent species of Rhizopogon . Mycologia 111: 225–234. [DOI] [PubMed] [Google Scholar]
- Nilsson, R. H. , Larsson K. H., Taylor A. F. S., Bengtsson‐Palme J., Jeppesen T. S., Schigel D., Kennedy P., et al. 2019. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 47(D1): D259–D264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovrebo, C. L. , Hughes K. W., and Halling R. E.. 2019. Three new species of Tricholoma from Costa Rica. Phytotaxa 392: 33–44. [Google Scholar]
- R Core Team . 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: Website http://www.R‐project.org/ [accessed 27 August 2020]. [Google Scholar]
- Raes, E. J. , Bodrossy L., van de Kamp J., Bissett A., Ostrowski M., Brown M. V., Waite A. M., et al. 2018. Oceanographic boundaries constrain microbial diversity gradients in the South Pacific Ocean. Proceedings of the National Academy of Sciences, USA 115(35): E8266–E8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S. E. , and Gladfelter A. S.. 2015. Nuclear autonomy in multinucleate fungi. Current Opinion in Microbiology 28: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, M. , Ellison C., Taylor J. W., and Glass N. L.. 2011. Nuclear and genome dynamics in multinucleate ascomycete fungi. Current Biology 21(18): R786–R793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosling, A. , Cox F., Cruz‐Martinez K., Ihrmark K., Grelet G. A., Lindahl B. D., Menkis A., and James T. Y.. 2011. Archaeorhizomycetes: Unearthing an ancient class of ubiquitous soil fungi. Science 333: 876–879. [DOI] [PubMed] [Google Scholar]
- RStudio Team . 2019. RStudio: Integrated development for R. RStudio, Boston, Massachusetts, USA. [Google Scholar]
- Schoch, C. L. , Wang Z., Townsend J. P., and Spatafora J. W.. 2009. Geoglossomycetes cl. nov., Geoglossales ord. nov. and taxa above class rank in the Ascomycota Tree of Life. Persoonia 22: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüβler, A. , Schwarzott D., and Walker C.. 2001. A new fungal phylum, the Glomeromycota: Phylogeny and evolution. Mycological Research 105: 1413–1421. [Google Scholar]
- Smith, M. E. , Trappe J. M., and Rizzo D. M.. 2006. Genea, Genabea and Gilkeya gen. nov.: Ascomata and ectomycorrhiza formation in a Quercus woodland. Mycologia 98: 699–716. [DOI] [PubMed] [Google Scholar]
- Stadler, M. , Lambert C., Wibberg D., Kalinowski J., Cox R. J., Kolařík M., and Kuhnert E.. 2020. Intragenomic polymorphisms in the ITS region of high‐quality genomes of the Hypoxylaceae (Xylariales, Ascomycota). Mycological Progress 19: 235–245. [Google Scholar]
- Sukarno, N. , Listiyowati S., Rahayu N., and Nara K.. 2019. Elaphomyces tropicalis sp. nov.: A new ectomycorrhizal fungus associated with dipterocarps from tropical Indonesia. Mycoscience 60: 83–88. [Google Scholar]
- Thiers, B. M. , and Halling R. E.. 2018. The Macrofungi Collection Consortium. Applications in Plant Sciences 6: e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Cruz, T. J. , Billingsley Tobias T. L., Almatruk M., Hesse C. N., Kuske C. R., Desirò A., Benucci G. M. N., et al. 2017. Bifiguratus adelaidae, gen. et sp. nov., a new member of Mucoromycotina in endophytic and soil‐dwelling habitats. Mycologia 109: 363–378. [DOI] [PubMed] [Google Scholar]
- Vadthanarat, S. , Raspé O., and Lumyong S.. 2018. Phylogenetic affinities of the sequestrate genus Rhodactina (Boletaceae), with a new species, R. rostratispora from Thailand. MycoKeys 29: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadthanarat, S. , Amalfi M., Halling R. E., Bandala V., Lumyong S., and Raspé O.. 2019. Two new Erythrophylloporus species (Boletaceae) from Thailand, with two new combinations of American species. MycoKeys 55: 29–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Větrovský, T. , Kolařík M., Žifčáková L., Zelenka T., and Baldrian P.. 2016. The rpb2 gene represents a viable alternative molecular marker for the analysis of environmental fungal communities. Molecular Ecology Resources 16: 388–401. [DOI] [PubMed] [Google Scholar]
- Vidal, J. M. , Alvarado P., Loizides M., Konstantinidis G., Chachuła P., Mleczko P., Moreno G., et al. 2019. A phylogenetic and taxonomic revision of sequestrate Russulaceae in Mediterranean and temperate Europe. Persoonia 42: 127–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vydryakova, G. A. , Van D. T., Shoukouhi P., Psurtseva N. V., and Bissett J.. 2012. Intergenomic and intragenomic ITS sequence heterogeneity in Neonothopanus nambi (Agaricales) from Vietnam. Mycology 3: 89–99. [Google Scholar]
- White, T. J. , Bruns T. D., Lee S. B., and Taylor J. W.. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In Innis M. A., Gelfand D. H., Sninsky J. J., and White T. J.eds.], PCR protocols: A guide to methods and applications, 315–322. Academic Press, Cambridge, Massachusetts, USA. [Google Scholar]
- Wickham, H. 2016. ggplot2: Elegant graphics for data analysis. Springer‐Verlag, New York, New York, USA. [Google Scholar]
- Willis, C. G. , Ellwood E. R., Primack R. B., Davis C. C., Pearson K. D., Gallinat A. S., Yost J. M., et al. 2017. Old plants, new tricks: Phenological research using herbarium specimens. Trends in Ecology and Evolution 32: 531–546. [DOI] [PubMed] [Google Scholar]
- Willis, K. J. [ed.]. 2018. State of the world’s fungi 2018 report. Royal Botanic Gardens Kew, Richmond, Surrey, United Kingdom. [Google Scholar]
- Wu, G. , Li Y. C., Zhu X. T., Zhao K., Han L. H., Cui Y. Y., Li F., et al. 2016. One hundred noteworthy boletes from China. Fungal Diversity 81: 25–188. [Google Scholar]
- Zalar, P. , Sybren de Hoog G. S., Schroers H. J., Frank J. M., and Gunde‐Cimerman N.. 2005. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie van Leeuwenhoek 87: 311–328. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Chen J., Lv Y., Gao C., and Guo S.. 2012. Mycena sp., a mycorrhizal fungus of the orchid Dendrobium officinale . Mycological Progress 11: 395–401. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Output file (.csv) for the BLAST analyses of the filtered Australian Microbiome Initiative (AusMic) sequences versus the National Center for Biotechnology Information (NCBI) and the National Herbarium of Victoria (MEL) data sets, showing the filtered operational taxonomic units (OTUs) (the AusMic identifiers in the first column and the sequences in the second column), the AusMic classification for each OTU (third column), the NCBI hits (fourth column) with percent identity (fifth column), and the MEL hits (sixth column) with percent identity (seventh column).
APPENDIX S2. Merged and omitted operational taxonomic unit (OTU) categories.
APPENDIX S3. Counts of Agaricomycete operational taxonomic units (OTUs) at familial and ordinal levels. The “freq” column shows the numbers of OTUs for the corresponding taxonomic classification in the “family” column, which also shows the same OTU count in parentheses. The “group_sum” column shows the total OTU counts for the corresponding orders.
APPENDIX S4. Latitudes (last column) with corresponding occurrences of Agaricomycetes indicated by Australian Microbiome Initiative (AusMic) taxonomic classifications, along with associated environmental sample IDs and operational taxonomic unit (OTU) counts. The “species” column contains 1263 unique classifications (this includes 219 unique determinations at ranks above species).
APPENDIX S5. FASTA file of the National Herbarium of Victoria (MEL) data.
Data Availability Statement
The AusMic data utilized in this study were downloaded from the BioPlatforms Australia data portal (https://data.bioplatforms.com/bpa/otu/ [accessed 14 March 2019]), using the ITS1FITS4_fungi amplicon filter (see the Methods for details on further processing). Our subsequent analyses using the AusMic data are available as Appendices S1–S4. The FASTA file containing the MEL data utilized in our study is included here as Appendix S5; the sequences were also deposited in GenBank (MT703045–MT703635).


