Abstract
Accurate interpretation and analysis of echocardiography is important in assessing cardiovascular health. However, motion tracking often relies on accurate segmentation of the myocardium, which can be difficult to obtain due to inherent ultrasound properties. In order to address this limitation, we propose a semi-supervised joint learning network that exploits overlapping features in motion tracking and segmentation. The network simultaneously trains two branches: one for motion tracking and one for segmentation. Each branch learns to extract features relevant to their respective tasks and shares them with the other. Learned motion estimations propagate a manually segmented mask through time, which is used to guide future segmentation predictions. Physiological constraints are introduced to enforce realistic cardiac behavior. Experimental results on synthetic and in vivo canine 2D+t echocardiographic sequences outperform some competing methods in both tasks.
Keywords: Echocardiography, Motion Tracking, Segmentation, Semi-Supervised, Deep Learning
1. INTRODUCTION
In 2018, coronary heart disease (CHD) accounted for 43.8% of deaths attributed to cardiovascular disease in the United States, making it the greatest cause of heart-related fatalities in the nation [1]. A widely used clinical technique for detecting CHD is visually assessing left ventricular (LV) wall motion abnormality using echocardiography by trained experts. However, this is inherently subjective due to inter-observer variability [2]. An objective, quantitative metric for analyzing LV deformation can reduce this variability and assist in the detection and diagnosis of CHD.
Motion estimation and segmentation of the myocardium both play crucial roles in the detection of wall motion abnormality. However, both of these tasks are traditionally treated as separate steps in deformation analysis. Many motion tracking algorithms rely on accurate segmentation in order to generate displacement fields [3, 4, 5, 6]. This becomes especially problematic in echocardiography due to the relatively low signal-to-noise ratio (SNR) inherent in ultrasound imaging which makes the task of segmentation challenging, thus affecting the accuracy of the displacement field and the overall LV deformation analysis. Furthermore, lack of ground truth data makes it difficult to train deep learning models for either motion tracking or segmentation tasks, and it is impractical to have an expert manually annotate the large amounts of data required for training these models.
Work in the computer vision domain suggests that the tasks of motion tracking and segmentation are closely related and that learning the information required to accomplish one task may provide complementary information in accomplishing the other. Cheng et al. proposed SegFlow, an end-to-end unified network that estimates optical flow and object segmentation [7]. Tsai et al. developed ObjectFlow, an algorithm that iteratively optimizes optical flow and segmentation in a multi-scale framework [8]. This approach of training the tasks concurrently has been successfully applied to 2D cardiac MR sequences. Qin et al. proposes a Siamese style recurrent spatial transformer network combined with a fully convolutional network to simultaneously learn both motion and segmentation [9]. Furthermore, there have been recent developments in an unsupervised deformable registration framework applied to MR images by Balakrishnan et al. that learns to minimize the difference between a transformed source frame and a target frame without the need for ground truth [10]. However, none of these methods directly address the added challenges imposed by inherent ultrasound properties and therefore cannot be directly applied to echocardiography. To our knowledge, there has been no success in applying these methods directly to ultrasound images.
In this paper, we propose a semi-supervised joint learning network that adopts the bridging architecture seen in [7] to track LV motion and predict LV segmentations in 2D+t echocardiographic sequences. Our method differs in that we introduce physiological constraints to regularize the predicted motion fields and segmentations to adhere to more realistic cardiac behaviors as seen in [6, 11, 12] as well as implement a semi-supervised learning framework adapted from [10].
2. METHODS
The proposed model architecture is shown in Figure 1. Our primary contributions can be summarized as follows: (1) Adapting an established joint learning framework for 2D+t echocardiographic sequences [7]. (2) Implementing a semi-supervised learning framework to reduce the need for heavily annotated data [10]. (3) Introducing physiological constraints to enforce realistic cardiac behavior [6, 11, 12].
Fig. 1.
Proposed joint architecture showing separate motion tracking and segmentation branches and the feature bridging during up-sampling
2.1. Motion Tracking
Inspired by the success of FlowNetCorr in tracking flow, we design a network that inputs two images (source and target frame) into separate, but identical convolutional streams which are combined via a correlation layer that effectively performs patch-matching in feature space [13]. This correlation map then follows an encoder-decoder architecture of U-Net [14] and a down-sampling and upsampling phase with skip links for feature concatenation. The network outputs a 2D displacement field that maps a source frame to a target frame.
Unlike the original FlowNetCorr model, which is trained in a supervised manner [13], our model is optimized in an unsupervised manner similar to Balakrishnan et al [10]. The output displacement field is used to transform the source frame via bilinear interpolation. The network seeks to minimize the mean square difference between this morphed source frame and the target frame. This loss function is defined as follows:
| (1) |
where I = (I1 … IN) is an echocardiographic sequence of N images, Ui is the displacement field that maps a source image I1 to a target Ii, and F = (I1,Ui)) is a bilinear interpolation operator that morphs I1 to Ii using Ui.
2.2. Segmentation
Our segmentation branch is a U-Net inspired convolutional network that begins with an encoding and down-sampling path that is followed by a decoding and up-sampling path with skip links [14]. The input is the target frame of the motion tracking branch. The source frame is manually segmented. The output displacement field from the motion tracking branch propagates the manual segmentation via bilinear interpolation as well. The network is optimized by minimizing the Dice loss of the propagated segmentation and the predicted segmentation of the target frame. This loss function is defined as follows:
| (2) |
where Y is the manually segmented mask of I1 and M = (M2 … MN) are the predicted masks for each image in the sequence.
2.3. Joint Model
Communication between the two branches is done by implementing a similar bridging technique used in SegFlow [7]. Learned features are joined bi-directionally during up-sampling by concatenating up-sampled features from each task with the other. This allows each branch to utilize complementary features learned from the other branch that it may not have learned by itself. A convolutional RNN is incorporated at the last layer to maintain information temporally, as seen in [9].
2.3.1. Physiological Regularization
To ensure spatial smoothness, we choose to enforce incompressibility and LV shape constraints. We enforce incompressibility by penalizing divergence in a similar manner to Parajuli et al. [6]. This term penalizes the presence of sources and sinks in the displacement field, which does not occur in real cardiac motion. We minimize the gradient of the displacement field, which would be maximized if divergence is present, as follows:
| (3) |
LV shape is enforced by incorporating anatomical priors as proposed by Otkay et al [12]. We train a convolutional autoencoder to learn non-linear representations of known LV masks. The predicted mask from our segmentation branch is then fed into this autoencoder and the feature differences between the known mask and the predicted mask are minimized as follows:
| (4) |
where f is the operation of the autoencoder.
In addition to spatial constraints, we incorporate a temporal constraint. Cardiac motion typically occurs in a cycle from diastole to systole and back again. To enforce this pattern, we add a periodicity constraint in a manner similar to Lu et al [11]. This is done by minimizing the temporal derivative of the displacement field over time, which should sum close to zero. This term is defined as follows:
| (5) |
Thus, the composite loss function of the joint network is defined as follows:
| (6) |
where the λ values are the weighting parameters of each term.
3. EXPERIMENTS AND RESULTS
We evaluated the performance of our proposed model on two 2D+t datasets: synthetic human echocardiographic sequences and in vivo canine models. The synthetic data were produced by Alessandrini et al. [15] and is comprised of seven 3D echocardiographic sequences generated by passing labeled masks of a real sequence through an electro-mechanical motion simulator with varying parameters and inputting scatter maps sampled from real recordings into an ultrasound simulation environment. The induced motion and generated masks were used as ground truth to evaluate our results.
In vivo data was acquired using a Philips iE33 (Philips Medical Systems, Andover, MA) scanner and a X7–2 probe on anesthetized open-chest canine under three physiological conditions: baseline, stenosis in the left anterior descending (LAD) artery, and LAD stenosis under pharmacologically induced low-dose stress using 5μg/kg/min of dobutamine [11]. Displacements calculated from implanted sonomicrometer crystals and manual segmentations were used as ground truth to evaluate our results. A total of seven procedures were done (total of 21 sequences). All procedures were approved under Institutional Animal Care and Use Committee policies.
The model was trained using leave-one-out cross validation per sequence. For each sequence, sixty 2D cross sectional short axis (SAX) slices were sampled from apex to base at each time point. Each slice was cropped to 128×128. Each sequence was comprised of roughly 20–30 time frames. In total, synthetic experiments were performed on 420 sequences, and in-vivo experiments were performed on 1260 sequences. Data augmentation was performed online and included random rotations, flips, and shears. Weights were initialized offline by training with available annotated data.
We evaluated the segmentation performance of our joint model against a Dynamic Appearance Model (DAM) [16] and our segmentation branch by itself (Seg Only) by comparing Dice coefficient (Dice) and Hausdorff Distance (HD). The results reported on Table 1 and Fig. 2 show that our proposed model outperforms the DAM and Seg Only model in both Dice and HD for both synthetic and in vivo data. Wilcoxon signed-rank test confirm significance with p < .05 against all segmentation results.
Table 1.
Dice similarity coefficient (Dice) and Hausdorff Distance (HD) calculated over each sequence
| Methods | Synthetic | In Vivo | ||
|---|---|---|---|---|
| Dice | HD (mm) | Dice | HD (mm) | |
| DAM | 0.91±0.03 | 3.00±0.33 | 0.85±0.02 | 3.49±0.30 |
| Seg Only | 0.88±0.03 | 3.60±0.41 | 0.78±0.07 | 3.75±0.84 |
| Our Model | 0.95±0.01 | 2.44±0.36 | 0.87±0.01 | 3.01±0.34 |
Fig. 2.
Illustration of in vivo segmentation predictions at baseline (red) and ground truth (yellow). Left to Right: DAM, Seg Only, Our Model
We evaluated the motion tracking performance of our model against other motion tracking algorithms such as Flow Network Tracking (FNT) [6], non-rigid Free Form Deformation (FFD) [17], and our motion branch by itself (Motion Only). We compare the root mean squared error (RMSE) of the the x-and-y displacement fields (Ux and Uy, respectively) to the ground truth. The results reported on Table 2 and Fig. 3 show that our proposed model outperforms the FNT, FFD, and Motion Only model for both synthetic and in vivo data. Wilcoxon signed-rank test confirms significance with p < 0.05 against all motion tracking results, except FNT when calculating Ux on synthetic data (p = 0.056).
Table 2.
Root Mean Squared Error (RMSE) for the x and y displacements (Ux, Uy) calculated over each sequence
| Methods | Synthetic | In Vivo | ||
|---|---|---|---|---|
| Ux (mm) | Uy (mm) | Ux (mm) | Uy (mm) | |
| FFD | 2.06±1.01 | 1.88±0.43 | 2.58±1.48 | 2.86±1.22 |
| FNT | 0.67±0.14 | 0.75±0.16 | 1.07±0.27 | 1.64±0.53 |
| Motion Only | 1.41±0.32 | 1.57±0.24 | 2.45±0.76 | 2.37±0.73 |
| Our Model | 0.61±0.13 | 0.69±0.14 | 1.01±0.34 | 0.79±0.22 |
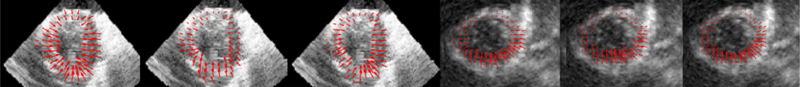
Fig. 3.
Illustration of motion estimation in studies with LAD stenosis (In Vivo) and LAD infarction (Synthetic): Left to Right: Ground Truth (In Vivo), FFD (In Vivo), Our Model (In Vivo), Ground Truth (Synthetic), FFD (Synthetic), Our Model (Synthetic)
4. CONCLUSION
In this paper, we present a semi-supervised joint learning method for motion tracking and segmentation in echocardiography through a novel integration of a previously established feature bridging technique [7] and a semi-supervised learning framework [10]. We expand on this network by introducing physiological constraints to enforce realistic cardiac behavior. This method reduces reliance on accurate segmentations that many motion tracking algorithms have while improving the performance of both tasks by simultaneously learning task similarities. We have shown that our proposed model outperforms competing methods in both motion tracking and segmentation. Future work will explore 3D+t and address the practical limitations of requiring initial segmentations.
5. ACKNOWLEDGEMENTS
We would like to acknowledge the technical assistance of the staff of the Yale Translational Research Imaging Center and Drs. Nabil Boutagy, Imran Alkhalil, Melissa Eberle, and Zhao Liu for assisting with the in vivo canine imaging studies.
6. REFERENCES
- [1].Benjamin EJ et al. , “Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association,” Circulation, 2018. [DOI] [PubMed] [Google Scholar]
- [2].Hoffmann R et al. , “Analysis of regional left ventricular function by cineventriculography, cardiac magnetic resonance imaging, and unenhanced and contrast-enhanced echocardiography: a multicenter comparison of methods,” J. Am. Coll. Cardiol, January 2006. [DOI] [PubMed] [Google Scholar]
- [3].Papademetris X et al. , “Estimation of 3-d left ventricular deformation from medical images using biomechanical models,” IEEE transactions on medical imaging, 2002. [DOI] [PubMed] [Google Scholar]
- [4].Shi P et al. , “Point-tracked quantitative analysis of left ventricular surface motion from 3-d image sequences,” IEEE transactions on medical imaging, 2000. [DOI] [PubMed] [Google Scholar]
- [5].Lin N et al. , “Generalized robust point matching using an extended free-form deformation model: application to cardiac images,” in Biomedical Imaging: Nano to Macro, 2004. IEEE International Symposium on IEEE, 2004. [Google Scholar]
- [6].Parajuli N et al. , “Flow network tracking for spatiotemporal and periodic point matching: Applied to cardiac motion analysis,” Medical Image Analysis, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cheng J et al. , “Segflow: Joint learning for video object segmentation and optical flow,” in IEEE International Conference on Computer Vision (ICCV), 2017. [Google Scholar]
- [8].Tsai Y et al. , “Video segmentation via object flow,” in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 2016, pp. 3899–3908. [Google Scholar]
- [9].Qin C et al. , “Joint learning of motion estimation and segmentation for cardiac MR image sequences,” CoRR, 2018. [Google Scholar]
- [10].Balakrishnan G et al. , “An unsupervised learning model for deformable medical image registration,” in The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 2018. [Google Scholar]
- [11].Lu A et al. , “Learning-based regularization for cardiac strain analysis with ability for domain adaptation,” CoRR, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Oktay O et al. , “Anatomically constrained neural networks (ACNN): application to cardiac image enhancement and segmentation,” CoRR, 2017. [DOI] [PubMed] [Google Scholar]
- [13].Fischer P et al. , “Flownet: Learning optical flow with convolutional networks,” CoRR, 2015. [Google Scholar]
- [14].Ronneberger O et al. , “U-net: Convolutional networks for biomedical image segmentation,” CoRR, 2015. [Google Scholar]
- [15].Alessandrini M et al. , “A pipeline for the generation of realistic 3d synthetic echocardiographic sequences: Methodology and open-access database,” IEEE transactions on medical imaging, 2015. [DOI] [PubMed] [Google Scholar]
- [16].Huang X et al. , “Contour tracking in echocardiographic sequences via sparse representation and dictionary learning,” Medical image analysis, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rueckert D et al. , “Nonrigid registration using free-form deformations: application to breast mr images,” IEEE transactions on medical imaging, 1999. [DOI] [PubMed] [Google Scholar]