Abstract
The de novo design of proteins that bind highly functionalized small molecules represents a great challenge. To enable computational design of binders, we developed a unit of protein structure—a van der Mer (vdM)—that maps the backbone of each amino acid to statistically preferred positions of interacting chemical groups. Using vdMs, we designed six de novo proteins to bind the drug apixaban; two bound with low and submicromolar affinity. X-ray crystallography and mutagenesis confirmed a structure with a precisely designed cavity that forms favorable interactions in the drug–protein complex. vdMs may enable design of functional proteins for applications in sensing, medicine, and catalysis.
The Anfinsen hypothesis states that a protein’s sequence encodes its tertiary structure and underlying function (1). Conversely, a protein’s tertiary structure encodes the possible sequences compatible with a particular function. De novo protein design has succeeded in the creation of proteins that fold to various targeted tertiary structures (structure to sequence) (2, 3). Nevertheless, it has been extremely challenging to design proteins that not only fold but also bind to complex small molecules (function and structure to sequence) (2–4). Use of algorithms optimized for packing apolar protein cores leads to difficulty when designing polar cavities required for binding hydrophilic molecules (5). Consequently, design of small-molecule–binding proteins has generally required recursive experimental screening and large libraries to engender function, mostly starting with natural proteins rather than de novo structures (Fig. 1A) (3, 4, 6–9). Here, we accomplish the reverse of the Anfinsen hypothesis by simultaneously designing structure and binding function from scratch, targeting a small-molecule drug with significant polarity and structural complexity. To do this, we developed a unit of local protein structure that directly links a tertiary structure to key interactions that engender tight and specific binding. These findings illuminate the principles underlying the emergence and evolution of complex function in proteins and provide a methodology for designing useful proteins.
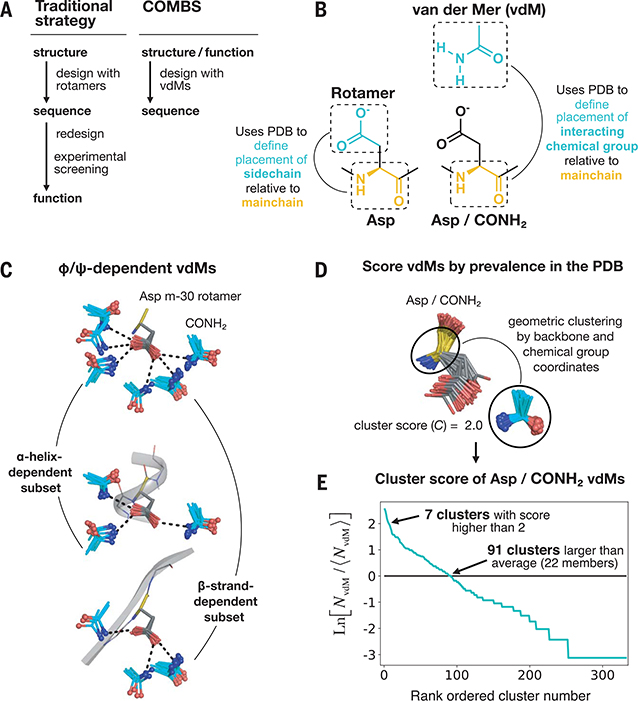
Fig. 1. A vdM is a structural unit relating chemical group position to the protein backbone.
(A) Workflow of a traditional protein design strategy versus that of COMBS. (B) Definition of a vdM. A chemical group is interacting if it is in van der Waals contact with the protein side chain or main chain. Like rotamers, vdMs are derived from a large set of high-quality protein crystal structures. A vdM of aspartic acid (Asp) and carboxamide (CONH2, cyan) is shown. (C) vdMs are φ, ψ, and rotamer dependent; this is illustrated by the top vdMs of the m-30 rotamer of Asp, clustered by location of CONH2 after exact superposition of main chain N, Cα, and C atoms. (D and E) We ranked vdMs by prevalence in the PDB, quantified by a cluster score C [the natural logarithm of the ratio of the number of members in a cluster (NvdM) to the average number of members in a cluster (⟨hNvdM⟩)]. The seventh-largest cluster of Asp/CONH2 vdMs is shown as an example in (D).
Targeted function and fold
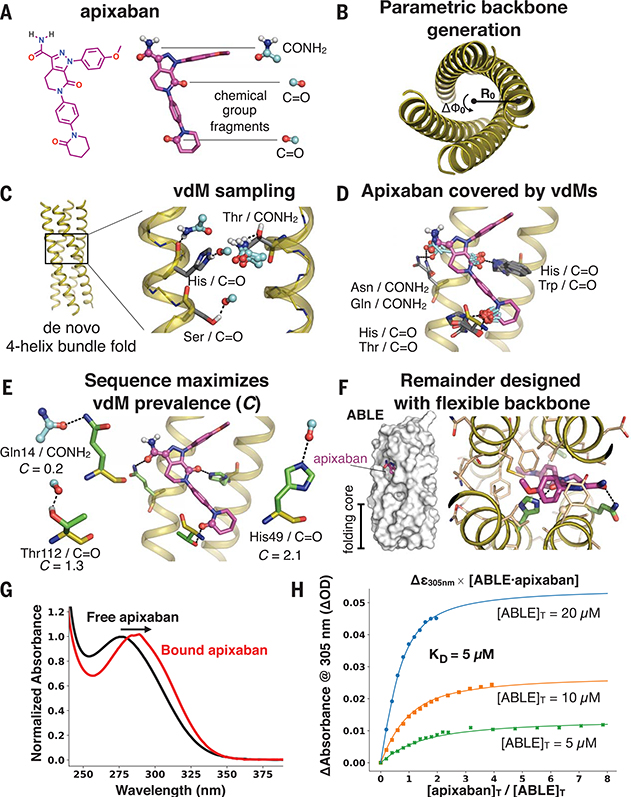
We targeted the factor Xa inhibitor apixaban, an organic compound with five rotatable bonds and eight heteroatoms. Our first objective was to compute a tertiary structure capable of cooperatively binding the polar groups of apixaban. Instead of repurposing natural binding proteins or folds that have been shown to bind a similar ligand, in this work, we use de novo four-helix bundles because they are mathematically parameterized (10, 11), designable (12), and share no similarity to the fold of factor Xa. Four-helix bundles generally do not bind small molecules and instead bind metal ions or metalloporphyrins by strong coordinate bonds (10, 13–16). However, four-helix bundles are tubular and can be designed to have high thermodynamic stability (11, 13) to compensate for the energetically demanding process of building binding cavities replete with buried polar functionality (17). Thus, the design of a de novo helical bundle that binds the drug apixaban critically tests the design method.
The van der Mer structural unit
The design of proteins relies on optimal packing of interior side chains in discrete conformations called rotamers (2, 3, 18–22). However, the design of ligand-binding proteins additionally requires side chains that interact favorably with the target small molecule. Previous design strategies approached this problem by computationally appending the target ligand to rotamers with idealized interaction geometries that—although composed of billions of conformations—sampled only a small fraction of the possible conformational space (6, 8, 23). These strategies rarely deliver submillimolar binders from the initial computational design, so subsequent steps rely on experimental random mutagenesis and screening of libraries.
We wondered how much of the vast, possible conformational space of protein–chemical group interactions is actually sampled in observed protein structures and if sampling interactions directly from this distribution might aid the design of high-affinity binders. Whereas previous analyses have focused on local side chain contacts with chemical groups (24), we sought a structural unit that directly maps backbone coordinates to chemical group locations, the link between the protein fold and binding function. We developed a unit of protein structure analogous to rotamers—a van der Mer (vdM)—that defines the placement of key chemical groups in the ligand relative to the backbone atoms of the contacting residue (Fig. 1B). vdMs are culled from a nonredundant set of protein structures by (i) identifying all residues of a certain type that interact with a particular chemical group, (ii) performing an all-by-all pairwise superposition of only the backbone and chemical group coordinates (side chains are not considered in the superposition, allowing some variation in their conformation), and (iii) geometric clustering with a tight root mean square deviation (RMSD) cutoff (0.5 Å). The resulting vdMs show backbone φ and ψ dependence (Fig. 1C) and capture compensatory effects of backbone and chemical group placement. Furthermore, single clusters may contain multiple rotamers (Figs. 1D and 6A and fig. S1), given that side-chain coordinates are not explicitly considered in clustering.
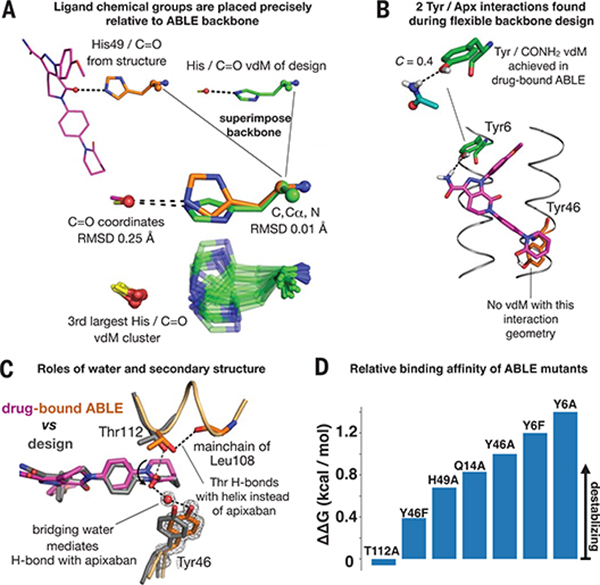
Fig. 6. Design inferences from the structure and function of ABLE.
(A) Exact sidechain positioning is not necessary for precise placement of ligand chemical groups relative to the mainchain. The placement of the C=O chemical group of apixaban relative to the backbone of residue 49 is exact (0.25 Å RMSD). The His49/C=O vdM from the design (green) (Fig. 3E) was superimposed onto His49 (orange) of the drug-bound ABLE structure through use of backbone atoms (N, Cα, C atoms, spheres). This backbone superposition places the C=O group of the original vdM precisely (0.25 Å RMSD) onto that of apixaban (purple) in the structure. The cluster describing the His C=O vdM, shown beneath, contains multiple rotamers of His that achieve the same placement of C=O relative to the position of the backbone. The rotamers of His49 in the structure and His from the original vdM are both observed in the cluster. (B) Flexible backbone sequence design (Fig. 3F) resulted in recruitment of two additional polar interactions with apixaban from Tyr6 and Tyr46. A Tyr6/CONH2 vdM is prevalent in the PDB, whereas the Tyr46/C=O interaction is not found in the database. (C) A water mediates an H-bond between Tyr46 and the C=O group of apixaban. Thr122 H-bonds the C=O of the helix backbone at residue 108. (D) Relative binding affinities of ABLE mutants with apixaban-PEG-FITC fluorophore by fluorescence anisotropy experiments (supplementary text and fig. S18).
The use of vdMs contrasts with procedures that place ligands at idealized locations relative to the terminal atoms of a side chain (6, 8, 23, 25), which results in vast numbers of ligand-rotamer combinations that might never occur in proteins. Instead, vdMs sample locations of chemical groups relative to the backbone that have been experimentally vetted to achieve favorable interactions. They also implicitly consider interactions with ordered or bulk water, which might influence their interaction geometries. Moreover, unlike ligand-appended and inverse rotamers used in earlier approaches (6, 8, 23, 25), vdMs may be derived from contacts with either main chain, side chain, or both in a multivalent interaction. Finally, the prevalence of a given vdM in the Protein Data Bank (PDB) can be used in scoring functions, similarly to scoring rotamers, which may assist automated selection of binding-site residues for design.
To maximize the number of observed protein–chemical group contacts, we created vdMs using the chemical groups of amino acids that constitute the protein (e.g., CONH2 of Gln and Asn and N-H and C=O of backbone amides). To avoid bias from local structure, we counted only the interactions that were distant in the linear polypeptide chain, as described in the supplementary materials. The set of chemical groups can also be expanded to include those from small-molecule drugs, metal ions, and cofactors, although these are not as pervasive in crystal structures.
We ranked vdMs by their prevalence in the PDB using a log-odds score, C (Fig. 1, D and E; fig. S2; and supplementary text). Although there are hundreds of vdMs associated with a given residue–chemical group combination (figs. S3 and S4), only a small fraction of vdMs are highly enriched in protein structures (C > 0). For example, only 91 Asp CONH2 vdMs have C values >0; these top vdMs map the locations of CONH2, relative to the backbone of an Asp residue, that are statistically preferred by proteins in the PDB (Fig. 1E and fig. S2D). This is on the order of the number of rotamers used for an amino acid during a typical protein design packing calculation (26). Thus, when combined with an efficient search algorithm, sampling protein–chemical group interactions with vdMs to design ligand-binding sites might be as expedient as sampling rotamers to pack a protein core. Furthermore, functionally relevant lower-probability rotamers may be included if contained in a high-scoring vdM.
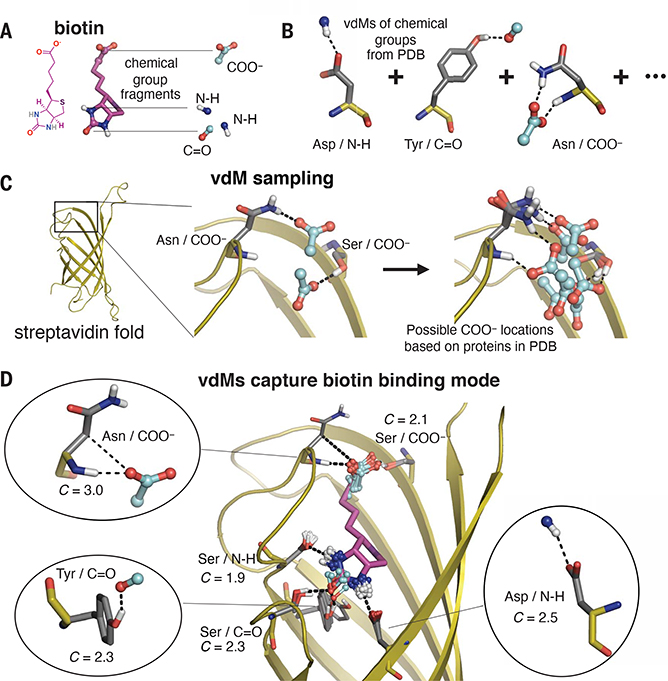
Proteins use the same set of 20 amino acids to fold as well as to recognize a vast array of highly functionalized ligands. We therefore hypothesized that the interaction modes used by amino acids to stabilize their tertiary structures would also be used to achieve tight binding of ligands, even those containing structurally distinct heterocyclic chemical groups. To test this hypothesis, we examined the streptavidin–biotin complex (Fig. 2). Using the natural sequence of streptavidin, we examined the positions of vdMs of N-H, C=O, and COO−, where these groups were derived from protein main chains and side chains. In each case, we observed that the side-chain interactions with biotin’s polar groups involved highly favorable vdMs, with enrichment scores of ~8-fold or greater (C > 2). The streptavidin sequence–fold pairing cooperatively positions highly favorable vdMs to cover each polar chemical group of biotin simultaneously.
Fig. 2. Prevalent vdMs describe the binding site of biotin in streptavidin.
(A and B) We constructed vdMs of the polar chemical groups of biotin by searching the PDB for protein interactions with the (i) backbone amide nitrogen (N-H), (ii) backbone carbonyl or carbonyl from Asn or Gln side chains (C=O), and (iii) carboxylate of Asp or Glu side chains (COO−). (C) Using the native sequence of streptavidin, vdMs were sampled on the streptavidin backbone to generate possible locations for productive interactions with the chemical groups. Here, Asn and Ser vdMs of COO− are sampled at two positions of the backbone. (D) vdMs with chemical groups (cyan) that are nearest neighbors (0.6 Å RMSD) to those of biotin in its binding site are overlaid on top of biotin (purple).
Our analysis of the streptavidin–biotin complex suggests that binding sites can be designed by considering folds that position vdMs to collectively bind the distinct chemical groups found in a target small-molecule ligand. Moreover, the vdMs of the binding site should be maximally prevalent in the PDB. We developed a search algorithm, called Convergent Motifs for Binding Sites (COMBS), to discover favorable poses of a ligand that satisfy these criteria.
De novo design strategy
Our design strategy consists of several hierarchic steps, which prioritize the most essential and difficult features to avoid sampling regions in sequence and structure space with little chance of success (fig. S5). First, we define the chemical groups within the small molecule that will be targeted. We initially focus on polar chemical groups, which are the most challenging to dehydrate but must be satisfied with H-bonds to achieve high affinity and specificity (27). Second, we choose a designable protein fold and create an ensemble of backbones with geometries that are consistent with the known plasticity of the fold. Next, for each backbone we use COMBS to identify members of the backbone ensemble that can position vdMs to collectively engage each of the targeted chemical groups of the small molecule. In this way, the binding of the desired ligand dictates the precise backbone geometry. Having discovered candidate backbones and binding sites, the design is completed by engineering a tightly packed folding core that supports the vdM-derived keystone interactions in the binding site (13). In this step, we constrain the keystone interactions and use flexible backbone design (13, 26) to pack additional residues within the binding site while simultaneously packing the protein core.
We focused on apixaban’s carboxamide (both the C=O and -NH2), as well as two additional carbonyls (Fig. 3A). (Other groups that were internally H-bonded or easily dehydrated were not initially targeted.) We created a set of vdMs of carboxamide (CONH2 from Asn and Gln side chains) and carbonyl [C=O from the protein backbone (supplementary text)] and used these vdMs to discover preferred CONH2 and C=O binding locations within a set of 32 mathematically generated de novo polyglycine backbones (10, 28) (Fig. 3, B and C; fig. S2; and table S1). For each of the mathematically generated backbones, we placed apixaban in the protein interior by using a separate set of vdMs with apixaban superimposed onto the chemical group of the vdM. For example, the CONH2 of apixaban can be superimposed on the CONH2 of a vdM, uniquely defining the position of apixaban in the binding site. Apixaban’s conformation in this step was fixed in a low-energy conformer found in its cocrystal structure with factor Xa (PDB code 2p16) (Fig. 3A and fig. S6; extension to multiple conformers is discussed in the supplementary text and illustrated in fig. S7). vdMs that cover the remaining C=O groups of the placed ligand, as well as additional vdMs to the carboxamide, were then queried in the nearby space (Fig. 3D). We chose binding poses by maximizing the PDB prevalence of sterically compatible vdMs (∑C) (Fig. 3E).
Fig. 3. Apixaban-binding helical bundle (ABLE) design strategy.
(A to F) Steps of the design process. (A) We targeted simultaneous engagement of two carbonyls (C=O) and the carboxamide (CONH2) of apixaban. (B) We computationally generated a set of 32 designable four-helix bundle folds based on a mathematical parameterization. (C) vdM sampling of CONH2 and C=O allowed us to enumerate statistically preferred locations of these chemical groups relative to the backbone. (D) We used a precomputed set of vdMs with apixaban superimposed by one of its chemical groups to position apixaban within the bundle, such that it was guaranteed to have at least one vdM that accommodates its position. Chemical groups of vdMs that overlap with those of apixaban are found by a nearest-neighbors lookup. Multiple vdMs contributing from one residue position are possible, e.g., His/C=O and Trp/C=O vdMs, and can be used in separate designs. (E) Specific choices of vdMs for each chemical group of the ligand were made by maximizing the use of highly enriched vdMs in the binding site (high C score) (Fig. 1, D and E). Final ligand positions and interactions for the six experimentally characterized designs were chosen by maximizing both C and the burial of the apolar surface area of apixaban. The vdMs chosen to comprise the binding site of ABLE are shown along with their cluster scores. (F) The location of apixaban and its vdM-derived interactions with the protein are constrained in a subsequent flexible backbone sequence design protocol. (G) The electronic absorbance spectrum of apixaban is red-shifted upon binding to ABLE. The black spectrum shows apixaban (4 μM) in buffer containing 50 mM NaPi, 100 mM NaCl (pH 7.4). The red spectrum is the difference of the absorbance spectrum of ABLE alone (20 μM) and the spectrum of ABLE (20 μM) with apixaban (4 μM). The spectra were normalized to the peak maximum for comparison. These experiments were facilitated by the high extinction coefficient of apixaban and the lack of Trp in ABLE. (H) Global fit of a single-site binding model to the absorbance changes at 305 nm upon titration of apixaban into 5, 10, and 20 μM solutions of ABLE. The KD from the fit is 5 (± 1) μM, which was confirmed by fluorescence polarization competition experiments (supplementary materials).
Side chains from vdMs in six selected binding poses were fixed, and their H-bonding interactions with apixaban were constrained in all subsequent steps of sequence design performed within the Rosetta modeling suite. After insertion of interhelical loops, we used a flexible backbone design protocol (13) (Fig. 3F) to compute the hydrophobic core while simultaneously completing the packing of the binding site. For some designs, new polar interactions were recruited during this step, as were Gly residues, which are known to interact favorably with aromatic groups (27). The use of small residues to make hydrophobic contacts minimizes the number of large, apolar side chains that might lead to nonspecific binding or hydrophobic collapse in the absence of ligand.
Description of designs and biophysical characterization
We designed six proteins of varying length, topology, ligand position, ligand burial, and keystone interactions (fig. S8). By contrast to factor Xa, which engages polar groups of apixaban through main-chain amides in loops (fig. S6), the designs interact with apixaban using predominantly side chains in helices. The six designs were well-expressed in bacteria, and each was helical based on far UV circular dichroism spectroscopy (fig. S9). Proton nuclear magnetic resonance (NMR) showed that two designs, ABLE (apixaban-binding helical bundle) and LABLE (longer ABLE), bound apixaban (fig. S10). These two designs had the same orientation of apixaban within the bundle and shared the same vdM-derived keystone interactions (Fig. 3E and fig. S8). For example, they shared a buried, high-scoring His/C=O vdM (8-fold enrichment, C = 2.1) (Fig. 3E). However, ABLE and LABLE differed in length (125 vs. 165 residues), topology, and loop geometry and shared only 22% sequence homology.
Binding of apixaban to ABLE restricts the drug’s conformation, resulting in a red shift of its electronic absorbance spectrum (Fig. 3G). Spectral titrations and fluorescence polarization competition experiments showed that ABLE and LABLE bind apixaban with a dissociation constant (KD) of 5 (± 1 [SEM]) μM and 0.6 (± 0.1) μM, respectively (Figs. 3H and 4D; figs. S11 and S12). Although LABLE showed a dispersed two-dimensional 1H-15N heteronuclear single-quantum coherence spectrum by NMR (fig. S13), indicative of a well-structured protein, it failed to crystallize in a sparse matrix screen, and so we focused our attention on characterization of ABLE. ABLE is monomeric in solution (fig. S14) and highly stable to heat denaturation (melting temperature of >95°C), despite the inclusion of three Gly and a polar His within its core (fig. S15).
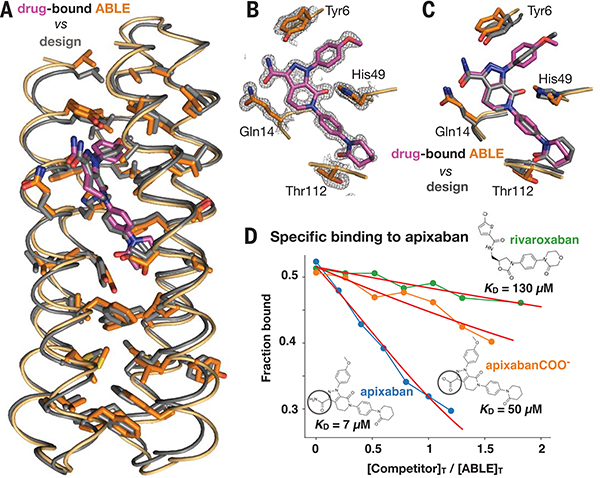
Fig. 4. The structure of apixaban-bound ABLE agrees with the design.
(A) Superposition of backbone Cα atoms of structure (protein in orange, apixaban in purple) and design (gray; 0.7 Å RMSD), showing side chains of amino acids in the protein core. (B) ABLE’s binding site from the structure (1.3-Å resolution), showing vdM-derived interactions with apixaban (purple). The 2mFo-DFc composite omit map is contoured at 1.5 σ. The map was generated from a model that omitted coordinates of apixaban. The protein backbone of these residues is shown in cartoon format. (C) Overlay of designed interactions (gray), after the designed model was superimposed onto the Cα atoms of the structure (protein in orange, apixaban in purple). (D) Fluorescence anisotropy competition experiments (485-nm excitation, 528-nm emission) showed that ABLE binds apixaban specifically. The bound fluorophore apixaban–polyethylene glycol–fluorescein isothiocyanate (apixaban-PEG-FITC) (supplementary text and fig. S9) is dislodged by addition of competing ligand. Anisotropy was converted to the fraction bound by use of a one-site binding model (supplementary text). The ABLE concentration was 20 μM, and the apixaban-PEG-FITC concentration was 25 nM in buffer containing 50 mM NaPi, 100 mM NaCl (pH 7.4). Apixaban COO− is identical to apixaban except that it contains a carboxylate instead of a carboxamide (circled). Rivaroxaban is another inhibitor that also binds tightly to factor Xa by using the same binding mode as apixaban but shows only very weak binding to ABLE. Fits to a competitive binding model are shown in red. KD values: rivaroxaban, 130 (± 10) μM; apixaban COO−, 50 (± 5) μM; apixaban, 7 (± 2) μM.
Structures of apixaban-bound and drug-free ABLE
ABLE readily crystalized with apixaban and diffracted to 1.3-Å resolution. Two very closely related monomers were observed in the asymmetric unit (fig. S16); apixaban is bound to both monomers, as expected for a specific, high-affinity complex. The structure of the drug-bound protein is in excellent agreement with the design (Cα RMSD of 0.7 Å) (Fig. 4). The rotamers of the core residues of ABLE, including the binding-site residues, overwhelmingly agree with the design model. Superimposing by all heavy atoms of core amino acids, including apixaban, gives an RMSD of 0.98 Å. ABLE buries almost all available apolar surface area (504 Å2) of apixaban, and it also forms most polar interactions included in the design (Fig. 4, B and C). Apixaban’s conformation is close to that used in the design (0.6 Å heavy atom RMSD), with small deviations that bring it closer to a quantum mechanically optimized geometry (fig. S17). The rigid body translation between apixaban’s center of mass in the designed versus that in observed structures is only 0.2 Å, with a rigid body rotation of 6°. The bespoke binding site is specific for apixaban, as shown by fluorescence polarization competition experiments (Fig. 4D), which indicated that ABLE binds apixaban 20-fold more tightly than a similar factor Xa inhibitor, rivaroxaban.
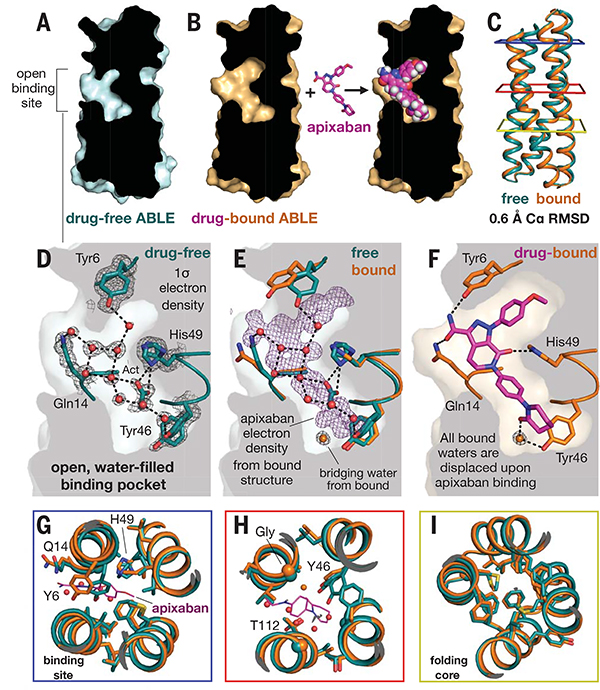
To assess the extent of preorganization of the protein, we also solved the drug-free structure to 1.3-Å resolution (Fig. 5). The structure shows an open, preorganized binding pocket, with an overall Cα RMSD of 0.65 Å to the apixaban–ABLE complex. The unoccupied binding site is solvated by nine ordered water molecules plus an acetate from the buffer (Fig. 5D). Binding of apixaban displaces ordered solvent from this site, suggesting a release of local frustration upon binding. The pocket has a 480-Å2 solvent-exposed surface area, which expands by 40% to accommodate the drug (680 Å2). The drug-free protein has nearly identical rotamers to that of the drug-bound protein throughout the core and binding site (Fig. 5, G to I). Unliganded ABLE shows two alternate rotamers for several of the residues that form H-bonds to apixaban (e.g., Tyr46 and His49); binding of apixaban selects one each of these alternate rotamers. Thus, like many natural proteins (29), ABLE has a limited degree of flexibility, which is reduced upon ligand binding, and the binding event appears to trade configurational entropy for enthalpically favorable interactions.
Fig. 5. Drug-free ABLE has a preorganized structure with an open binding site competent for binding.
(A) A slice through a surface representation of the 1.3-Å resolution structure of unliganded ABLE shows an open binding cavity. (B) Same slice, shown for the structure of apixaban-bound ABLE. (C) The Cα atom backbone superposition of unliganded and liganded ABLE. Colored squares surrounding the structure correspond to panels in (G), (H), and (I), looking down from the top. (D) The binding site of drug-free ABLE shows nine buried, crystallographic waters (red spheres, occupancy > 0.9) involved in an extensive H-bonded network with binding-site residues Tyr6, Gln14, Tyr46, and His49. The 2mFo-DFc electron density map of drug-free ABLE is contoured at 1 σ. An acetate (Act) group from the crystallization condition H-bonds with His49. His49 and Tyr46 are observed with alternate rotamers. (E) Same view as in (D) but with the addition of the corresponding residues from the apixaban-bound structure, after an all-Cα-atom backbone superposition. The 1-σ 2mFo-DFc electron density (purple) of apixaban from the drug-bound structure shows where the crystallographic waters bind in the ligand-free structure relative to the bound structure. A water (shown as an orange sphere) mediates the H-bond between Tyr46 and apixaban. This water is not observed in the unliganded structure. (F) Binding of apixaban in the drug-bound structure displaces all of the nine buried waters in the drug-free structure. Stick renderings, as well as the surface background, show the binding site of the ABLE-apixaban complex. (G and H) Binding-site overlay of liganded (orange, apixaban purple) and unliganded (cyan) ABLE shows preorganized rotamers. (I) The remote folding core contains identical rotamers in drug-free and drug-bound ABLE, predisposing the drug-free protein for binding.
Insights from the structure and function of ABLE
Two of the three keystone interactions identified by COMBS contribute appreciably to binding affinity. Substitution of His49 or Gln14 with alanine individually decreases affinity by approximately 1 kcal/mol (~3-fold) (Fig. 6D and fig. S18). Gln14 was observed in its intended rotamer, whereas His49 occupied an alternate rotamer that nevertheless maintained the intended position of apixaban’s carbonyl relative to the main chain (Fig. 6A). Indeed, the cluster describing this His/C=O vdM contains multiple His rotamers, each capable of achieving identical placements of C=O relative to the main chain. Thus, we observed vdM convergence, even amidst rotamer divergence.
We also examined the structural consequences of substituting His49 with Ala by solving the crystal structure of the unliganded His49→Ala (H49A) mutant protein (fig. S19). Although the structures of drug-free ABLE and drug-free H49A are similar (Cα RMSD = 1.2 Å), the residues that surround His49 show rotameric differences in the absence of this side chain; released from the restraints of tight packing, they instead adopt their preferred rotamers. The structure illustrates that global packing of core residues supports the positioning of a key functional group, even when this requires local frustration at individual sites.
Substitution of the third keystone residue, Thr112, with Ala resulted in little change in affinity (Fig. 6D). In the complex, its side chain did not form the intended H-bond to apixaban but instead formed an intrahelical H-bond to a backbone carbonyl (Fig. 6C). The intended Thr/C=O vdM is favored in the backbone-independent vdM library used in the design of ABLE, but it is disfavored in a backbone-dependent vdM library. The lack of engagement with apixaban’s carbonyl resulted in some disorder of the terminal oxopiperidine, which has higher b-factors and two alternate conformations (related by a 180° ring flip) in the structure (fig. S16). Thus, backbone-dependent vdM libraries should be used in future applications.
Flexible backbone sequence design of ABLE recruited two Tyr residues that interact with apixaban (Fig. 6B). One of these interactions was represented in the vdM database (Tyr6/CONH2, C = 0.4), but the other (Tyr46/C=O) was not. The structure of drug-bound ABLE confirmed the H-bond of Tyr6/CONH2 (Fig. 4C), but an unanticipated water enters the binding site to mediate an H-bond between apixaban and Tyr46 (Fig. 6C). Furthermore, substitution of Tyr6 with Phe or Ala was more destabilizing than the same substitutions for Tyr46, tracking with prevalence in the PDB. Thus, vdMs can be used to filter and rank interactions obtained using a variety of computational methods (30).
Finally, we wondered if ab initio folding predictions (26) might distinguish between successful versus unsuccessful designs. Of the six designs, only two—ABLE and LABLE—were predicted by folding simulations to maintain uncollapsed binding sites (fig. S20). Moreover, the lowest-energy models predicted from ab initio folding simulations of ABLE’s sequence largely agreed with the crystallographic structure (fig. S20A). Thus, ab initio folding may be useful as a screen to ensure that designs maintain an open, preorganized site. These results emphasize the degree to which the folding and binding problems are intimately coupled.
Conclusion
Previously, the design of de novo proteins that bind in a shape-selective manner to rigid, flat, hydrophobic dyes or lipidic metabolites had been possible, but binding flexible molecules replete with polar atoms has been more challenging (4, 8, 31–33). Natural proteins bind highly functionalized ligands by first accruing the ability to weakly bind fragments within the context of a particular fold (34–36). To mimic this process, we developed the vdM structural unit to directly link the protein fold to statistically preferred binding modes of chemical groups. We sampled vdMs on the backbone of a designable four-helix bundle to create constellations of chemical groups that, when matched with the shape of apixaban, defined the binding site. This contrasts with previous approaches that search for positional matching of whole ligands, sampled using idealized interaction geometries. Such approaches are highly sensitive to small changes in the interaction geometries and thus require an enormous amount of sampling to discover possible binding solutions, many of which may contain interactions not observed in the PDB.
vdMs sample from the experimentally vetted distribution of observed protein structures. vdMs are surprisingly sparse and discrete (Fig. 1E and figs. S3 and S4), and they enable facile sampling of sequence space to discover convergent combinations of keystone interactions (supplementary text and fig. S2). We consider only the backbone and the orientation of the pendant chemical group, which obviates the need to enumerate a large ensemble of ligand-appended rotamers for each amino acid type at each position of the sequence. We focused here on simple, fully de novo scaffolds rather than redesigning the specificity of natural ligand-binding proteins, because we wished to address the challenge of designing function entirely from scratch. Indeed, ABLE shares no sequence homology to any known proteins (BLAST E value of <0.42 against the nonredundant protein sequence database nr). We used only prevalence to rank vdMs and choose binding sites, but we suspect the true power of vdMs may lie in higher-order correlations of the interactions.
COMBS and vdMs can now be used for a variety of protein engineering applications and in full partnership with experimental optimization strategies for exploring sequence space. We anticipate that vdMs can also be used to predict chemical group hot spots of proteins with fixed sequence. vdMs may also enable design of protein-protein interfaces in a self-consistent manner. Finally, because vdMs sample from the distribution of evolved interaction geometries observed in protein structures, it is tempting to view the chemical group constellations constructed by vdMs as a structural hypothesis of the evolutionary path to acquire binding within the context of a given fold.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Jo for synthesis of the apixaban-FITC conjugate used in fluorescence polarization experiments, and we thank Y. Wu for performing NMR experiments. We are also grateful to E. Weiss for suggesting we target the drug apixaban.
Funding: N.F.P. and W.F.D. acknowledge research support from grants from NIH (R35 GM122603), NSF (1709506), and the U.S. Air Force Office of Scientific Research (FA9550-19-1-0331). N.F.P. acknowledges support from NIH (4 T32 HL 7731-25 and K99GM135519). ABLE structures were solved using the NE-CAT 24-ID-E beamline (GM124165) and an Eiger detector (OD021527) at the APS (DE-AC02-06CH11357). The structure of the H49A mutant was solved at the 8.3.1 beamline (R01 GM124149 and P30 GM124169) of the Advanced Light Source (DE-AC02-05CH11231).
Footnotes
Competing interests: N.F.P. and W.F.D. are inventors on a provisional patent application submitted by the University of California, San Francisco, for the design, composition, and function of the proteins in this study.
Data and materials availability: Computational code and design scripts are available in the supplementary materials and at Zenodo (37). Coordinates and data files of ABLE structures have been deposited to the PDB with accession codes 6W6X (drug-free ABLE), 6W70 (apixaban-bound ABLE), 6X8N (H49A ABLE mutant). Materials are available from the authors on request. The plasmid of ABLE is available from Addgene (no. 158627).
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/369/6508/1227/suppl/DC1
View/request a protocol for this paper from Bio-protocol.
REFERENCES AND NOTES
- 1.Anfinsen CB, Science 181, 223–230 (1973). [DOI] [PubMed] [Google Scholar]
- 2.Korendovych IV, DeGrado WF, Q. Rev. Biophys 53, e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhlman B, Bradley P, Nat. Rev. Mol. Cell Biol 20, 681–697 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dou J et al. , Protein Sci. 26, 2426–2437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcos E et al. , Science 355, 201–206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tinberg CE et al. , Nature 501, 212–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day AL et al. , Protein Eng. Des. Sel 31, 375–387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou J et al. , Nature 561, 485–491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barros EP et al. , J. Chem. Theory Comput 15, 5703–5715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoryan G, DeGrado WF, J. Mol. Biol 405, 1079–1100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang P-S et al. ,Science 346, 481–485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szczepaniak K, Lach G, Bujnicki JM, Dunin-Horkawicz S, J. Struct. Biol 188, 123–133 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Polizzi NF et al. , Nat. Chem 9, 1157–1164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhys GG et al. , J. Am. Chem. Soc 141, 8787–8797 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Reig AJ et al. , Nat. Chem 4, 900–906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupas AN, Bassler J, Dunin-Horkawicz S, in Fibrous Proteins: Structures and Mechanisms, Parry DAD, Squire JM, Eds. (Springer, Cham, 2017), pp. 95–129. [Google Scholar]
- 17.Lombardi A, Pirro F, Maglio O, Chino M, DeGrado WF, Acc. Chem. Res 52, 1148–1159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desjarlais JR, Handel TM, Protein Sci. 4, 2006–2018 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janin J, Wodak S, Levitt M, Maigret B, J. Mol. Biol 125, 357–386 (1978). [DOI] [PubMed] [Google Scholar]
- 20.McGregor MJ, Islam SA, Sternberg MJE, J. Mol. Biol 198, 295–310 (1987). [DOI] [PubMed] [Google Scholar]
- 21.Ponder JW, Richards FM, J. Mol. Biol 193, 775–791 (1987). [DOI] [PubMed] [Google Scholar]
- 22.Dahiyat BI, Mayo SL, Protein Sci. 5, 895–903 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lassila JK, Privett HK, Allen BD, Mayo SL, Proc. Natl. Acad. Sci. U.S.A 103, 16710–16715 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh J, Thornton JM, Atlas of Protein Side-Chain Interactions (Oxford Univ. Press, 1992). [Google Scholar]
- 25.Zanghellini A et al. , Protein Sci. 15, 2785–2794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J, Biochemistry 49, 2987–2998 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira de Freitas R, Schapira M, MedChemComm 8, 1970–1981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.North B, Summa CM, Ghirlanda G, DeGrado WF, J. Mol. Biol 311, 1081–1090 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Williams DH, Stephens E, O’Brien DP, Zhou M, Angew. Chem. Int. Ed 43, 6596–6616 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Tan SK et al. , Biochemistry 58, 3251–3259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas F et al. , ACS Synth. Biol 7, 1808–1816 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Park J et al. , eLife 8, e47839 (2019).31854299 [Google Scholar]
- 33.Glasgow AA et al. , Science 366, 1024–1028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tokuriki N, Tawfik DS, Science 324, 203–207 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Stout TJ, Sage CR, Stroud RM, Structure 6, 839–848 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Keedy DA et al. , eLife 7, e36307 (2018).29877794 [Google Scholar]
- 37.Polizzi N, npolizzi/combs_pub: Combs, Version v0.0.1, Zenodo; 10.5281/zenodo.3910780. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.