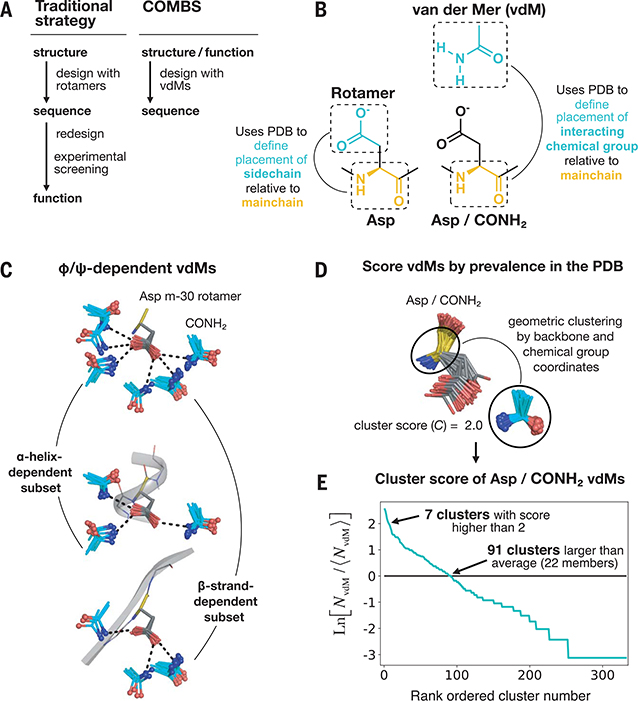
Fig. 1. A vdM is a structural unit relating chemical group position to the protein backbone.
(A) Workflow of a traditional protein design strategy versus that of COMBS. (B) Definition of a vdM. A chemical group is interacting if it is in van der Waals contact with the protein side chain or main chain. Like rotamers, vdMs are derived from a large set of high-quality protein crystal structures. A vdM of aspartic acid (Asp) and carboxamide (CONH2, cyan) is shown. (C) vdMs are φ, ψ, and rotamer dependent; this is illustrated by the top vdMs of the m-30 rotamer of Asp, clustered by location of CONH2 after exact superposition of main chain N, Cα, and C atoms. (D and E) We ranked vdMs by prevalence in the PDB, quantified by a cluster score C [the natural logarithm of the ratio of the number of members in a cluster (NvdM) to the average number of members in a cluster (⟨hNvdM⟩)]. The seventh-largest cluster of Asp/CONH2 vdMs is shown as an example in (D).