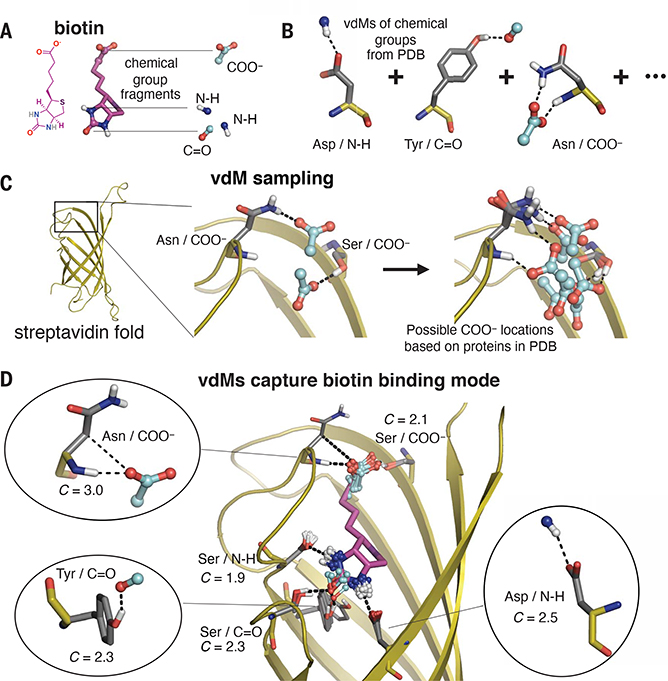
Fig. 2. Prevalent vdMs describe the binding site of biotin in streptavidin.
(A and B) We constructed vdMs of the polar chemical groups of biotin by searching the PDB for protein interactions with the (i) backbone amide nitrogen (N-H), (ii) backbone carbonyl or carbonyl from Asn or Gln side chains (C=O), and (iii) carboxylate of Asp or Glu side chains (COO−). (C) Using the native sequence of streptavidin, vdMs were sampled on the streptavidin backbone to generate possible locations for productive interactions with the chemical groups. Here, Asn and Ser vdMs of COO− are sampled at two positions of the backbone. (D) vdMs with chemical groups (cyan) that are nearest neighbors (0.6 Å RMSD) to those of biotin in its binding site are overlaid on top of biotin (purple).