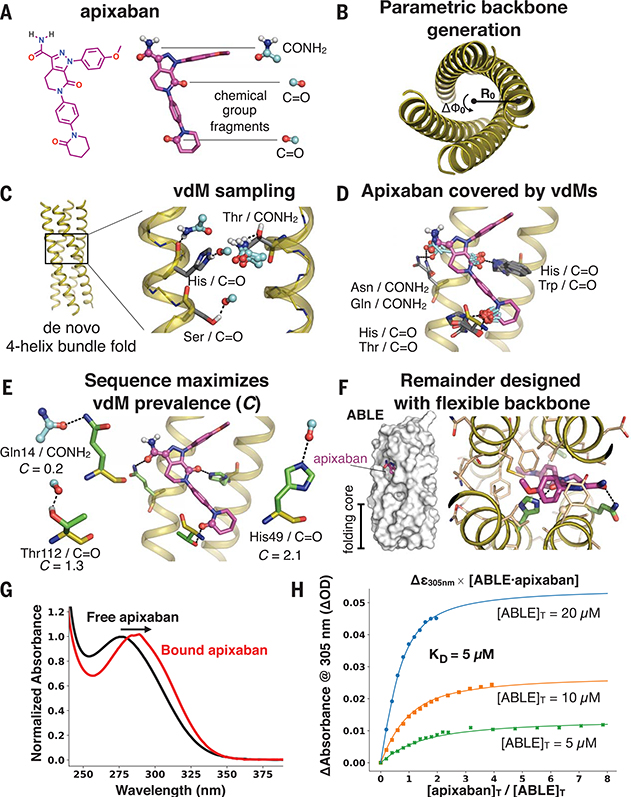
Fig. 3. Apixaban-binding helical bundle (ABLE) design strategy.
(A to F) Steps of the design process. (A) We targeted simultaneous engagement of two carbonyls (C=O) and the carboxamide (CONH2) of apixaban. (B) We computationally generated a set of 32 designable four-helix bundle folds based on a mathematical parameterization. (C) vdM sampling of CONH2 and C=O allowed us to enumerate statistically preferred locations of these chemical groups relative to the backbone. (D) We used a precomputed set of vdMs with apixaban superimposed by one of its chemical groups to position apixaban within the bundle, such that it was guaranteed to have at least one vdM that accommodates its position. Chemical groups of vdMs that overlap with those of apixaban are found by a nearest-neighbors lookup. Multiple vdMs contributing from one residue position are possible, e.g., His/C=O and Trp/C=O vdMs, and can be used in separate designs. (E) Specific choices of vdMs for each chemical group of the ligand were made by maximizing the use of highly enriched vdMs in the binding site (high C score) (Fig. 1, D and E). Final ligand positions and interactions for the six experimentally characterized designs were chosen by maximizing both C and the burial of the apolar surface area of apixaban. The vdMs chosen to comprise the binding site of ABLE are shown along with their cluster scores. (F) The location of apixaban and its vdM-derived interactions with the protein are constrained in a subsequent flexible backbone sequence design protocol. (G) The electronic absorbance spectrum of apixaban is red-shifted upon binding to ABLE. The black spectrum shows apixaban (4 μM) in buffer containing 50 mM NaPi, 100 mM NaCl (pH 7.4). The red spectrum is the difference of the absorbance spectrum of ABLE alone (20 μM) and the spectrum of ABLE (20 μM) with apixaban (4 μM). The spectra were normalized to the peak maximum for comparison. These experiments were facilitated by the high extinction coefficient of apixaban and the lack of Trp in ABLE. (H) Global fit of a single-site binding model to the absorbance changes at 305 nm upon titration of apixaban into 5, 10, and 20 μM solutions of ABLE. The KD from the fit is 5 (± 1) μM, which was confirmed by fluorescence polarization competition experiments (supplementary materials).