Summary
Epigenomic approaches in cells affected in CVDs can be exploited to understand the function of genetic polymorphisms at cis-regulatory elements and crosstalk between enhancers and lncRNAs associated with disease susceptibility and progression. The reversible nature of epigenetics provides opportunities for the development of novel therapeutic strategies for CVD.
Keywords: Epigenetics, enhancer, long non-coding RNAs, circular RNA, cardiovascular disease
Introduction
Cardiovascular disease (CVD) is a chronic inflammatory disease and accounts for over 50% of deaths worldwide. Major risk factors such as obesity, diabetes, hyperlipidemia and hypertension promote production of inflammatory mediators which lead to monocyte-endothelial cell (EC) adhesion, macrophage foam cell formation and vascular smooth muscle cell (VSMC) proliferation and migration in the blood vessel [1]. These events promote atherosclerosis and plaque development to contribute to CVD. Despite significant progress in understanding the signaling and transcriptional mechanisms and determinants of disease susceptibility and progression of CVD, present therapies show limited impact. Although there is strong evidence that genetics plays a major role in CVDs [2], the majority of genetic loci highly associated with CVD are in non-coding regions of the genome, implying novel regulatory mechanisms remain to be characterized. Epigenetic changes are able to regulate gene expression at proximal and distal regulatory regions without altering the underlying DNA sequence. Accordingly, there is increased attention on epigenetic mechanisms during development and in chronic disease states like CVDs. Additionally, because, unlike genetics, epigenetics can be reversible, it may have therapeutic implications. This review covers the latest insights into the epigenetic mechanisms that impact CVDs.
Epigenetic Processes and CVD
Epigenetic processes including DNA methylation (DNAme), histone post-translational modifications (PTMs) and long non-coding RNAs (lncRNAs) modulate chromatin structure and regulate the function of cis-regulatory elements such as promoters, enhancers and insulators to alter gene activity [3–5]. High throughput epigenomic approaches revealed the presence of specific histone modifications at these cis-regulatory elements. Active promoters show increased levels of histone H3 lysine 4 tri-methylation (H3K4me3) and H3/H4K acetylation (H3/H4Kac) and inactive promoters with H3K9me3/ H3K27me3, as well as cytosine DNAme which is generally observed at CpG islands [6]. Enhancers are enriched with binding sites for multiple transcription factors (TFs), H3K27ac, and coactivator p300. Clusters of enhancers, called super enhancers (SEs) are enriched with the coactivator bromodomain-containing protein 4 (BRD4), mediator proteins and TFs [7, 8]. Enhancers/SEs can be located nearby or several 100 kb away from target promoters and regulate tissue and lineage-specific genes. Recent data suggest these epigenetic processes are fine-tuned by lncRNAs (defined as >200 nucleotide long transcripts without protein coding potential) via direct interaction with cis-regulatory elements, or through interactions with key RNA binding proteins and chromatin modifiers [3, 9]. They can interact with enhancers and regulate gene expression via cis or in trans mechanisms. Several CVD susceptibility loci have mapped to enhancers/SE and lncRNAs [10, 11] implying that epigenetic mechanisms involving enhancers and lncRNAs impact the development and progression of CVD (Fig. 1). A summary of lncRNAs and their phenotypes is provided in Table 1.
Fig.1. Role of transcriptional regulatory elements (enhancers) and long non-coding RNAs (lncRNAs) in cardiovascular disease.
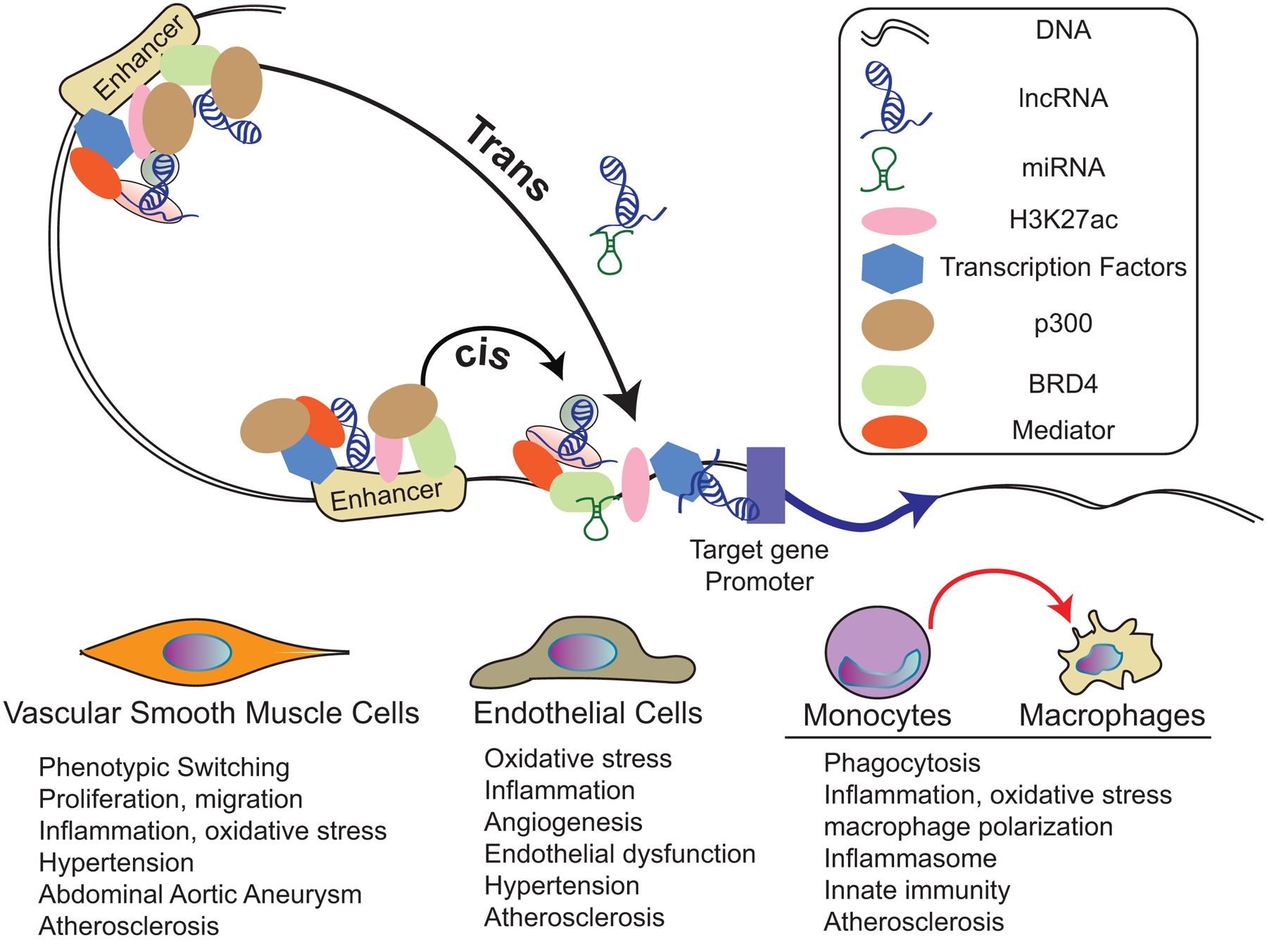
LncRNAs and enhancers regulate target gene expression alone or in combination. Enhancers are characterized by the presence of H3K27ac, transcription factors, and coactivators such as mediator, p300 and BRD4. LncRNAs and enhancers control nearby (cis) or distal (trans) gene expression. LncRNAs can also function as competing endogenous RNAs for some miRNAs. Together, these regulatory elements play important roles in epigenetic mechanisms controlling the expression of genes mediating the functions of vascular smooth muscle cells, endothelial cells, and monocytes/macrophages. Dysregulation of epigenetic mechanisms and related genes in these cells can lead to the pathological effects (schematically indicated under each cell-type) that can lead to cardiovascular diseases like hypertension and atherosclerosis.
Table 1:
LncRNAs and their biological effects in the vascular system.
| LncRNA | Cell Type | Phenotype | Putative human Ortholog | Ref. # |
|---|---|---|---|---|
| Giver | VSMC | Inflammation, oxidative stress, proliferation and hypertension | Yes | [15] |
| Lnc–Ang362 | VSMC | Proliferation | Unknown | [18] |
| H19 | VSMC | Abdominal aortic aneurysm | Yes | [19] |
| NEAT1 | VSMC | VSMC phenotypic switching | Yes | [16] |
| SMILR | VSMC | VSMC proliferation and cell cycle progression | Yes | [17] |
| SENCR | HUVEC | Stabilization of EC adherens junctions | Yes | [27] |
| LEENE | HUVEC | eNOS regulation and endothelial homeostasis | Yes | [28] |
| GATA6–AS | HUVEC | Regulates EC gene expression | Yes | [30] |
| ANRIL | HUVEC | EC phenotype | Yes | [31] |
| AF131217.1 | HUVEC | Inflammation | Yes | [32] |
| Dnm3os | Monocytes/macrophages | Inflammatory gene expression and phagocytosis | Yes | [36] |
| E330013P06 | Monocytes/macrophages | Modified LDL uptake and inflammatory gene expression | Yes | [37] |
| NTT | Monocytes | Monocyte and macrophage differentiation | Yes | [38] |
| MALAT1 | Macrophages | Regulates inflammation and promotes atherosclerosis | Yes | [39] |
| NEAT1 | Macrophages | Inflammasome activation | Yes | [40] |
| MeXis | Macrophages | Cholesterol efflux and atherosclerosis | Yes | [41] |
| CHROME | Hepatocytes/macrophages | Cholesterol homeostasis | Yes | [42] |
VSMC = vascular smooth muscle cells; HUVEC = human umbilical vein endothelial cells; EC = endothelial cells. This list represents a selection from recent studies, but does not cover all published studies.
Epigenetic mechanisms involved in VSMC dysfunction
Growth factors, cytokines and diabetic stimuli induce phenotypic switching (contractile to synthetic state) of VSMC to promote inflammatory gene expression, proliferation and migration, all key events in the pathogenesis of atherosclerosis. Phenotypic switching is tightly regulated by TFs such as serum response factor (SRF) via binding with CARG boxes at the contractile gene promoters. This process is fine-tuned by permissive histone modifications (H3/H4Kac and H3K4me2) which increase chromatin access to SRF in the contractile state. But, under pathological conditions, inhibition of such epigenetic mechanisms represses contractile genes [12]. H3K9/14ac and repressive H3K9me3 also regulate TNF-α and Angiotensin II (AngII)-induced inflammatory genes in VSMC and these epigenetic mechanisms are further modified by hyperglycemia and oxidized lipids to augment inflammation [13]. Recently, chromatin-immunoprecipitation coupled to next generation sequencing (ChIP-seq) with H3K27ac and BRD4 antibodies showed that AngII treatment reshapes enhancer and SE repertoires in rat VSMC [10]. These enhancers/SEs were enriched with binding sites for TFs regulated by AngII type 1 receptor, like AP1. Their deletion using CRISPR-Cas9 editing altered the expression of genes involved in growth factor signaling and atherosclerosis. Furthermore, treatment with the BRD4 inhibitor JQ1 blocked AngII-induced gene expression in VSMC, and ameliorated hypertension, medial hypertrophy and inflammation in AngII-infused mice. These enhancers/SE also harbor SNPs associated with cardiovascular traits/disease [10]. BET bromodomain inhibition with JQ1 also conferred protection against heart failure [14], suggesting these inhibitors of enhancer regulatory events could be promising therapies for various cardiac and CVDs. Further studies are needed to understand the function of such cis-regulatory elements and related novel epigenetic processes mediating VSMC dysfunction in CVD.
Role of lncRNAs in VSMC dysfunction
Increasing evidence shows that lncRNAs promote oxidative stress, proliferation, inflammatory genes and phenotypic switching of VSMC [15–17]. AngII-induced lncRNAs were identified in VSMC using RNA-seq and ChIP-seq with H3K4me3 [18]. One of these novel lncRNAs, GIVER is regulated by AngII-induced Nuclear Receptor Subfamily4 GroupA Member3 (NR4A3) in rat and human VSMC [15*]. Giver increases inflammation, oxidative stress, and proliferation in rat VSMC via interaction with Non-POU domain-containing octamer-binding protein and epigenetic mechanisms. Moreover, GIVER and NR4A3 are upregulated in hypertensive patients and these changes are attenuated in patients taking anti-hypertensive drugs [15], thus suggesting a potential role for GIVER in hypertension. Interestingly, another AngII-induced lncRNA H19 mediates progression of abdominal aortic aneurysm (AAA) in AngII-infused ApoE−/− mice and in a pig model of AAA. In VSMC, H19 upregulates HIF-1α through SP1 TF in the nucleus. Conversely, in the cytoplasm its association with HIF-1α promotes p53 stability and increases VSMC apoptosis [19]. The nuclear lncRNA NEAT1 regulates VSMC phenotypic switching in vitro and in vivo models of vascular injury [16]. Mitogens and vascular injury upregulate NEAT1, which sequesters WDR5 from the promoters of contractile genes and promotes chromatin compaction, possibly via epigenetic mechanisms, leading to their downregulation and increased proliferation and migration [16**]. Another human VSMC-specific lncRNA, SMILR, is induced by IL-1β and PDGF. SMILR promotes VSMC proliferation and regulates cell cycle progression genes via interaction with CENPF and Staufen1 [17**]. SMILR and its target genes are upregulated in unstable atherosclerotic plaques from carotid endarterectomy and in a pathological ex vivo vein graft model. Moreover, siRNA-mediated SMILR knockdown reduced VSMC proliferation in the ex vivo vein graft model [17]. Crosstalk between epigenetic modulators such as ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) and non-coding miR-145 have also been demonstrated in growth factor-induced VSMC dedifferentiation implicated in restenosis. These studies revealed that inhibition of miR-145 upregulates UHRF1, which regulates DNAme and repressive histone modification H3K27me3 at key VSMC differentiation genes [20], suggesting regulation of chromatin modifiers by miRNAs provides another layer of complexity to VSMC dysfunction. Together, it is clear that noncoding RNAs like lncRNAs can influence VSMC functions related to CVDs acting via RNA binding proteins and other epigenetic mechanisms.
Epigenetic mechanisms involved in EC dysfunction
EC dysfunction induced by shear stress, hyperlipidemia or metabolic insults promotes monocyte adhesion and inflammation. Changes in epigenetic histone modifications that mediate gene activation and repression have been reported in regulating EC-specific genes that promote oxidative stress and inflammation. Nitric oxide synthase 3 (eNOS) is specifically expressed in ECs and protects the endothelium by producing anti-inflammatory and vasodilatory nitric oxide. This EC-specific expression of eNOS was found to be associated with increased permissive histone modifications promoting open chromatin (accessibility) at the gene promoter. In contrast, chromatin at eNOS locus is compacted in cells other than ECs, thus limiting eNOS expression [21]. Genomewide approaches revealed the role of enhancers/SE in the maintenance of endothelial lineage and key EC functions in blood vessels. In human umbilical vein ECs (HUVEC), ETS related TF ERG maintained endothelial lineage genes by altering the enhancer/SE landscape [22**]. Interestingly, these SEs harbor CVD-related SNPs in regions occupied by ERG, supporting its importance in development and disease [22]. Notch1 signaling activated by Bone morphogenetic protein receptor type 2 (BMPR2) regulates epigenetic mechanisms that maintain EC integrity and homeostasis [23, 24]. After vascular injury, EC-VSMC contact activates BMPR2 and Notch1 signaling and upregulates phosphofructokinase PFKFB3 (fructose-2,6-bisphosphatase 3). PFKFB3 promotes glycolysis and increases acetyl-CoA, which is utilized by p300 to enrich H3K27ac at SEs and upregulate Notch1 target genes involved in EC regeneration. These data demonstrate an interesting link between metabolism and epigenetic mechanisms in vascular homeostasis [23**]. Disruption of such epigenetic mechanisms could lead to CVDs like hypertension. Together, these reports illustrate the importance of histone PTMs at distal transcriptional regulatory elements (enhancers) to maintain vascular tone in the arterial wall. Recently, high-resolution promoter capture Hi-C analysis of human induced pluripotent stem cells (iPSCs)-derived cardiomyocytes revealed about 2000 CVD-associated SNPs were linked to 347 genes and the majority of SNPs interacted with distal genes. This highlights the importance of long-range enhancer-promoter interactions in regulating genes associated with CVDs [25*]. The remarkable advances in such combinatorial chromatin conformation and structure technologies can be exploited to enhance our understanding of epigenomic mechanisms regulating the precise function of cis-regulatory elements and lncRNA loci driving the expression of genes associated with CVD [26].
Role of lncRNAs in EC dysfunction
Several studies described the role of lncRNAs induced by laminar shear stress and metabolic insults in EC function and CVD [27–29]. For instance, SENCR, identified as a shear-induced lncRNA, was upregulated in shear stressed aortas of SENCR-expressing humanized mice [27]. Mechanistically, SENCR promotes EC adherens junction formation by interaction with cytoskeletal-associated protein 4 [27]. LEENE is a flow-responsive enhancer associated lncRNA and regulates eNOS via increasing RNA Pol II recruitment [28]. Hypoxia-induced lncRNA GATA6-AS was reported as a negative modulator of epigenetic regulator LOXL2 [30]. GATA6-AS regulates endothelial gene expression by modulation of histone methylation [30]. More recently, lncRNA ANRIL was reported to control genes that promote an EC phenotype associated with CVD [31]. In addition, the laminar shear stress-induced lncRNA AF131217.1 suppresses inflammation by acting as a competing endogenous RNA (ceRNA) for miR-128–3p and increases expression of atheroprotective KLF4 [32]. Several other lncRNAs (Braveheart, Fendrr, Carmen,Miat, Alien, H19) showed specific functions in the cardiac atrium and ventricle, suggesting temporal and tissue-dependent roles during cardiac development [33]. Thus, lncRNAs have distinct roles in the ECs and can be potential therapeutic targets to ameliorate EC dysfunction in CVDs.
Epigenetic mechanisms in monocytes/macrophages
Apart from VSMCs and ECs, monocytes and macrophages also play important roles in CVD by promoting inflammation, lipid uptake and apoptosis. Monocyte to macrophage differentiation, lipid uptake, and macrophage polarization into pro-inflammatory activated phenotypes are critical in the development of CVDs. The role of epigenetic mechanisms in regulating such processes has been reviewed recently [34, 35].
Role of lncRNAs in monocytes and macrophages
Epigenetic factors, including lncRNAs, in monocytes and macrophages have been implicated in the development of inflammatory phenotypes [34]. Numerous lncRNAs are upregulated in macrophages from diabetic mice such as E330013P06 and Dnm3os [36*, 37]. Importantly, these lncRNAs are conserved in humans and upregulated in monocytes from individuals with type 2 diabetes [36, 37]. E330013P06 and Dnm3os regulate phagocytosis in macrophages and induce an inflammatory phenotype [36, 37]. Mechanistically, Dnm3os, which is associated with chromatin, interacts with the nuclear protein nucleolin. Under normal glucose homeostasis conditions, nucleolin inhibits Dnm3os actions at inflammatory gene promoters. However, in diabetes, upregulation of Dnm3os and downregulation of nucleolin disrupts this interaction, allowing Dnm3os to increase promoter H3K9ac and inflammatory gene expression [36]. Dnm3os is also increased in macrophages from diabetic Apoe−/− mice, hinting at a possible role in diabetic vascular disease [36]. A monocyte-specific lncRNA, NTT, is involved in monocyte and macrophage differentiation and in the pathogenesis of rheumatoid arthritis. NTT is upregulated in peripheral blood mononuclear cells from individuals with rheumatoid arthritis and regulates the adjacent gene PBOV1 by interacting with HnRNP-U [38]. LncRNAs also interact with miRNAs to regulate key immune functions. Loss of lncRNA MALAT1 is associated with atherosclerotic lesions in high-fat fed (Apoe−/− Malat1−/−) mice and in human atherosclerotic plaques [39**]. MALAT1 promotes an anti-inflammatory phenotype through interaction with miR-503 and decreased MALAT1 is associated with advanced plaques and worse prognosis in people [39]. The function of lncRNAs as miRNA sponges is largely limited to cytoplasmic lncRNAs. It is thus unclear how nuclear MALAT1 regulates miR-503 function. More recently, NEAT1 was shown to be a key regulator of inflammasomes in mouse macrophages through association with NLRP3, NLRC4 and AIM2 [40], further supporting the role of lncRNAs in innate immunity. Furthermore, lncRNA MeXis was shown to regulate expression of Abca1 and cholesterol efflux in macrophages [41**]. Recently a primate-specific lncRNA CHROME was also shown to regulate cholesterol efflux and HDL biogenesis by interfering with actions of key miRNAs and altering expression of ABCA1 in hepatocytes and macrophages [42**]. These studies further demonstrate the importance of lncRNAs in cholesterol metabolism and atherosclerosis.
Epigenetic crosstalk between lncRNAs, miRNAs and enhancers
Enhancers, lncRNAs and miRNAs, alone or in combination, play central roles in gene regulation via epigenetic crosstalk [26, 43]. Recently, several lncRNA and nearby protein coding gene (mRNA) pairs that are coordinately regulated were identified in IL-1β treated ECs. Interestingly, these pairs were divergently transcribed by shared epigenetic mechanisms and were located within the same transcriptional activation domain (TAD). One of them, lncRNA-CCL2, regulated nearby gene CCL2 via interaction with RNA-binding proteins. Inhibitors of BRD4, NF-κB and p300 blocked regulation of CCL2, demonstrating interaction between lncRNAs, enhancers and epigenetic mechanisms. Furthermore, lncRNA-CCL2 and CCL2 were upregulated in atherosclerosis, demonstrating functional significance of lncRNA-mRNA networks in CVDs [44*]. Another lncRNA, lncEGF7OS is expressed from the opposite strand of the EGFL7/miR-126 in human ECs and is regulated by TF ETS via a bidirectional promoter. LncEGF7OS regulates angiogenesis via interaction with TF Max and regulates H3K27ac at the EGFL/miR-126 promoter and enhancer. CRISPR-Cas9-mediated deletion of this locus suppressed angiogenesis, suggesting the possible therapeutic potential of lncEGFL7OS [45**]. In VSMCs, lnc-Ang383, and an overlapping enhancer, control expression of the proximate gene Ramp3, which promotes VSMC dysfunction [10]. Furthermore, treatment with SE inhibitor JQ1 or CRISPR-mediated deletion of lncRNA associated enhancer abrogates expression of Ramp3 and distal inflammatory genes in VSMCs. These studies suggest a close relationship between enhancers and noncoding RNAs, but further studies are required to understand the subtle crosstalk between them in gene regulation. [26]. CRISPR-Cas9 genome editing will be useful to systematically understand the intercommunication between these regulators in the vascular system [46].
Circular RNA (circRNA) and circulating lncRNA in CVD
Another class of noncoding RNAs, circRNAs, are recently implicated in CVD [47]. Bioinformatics approaches identified several circRNAs in human and murine VSMCs. These circRNAs specifically bind to different miRNAs in VSMC. VSMC Circ-Lrp6 displays numerous miRNA-145 binding sites that are conserved between mice and humans. Both Circ-Lrp6 and miR-145 localized in the P-bodies and were expressed in stenotic human arteries. Functional studies revealed that Circ-Lrp6 regulates VSMC phenotypic switching by modulating miR-145 [47]. Changes in circulating lncRNAs in plasma were assessed for diagnostic and therapeutic purposes and could complement ongoing efforts examining circulating noncoding RNAs [48, 49]. The circulating levels of lncRNA LIPCAR were elevated in the plasma of heart failure (HF) patients and was associated with hospitalization and poor outcomes [48]. In contrast, another study did not find increases in circulating levels of lncRNAs (including LIPCAR) and circRNAs were undetectable during myocardial injury [49]. Further studies are needed to establish the functions of circRNAs and circulating lncRNAs in cardiac injury and CVDs.
Conclusions
Contemporary findings pertaining to the role of transcriptional regulatory elements and related epigenetic mechanisms in prompting CVD are reviewed. Given the strong influence of lifestyles and environment in the development of CVDs, clearly epigenetic mechanisms are likely to play a major role. Apart from changes at enhancers, numerous lncRNAs are found associated with CVD, however, more work is required to determine their functionality and therapeutic usage. In general, lncRNA are less conserved across species, making it difficult to use mice for evaluating their in vivo functional roles. More mechanistic studies including CRISPR-Cas9 genome editing and genome-wide profiling with emerging technologies are required to precisely define the mode of actions of these epigenetic players in CVD. Single-cell RNA, ChIP- and ATAC-sequencing [50] may also assist in demonstrating cell-type specific roles of specific lncRNAs and enhancers in CVDs. Finally, in CVD, the biomarker and therapeutic capacity of enhancers and lncRNAs warrant exploration, including epigenetic therapies. Epigenetics can co-operate with genetics to impact the individual differences in disease susceptibility, development and response to treatment. Understanding epigenetic variations could also aid precision medicine strategies for optimized individual treatment [51].
Purpose of review.
Hyperlipidemia, hypertension, diabetes and related metabolic disorders increase the risk for cardiovascular disease (CVD). Despite significant progress in the identification of key mechanisms and genetic polymorphisms linked to various CVDs, the rates of CVDs continue to escalate, underscoring the need to evaluate additional mechanisms for more effective therapies. Environment and lifestyle changes can alter epigenetic mechanisms mediated by histone modifications and long non-coding RNAs (lncRNAs) which play important roles in gene regulation. This review summarizes recent findings on the role of epigenetic mechanisms in CVD.
Key points.
Recent findings highlighting the role of transcriptional regulatory elements including enhancers and lncRNAs (epigenetic mechanisms) in the regulation of CVD development and progression.
LncRNAs, either alone or in combination with enhancers or miRNAs, regulate key phenotypes in CVDs by modulating gene expression in cis and trans by epigenetic mechanisms.
Epigenetic regulators, including enhancers, and lncRNAs may be useful as novel biomarkers and therapeutic targets in CVDs.
Recent findings.
Recent studies identified dysregulated histone modifications and chromatin modifying proteins at cis-regulatory elements, including enhancers/super-enhancers, mediating the expression of genes associated with CVD in vascular and immune cells in response to growth factors and inflammatory mediators. Several lncRNAs have also been reported to contribute to pathological gene expression via cis and trans mechanisms involving interactions with nuclear proteins, co-operation with enhancers/super enhancers and acting as microRNA sponges.
Funding:
We gratefully acknowledge funding from the National Institutes of Health (NIDDK and NHLBI), the American Diabetes Association, the American Heart Association, and the Wanek Family Project for Type 1 Diabetes at the City of Hope.
Footnotes
Conflicts of interest: None
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Geovanini GR, Libby P. Atherosclerosis and inflammation: overview and updates. Clin Sci (Lond). 2018;132:1243–52. [DOI] [PubMed] [Google Scholar]
- 2.Musunuru K, Kathiresan S. Genetics of Common, Complex Coronary Artery Disease. Cell. 2019;177:132–45. [DOI] [PubMed] [Google Scholar]
- 3.Long Y, Wang X, Youmans DT, et al. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20:590–607. [DOI] [PubMed] [Google Scholar]
- 5.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato M, Natarajan R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol. 2019;15:327–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. [DOI] [PubMed] [Google Scholar]
- 9.Sallam T, Sandhu J, Tontonoz P. Long Noncoding RNA Discovery in Cardiovascular Disease: Decoding Form to Function. Circ Res. 2018;122:155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S, Senapati P, Chen Z, et al. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nat Commun. 2017;8:1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jae N, Heumuller AW, Fouani Y, et al. Long non-coding RNAs in vascular biology and disease. Vascul Pharmacol. 2019;114:13–22. [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Leslie KL, Martin KA. Epigenetic regulation of smooth muscle cell plasticity. Biochim Biophys Acta. 2015;1849:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy MA, Zhang E, Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia. 2015;58:443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan Q, McMahon S, Anand P, et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci Transl Med. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das S, Zhang E, Senapati P, et al. A Novel Angiotensin II-Induced Long Noncoding RNA Giver Regulates Oxidative Stress, Inflammation, and Proliferation in Vascular Smooth Muscle Cells. Circ Res. 2018;123:1298–312. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This paper demonstrates Angiotensin II-induced novel lncRNA GIVER is regulated by the nuclear receptor Nr4a3 and promotes oxidative stress and inflammation in vascular smooth muscle cells via epigenetic mechanisms. Increased expression of a human ortholog correlates with human hypertension.
- 16.Ahmed ASI, Dong K, Liu J, et al. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2018;115:E8660–E7. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study demonstrates key of role of lncRNA NEAT1 in VSMC phenotypic switching via epigenetic repression of contractile genes.
- 17.Mahmoud AD, Ballantyne MD, Miscianinov V, et al. The Human-Specific and Smooth Muscle Cell-Enriched LncRNA SMILR Promotes Proliferation by Regulating Mitotic CENPF mRNA and Drives Cell-Cycle Progression Which Can Be Targeted to Limit Vascular Remodeling. Circ Res. 2019;125:535–51. [DOI] [PMC free article] [PubMed] [Google Scholar]; **In this paper, the authors demonstrate novel mechanisms by which human VSMC specific lncRNA SMILR regulates cell cycle progression. SMILR knockdown in ex vivo graft pathological model reduces proliferation, suggesting its therapeutic potential.
- 18.Leung A, Trac C, Jin W, et al. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res. 2013;113:266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li DY, Busch A, Jin H, et al. H19 Induces Abdominal Aortic Aneurysm Development and Progression. Circulation. 2018;138:1551–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elia L, Kunderfranco P, Carullo P, et al. UHRF1 epigenetically orchestrates smooth muscle cell plasticity in arterial disease. J Clin Invest. 2018;128:2473–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirodkar AV, Marsden PA. Epigenetics in cardiovascular disease. Curr Opin Cardiol. 2011;26:209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalna V, Yang Y, Peghaire CR, et al. The Transcription Factor ERG Regulates Super-Enhancers Associated With an Endothelial-Specific Gene Expression Program. Circ Res. 2019;124:1337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study demonstrate the role of transcription factor ERG in lineage specific gene regulation by enhancers and super enhancers in endothelial homeostasis. SNPs at ERG super enhancers are associated with CVD.
- 23.Miyagawa K, Shi M, Chen PI, et al. Smooth Muscle Contact Drives Endothelial Regeneration by BMPR2-Notch1-Mediated Metabolic and Epigenetic Changes. Circ Res. 2019;124:211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]; **In this study, the authors show that SMC-EC interaction activates BMPR2-Notch1 signaling which leads to increase in glycolysis and activation of enhancers involved in EC proliferation and homeostasis. This study links metabolic changes with epigenetic mechanisms.
- 24.Schlereth K, Weichenhan D, Bauer T, et al. The transcriptomic and epigenetic map of vascular quiescence in the continuous lung endothelium. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montefiori LE, Sobreira DR, Sakabe NJ, et al. A promoter interaction map for cardiovascular disease genetics. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study identified interaction of several CVD associated SNPs with target genes in endothelial cells, highlighting the significance of long-range chromatin interactions.
- 26.Wang KC, Chang HY. Epigenomics: Technologies and Applications. Circ Res. 2018;122:1191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyu Q, Xu S, Lyu Y, et al. SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc Natl Acad Sci U S A. 2019;116:546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Y, Ajami NE, Huang TS, et al. Enhancer-associated long non-coding RNA LEENE regulates endothelial nitric oxide synthase and endothelial function. Nat Commun. 2018;9:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monteiro JP, Bennett M, Rodor J, et al. Endothelial function and dysfunction in the cardiovascular system: the long non-coding road. Cardiovasc Res. 2019;115:1692–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann P, Jae N, Knau A, et al. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat Commun. 2018;9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho H, Shen GQ, Wang X, et al. Long noncoding RNA ANRIL regulates endothelial cell activities associated with coronary artery disease by up-regulating CLIP1, EZR, and LYVE1 genes. J Biol Chem. 2019;294:3881–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Q, Meng Q, Qi M, et al. Shear-Sensitive lncRNA AF131217.1 Inhibits Inflammation in HUVECs via Regulation of KLF4. Hypertension. 2019;73:e25–e34. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Padilla C, Dominguez JN, Aranega AE, et al. Differential chamber-specific expression and regulation of long non-coding RNAs during cardiac development. Biochim Biophys Acta Gene Regul Mech. 2019;1862:194435. [DOI] [PubMed] [Google Scholar]
- 34.Davis FM, Gallagher KA. Epigenetic Mechanisms in Monocytes/Macrophages Regulate Inflammation in Cardiometabolic and Vascular Disease. Arterioscler Thromb Vasc Biol. 2019;39:623–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoeksema MA, Glass CK. Nature and nurture of tissue-specific macrophage phenotypes. Atherosclerosis. 2019;281:159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das S, Reddy MA, Senapati P, et al. Diabetes Mellitus-Induced Long Noncoding RNA Dnm3os Regulates Macrophage Functions and Inflammation via Nuclear Mechanisms. Arterioscler Thromb Vasc Biol. 2018;38:1806–20. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This report showed that lncRNA Dnm3os is upregulated in macrophages from diabetic mice and in monocytes from diabetic human subjects. Mechanistic studies showed that dysregulation of Dnm3os interaction with the nucleolin under diabetic conditions promotes macrophage inflammatory phenotype.
- 37.Reddy MA, Chen Z, Park JT, et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes. 2014;63:4249–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang CA, Li JP, Yen JC, et al. lncRNA NTT/PBOV1 Axis Promotes Monocyte Differentiation and Is Elevated in Rheumatoid Arthritis. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cremer S, Michalik KM, Fischer A, et al. Hematopoietic Deficiency of the Long Noncoding RNA MALAT1 Promotes Atherosclerosis and Plaque Inflammation. Circulation. 2019;139:1320–34. [DOI] [PubMed] [Google Scholar]; **This study showed MALAT1 regulates inflammation via miR-503 and its deficiency promotes atherosclerosis in Apoe−/− mice.
- 40.Zhang P, Cao L, Zhou R, et al. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;10:1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sallam T, Jones M, Thomas BJ, et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. 2018;24:304–12. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study showed lncRNA MeXis regulates LXR-dependent expression of Abca1 and cholesterol efflux.
- 42.Hennessy EJ, van Solingen C, Scacalossi KR, et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat Metab. 2019;1:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study demonstrated the function of a primate-specific lncRNA CHROME in ABCA1 expression and cholesterol homeostasis.
- 43.Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. 2019. [DOI] [PubMed] [Google Scholar]
- 44.Khyzha N, Khor M, DiStefano PV, et al. Regulation of CCL2 expression in human vascular endothelial cells by a neighboring divergently transcribed long noncoding RNA. Proc Natl Acad Sci U S A. 2019;116:16410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This report showed regulation of inflammation in endothelial cells by a network of neighboring mRNA-lncRNA pairs in association with super enhancers.
- 45.Zhou Q, Yu B, Anderson C, et al. LncEGFL7OS regulates human angiogenesis by interacting with MAX at the EGFL7/miR-126 locus. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study showed the utility of CRISPR-Cas9 genome editing approach in determining the role of lncRNA-enhancer interactions in angiogenesis.
- 46.Engreitz J, Abudayyeh O, Gootenberg J, et al. CRISPR Tools for Systematic Studies of RNA Regulation. Cold Spring Harb Perspect Biol. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall IF, Climent M, Quintavalle M, et al. Circ_Lrp6, a Circular RNA Enriched in Vascular Smooth Muscle Cells, Acts as a Sponge Regulating miRNA-145 Function. Circulation research. 2019;124:498–510. [DOI] [PubMed] [Google Scholar]
- 48.Santer L, Lopez B, Ravassa S, et al. Circulating Long Noncoding RNA LIPCAR Predicts Heart Failure Outcomes in Patients Without Chronic Kidney Disease. Hypertension. 2019;73:820–8. [DOI] [PubMed] [Google Scholar]
- 49.Schulte C, Barwari T, Joshi A, et al. Comparative Analysis of Circulating Noncoding RNAs Versus Protein Biomarkers in the Detection of Myocardial Injury. Circ Res. 2019;125:328–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirka RC, Wagh D, Paik DT, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leopold JA, Loscalzo J. Emerging Role of Precision Medicine in Cardiovascular Disease. Circ Res. 2018;122:1302–15. [DOI] [PMC free article] [PubMed] [Google Scholar]


