Abstract
The COVID-19 pandemic caused by SARS-CoV-2 has now emerged as a global health problem and is responsible for high mortality and morbidity. The SARS-CoV-2 main protease (Mpro) emerged as a promising drug target because of its essential role in the processing of polyproteins, which is translated from viral RNA. The present study reports a designed novel hybrid antiviral molecule (VTRRT-13.V2.1) against SARS-CoV2 main protease. A series of different combinations of hybrid antiviral were generated from nonspecific antiviral molecules currently used to control COVID-19. To enhance the specificity of the designed hybrid antiviral molecule, the core pocket region of the active site of Mpro protein was targeted. In-silico screening, molecular mechanics, molecular dynamics simulation (MDS) analysis identified a hybrid VTRRT-13.V2 molecule. Retrosynthetic analysis and combinatorial synthesis generated 1000 analogs of VTRRT-13.V2 molecules. Docking, molecular mechanics, and MDS analysis selected VTRRT-13.V2.1 as a possible inhibitor for SARS-CoV2 main protease. Comparative analysis of all the results showed that VTRRT-13.V2.1 have the highest docking Glide score (-12.28 kcal/mol) and best binding energy (-52.23 kcal/mol) as compared to the other hybrid constructs such as VTRRT-13.V2 (-9.47 and -47.36 kcal/mol), VTRRT-13 (-8.9 and -47.55 kcal/mol), and current antiviral investigated. The mutational sensitivity screening showed that binding residues of Mpro are not present in mutation hotspots. It was also observed that VTRRT-13.V2.1 does not have any human off-targets. SARS-CoV2 main protease is essential for the survival of this virus; hence, a designed novel hybrid antiviral molecule (VTRRT-13.V2.1) might be useful to control the infection of COVID-19 infection.
Keywords: Bioinformatics, Microbiology, Infectious disease, Medical microbiology, Hybrid antiviral molecule, SARS-CoV2 main protease, In-silico designing, Molecular dynamics simulation, Retro-combinatorial synthesis
Bioinformatics; Microbiology; Infectious disease, Medical Microbiology; Hybrid Antiviral molecule; SARS-CoV2 main protease; In-silico designing; Molecular Dynamics Simulation; Retro-combinatorial synthesis
1. Introduction
A novel coronavirus caused an outbreak of the pulmonary disease in Wuhan city, and now it has spread globally (Zhou et al., 2020) (Wu et al., 2020). The virus is identified as SARS-CoV-2 (Gorbalenya et al., 2020) due to 82% similarity in RNA genome with SARS coronavirus (SARS-CoV), and classified in clade b of the genus Betacoronavirus (Wu et al., 2020) (Zhou et al., 2020). The disease by this pathogen SARS-CoV-2 is called COVID-19. On March 11, 2020, the world health organization (WHO) declared the outbreak a pandemic. On July 27, 2020, there were >1,60,00,000 cumulative cases globally and >6,40,000 death with >5% mortality rate in outcome cases. There are different targets in the SARS-CoV2, but the best-characterized drug target is the main protease (Mpro) among coronaviruses (Zhang et al., 2020a) (Anand et al., 2003). This protease is essential for processing the polyproteins that are translated from virus RNA (Hilgenfeld, 2014). Mpro works with more than 11 cleavage sites on the large polyprotein 1ab (replicase 1ab, ~790kDa), and its recognition sequence is Leu-Gln∗-(Ser, Ala, Gly) where ∗ is cleavage site. Inhibiting, this protease would block viral replication. No human homolog of this protease is known; hence it may be non-toxic to humans. Ritonavir, Lopinavir, and Remdesivir have been used to control other coronaviruses (Sheahan et al., 2020). Ritonavir and lopinavir have been tested against the SARS-CoV (Chu et al., 2004). Lopinavir is an HIV-1 protease inhibitor (Reddy et al., 2007). Ritonavir and Lopinavir were found to be effective against coronavirus like MERS-CoV (Spanakis et al., 2014). Lopinavir-ritonavir with other antiviral molecules controls the SARS-CoV (Chu et al., 2004) and MERS-CoV (Arabi et al., 2018). Remdesivir (GS-5734) is a broad-spectrum antiviral nucleotide prodrug with potent antiviral activity against diverse RNA viruses like SARS-CoV, MERS-CoV (Sheahan et al., 2017). Recent clinical trials of Lopinavir-Ritonavir showed that it did not significantly accelerate clinical improvement, reduced mortality, or diminish throat viral RNA detection in patients with serious COVID-19 (Cao et al., 2020). Therefore, there is an urgent need to find an inhibitor for SARS-CoV2 main protease. Designing a new molecule takes a long time to develop it as effective antiviral, hence in the present study, a hybrid molecule VTRRT-13.V2.1 was designed from FDA approved antiviral molecules against SARS-CoV2 main protease (Mpro) using in-silico drug design, molecular mechanics, retrosynthetic analysis, combinatorial synthesis, and molecular dynamics simulation analysis.
2. Results
2.1. Generation of receptor grid for binding site of SARS-CoV2 main protease
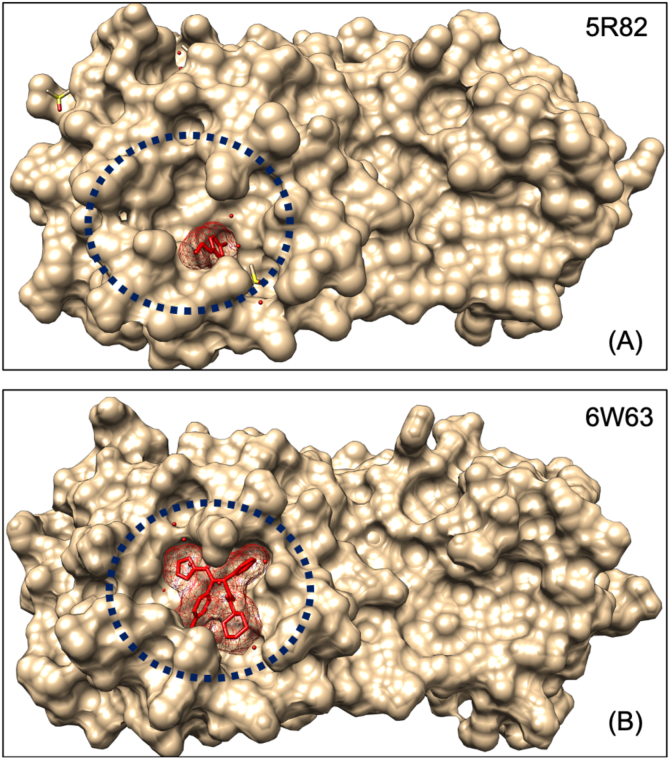
The PDB structures of SARS-CoV2 main protease (Mpro) complexed with a different ligand were taken from RCSB (PDB number 5R82, resolution 1.31Å; and PDB number 6W63, resolution 2.1Å). Binding site residues of the ligand in 5R82 were found to be deep in the active site (Figure 1A) and consist of His41, Met49, Gln189, Arg188, Asp187, His164, and Met165. Similarly, the binding site of the ligand in 6W63 was found to be wide (Figure 1B) and consisting of Thr25, Thr26, Leu27, His41, Cys44, Met49, Tyr54, Phe140, Leu,141, Asn142, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, Leu167, Pro168, Asp 187, Arg188, and Gln189. The ligand was removed from the crystal structure, and the unliganded structure was pre-processed, optimised, and minimised using the OPLS_2005 force field. Ligand binding site in the 5R82 is at the center (core pocket) of the active site of 6W63; hence, a molecule which is designed against the core binding site (binding pocket of ligand in 5R82) must have better chance to have good binding affinity and specificity with the active site of the SARS-CoV2 main protease (Figure 1). Two receptor grids were generated using core pocket residues of the active site, and complete binding site residues. The initial drug screening was started with a core-binding pocket, and the identified hybrid molecule was validated for the complete binding site or active site of SARS-CoV2 main protease.
Figure 1.
Binding site of the SARS-CoV2 main protease showing bound ligand. The ligand in PDB 5R82 is present in the deep and core binding pocket of active site while ligand of the PDB 6W63 is broad and covering most part of the active site of SARS-CoV2 main protease.
2.2. Prediction of antiviral activity of the designed hybrid antiviral molecule
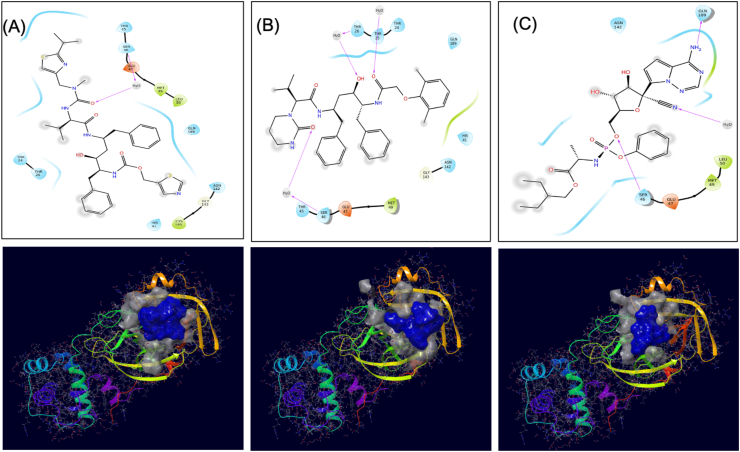
In the present work, antiviral Ritonavir, Lopinavir, and Remdesivir that have shown some antiviral effect on SARS-CoV2 as per currently available literature on 2 March 20, 2020, were selected. These antivirals were docked to the core receptor grid of SARS-CoV2 main protease using Glide XP docking. The interacting parts of these ligands were analysed (Figure 2). Different hybrid molecules were randomly created using various combinations of antiviral molecules by keeping important features of parent antiviral molecules (involved in its targets interactions) conserved. The designed hybrid antiviral molecules were in-silico predicted for structural based antiviral activity. The result presented as probability active (Pa Score) that varies from 0 to 1, and a score of >0.3 can be considered as active. A total of 20 hybrid molecules have predicted to possess the antiviral activity score (Pa) of 0.4 (40%). These 20 hybrid molecules, along with three-parent molecules, were investigated for a possible inhibitor for SARS-CoV2 main protease, and their smile formula, chemical name, and predicted antiviral score are listed in Table 1, and their 2D structure are shown in supplementary Table 1. These antiviral molecules have been prepared by LigPrep modules of Schrodinger for docking to the binding site of SARS-CoV2 main protease.
Figure 2.
Interaction diagram showing interacting amino acid residues and their docking pose in SARS-CoV2 main protease-Ritonavir complex (A), SARS-CoV2 main protease-Lopinavir complex (B), and SARS-CoV2 main protease-Remdesivir complex (C). Docking grid was generated using the core-binding pocket of the active site of this protein as per PDB 5R82.
Table 1.
Antiviral molecules and their hybrid molecules: Smile formula, IUPAC name and predicted antiviral score are shown for the molecules. Antiviral activity was predicted for the antiviral molecules and their hybrids based on Multi-level Neighborhoods of Atoms (MNA) descriptors. Antiviral activity score (Pa) varies between 0 to 1, where 1 is the best antiviral activity, and zero stand for no predicted antiviral activity.
| Compound | Smile Formula of molecule | Chemical Name | Antiviral score (Pa) |
|---|---|---|---|
| Remdesivir(R) | CCC(CC)COC(=O)C(C)NP(=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3N2N = CN = C3N)O)O)OC4 = CC = CC = C4 | 2-ethylbutyl 2-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)propanoate | 0.814 |
| Lopinavir(L) | CC1 = C(C(=CC = C1)C)OCC(=O)NC(CC2 = CC = CC = C2)C(CC(CC3 = CC = CC = C3)NC(=O)C(C(C)C)N4CCCNC4 = O)O | N-{5-[2-(2,6-dimethylphenoxy)acetamido]-4-hydroxy-1,6-diphenylhexan-2-yl}-3-methyl-2-(2-oxo-1,3-diazinan-1-yl)butanamide | 0.482 |
| Ritonavir(Rt) | CC(C)C1 = NC(=CS1)CN(C)C(=O)NC(C(C)C)C(=O)NC(CC2 = CC = CC = C2)CC(C(CC3 = CC = CC = C3)NC(=O)OCC4 = CN = CS4)O | ((1,3-thiazol-5-yl)methyl N-[3-hydroxy-5-(3-methyl-2-{[methyl({[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl})carbamoyl]amino}butanamido)-1,6-diphenylhexan-2-yl]carbamate | 0.602 |
| VTRL1 | CCC(CC)COC(=O)C(C)N([P](=O) (OC(C(N(C(COC1 = C(C)C=CC = C1C) = O)[H])CC2 = CC = CC = C2)O[H])OC3 = CC = CC = C3)[H] | 2-ethylbutyl 2-[({2-[2-(2,6-dimethylphenoxy)acetamido]-1-hydroxy-3-phenylpropoxy}(phenoxy)phosphoryl)amino]propanoate | 0.716 |
| VTRL2 | C1(C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H])CC(C(N(C(COC4 = C(C)C=CC = C4C) = O)[H])CC5 = CC = CC = C5)O[H] | N-[4-(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)-3-hydroxy-1-phenylbutan-2-yl]-2-(2,6-dimethylphenoxy)acetamide | 0.607 |
| VTRL3 | CCC(CC)COC(=O)C(N([P](=O) (OCCC(CC1 = CC = CC = C1)N(C(=O)C(C(C)C)N2CCCN(C2 = O)[H])[H])OC3 = CC = CC = C3)[H])[H] | 2-ethylbutyl 2-[({3-[3-methyl-2-(2-oxo-1,3-diazinan-1-yl)butanamido]-4-phenylbutoxy}(phenoxy)phosphoryl)amino]acetate | 0.636 |
| VTRL4 | C1(C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H])CC(CC4 = CC = CC = C4)N(C(=O)C(C(C)C)N5CCCN(C5 = O)[H])[H] | N-[1-(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)-3-phenylpropan-2-yl]-3-methyl-2-(2-oxo-1,3-diazinan-1-yl)butanamide | 0.446 |
| VTRL5 | CCC(CC)COC(=O)C(C)N(C(CC(C(N(C(COC1 = C(C)C=CC = C1C) = O)[H])CC2 = CC = CC = C2)O[H])CC3 = CC = CC = C3)[H] | 2-ethylbutyl 2-({5-[2-(2,6-dimethylphenoxy)acetamido]-4-hydroxy-1,6-diphenylhexan-2-yl}amino)propanoate | 0.478 |
| VTRL6 | [P](=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H]) (OC4 = CC = CC = C4)N([H])C(CC(C(N(C(COC5 = C(C)C=CC = C5C) = O)[H])CC6 = CC = CC = C6)O[H])CC7 = CC = CC = C7 | N-[5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)-3-hydroxy-1,6-diphenylhexan-2-yl]-2-(2,6-dimethylphenoxy)acetamide | 0.823 |
| VTRL7 | [P](=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H]) (OC4 = CC = CC = C4)C(CC(C(N(C(COC5 = C(C)C=CC = C5C) = O)[H])CC6 = CC = CC = C6)O[H])CC7 = CC = CC = C7 | (5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methyl phenyl {5-[2-(2,6-dimethylphenoxy)acetamido]-4-hydroxy-1,6-diphenylhexan-2-yl}phosphonate | 0.554 |
| VTRL8 | [P](=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H]) (OC4 = CC = CC = C4)CC(CC(C(N(C(COC5 = C(C)C=CC = C5C) = O)[H])CC6 = CC = CC = C6)O[H])CC7 = CC = CC = C7 | (5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methyl phenyl {2-benzyl-5-[2-(2,6-dimethylphenoxy)acetamido]-4-hydroxy-6-phenylhexyl}phosphonate | 0.579 |
| VTRL9 | N([P](=O) (OCC1C(CC(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])OC4 = CC = CC = C4)C(CC(C(N(C(COC5 = C(C)C=CC = C5C) = O)[H])CC6 = CC = CC = C6)O[H])CC7 = CC = CC = C7 | N-[5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3-hydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)-3-hydroxy-1,6-diphenylhexan-2-yl]-2-(2,6-dimethylphenoxy)acetamide | 0.827 |
| VTRL10 | [P](=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O)O) (OC4 = CC = CC = C4)N(C(=O)C(C(C)C)N5CCCN(C5 = O)[H])[H] | N-{[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}-3-methyl-2-(2-oxo-1,3-diazinan-1-yl)butanamide | 0.711 |
| VTRL11 | [P](=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H]) (OC4 = CC = CC = C4)N(C(COC5 = C(C)C=CC = C5C) = O)[H] | N-{[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}-2-(2,6-dimethylphenoxy)acetamide | 0.763 |
| VTRL12 | [P](=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H]) (OC)N([H])C(NC(COC4 = C(C)C=CC = C4C) = O)C(CC(CC5 = CC = CC = C5)N(C(=O)C(C)N6CCCN(C6 = O)[H])[H])O[H] | N-[5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methoxy](methoxy)phosphoryl}amino)-5-[2-(2,6-dimethylphenoxy)acetamido]-4-hydroxy-1-phenylpentan-2-yl]-2-(2-oxo-1,3-diazinan-1-yl)propanamide | 0.581 |
| VTRRT13 | [P](=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H]) (OC4 = CC = CC = C4)N(C(CC5 = CC = CC = C5)CC(C(CC6 = CC = CC = C6)N(C(=O)OCC7 = CN = CS7)[H])O[H])[H] | (1,3-thiazol-5-yl)methyl N-[5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)-3-hydroxy-1,6-diphenylhexan-2-yl]carbamate | 0.867 |
| VTRRT14 | CCC(CC)COC(=O)C(C)N[P](=O) (OCC(N(C(C(N(C(N(CC1 = CSC(=N1)C(C)C)C) = O)[H])C(C)C) = O)[H])CC2 = CC = CC = C2)OC3 = CC = CC = C3 | 2-ethylbutyl 2-({[2-(3-methyl-2-{[methyl({[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl})carbamoyl]amino}butanamido)-3-phenylpropoxy](phenoxy)phosphoryl}amino)propanoate | 0.628 |
| VTRRT15 | N([P](=O) (OCC1C(C(C(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H])O[H])OC4 = CC = CC = C4) (C(C(CC(N(C(CN(C(N(CC5 = CSC(=N5)C(C)C)[H]) = O)[H]) = O)[H])CC6 = CC = CC = C6)O[H])CC7 = CC = CC = C7)[H] | N-[5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)-4-hydroxy-1,6-diphenylhexan-2-yl]-2-[({[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)amino]acetamide | 0.734 |
| VTRRT16 | [P](=O) (OCC1C(CC(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H]) (OC4 = CC = CC = C4)N(C(C(CC(N(C(C(N(C(N(CC5 = CSC(=N5)C(C)C)C) = O)[H])C) = O)[H])CC6 = CC = CC = C6)O[H])CC7 = CC = CC = C7)[H] | N-[5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3-hydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)-4-hydroxy-1,6-diphenylhexan-2-yl]-2-{[methyl({[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl})carbamoyl]amino}propanamide | 0.746 |
| VTRRT17 | [P](=O) (OCC1C(CC(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N)O) (OC4 = CC = CC = C4)N(C(C(CC(N(C(C(N(C(N(CC5 = CSC(=N5)C(C)C)C) = O)[H])C(C)C) = O)[H])CC6 = CC = CC = C6)O[H])CC7 = CC = CC = C7)[H] | N-[5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3-hydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)-4-hydroxy-1,6-diphenylhexan-2-yl]-3-methyl-2-{[methyl({[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl})carbamoyl]amino}butanamide | 0.765 |
| VTRRT18 | [P](=O) (OCC1C(CC(O1) (C#N)C2 = CC = C3[N]2N = CN = C3N([H])[H])O[H]) (OC4 = CC = CC = C4)N(C(CC5 = CC = CC = C5)CC(C(CC6 = CC = CC = C6)N(C(=O)OCC7 = CN = CS7)[H])O[H])[H] | (1,3-thiazol-5-yl)methyl N-[5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3-hydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)-3-hydroxy-1,6-diphenylhexan-2-yl]carbamate | 0.870 |
| VTLRT19 | N(C(CC1 = CC = CC = C1)C(CC(CC2 = CC = CC = C2)N(C(=O)C(C(C)C)N3CCCN(C3 = O)[H])[H])O[H]) (C(=O)OCC4 = CN = CS4)[H] | (1,3-thiazol-5-yl)methyl N-{3-hydroxy-5-[3-methyl-2-(2-oxo-1,3-diazinan-1-yl)butanamido]-1,6-diphenylhexan-2-yl}carbamate | 0.611 |
| VTLRT20 | CC1 = C(C(=CC = C1)C)OCC(=O)N(C(C(CC(N(C(C(N(C(N(CC2 = CSC(=N2)C(C)C)C) = O)[H])C(C)C) = O)[H])CC3 = CC = CC = C3)O[H])CC4 = CC = CC = C4)[H] | N-{5-[2-(2,6-dimethylphenoxy)acetamido]-4-hydroxy-1,6-diphenylhexan-2-yl}-3-methyl-2-{[methyl({[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl})carbamoyl]amino}butanamide | 0.498 |
| VTRRT13-V1 | [H]OC(CC(CC1 = CC = CC = C1)N([H])P(=O) (OCC1OC(C#N) (C(O[H])C1O[H])C1 = CC = C2N1N = CN = C2N([H])[H])OC1 = CC = CC = C1)C(CC1 = CC = CC = C1)N([H])C(O) = O | [5-({[(5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-3,4-dihydroxyoxolan-2-yl)methoxy](phenoxy)phosphoryl}amino)-3-hydroxy-1,6-diphenylhexan-2-yl]carbamic acid | 0.846 |
| VTRRT13-V2 | [H]NC(CC1 = CC = CC = C1)C(O[H])CC(CC1 = CC = CC = C1)N([H])P(=O) (OCC1OC(C#N) (C(O[H])C1O[H])C1 = CC = C2N1N = CN = C2N([H])[H])OC1 = CC = CC = C1 | 5-[({[(5-amino-4-hydroxy-1,6-diphenylhexan-2-yl)amino](phenoxy)phosphoryl}oxy)methyl]-2-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-3,4-dihydroxyoxolane-2-carbonitrile | 0.847 |
2.3. Docking via Glide's Extra Precision XP mode identified that a hybrid antiviral molecule binds to the core-binding pocket of SARS-CoV2 main protease
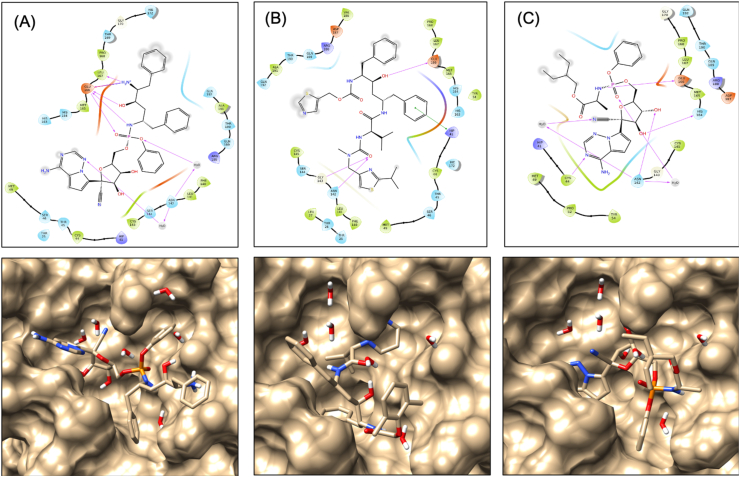
A docking study was performed using Glide's Extra Precision XP mode. XP docking identifies that all hybrid and parent molecules except VTRRT-17, docked to the core binding site of SARS-CoV2 main protease with favourable docking score, and their docking score has a significant difference. The results are shown in Table 2. It was also observed that hybrids molecules have better docking score than the parent molecules, that signifies the design of hybrid molecules. Among all the molecules, hybrid antiviral VTRL-12, VTRL-6, VTRRT-13 showed the best docking as compared to other hybrids and parent antiviral molecules. The interacting residues of the core-binding pocket of SARS-CoV2 main protease with three best docked hybrid antiviral molecules along with their docked poses are shown in Figure 3.
Table 2.
Result showing outcome of GLIDE molecular docking in XP mode and Binding free energies result from Prime analysis using the MMGBSA approach. The docking was performed using Grid of core binding pocket residues of the active site of SARS-CoV2 main protease (as per PDB ID 5R82).
| Antiviral molecules and their hybrids | Docking Score in Kcal/mol (XP docking result) | Binding Gibbs Free energy change in Kcal/mol (MMGBSA result) |
|---|---|---|
| VTRL12 | -5.69 | -28.85 |
| VTRL6 | -5.33 | -9.53 |
| VTRRT13 | -5.08 | -30.30 |
| VTRRT18 | -4.85 | -14.26 |
| VTRL10 | -4.28 | -17.62 |
| VTRL2 | -4.11 | -13.17 |
| Remdesivir | -4.00 | -13.58 |
| VTRL4 | -3.90 | -7.73 |
| VTRL8 | -3.66 | -24.86 |
| VTRL9 | -3.52 | -2.98 |
| Lopinavir | -3.40 | -8.34 |
| VTRL11 | -3.40 | -13.15 |
| VTRL1 | -3.19 | -6.88 |
| VTLRT19 | -2.90 | -0.21 |
| VTRL7 | -2.69 | 1.80 |
| VTRRT14 | -2.61 | -2.37 |
| VTRL5 | -2.50 | 0.40 |
| VTRRT16 | -2.47 | -6.84 |
| VTRL3 | -2.40 | -13.27 |
| VTRL5 | -2.28 | -9.35 |
| VTRRT15 | -2.12 | -14.52 |
| Ritonavir | -2.12 | -0.77 |
| VTLRT20 | -1.66 | -8.06 |
| VTRRT17 | 0.52 | -12.14 |
Figure 3.
Interaction diagram showing interacting amino acid residues and their docking pose in SARS-CoV2 main protease-VTRRT13 complex (A), SARS-CoV2 main protease-VTRL12 complex (B), and SARS-CoV2 main protease-VTRL6 complex (C). Docking grid was generated using the core-binding pocket of the active site of this protein as per PDB 5R82.
2.4. Gibbs free energy calculation confirms the binding of hybrid antiviral molecules with the core-binding pocket of SARS-CoV2 main protease
All the possible leads were underdone MMGBSA (molecular mechanics generalized born and surface area solvation) analysis to calculate the Gibbs free energy of binding. The result of this analysis is listed in Table 2. It was found that all the molecules have favorable Gibbs free energy with the core binding pocket of SARS-CoV2 main protease, except VTRL-7 and VTRL-5. Among all the molecules, VTRRT-13 showed the best binding energy (-30.3 kcal/mol) followed by VTRL-12 (-28.9 kcal/mol). It further confirms that hybrid antiviral molecules have more favorable Gibbs free energy as compared to parent antiviral such as lopinavir and ritonavir. Less favorable Gibbs free energy explain the possible reason for the failure of these drugs to treat COVID-19 patients.
2.5. Molecular dynamics simulation analysis confirms the interaction of VTRRT-13 with the core-binding region of SARS-CoV2 main protease
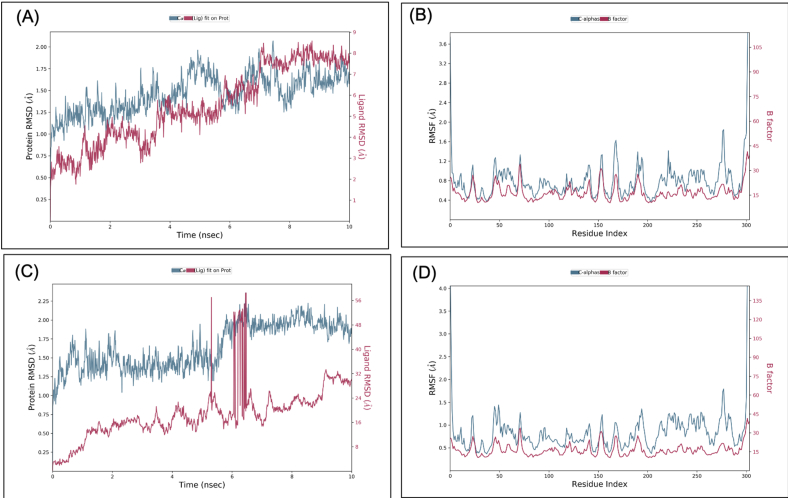
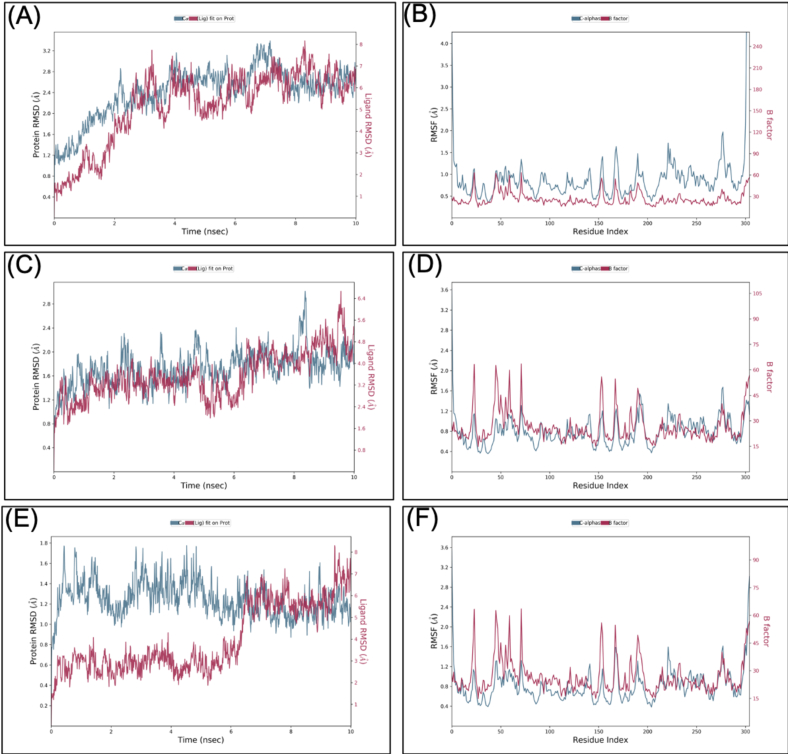
Molecular dynamics simulation (MDS) was performed for Mpro-VTRRT-13 complex and Mpro-VTRL-12 complex with 10ns simulation time, and results were analysed for RMSD, RMSF, ligand stability, bonding, etc. RMSD calculation showed that the Mpro-VTRRT-13 complex is more stable throughout simulation as compared to the Mpro-VTRL-12 complex (Figure 4). RMSF analysis showed that the protein has the RMSF less than 1.5Å for both the complexes that showed less fluctuation in protein conformation hence a stable structure. Interaction between the core-binding pocket of SARS-CoV2 main protease with hybrid molecule VTRRT-13 involves Thr24, Thr26, His41, Asn119, Asn142, Glu166, Gln189, and at least four contacts always exist more than 30% simulation time (Figure 5). In contrast, the Mpro-VTRL-12 complex has no contact in the range of >30% simulation time that showed its weak interaction with the core binding pocket of SARS-CoV2 main protease. Similarly, ligand RMSF of the Mpro-VTRRT-13 complex was found to have less change after binding with SARS-CoV2 main protease (data not shown) and possess low RMSD as compared to the Mpro-VTRL-12 complex that further confirms better and stable interaction of VTRRT-13 with SARS-CoV2 main protease. Hence, MDS analysis confirms that VTRRT-13 hybrid antiviral molecule is better at targeting the core binding pocket of SARS-CoV2 main protease. During MDS, it was observed that RMSD of VTRRT-13 is below 3Å except for some parts (data not shown); hence, VTRRT-13 was modified by deleting the atoms with high RMSD. Two variants VTRRT-13.V1 and VTRRT-13.V2, were created. The docking was performed with the core binding pocket, and the result showed that VTRRT-13.V2 have good docking score and negative binding energy. Hence it was included in further study.
Figure 4.
Root-mean-square deviation (RMSD) and Root mean square fluctuations (RMSF) result of the SARS-CoV2 main protease-VTRRT-13 complex (A & B), SARS-CoV2 main protease-VTRL-12 complex (C and D). The RMSD and RMSF were generated from Molecular dynamics analysis.
Figure 5.
Interacting residues and the interacting fraction of the hybrid molecule VTRRT-13 (A & B) and VTRL-12 (C & D) during molecular dynamic simulation protein-ligand complex.
2.6. Docking and molecular mechanics analysis confirms the binding interaction of hybrid lead with the binding site of SARS-CoV2 main protease
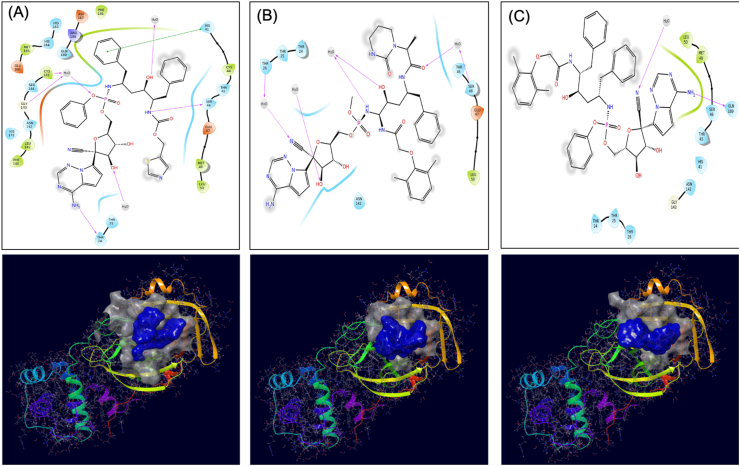
All the four hybrid molecules (VTRRT-13, VTRL-12, VTRL-6, VTRRT-13.V2) and parent molecules (ritonavir, lopinavir, and remdesivir) were further docked to the complete binding site of SARS-CoV2 main protease (as per PDP 6W63). The binding energy of the docked complexes was calculated using the molecular mechanics approach. The results are presented in Table 3 and Figure 6. The result showed that VTRRT-13.V2 has the best docking with (-9.4) and good binding energy (-47.36Kcal/mol) as compared to other antiviral and hybrid molecules tested. This further signifies our approach to design the drug targeted to the core region of the active site that helps to design an inhibitor for the active site of any protein. Figure 6A also showed that hybrid antiviral VTRRT-13.V2 covers most of the binding site of SARS-CoV2 main protease as compared to their parent antiviral molecules i.e, ritonavir, and remdesivir.
Table 3.
Result showing the outcome of GLIDE molecular docking in XP mode and Binding free energies result from Prime analysis using the MMGBSA approach. The docking was performed using Grid of the active site of residues of SARS-CoV2 main protease (as per PDB ID 6W63).
| Antiviral molecules and their hybrids | Docking Score in Kcal/mol (XP docking result) | Binding Gibbs Free energy change (in Kcal/mol) (MMGBSA result) |
|---|---|---|
| VTRRT13-V2 | -9.47 | -47.36 |
| VTRRT-13 | -8.90 | -47.55 |
| Ritonavir | -8.19 | -47.43 |
| Lopinavir | -8.17 | -37.04 |
| VTRL-12 | -7.62 | -32.96 |
| Remdesivir | -7.29 | -36.52 |
| VTRL6 | -7.04 | -33.45 |
| VTRRT13-V2.1 | -12.28 | -52.23 |
Figure 6.
Interaction diagram showing interacting amino acid residues and their docking pose in the active site of SARS-CoV2 main protease-VTRRT-13.V2 complex (A), SARS-CoV2 main protease-Ritonavir complex (B), and SARS-CoV2 main protease-Remdesivir complex (C). Docking grid used generated using complete active site residues of this protein as per PDB 6W63.
2.7. MDS analysis confirms VTRRT-13.V2 have better interaction with SARS-CoV2 main protease than other tested hybrids
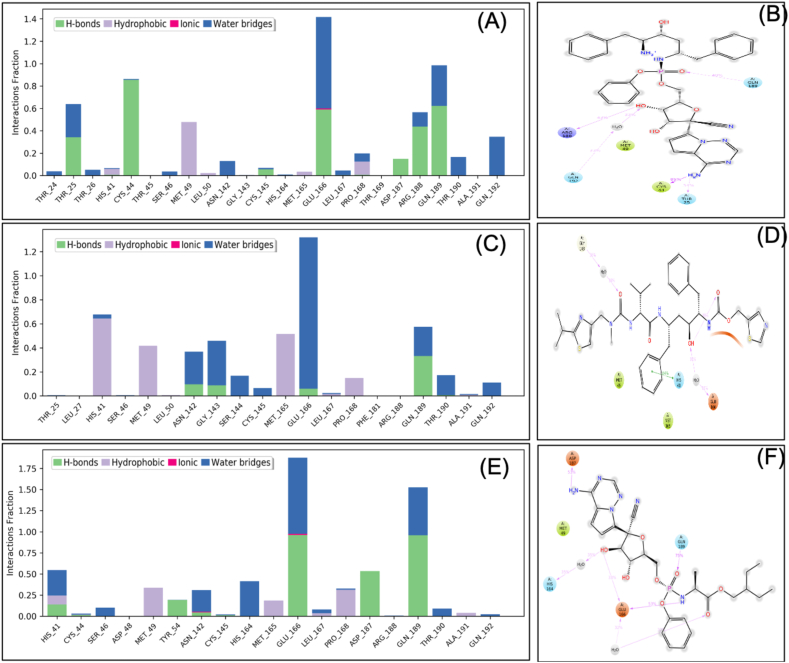
VTRRT-13.V2 is a hybrid of ritonavir and remdesivir; hence MDS was performed for docked complexes of the complete binding site of SARS-CoV2 main protease with VTRRT-13.V2, ritonavir, and remdesivir. MDS was conducted up to 10ns, and results were analysed for RMSD, RMSF, ligand stability, bonding etc. RMSD calculation showed that the Mpro-VTRRT-13.V2 complex is stable throughout simulation with protein RMSD value less than 2.5 Å at 10ns (Figure 7). RMSF analysis showed that the Mpro-VTRRT-13.V2 complex has the RMSF less than 1.5Å that showed a stable complex structure. Interaction between SARS-CoV2 main protease and VTRRT-13.V2 involves Thr25, Cys44, Met49, Glu166, Arg188, Gln189, Gln192, and at least five contacts always exist more than 30% simulation time (Figure 8). Hydrogen bonding and ionic interaction are considered very important for ligand binding and have a strong influence on drug specificity, metabolism, and absorption. The MDS result showed that VTRRT-13.V2 form five hydrogen bonds with bond length less than 2.5Å (>30% simulation time) that showed specific interaction of VTRRT-13.V2 with SARS-CoV2 main protease. Ritonavir forms one hydrogen bond, while Remdesivir forms three hydrogen bonds in the same simulation time. This confirms that the hybrid VTRRT-13.V2 antiviral molecule has more specific interaction than its parent molecules. In addition, MDS analysis of the ligand showed that VT-RRT-13.V2 has less RMSD, a stable radius of gyration (measure stability of ligand during simulation), less intramolecular hydrogen bonding than ritonavir, which further supports better interaction of VTRRT-13.V2 with SARS-CoV2 main protease. Hence, MDS analysis confirms that VTRRT-13.V2 hybrid antiviral molecule may act as a better inhibitor for SARS-CoV2 main protease.
Figure 7.
Root-mean-square deviation (RMSD) and Root mean square fluctuations (RMSF) result of the SARS-CoV2 main protease-VTRRT-13.V2 complex (A & B), SARS-CoV2 main protease-Ritonavir complex (C and D), and SARS-CoV2 main protease-Remdesivir complex (E and F). The RMSD and RMSF were generated from Molecular dynamics analysis.
Figure 8.
Interacting residues and the interacting fraction of the hybrid molecule VTRRT-13-V2 (A & B), Ritonavir (C & D), and Remdesivir (E and F) during molecular dynamic simulation protein-ligand complex. Types of bonding involved between protein and ligands are shown in different colors.
2.8. Retrosynthetic analysis, combinatorial synthesis, and MDS analysis identified VTRRT-13.V2.1 as a possible inhibitor for SARS-CoV2 main protease
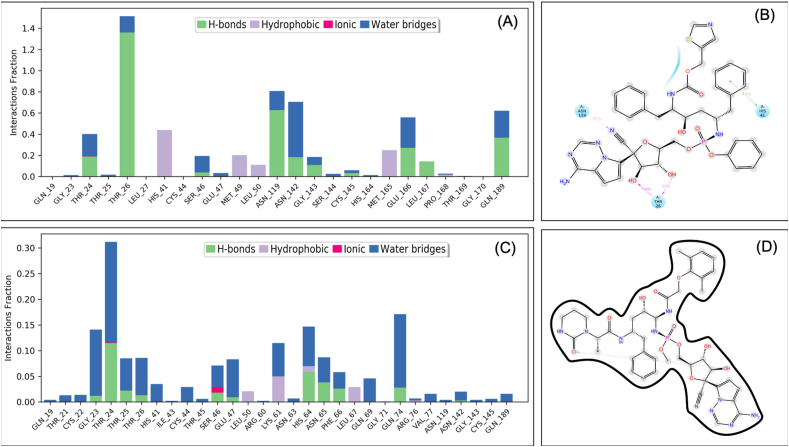
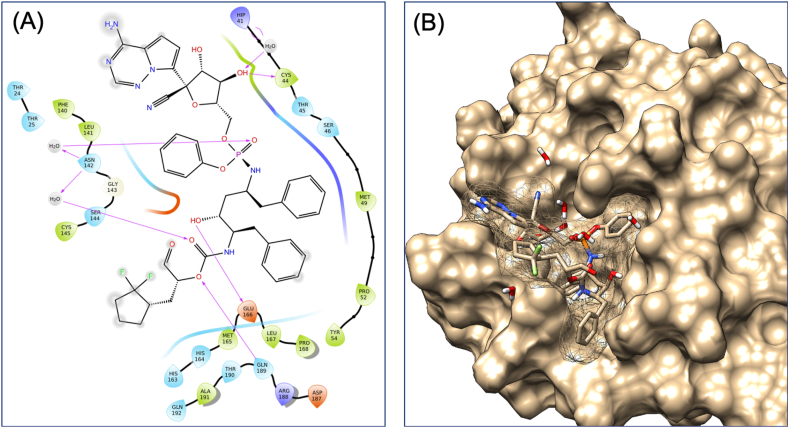
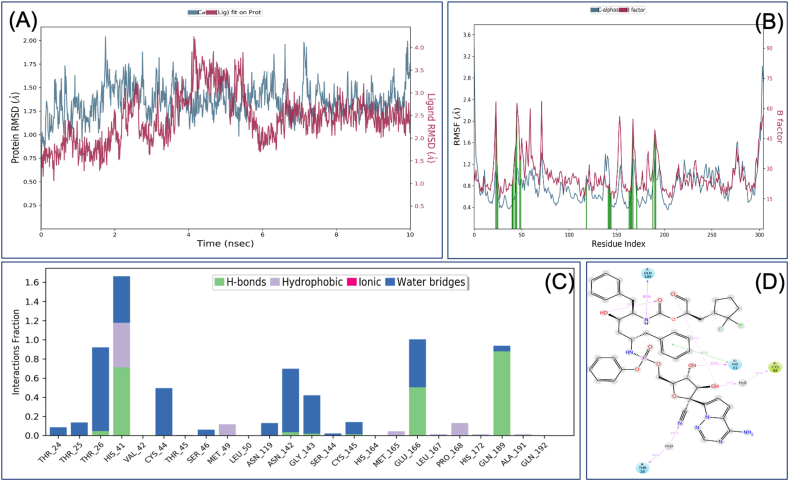
Retrosynthetic analysis and combinatorial synthesis produces synthetically tractable lead-like 1000 products via more than 100 modifications of VTRRT-13.V2. Docking and binding energy calculation of 1000 products with Mpro protein, identified VTRRT-13.V2.1 as best docked molecule (-12.28) with the binding energy of -52.23 kcal/mol (Figure 9). To further confirm the interaction, the MDS analysis of protein-ligand complex was performed, and the result showed that RMSD of protein is < 2.0 Å and ligand in the complex is < 2.5 Å that showed a very stable complex (Figure 10). All the fluctuation seen in the RMSD calculations is within <3Å that showed that this complex is stable. Similarly, RMSF analysis showed that the most part of the protein has the RMSF <2.5Å that showed less fluctuation in protein conformation under complex conditions. Interaction between VTRRT-13.V2.1 and SARS-CoV2 main protease complex involves Thr26, His41, Cys44, Asn142, Gly143, Glu166, Gln189, and at least seven contacts always exist more than 30% simulation time (Figure 10D). It is also noted in MDS analysis that interaction of VT-RRT-13.V2.1 with the Thr26, His41, and Glu166 exists at 100% simulation time. This further confirms its strong interactions between VTRRT-13.V2.1 and SARS-CoV2 main protease. VTRRT-13.V2.1 can be synthesised by curtius-2 and ester-hydrolysis-2 reaction using two reagents i.e 2R,3R,5S)-5-{[(S)-{[(2S,3R,4R,5S)-3-(acetyloxy)-5-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-5-cyano-4-hydroxyoxolan-2-yl]-methoxy}(phenoxy)phosphoryl]amino}-2-benzyl-3-hydroxy-6-phenylhexanoic acid, and 3-(2,2-difluorocyclopentyl) 2-hydroxypropanal.
Figure 9.
Interaction diagram showing interacting amino acid residues (A) and their docking pose (B) in SARS-CoV2 main protease-VTRRT-13.V2.1 complex.
Figure 10.
Root-mean-square deviation (A) and Root mean square fluctuations (B), Interacting residues (C), and the interacting fraction (D) during molecular dynamics simulation analysis of SARS-CoV2 main protease-VTRRT-13.V2.1 complex.
2.9. Designed VTRRT13.V2.1 has favourable ADMET properties
ADMET (absorption, distribution, metabolism, excretion, and toxicity) analysis of VTRRT-13.V2.1 showed that most of its ADMET parameters are in the acceptable limits (Table 4) that further support the designed lead. QPlogPw, QPlogPo/w, QPlogS showed that VTRRT13-V2.1 is water-soluble. It has also seen that it possesses >35% oral absorption that showed its good uptake. The availability of free drugs in blood is essential to cross the membrane and binding to molecular targets. The QPlogKhasa showed that the VTRRT13-V2.1 has very low interaction with plasma proteins.
Table 4.
Result showing the outcome of ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity)analysis of VTRRT13-V2.1. The prediction was performed using QikProp analysis.
| ADMAT Properties | Value for VTRRT13-V2.1 |
|---|---|
| mol MW | 917.901 |
| donorHB | 7.0 |
| accptHB | 20.85 |
| QPlogPw | 32.712 |
| QPlogPo/w | 3.508 |
| QPlogS | -5.266 |
| QPlogBB | -3.526 |
| QPlogKp | -2.466 |
| QPlogKhsa | -0.246 |
| Oral Absorption (%) | 35.376 |
| Rule of Five | 3 |
| QPlogHERG | -7.016 |
| QPPCaco | 31.284 |
2.10. Mutational profiling of binding site residues of SARS-CoV2 main protease
Mutational sensitivity analysis of the binding site residues (Thr26, His41, Cys44, Asn142, Gly143, Glu166, and Gln189) of Mpro involved in the interaction with VTRRT13-V2.1 showed ΔΔGpred values close to zero that suggest less chance of mutations at these sites. Similarly, analysis of modifications (replacements) in CoV-GLUE database showed no mutation hot spot involving these binding residues and only single or double mutation present in some residues i.e. Thr26 (No mutation), His41 (H41P), Cys44 (No mutations), Asn142 (N142L/S), Gly143(Q143S), Glu166 (Q166K) and Gln189 (Q189K). Hence viral protease has less change for escape mutation that further enhances the significance of the designed inhibitor.
2.11. Interaction of VTRRT13-V2.1 with the main protease of cross-species i.e SARS-CoV1 and MERS-CoV
To further evaluate the interaction of VTRRT13-V2.1 with cross species protease like the main protease of SARS-CoV1 and MERS-CoV, docking and MMGBSA analysis were performed. The result showed the lead inhibitor docked to both SARS-CoV1 (-6.5 kcal/mol) and MERS-CoV (-8.5 kcal/mol). It also showed favourable binding energy with SARS-CoV1(-56.13 kcal/mol) but unfavourable binding energy with MERS-CoV (10.07 kcal/mol). This suggests VTRRT13-V2.1 can also bind to SARS-CoV1 but not to MERS-CoV.
2.12. VTRRT13-V2.1 has no human off-targets and no cytotoxicity in the cell line
Presence of lead's off-targets in human may reduce the efficacy and causes cytotoxicity. Hence, human off targets and cytotoxicity of VTRRT13.V2.1 in cell lines were predicted. The result showed that no human off-targets except arachidonate 5-lipooxygeanse (with less probability i.e 0.064). Similarly, the predicted cytotoxicity of the VTRRT13-V2.1 has been demonstrated that it is non-cytotoxic to tumor and non-tumor cell lines (at Pa> 0.7). This further enhances the possibility of the development of VTRRT13.V2.1 as a possible inhibitor against the main protease of SARS-CoV2.
3. Discussion
The emergence of COVID-19 caused by SARS-CoV2 has caused global pandemics. Currently, there is no FDA approved treatment for infection caused by SARS-CoV2. Lopinavir, an HIV type 1 Aspartate protease inhibitor, having activity against other coronaviruses like SARS-CoV (Chu et al., 2004). Ritonavir is combined with lopinavir to increase its plasma half-life through the inhibition of Cytochrome P450. The result showed that lopinavir-ritonavir with ribavirin and interferon alfa resulted in survival in MERS-CoV infection (Kim et al., 2016). Remdesivir (GS-5734) is a broad-spectrum antiviral nucleotide prodrug with potent antiviral activity against diverse RNA viruses like SARS-CoV, MERS-CoV (Sheahan et al., 2017). Ritonavir, lopinavir, and remdesivir has been used to control other coronaviruses (Sheahan et al., 2020). Remdesivir (GS-5734) is currently used with lopinavir to control COVID-19 without knowing its targets in SARS-CoV (Choy et al., 2020). Recent clinical trials showed that broad-spectrum antiviral lopinavir and ritonavir alone were ineffective against COVID-19 (Cao et al., 2020). Hence, in the present study, several hybrid molecules were designed based on the lopinavir, ritonavir, and remdesivir that are FDA approved antiviral molecules.
In the present study, a novel approach is developed to design an inhibitor against SARS-CoV2 main protease by targeting the core pocket of protein before its complete active site. Here, two PDB structure of SARS-CoV2 main protease (5R82, and 6W63) were taken. We have selected these two PDB structures because they only differ in the presence of the ligand molecule, but their SARS-CoV2 main protease is the same. The study started with screening antiviral molecules for a core-binding pocket of 5R82. This PDB structure has a good resolution (RMSD 1.31Å) that may further strengthen the outcome of this study. During analysis of the ligand molecules in the PDB structures, it was found that 5R82 has a 6-(ethylamino)pyridine-3-carbonitrile, which fits deep into the active site hence binding to core pocket of this protein. Docking grid was designed at the binding sites of this ligand. Similarly, 6W63 having an N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide occupy a large part of the active site hence second grid was generated at the binding site of this molecule.
The study aims to design specific hybrid molecules against the core binding pocket (binding site of a ligand in 5R82) that have a better chance to have a good binding affinity with the active site of the SARS-CoV2 main protease with minimum non-specificity. Our in-silico drug design, molecular mechanics, and molecular dynamic simulation analysis identify VTRRT-13 as a lead that has good interaction with the core binding pocket of the active site of SARS-CoV2 main protease. The MDS analysis showed that the terminal atoms of this hybrid inhibitor were unstable and having no interaction with the core binding pocket of the active site of SARS-CoV2 main protease; hence, VTRRT-13.V2 was modified. Further docking analysis and molecular mechanic analysis identifies VTRRT-13.V2 as a lead that has good interaction with the core-binding pocket of the active site of SARS-CoV2 main protease.
Further, VTRRT-13.V2, VTRRT-13, VTRL-12, VTRL-6, lopinavir, ritonavir, and remdesivir were taken and docked them into the complete binding site of the SARS-CoV2 main protease. It was found that VTRRT-13.V2 has very good interaction with the binding site of SARS-CoV2 main protease. VTRRT-13.V2 is a hybrid of ritonavir and remdesvir; hence these three complexes were analyzed with molecular dynamics simulation. The MDS result showed that VTRRT-13V2 form a stable complex and produces more directional interaction (hydrogen bonding) than ritonavir and remdesivir. This hybrid VTRRT-13V2 is formed from two FDA approved drugs hence have more chance to be active in in-vivo experiments. The VTRRT-13.V2 is chemically 5-[({[(5-amino-4-hydroxy-1,6-diphenylhexan-2-yl)amino](phenoxy)phosphoryl}oxy)methyl] -2-{4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl}-3,4-dihydroxyoxolane-2-carbonitrile.
Lead optimization required the design, synthesis, and profiling of thousands of leads analogs before clinical candidate nomination that takes a longer time. Hence, to further enhance the efficacy of the lead molecule, 1000 analogs of VTRRT-13.V2 were generated using retrosynthetic analysis and combinatorial synthesis. In-silico docking, molecular mechanics, molecular dynamics analysis study, suggested VTRRT-13.V2.1 as a possible inhibitor of the SARS-CoV2 main protease. MDS analysis also showed that VTRRT-13.V2.1 has a very strong interaction with SARS-CoV2 main protease via interaction with Thr26, His41, and Glu166 at 100% simulation time. This molecule is identified as (2R)-1-[(1S)-2,2-difluorocyclopentyl]-3-oxopropan-2-ylN-[(2R,3R,5S)-5-{[(S){[(2S,3R,4R,5S)-5-{4-aminopyrrolo [2,1-f] [1,2,4] triazine-7-yl}-5-cyano-3,4dihydroxyoxolan-2-yl]methoxy} (phenoxy) phosphoryl]amino}-3-hydroxy-1,6-diphenylhexan-2-yl]carbamate.
Comparative analysis of all the results showed that VTRRT-13.V2.1 have the highest docking G-score (-12.28) and best binding energy (-52.23 kcal/mol) as compared to the other hybrid constructs investigated such as VTRRT-13.V2 (-9.47 and -47.36 kcal/mol), VTRRT-13, and their parent molecules like ritonavir, lopinavir, and remdesivir. It was also observed that VTRRT13.V2.1 interacts with Thr26, His41, and Glu166 throughout the simulation time. It is also reported that His41 and Glu166 are also involved in the interaction of this protease with Ketoamide 13a inhibitor (Zhang et al., 2020b). To further evaluate the designed inhibitor, the ADMET analysis was performed that showed most of the ADMET properties are favorable. Mutational escape is one of the mechanisms used by viruses to escape the antiviral therapy hence mutation possibility of binding residues of Mpro involved in the interaction with VTRRT13-V2.1 was investigated. It was observed that there is less chance of mutations at binding residues and not present in mutation hot spots. The designed lead does not have any human off-target and less predicted cytotoxicity. The lead also interacts with the main protease of SARS-CoV1 but not to MERS-CoV that further signifies the use of designed lead.
Therefore, the present In-silico design, molecular mechanics, retrosynthetic analysis, combinatorial synthesis, molecular dynamic simulation-based analysis design a novel hybrid antiviral VTRRT-13.V2.1 against SARS-CoV2 main protease. The designed hybrid molecule needs to be synthesized and experimentally validated with purified SARS-CoV2 main protease, and requires in-vivo validation in SARS-CoV2, animal, and human model before use as therapeutics.
4. Methods
4.1. Retrieval of the structure of SARS-CoV2 main protease and preparation
The PDB structure of SARS-CoV2 main protease is available hence retrieved from RCSB (PDB number 5R82, resolution 1.31Å; PDB 6W63, resolution 2.1Å). Similarly, the SARS-CoV1 main protease (PDB number 5N19, Resolution 1.62), and MERS-CoV main protease (PDB number 5WKK, Resolution 1.55) were also retrieved from RCSB. Ligand interacting residue of the PDB structure was determined using maestro. The ligand was removed, and the unliganded PDB structure was pre-processed by assigning bond orders, adding hydrogens, creating zero-order bonds for metals, creating disulphide bonds, optimised for water orientation and pH, and minimised to 0.3RMSD using OPLS_2005 force field. Receptor grid was generated using the binding residues of the ligand as per our published protocol (Tiwari et al., 2019).
4.2. Designing of the hybrid antiviral molecule
Several hybrid combinations were generated using different parts of ritonavir, lopinavir, and remdesivir. The designed combinations were in-silico predicted for antiviral activity using the PASS online server as per our published method (Tiwari et al., 2018a). The hybrid antiviral molecules were prepared using LigPrep modules of the Schrodinger as per our published protocol (Tiwari et al., 2018b).
4.3. Docking in XP mode
All the prepared ligands were docked to the docking grid of the main protease of SARS-CoV2, SARS-CoV1 and MERS-CoV in XP (Extra Precision) mode as per our published protocol (Verma and Tiwari, 2018).
4.4. Molecular mechanics/generalized born surface area (MM-GBSA) calculations of selected library compounds
To get more accurate interaction, XP selected compounds were further subjected to molecular mechanics with generalized born surface area (MM-GBSA) calculations using Prime module of Schrodinger as per our published methods (Tiwari et al., 2019).
4.5. Molecular dynamics simulation (MDS) analysis
MDS was performed using Desmond modules of the Schrodinger 2019-4 as per published methods (Wright et al., 2020). The protein-ligand complex was prepared and subjected to the system builder using the OPLS3e force field. The system was built for the protein-ligand complex using the TIP3P solvent model; sodium ion was added to make charge neutral, 0.15M NaCl was added to make the system close to the natural system. The simulation was run for 10ns, with 10ps trajectory recording intervals. System energy was set to be 1.2, and the ensemble class used was NPT. The simulation was set to run at 300k at 1.01325bar. The option to relax system before simulation was selected. The simulated system was analysed for the simulation interaction diagram.
4.6. Retrosynthetic analysis and combinatorial synthesis
To produce synthetically tractable lead-like compounds, retrosynthetic analysis, and combinatorial synthesis was performed for 'VTRRT-13.V2′ hybrid antiviral molecule as per published method using PathFinder (Konze et al., 2019). In-silico enumeration provides a more exhaustive exploration of available chemical space than traditional empirical SAR studies. PathFinder can incorporate more than 100 reactions that are required for the molecular scaffolds and drug discovery (Bemis and Murcko, 1996). A total of 100 number of pathways are investigated to produce 10000 products. The top 10% product that are similar to the input molecule VTRRT-13.V2 were selected for further analysis. The selected 1000 products were analysed by docking, molecular mechanics, and molecular dynamics simulations.
4.7. ADMET analysis
Absorption, distribution, metabolism, excretion, and toxicity analysis were carried out for the selected lead VTRRT-13.V2.1 using QikProp analysis as per our published protocol (Tiwari et al., 2020).
4.8. Mutational sensitivity profiling and analysis of experimental mutation data
Mutational sensitivity profiling of the Mpro was performed using the MAESTROweb as per the published method (Laimer et al., 2016). The amino acid sequence of Mpro was further analysed for any mutation which was originally present in the SARS-CoV2 virus using the CoV-GLUE database.
4.9. Identification of human off-targets of designed lead
The human off targets of the designed lead was predicted by Swiss Target Prediction using the published protocol (Tiwari et al., 2018a).
4.10. Prediction of cytotoxicity of designed lead on cell lines
In-silico cytotoxicity screening was performed using online CLC-Pred (Cell line Cytotoxicity Predictor) as per published protocol (Lagunin et al., 2018).
Declarations
Author contribution statement
Vishvanath Tiwari: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The author declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
VT would like to thank the Central University of Rajasthan for providing Schrodinger suite. VT also want to thank Dr. Monalisa Tiwari for proof reading the revised manuscript.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary Table 1
References
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Arabi Y.M., Alothman A., Balkhy H.H., Al-Dawood A., AlJohani S., Al Harbi S., Kojan S., Al Jeraisy M., Deeb A.M., Assiri A.M. Treatment of Middle East Respiratory Syndrome with a combination of lopinavir-ritonavir and interferon-beta1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials. 2018;19:81. doi: 10.1186/s13063-017-2427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis G.W., Murcko M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.T., Yin-Lam Wong A., Kaewpreedee P., Sia S.F., Chen D., Yan Hui K.P., Wing Chu D.K., Wai Chan M.C., Pak-Hang Cheung P., Huang X. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S., Kao R.Y., Poon L.L., Wong C.L., Guan Y. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U.J., Won E.J., Kee S.J., Jung S.I., Jang H.C. Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome. Antivir. Ther. 2016;21:455–459. doi: 10.3851/IMP3002. [DOI] [PubMed] [Google Scholar]
- Konze K.D., Bos P.H., Dahlgren M.K., Leswing K., Tubert-Brohman I., Bortolato A., Robbason B., Abel R., Bhat S. Reaction-based enumeration, active learning, and free energy calculations to rapidly explore synthetically tractable chemical space and optimize potency of cyclin-dependent kinase 2 inhibitors. J. Chem. Inf. Model. 2019;59:3782–3793. doi: 10.1021/acs.jcim.9b00367. [DOI] [PubMed] [Google Scholar]
- Lagunin A.A., Dubovskaja V.I., Rudik A.V., Pogodin P.V., Druzhilovskiy D.S., Gloriozova T.A., Filimonov D.A., Sastry N.G., Poroikov V.V. CLC-Pred: a freely available web-service for in silico prediction of human cell line cytotoxicity for drug-like compounds. PloS One. 2018;13 doi: 10.1371/journal.pone.0191838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimer J., Hiebl-Flach J., Lengauer D., Lackner P. MAESTROweb: a web server for structure-based protein stability prediction. Bioinformatics (Oxford, England) 2016;32:1414–1416. doi: 10.1093/bioinformatics/btv769. [DOI] [PubMed] [Google Scholar]
- Reddy G.S., Ali A., Nalam M.N., Anjum S.G., Cao H., Nathans R.S., Schiffer C.A., Rana T.M. Design and synthesis of HIV-1 protease inhibitors incorporating oxazolidinones as P2/P2' ligands in pseudosymmetric dipeptide isosteres. J. Med. Chem. 2007;50:4316–4328. doi: 10.1021/jm070284z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schafer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanakis N., Tsiodras S., Haagmans B.L., Raj V.S., Pontikis K., Koutsoukou A., Koulouris N.G., Osterhaus A.D., Koopmans M.P., Tsakris A. Virological and serological analysis of a recent Middle East respiratory syndrome coronavirus infection case on a triple combination antiviral regimen. Int. J. Antimicrob. Agents. 2014;44:528–532. doi: 10.1016/j.ijantimicag.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari M., Panwar S., Kothidar A., Tiwari V. Rational targeting of Wzb phosphatase and Wzc kinase interaction inhibits extracellular polysaccharides synthesis and biofilm formation in Acinetobacter baumannii. Carbohydr. Res. 2020;492:108025. doi: 10.1016/j.carres.2020.108025. [DOI] [PubMed] [Google Scholar]
- Tiwari V., Meena K., Tiwari M. Differential anti-microbial secondary metabolites in different ESKAPE pathogens explain their adaptation in the hospital setup. Infect. Genet. Evol. 2018;66:57–65. doi: 10.1016/j.meegid.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Tiwari V., Patel V., Tiwari M. In-silico screening and experimental validation reveal L-Adrenaline as anti-biofilm molecule against biofilm-associated protein (Bap) producing Acinetobacter baumannii. Int. J. Biol. Macromol. 2018;107:1242–1252. doi: 10.1016/j.ijbiomac.2017.09.105. [DOI] [PubMed] [Google Scholar]
- Tiwari V., Rajeswari M.R., Tiwari M. Proteomic analysis of iron-regulated membrane proteins identify FhuE receptor as a target to inhibit siderophore-mediated iron acquisition in Acinetobacter baumannii. Int. J. Biol. Macromol. 2019;125:1156–1167. doi: 10.1016/j.ijbiomac.2018.12.173. [DOI] [PubMed] [Google Scholar]
- Verma P., Tiwari V. Targeting outer membrane protein component AdeC for the discovery of efflux pump inhibitor against AdeABC efflux pump of multidrug resistant acinetobacter baumannii. Cell Biochem. Biophys. 2018;76:391–400. doi: 10.1007/s12013-018-0846-5. [DOI] [PubMed] [Google Scholar]
- Wright W.C., Chenge J., Wang J., Girvan H.M., Yang L., Chai S.C., Huber A.D., Wu J., Oladimeji P.O., Munro A.W. Clobetasol propionate is a heme-mediated selective inhibitor of human Cytochrome P450 3A5. J. Med. Chem. 2020;63:1415–1433. doi: 10.1021/acs.jmedchem.9b02067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020 doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, NY) 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1