Abstract
COVID-19 pandemic has underlined the impact of emergent pathogens as a major threat to human health. The development of quantitative approaches to advance comprehension of the current outbreak is urgently needed to tackle this severe disease.
Considering different starting times of infection, mathematical models are proposed to represent SARS-CoV-2 dynamics in infected patients. Based on the target cell limited model, the within-host reproductive number for SARS-CoV-2 is consistent with the broad values of human influenza infection. The best model to fit the data was including immune cell response, which suggests a slow immune response peaking between 5 to 10 days post-onset of symptoms. The model with the eclipse phase, time in a latent phase before becoming productively infected cells, was not supported. Interestingly, model simulations predict that SARS-CoV-2 may replicate very slowly in the first days after infection, and viral load could be below detection levels during the first 4 days post infection.
A quantitative comprehension of SARS-CoV-2 dynamics and the estimation of standard parameters of viral infections is the key contribution of this pioneering work. These models can serve for future evaluation of control theoretical approaches to tailor new drugs against COVID-19.
Keywords: COVID-19, SARS-CoV-2, Mathematical Modelling, Viral Kinetics, Within-Host, Immune responses
1. INTRODUCTION
Epidemics by infectious pathogens are a major threat to humankind. The year 2020 has uncovered one of the biggest pandemics in history, the novel coronavirus SARS-CoV-2 that was first reported in Wuhan, Hubei Province, China in December 2019 (CDC, 2020). While China did a large effort to shrink the outbreak, COVID-19 developed into a pandemic in more than 210 countries moving the epicentre from China to Europe and consequently to America(CDC, 2020). Several countries are planning to relax the strict social distancing regulations. Nevertheless, epidemic rebound risks are latent(Lopez, Rodo, 2020, Ricardo-Azanza, Vargas-Hernandez, 2020).
Coronaviruses are found in different species of animals (e.g. bats and camels) and can evolve to infect humans by droplets from coughing or sneezing. In February 2003, the Severe Acute Respiratory Syndrome (SARS-CoV) was reported in Asia resulting in 8422 cases with a case-fatality rate of 11% (CDC, 2020). Later, in 2012, the Middle East respiratory syndrome (MERS-CoV) was identified in Saudi Arabia with about 2506 cases, killing 862 between 2012 and 2020 (CDC, 2020). Metagenomics studies previous to the COVID-19 outbreak envisaged the possibility of future threats due to the identification of several sequences closely related SARS-like viruses circulating in the Chinese bat populations (He, Zhang, Xu, Yang, Yang, Feng, Xia, Zhou, Zhen, Feng, Guo, Zhang, Tu, 2014, Menachery, Yount, Debbink, Agnihothram, Gralinski, Plante, Graham, Scobey, Ge, Donaldson, Randell, Lanzavecchia, Marasco, Shi, Baric, 2015).
So far, no vaccine or antiviral drug is likely to be available soon. Either monoclonal antibody or vaccine approaches have failed to neutralize and protect from coronavirus infections (Menachery et al., 2015). Therefore, individual behaviour (e.g early self-isolation and social distancing), as well as preventive measures such as hand washing and covering when coughing are critical to control the spread of COVID-19 (Anderson, Heesterbeek, Klinkenberg, Hollingsworth, 2020, Hernandez-Mejia, Hernandez-Vargas, 2020, Prather, Wang, Schooley, 2020). Additionally to these measures, several travel restrictions and quarantines have taken place in many countries around the globe.
Epidemiological models have highlighted that social distancing interventions to mitigate the epidemic is a key aspect (Anderson, Heesterbeek, Klinkenberg, Hollingsworth, 2020, Lopez, Rodo, 2020, Hernandez-Mejia, Hernandez-Vargas, 2020, Ricardo-Azanza, Vargas-Hernandez, 2020). Nevertheless, there are many epidemiological unknowns with the COVID-19 pandemic (Anderson et al., 2020). The case fatality rate for COVID-19 is about 0.3-1% (CDC, 2020). However, adjusted estimations by Baud et al. (2020) indicates that the COVID-19 mortality rate could be as high as 20% in Wuhan. In its early stages, the epidemic has doubled in size every 7.4 days (Li et al., 2020). Moreover, the basic reproductive number was estimated to be 2.2 (95% CI, 1.4 to 3.9) (Li et al., 2020). Based on the relatively long incubation period for COVID-19, about 5–6 days, (Anderson, Heesterbeek, Klinkenberg, Hollingsworth, 2020, CDC) suggested a considerable pre-symptomatic infectiousness.
While there are many mathematical models developed at epidemiological level for COVID-19 to discuss the transmission of SARS-CoV-2 and de-confinement strategies (Anderson, Heesterbeek, Klinkenberg, Hollingsworth, 2020, Ferretti, Wymant, Kendall, Zhao, Nurtay, Abeler-Dörner, Parker, Bonsall, Fraser, 2020, Lopez, Rodo, 2020, Hernandez-Mejia, Hernandez-Vargas, 2020, Peng, Yang, Zhang, Zhuge, & Hong, Ricardo-Azanza, Vargas-Hernandez, 2020), there are too few models at within-host level to understand SARS-CoV-2 replication cycle, interactions with the immune system, and drug effects (Du, Yuan, 2020, Ejima, Kim, Ito, Iwanami, Ohashi, Koizumi, Watashi, Bento, Aihara, Iwami, 2020, Gonçalves, Bertrand, Ke, Comets, de Lamballerie, Malvy, Pizzorno, Terrier, Calatrava, Mentré, Smith, Perelson, Guedj, 2020, Goyal, Cardozo-Ojeda, Schiffer, 2020, Su, Ejima, Ito, Iwanami, Ohashi, 2020, Wang, Heiland, Craig, Davis, Ford Versypt, Jenner, Ozik, Collier, Cockrell, Becker, An, Glazier, Narayanan, Smith, Macklin, 2020). Among different model structures to represent viral dynamics, the target cell limited model has served to represent several diseases such as HIV (Hernandez-Vargas and Middleton (2013); Perelson and Ribeiro (2013); Pinkevych et al. (2016); Rong and à (2009)), Hepatitis (Graw and Perelson (2015); Reluga, Dahari, and Perelson (2009)), Ebola (Nguyen, Binder, Boianelli, Meyer-Hermann, and Hernandez-Vargas (2015); Nguyen and Hernandez-Vargas (2017)), influenza (Baccam, Beauchemin, Macken, Hayden, and Perelson (2006); Handel, Longini, and Antia (2007); Hernandez-Vargas et al. (2014b); Pawelek, Dor, Salmeron, and Handel (2016)), among many others. A detailed reference for viral modelling can be found in Hernandez-Vargas (2019). Very recent data from infected patients with COVID-19 has enlightened the within-host viral dynamics. Zou et al. (2020) presented the viral load in nasal and throat swabs of symptomatic patients. Interestingly, SARS-CoV-2 replication cycles may last longer than flu, about 10 days or more after the incubation period (Anderson, Heesterbeek, Klinkenberg, Hollingsworth, 2020, Zou, Ruan, Huang, Liang, Huang, Hong, Yu, Kang, Song, Xia, Guo, Song, He, Yen, Peiris, Wu, 2020). Here, we contribute to the mathematical study of SARS-CoV-2 dynamics at the within-host level based on data presented by Wölfel et al. (2020).
2. MODELLING SARS-CoV-2 DYNAMICS IN THE HOST
Using ordinary differential equations (ODEs), different mathematical models are presented to adjust the viral kinetics reported by Wölfel et al. (2020) in patients with COVID-19. ODEs are solved using the MATLAB library ode45, which is considered for solving non-stiff differential equations (Mathworks, 2020).
Note that the viral load (Wölfel et al., 2020) was sampled from throat swab cultures and measured in copies/ml, g Swab, at Log10 scale. The clinical data set of 9 individuals is from Wölfel et al. (2020). Due to close contact with index cases and an initial diagnostic test before admission, patients were hospitalized in Munich (Wölfel et al. (2020)). Viral load kinetics were reported in copies/ml per the whole swab for 9 individual cases. All samples were taken about 2 to 4 days post symptoms. Further details can be found in Wölfel et al. (2020).
Parameter identification aims to estimate model parameters based on minimizing the error between model prediction and experimental data. The viral load is measured in Log10 scales, thus, parameter fitting is performed using the following cost function:
Definition 1. Root Mean Squared Logarithmic Error (RMSLE)
This is the difference on Log10 scales between the model output (), and the experimental measurement (yi). This writes as follows
(1) where n is the number of measurements.
Turn out that the minimization of a cost function implies a nonlinear optimization problem, complex, with several variables, and multiple minima. This complexity can be tackled using evolutionary optimization algorithms such as the Differential evolution (DE) algorithm (Storn & Price, 1997). Note that several optimization solvers were considered in previous modeling work (Hernandez-Vargas et al., 2014b), including both deterministic (fmincon Matlab routine) and stochastic (e.g Genetic and Annealing algorithm) methods. Simulation results revealed that the DE global optimization algorithm is robust to initial guesses of parameters than other mentioned methods (Torres-Cerna, Alanis, Poblete-Castro, Bermejo-Jambrina, & Hernandez-vargas, 2016).
Note that multiple models can provide the same fit with observed experimental data. Thus, it becomes necessary to choose between different models. The standard approach to model selection is first estimate all model parameters from the data, then select the model with the best-fit error and some penalties on model complexity. A very used model selection criteria is defined next.
Definition 2. Akaike information criterion AIC
The corrected (AIC) writes as follows:
(2) where N is the number of data points, M is the number of unknown parameters and RSS is the residual sum of squares obtained from the fitting routine. AIC is used here to compare the goodness-of-fit for models that evaluate different hypotheses (Burnham & Anderson, 2002). A lower AIC value means that a given model describes the data better than other models with higher AIC values. Small differences in AIC scores (e.g. < 2) are not significant (Burnham & Anderson, 2002).
Redundant parametrization in models provides difficulties to estimate the parameters uniquely. Ambiguous parameters θsub ⊂ θ may be varied without changing the output y resulting in constant values for the cost function to minimize e.g residuals. This is particularly important for biological systems, where a large variability is presented from one host to another host, limiting the prediction value of mathematical models and estimated parameters (Xia & Moog, 2003).
Definition 3. Identifiability Xia & Moog, 2003 —
A mathematical model is identifiable if θ can be uniquely determined from the measurable output y(t); otherwise, the system is unidentifiable.
Note that a mathematical model is algebraically identifiable may still be practically non-identifiable if the amount and quality of the data are insufficient and the data manifest large deviations. The computational approach by Raue et al. (2009) exploits the profile likelihood to determine identifiability and is considered here. This methodology can detect both structurally and practically non-identifiable parameters (Hernandez-Vargas, 2019).
2.1. Target cell limited model for SARS-CoV-2
The mathematical model used here to represent SARS-CoV-2 dynamics is based on the target cell-limited model (Ciupe, Heffernan, 2017, Hernandez-Vargas, 2019, Perelson, 2002), which writes as follows:
| (3) |
| (4) |
| (5) |
Equation (3) represents the dynamics of susceptible cells (U) while the equation (4) represents the dynamics of infected cells (I). Note that SARS-CoV-2 can replicate in a variety of cell types, including epithelial cells. In fact, SARS-CoV-2 infection may promote the induction of endotheliitis in several organs as a direct consequence of viral involvement (Varga et al., 2020).
Viral dynamics are represented by (5). Viral particles (V) infect susceptible cells with a rate β ((copies/ml)1 day1). Once cells are productively infected, they release virus at a rate p (copies/ml day1 cell1) and virus particles are cleared with a rate c (day1). Infected cells are cleared at rate δ (day1) as a consequence of cytopathic viral effects and immune responses.
It is still debatable in the literature which compartments SARS-CoV-2 can infect, however, there is a common agreement that the infection mainly takes place in respiratory epithelial cells (Tyrrell & Myint, 1996). Previous mathematical modelling work for human influenza infection has computed the number of target cells in an adult, U(0), is about 4 × 108 cells (Baccam et al., 2006). Initial values for infected cells (I(0)) are taken as zero. Note that V(0) cannot be measured as it is below detectable levels (about 100 copies/ml) (Wölfel et al., 2020). Previous modelling work has suggested using half of the detection levels (less than 50 copies/ml) (Hernandez-Vargas, Wilk, Canini, Toapanta, Binder, Uvarovskii, Ross, Guzman, Perelson, Meyer-Hermann, 2014b, Thiebaut, Guedj, Jacqmin-Gadda, Chene, Trimoulet, Neau, Commenges, 2006). Here, using a regression model, the initial viral concentration V(0) was estimated to be about 0.31 copies/ml.
Remark 1
SARS-CoV-2 kinetics are measured after the onset of symptoms (Wölfel et al., 2020). However, it is unknown when the initial infection took place. Patients infected with MERS-CoV in Oh et al. (2016) showed that the virus peaked during the second week of illness, which indicated that the median incubation period was 7 days (range, 2 to 14) (Oh et al., 2016). The incubation period for SARS-CoV-2 has a median time of 2-5 days from exposure to symptoms onset (Lauer et al., 2020). Therefore, for simulation purposes, we explore different scenarios of initial infection day (ti), that is, -7, -3 days before the onset of symptoms.
Definition 4
Infectivity. This is the ability of a pathogen to establish an infection (Diekmann, Heesterbeek, & Metz, 1990). To quantify infectivity, the within-host reproductive number (R 0) is employed to compute the expected number of secondary infections produced by an infected cell (Heffernan, Smith, & Wahl, 2005). When R 0 < 1, one infected cell would infect less than one cell. Thus, the infection would be cleared from the population. Otherwise, if R 0 > 1, the pathogen would be able to invade the target cell population. For the model (3)-(5), the reproductive number is
(6)
Remark 2
Using only the viral titters for parameter fitting procedures in the target cell limited model would result in identifiability problems (Nguyen, Klawonn, Mikolajczyk, & Hernandez-Vargas, 2016). That is, parameter values are rescaled and consequently the validity of parameters would be doubtful to extract biological meaning. As the viral clearance is attributed to a process not directly related to the immune system or viral particle per se, the viral clearance parameter (c) is fixed here with previous estimates in humans, e.g approximately 2.4 for influenza and HIV (Baccam, Beauchemin, Macken, Hayden, Perelson, 2006, Hernandez-Vargas, Middleton, 2013).
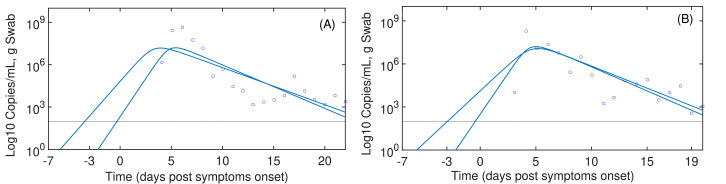
Assuming the day of infection closer to the post symptom onset (pso), day 0, numerical results show high reproductive numbers (R 0) and higher infection rates (β) as presented in Table 1 . Considering the Remark 1, it is assumed the initial day of infection is -7 or -3 pso, therefore, the rate of infection (β) would be slow but associated with a high replication rate (p). Note that individual parameter values should not be interpreted because of identifiability problems. Fig. 1 reveals that viral replication is below detectable levels from 3 to 4 days post-infection. Independently of the starting infection time (ti), numerical results at the Table 1 reveal high reproductive numbers (more than 4.91), implying that SARS-CoV-2 would invade most of the susceptible target cells.
Remark 3
An additional state known as the “Eclipse Phase” has been proposed by Beauchemin et al. (2008); Holder et al. (2011) to represent the time frame of the infection more adequately. Newly infected cells spend time in a latent phase (E) before becoming productively infected cells (I), this can be written as follows:
(7)
(8)
(9)
(10) Cells in the eclipse phase (E) can become productively infected at rate k. For SARS-CoV-2, we found that the eclipse phase model (AIC ≈ 34) does not improve the fitting respect to the target cell model. This can be attributed to identifiability problems as we only have data for the viral titter (Nguyen et al., 2016).
Table 1.
Estimations for the target cell model (3)-(5). The “Mean” arrow represents the average of parameter fitting results to the data sets of 9 patients from Wölfel et al. (2020). The 95% confidence intervals (95% CI) of the parameters is computed based on the fitting results for the 9 patients. The parameter c is fixed in 2.4.
| ti | β | δ | p | R0 | AIC | |
|---|---|---|---|---|---|---|
| -3 | Mean | 4.71 | 1.07 | 3.07 | 22.53 | 11.64 |
| 95% CI | [0.075-21.3] | [0.71-1.91] | [0.2-360] | [9.13-70] | [6.81-20.91] | |
| -7 | Mean | 1.58 | 1.04 | 5.36 | 13.51 | 7.59 |
| 95% CI | [0.03-13.5] | [0.61-2.01] | [0.2-362] | [4.5-45.12] | [4.91-20.8] |
Fig. 1.
Target cell model for SARS-CoV-2. Continuous line are simulation based on the taget cell model (3)-(5). Blue circles represents the data from Wölfel et al. (2020). The most complete data sets to represent the exponential viral growth in Wölfel et al. (2020) were for the patient A in panel (A) and the patient B in panel (B), respectively. Infection time was assumed at -7 and -3 days post symptoms onset.
2.2. Model for SARS-CoV-2 and its immune response
Previous modelling studies have acknowledged the relevance of the immune T-cell response to clear influenza virus (Hancioglu, Swigon, Clermont, 2007, Hernandez-Vargas, Wilk, Canini, Toapanta, Binder, Uvarovskii, Ross, Guzman, Perelson, Meyer-Hermann, 2014b, Lee, Topham, Park, Hollenbaugh, Treanor, Mosmann, Jin, Ward, Miao, Holden-Wiltse, Others, 2009, Miao, Hollenbaugh, Zand, Holden-Wiltse, Mosmann, Perelson, Wu, Topham, 2010, Pawelek, Huynh, Quinlivan, Cullinane, Rong, Perelson, 2012, Saenz, Quinlivan, Elton, MacRae, Blunden, Mumford, Daly, Digard, Cullinane, Grenfell, McCauley, Wood, Gog, 2010). Here, we adapted a minimalistic model derived by Almocera, Nguyen, and Hernandez-Vargas (2018); Boianelli et al. (2015) to represent the interaction between influenza and immune response dynamics. The model assumes that the virus (V) level induces the proliferation of T cells (T) as follows:
| (11) |
| (12) |
Equation (11) refers to SARS-CoV-2 dynamic. Viral replication is modelled with a logistic function with a maximum carrying capacity K and a replication rate p. K is the maximum viral load for each of the patients in Wölfel et al. (2020). The initial viral concentration V(0) is 0.31 copies/ml. The virus is cleared at a rate c, which is considered as in the Remark 2. The term cTVT represents the rate of killing infected cells by the immune response.
Equation (12) represents the T cell response against SARS-CoV-2. T cell homoeostasis is represented by where T(0) is the initial number of T cells and δT is the half life of T cells. It is assumed T(0) is about 106 cells. The steady state condition must be satisfied to guarantee the T cell homeostatic value in the absence of viral infection. The half life of T cells is approximately 4-34 days (McDonagh & Bell, 1995), therefore we take . T cells can proliferate at a rate r. It is assumed that the activation of T cell proliferation by the virus follows a log-sigmoidal form with half saturation constant kT. The coefficient m relates to the width of the sigmoidal function. While different values of m were tested, rendered the best fit.
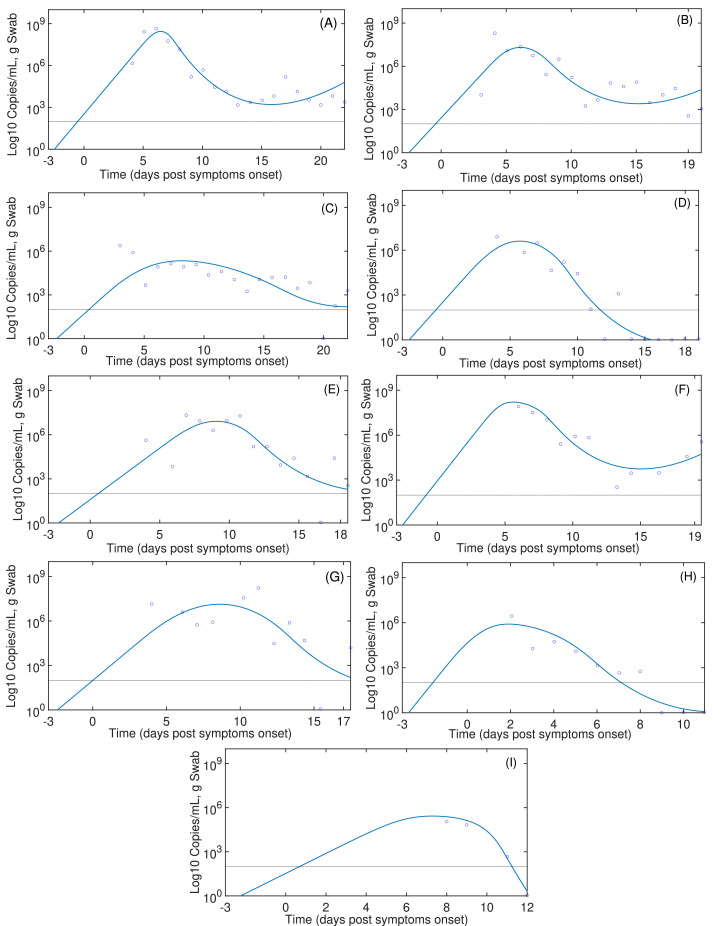
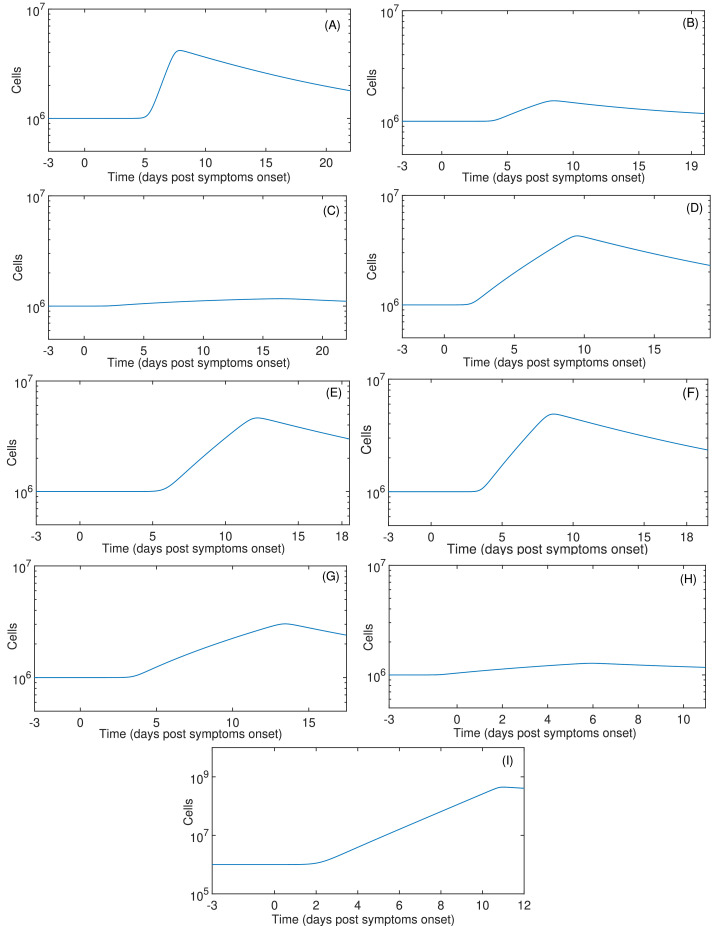
The summary of fitting procedures at dpso is presented in Table 2 , which highlights that the mean of AICs values of the model (11)-(12) are smaller than those presented in the target cell limited model. Results of parameter fitting are portrayed in Fig. 2 . Independently of the starting infection day, the immune response by T cells peaks between 5 to 10 dpso as shown in Fig. 3 . Note that this model represents viral clearance only for patients D, H and I. In the other patients the viral clearance was not reported in the data (Wölfel et al., 2020).
Table 2.
Estimations for model with immune system (11)-(12) using experimental data from Wölfel et al. (2020) assuming and infection time -3 dpso.
| Patient | r | cT | p | kT | AIC |
|---|---|---|---|---|---|
| A | 0.794 | 1.58 | 6.31 | 7.94 × 107 | -1.33 |
| B | 0.126 | 7.94 | 12.58 | 1.99 × 106 | 11.43 |
| C | 0.020 | 1.58 | 19.95 | 1.58 × 103 | 14.28 |
| D | 0.251 | 1.58 | 6.31 | 3.16 × 104 | 7.16 |
| E | 0.316 | 1.00 | 5.01 | 5.01 × 105 | 16.04 |
| F | 0.398 | 1.26 | 6.31 | 1.00 × 107 | 7.07 |
| G | 0.158 | 1.99 | 6.31 | 7.94 × 104 | 24.59 |
| H | 0.050 | 2.52 | 31.62 | 2.52 × 103 | 10.23 |
| I | 0.794 | 1.58 | 3.98 | 1.00 × 103 | -42.49 |
| Mean | 0.194 | 1.89 | 8.57 | 1.26 × 105 | 5.22 |
| 95% CI | [0.05-0.79] | [1-15.8] × 10 | [5.01-12.58 ] | [1.58-2000] × 103 | [-1.33-24.59] |
Fig. 2.
SARS-CoV-2 dynamics with immune responses. Continuous lines are the simulations based on (11)-(12). Blue circles represent the data from Wölfel et al. (2020). Infection time was assumed at -3 days post symptom onset.
Fig. 3.
T cell immune response dynamics. Continuous lines are the simulations based on (11)-(12). Infection time was assumed at -3 days post symptom onset.
3. CONTROL INSIGHTS INTO COVID-19 TREATMENT
The novel coronavirus SARS-CoV-2 first reported in Wuhan in December 2019 has paralysed our societies, leading to self-isolation and quarantine for several days. Indeed, COVID-19 is a major threat to humans, with alarming levels of spread and death tolls, in particular on the elderly. COVID-19 is the first pandemic after the H1N1 ”swine flu” in 2009 (CDC, 2020). While many mathematical models have concentrated on the epidemiological level predicting how SARS-CoV-2 would spread, this paper aims to model SARS-CoV-2 dynamics at the within-host level to quantify SARS-CoV-2 infection kinetics in humans.
Data from Oh et al. (2016) showed that MERS-CoV levels peak during the second week with a median value of 7.21 (log10 copies/mL) in the severe patient group, and about 5.54 (log10 copies/mL) in the mild group. For SARS, the virus peaked at 5.7 (log10 copies/mL) between 7 to 10 days after onset (Peiris et al., 2003). For COVID-19, the viral peak was approximately 8.85 (log10 copies/mL) before 5 dpso (Wölfel et al., 2020). Liu et al. (2020) found that patients with severe disease reported a mean viral load on admission 60 times higher than mild disease cases. Additionally, high viral levels persisted in severe patients for 12 days after onset (Liu et al., 2020).
The reproductive number for human influenza ranges from 3.5 - 75 (Baccam, Beauchemin, Macken, Hayden, Perelson, 2006, Hernandez-Vargas, Colaneri, Middleton, 2014a), which is consistent with the values reported here for COVID-19. Interestingly, both of our models when fitted to the data set of patient A predict that the virus could replicate below detection levels for the first 4 dpi. This may explain why infected patients with SARS-CoV-2 would take from 2-14 dpi to exhibit symptoms.
The model with immune system (Fig. 3) highlights that the T cell response is slowly mounted against SARS-CoV-2 (Anderson et al., 2020). Thus, the slow T cell response may promote low inflammation levels during the first days post infection (Hernandez-Vargas et al., 2014a), which might be a reason to the observations during COVID-19 pandemic of the detrimental outcome on French patients that used non-steroidal anti-inflammatory drugs (NADs) such as ibuprofen. However, so far, there is not any conclusive clinical evidence on the adverse effects by NADs on SARS-CoV-2 infected patients.
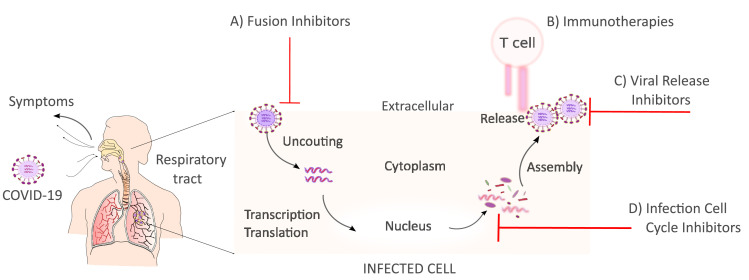
Accelerating therapeutic and prophylactic medication against SARS-CoV-2 is unprecedented. Among the different medical strategies, antivirals are central in inhibiting one or several parts of the viral cycle, see Fig. 4 . Network proximity analyses of drug targets by Zhou et al. (2020) prioritized 16 potential anti-SARS-CoV-2 drugs among them melatonin, mercaptopurine, and sirolimus. Additionally, Zhou et al. (2020) identified three potential drug combinations (e.g sirolimus plus dactinomycin, mercaptopurine plus melatonin, and toremifene plus emodin). Based on a rhesus macaque model of MERS-CoV infection, (de Wit et al., 2020) revealed that early remdesivir treatment may provide a clear clinical benefit with a decrease virus replication in the lungs.
Fig. 4.
SARS-CoV-2 Replication Cycle. After the binding to receptors of the host cell, the virus RNA is uncoated in the cytoplasm. Then, transcription/translation processes take place to generate new viral RNA material and proteins. Virus assembly occurs within vesicles followed by virus release. Once the virus is released can infect other cells.
Several stages would take place to optimize protocols for the different drugs pointed out against SARS-CoV-2. Mathematical models can help to evaluate in silico the potential of hypothetical drugs. For instance, mathematical terms to represent antiviral effects in the model (3)-(5) writes as follow
| (13) |
| (14) |
| (15) |
where uh and ur would represent the effect of inhibiting the replication cycle and the release of virus, respectively. In this direction, (Gonçalves et al., 2020) found that in order to reduce the peak viral load by more than 2 logs, drug efficacy needs to be greater than 80% if treatment is administered after symptom onset. In a similar direction, the modelling work by Goyal et al. (2020) predicted that to lower viral area under the curve therapies need to be given before the SARS-CoV-2 viral load peaks.
Additionally, mathematical models have served to investigate the effect of immune therapies such as in vivo neutralization of pro-inflammatory cytokines during secondary streptococcus pneumoniae infection post influenza infection (Sharma-Chawla et al., 2019). In the context of COVID-19, our model with immune response (11)-(12) could integrate a term to represent antiviral effects (ur) as well as immune modulation (um) to promote the proliferation of T cells, this would be represented as follows:
| (16) |
| (17) |
In other viral infections such as Ebola Nguyen and Hernandez-Vargas (2017), monoclonal antibodies (mAbs) played an important role to inhibit viral replication, e.g the term while increasing viral clearance e.g cV. Another way to suppress SARS-CoV-2 replication is when in co-infections with other viruses because of SARS-CoV-2 has a slower growth rate than the other viruses examined in Pinky and Dobrovolny (2020). Our parameters fitted with the data sets from Wölfel et al. (2020) support also the idea of a slow replication by SARS-CoV-2.
The humoral response against SARS-CoV-2 is urgently needed to evaluate the protection to reinfections. A longitudinal study in rhesus monkeys by Bao et al. (2020) uncovered that infected monkeys presented viral replication at 7 days post-infection (dpi). A significant increase of specific IgG was detected at 14, 21 or 28 dpi. Infected monkeys were re-challenged after specific antibody tested positively and symptoms vanished. Monkeys with re-exposure presented no recurrence of COVID-19, highlighting that protection can be presented to subsequent exposures. Regarding antiviral drugs, Remdesivir treatment has shown a good prophylactic effect during the first 24 hours post MERS-CoV infection in a non-human primate model (de Wit et al., 2020). Furthermore, several benefits were reported for treatment if provided during 12 hours post MERS-CoV infection (de Wit et al., 2020). Our study here mainly addressed T cell responses, therefore, future modelling attempts should be directed to establish a more detailed model of antibody production and cross-reaction (Hernandez-Mejia & Hernandez-Vargas, 2020) as well as in silico testing of different antivirals (Hernandez-Mejia, Alanis, Hernandez-Gonzalez, Findeisen, & Hernandez-vargas, 2019).
There are technical limitations in this study that need to be highlighted. The data for SARS-CoV-2 kinetics in Wölfel et al. (2020) is at the onset of symptoms. This is a key aspect that can bias parameter estimation as mathematical models initiate on the day of infection. In fact, we could miss viral dynamics at the onset of symptoms as well as the SARS-CoV-2 viral load peak. For example, from throat samples in Rhesus macaques infected with SARS-CoV-2, two peaks were reported on most animals at 1 and 5 dpi (Shan, 2020).
In a more technical aspect using only viral load on the target cell limited model to estimate parameters may lead to identifiability problems (Miao, Xia, Perelson, Wu, 2011, Nguyen, Hernandez-Vargas, 2015, Nguyen, Klawonn, Mikolajczyk, Hernandez-Vargas, 2016, Xia, 2003). Thus, our parameter values should be taken with caution when parameter quantifications are interpreted to address within-host mechanisms. For the model with the immune system, there is not data confrontation with immune response predictions, thus, new measurements on cytokines and T cell responses would uncover new information.
The race to develop the first vaccine to tackle COVID-19 has started with the first clinical trial just 60 days after the genetic sequence of the virus. Modelling work developed in this paper paves the way for future mathematical models of COVID-19 to reveal prophylactic and therapeutic interventions at multi-scale levels (Almocera, Hernandez-Vargas, 2019, Feng, Cen, Zhao, Velasco-Hernandez, 2015, Feng, Velasco-Hernandez, Tapia-Santos, Leite, 2012, Handel, Rohani, 2015, Nguyen, Mikolajczyk, Hernandez-Vargas, 2018, Parra-Rojas, Nguyen, Hernandez-Mejia, Hernandez-Vargas, 2018). Further insights into immunology and pathogenesis of SARS-CoV-2 will help to improve the outcome of this and future pandemics.
Ultimately, previous modelling efforts here and from others (Gonçalves, Bertrand, Ke, Comets, de Lamballerie, Malvy, Pizzorno, Terrier, Calatrava, Mentré, Smith, Perelson, Guedj, 2020, Goyal, Cardozo-Ojeda, Schiffer, 2020, Pinky, Dobrovolny, 2020) to represent SARS-CoV-2 could be further extended with control theoretical approaches such as optimal control and model predictive control to schedule drug candidate as well as immune modulators. Control theory in cooperation with Pharmacokinetic (PK)/Pharmacodynamic (PD) modelling have served for optimizing therapies in HIV and influenza infection (Chang, Astolfi, 2008, Hernandez-Mejia, Alanis, Hernandez-Gonzalez, Findeisen, Hernandez-vargas, 2019, Rivadeneira, Caicedo, Ferramosca, Gonzalez, 2017, Rivadeneira, Moog, Stan, Brunet, Raffi, Ferré, Costanza, Mhawej, Biafore, Ouattara, Ernst, Fonteneau, Xia, 2014).
CRediT authorship contribution statement
Esteban A. Hernandez-Vargas: Conceptualization, Formal analysis, Writing - original draft. Jorge X. Velasco-Hernandez: Writing - original draft.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
This research was funded by the Universidad Nacional Autonoma de Mexico (UNAM), CONACYT, and the Alfons und Gertrud Kassel-Stiftung. JXVH acknowledges support form grant PAPIIT-UNAM IN115720 and DGAPA-PAPIIT-IV100220.
References
- Almocera A., Nguyen V., Hernandez-Vargas E. Multiscale model within-host and between-host for viral infectious diseases. Journal of Mathematical Biology. 2018;77(4):1035–1057. doi: 10.1007/s00285-018-1241-y. [DOI] [PubMed] [Google Scholar]
- Almocera A.E.S., Hernandez-Vargas E.A. Coupling multiscale within-host dynamics and between-host transmission with recovery (SIR) dynamics. Mathematical Biosciences. 2019;309(August 2018):34–41. doi: 10.1016/j.mbs.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? The Lancet. 2020 doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccam P., Beauchemin C., Macken C.a., Hayden F.G., Perelson A.S. Kinetics of influenza A virus infection in humans. Journal of virology. 2006;80(15):7590–7599. doi: 10.1128/JVI.01623-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J.…Qin C. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.03.13.990226. [DOI] [Google Scholar]; p. 2020.03.13.990226
- Baud D., Qi X., Nielsen-Saines K., Musso D., Pomar L., Favre G. Real estimates of mortality following COVID-19 infection. The Lancet Infectious Diseases. 2020;3099(20):30195. doi: 10.1016/s1473-3099(20)30195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C.A.A., McSharry J.J., Drusano G.L., Nguyen J.T., Went G.T., Ribeiro R.M., Perelson A.S. Modeling amantadine treatment of influenza A virus in vitro. Journal of Theoretical Biology. 2008;254(2):439–451. doi: 10.1016/j.jtbi.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boianelli A., Nguyen V.K., Ebensen T., Schulze K., Wilk E., Sharma N.…Hernandez-Vargas E.A. Modeling Influenza Virus Infection: A Roadmap for Influenza Research. Viruses. 2015;7(10):5274–5304. doi: 10.3390/v7102875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham K.P., Anderson D.R. Springer Science & Business Media; 2002. Model selection and multimodel inference: a practical information-theoretic approach. [Google Scholar]
- CDC (2020). Coronavirus diseases (COVID-2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/.
- Chang H., Astolfi A. Control of HIV Infection Dynamics. IEEE Control Systems Magazine. 2008;28(2):28–39. [Google Scholar]
- Ciupe S.M., Heffernan J.M. In-host modeling. Infectious Disease Modelling. 2017;2(2):188–202. doi: 10.1016/j.idm.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann O., Heesterbeek J.A., Metz J.A. On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. Journal of Mathematical Biology. 1990;28(4):365–382. doi: 10.1007/BF00178324. [DOI] [PubMed] [Google Scholar]
- Du S.Q., Yuan W. Mathematical Modeling of Interaction between Innate and Adaptive Immune Responses in COVID-19 and Implications for Viral Pathogenesis. Journal of Medical Virology. 2020:0–2. doi: 10.1002/jmv.25866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejima K., Kim K.S., Ito Y., Iwanami S., Ohashi H., Koizumi Y.…Iwami S. Inferring Timing of Infection Using Within-host SARS-CoV-2 Infection Dynamics Model: Are ”Imported Cases” Truly Imported? medRxiv. 2020;4297 doi: 10.1101/2020.03.30.20040519. [DOI] [Google Scholar]
- Feng Z., Cen X., Zhao Y., Velasco-Hernandez J.X. Coupled within-host and between-host dynamics and evolution of virulence. Mathematical Biosciences. 2015;270:204–212. doi: 10.1016/j.mbs.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Feng Z., Velasco-Hernandez J., Tapia-Santos B., Leite M.C.A. A model for coupling within-host and between-host dynamics in an infectious disease. Nonlinear Dynamics. 2012;68(3):401–411. doi: 10.1007/s11071-011-0291-0. [DOI] [PubMed] [Google Scholar]
- Ferretti L., Wymant C., Kendall M., Zhao L., Nurtay A., Abeler-Dörner L.…Fraser C. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science (New York, N.Y.) 2020;6936(March):1–13. doi: 10.1126/science.abb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves A., Bertrand J., Ke R., Comets E., de Lamballerie X., Malvy D.…Guedj J. Timing of antiviral treatment initiation is critical to reduce SARS-Cov-2 viral load. medRxiv. 2020 doi: 10.1101/2020.04.04.20047886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A., Cardozo-Ojeda E., Schiffer J.T. Potency and timing of antiviral therapy as determinants of duration of SARS CoV-2 shedding and intensity of inflammatory response. medRxiv. 2020 doi: 10.1101/2020.04.10.20061325. [DOI] [PMC free article] [PubMed] [Google Scholar]; p. 2020.04.10.20061325
- Graw F., Perelson A.S. Modeling Viral Spread. Annual Review of Virology. 2015;(July):1–18. doi: 10.1146/annurev-virology-110615-042249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancioglu B., Swigon D., Clermont G. A dynamical model of human immune response to influenza A virus infection. Journal of Theoretical Biology. 2007;246(1):70–86. doi: 10.1016/j.jtbi.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Handel A., Longini I.M., Antia R. Neuraminidase inhibitor resistance in influenza: Assessing the danger of its generation and spread. PLoS Computational Biology. 2007;3(12):2456–2464. doi: 10.1371/journal.pcbi.0030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel A., Rohani P. Crossing the scale from within-host infection dynamics to between-host transmission fitness: a discussion of current assumptions and knowledge. Philosophical Transactions of the Royal Society B: Biological Sciences. 2015;370(1675):20140302. doi: 10.1098/rstb.2014.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang Y., Xu L., Yang W., Yang F., Feng Y.…Tu C. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. Journal of virology. 2014;88(12):7070–7082. doi: 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan J., Smith R., Wahl L. Perspectives on the basic reproductive ratio. Journal of The Royal Society Interface. 2005;2(4):281–293. doi: 10.1098/rsif.2005.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Mejia G., Alanis A.Y., Hernandez-Gonzalez M., Findeisen R., Hernandez-vargas E.A. Passivity-based Inverse Optimal Impulsive Control for Influenza Treatment in the Host. IEEE Transactions on Control Systems Technology. 2019:1–12. [Google Scholar]
- Hernandez-Mejia G., Hernandez-Vargas E.A. Uncovering antibody cross-reaction dynamics in influenza A infections. Bioinformatics. 2020 doi: 10.1101/2020.01.06.896274. [DOI] [PubMed] [Google Scholar]; p. 2020.01.06.896274
- Hernandez-Mejia G., Hernandez-Vargas E.A. When is SARS-CoV-2 in your shopping list? Mathematical Biosciences. 2020;328:1–7. doi: 10.1016/j.mbs.2020.108434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Vargas E.A. 1st. ELSEVIER Academic Press; London: 2019. Modeling and Control of Infectious Diseases: with MATLAB and R. [Google Scholar]
- Hernandez-Vargas E.A., Colaneri P., Middleton R.H. Switching Strategies to Mitigate HIV Mutation. IEEE Transactions on Control Systems Technology. 2014;(1):1–6. [Google Scholar]
- Hernandez-Vargas E.A., Middleton R.H. Modeling the three stages in HIV infection. Journal of Theoretical Biology. 2013;320:33–40. doi: 10.1016/j.jtbi.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Hernandez-Vargas E.A., Wilk E., Canini L., Toapanta F.R., Binder S.C., Uvarovskii A.…Meyer-Hermann M. Effects of aging on influenza virus infection dynamics. Journal of Virology. 2014;88(8):4123–4131. doi: 10.1128/JVI.03644-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder B.P., Simon P., Liao L.E., Abed Y., Bouhy X., Beauchemin C.A.A., Boivin G. Assessing the in vitro fitness of an oseltamivir-resistant seasonal A/H1N1 influenza strain using a mathematical model. PloS one. 2011;6(3):e14767. doi: 10.1371/journal.pone.0014767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R.…Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Annals of Internal Medicine. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.Y., Topham D.J., Park S.Y., Hollenbaugh J., Treanor J., Mosmann T.R.…Others Simulation and prediction of the adaptive immune response to influenza A virus infection. Journal of virology. 2009;83(14):7151–7165. doi: 10.1128/JVI.00098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y.…Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. New England Journal of Medicine. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]; p. NEJMoa2001316
- Liu Y., Yan L.-M., Wan L., Xiang T.-X., Le A., Liu J.-M.…Zhang W. Viral dynamics in mild and severe cases of COVID-19. The Lancet Infectious Diseases. 2020;0(0) doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez L., Rodo X. Technical Report. 2020. The end of the social confinement in Spain and the COVID-19 re-emergence risk. [DOI] [PubMed] [Google Scholar]
- Mathworks (2020). Solve nonstiff differential equations — medium order method - MATLAB ode45. https://www.mathworks.com/help/matlab/ref/ode45.html.
- McDonagh M., Bell E.B. The survival and turnover of mature and immature CD8 T cells. Immunology. 1995;84(4):514–520. [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A.…Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nature Medicine. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H., Hollenbaugh J.A., Zand M.S., Holden-Wiltse J., Mosmann T.R., Perelson A.S.…Topham D.J. Quantifying the Early Immune Response and Adaptive Immune Response Kinetics in Mice Infected with Influenza A Virus. Journal of Virology. 2010;84(13):6687–6698. doi: 10.1128/JVI.00266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H., Xia X., Perelson A.S., Wu H. On Identifiability of Nonlinear Ode Models and Applications in Viral Dynamics. SIAM review. Society for Industrial and Applied Mathematics. 2011;53(1):3–39. doi: 10.1137/090757009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.K., Binder S.C., Boianelli A., Meyer-Hermann M., Hernandez-Vargas E.A. Ebola virus infection modeling and identifiability problems. Frontiers in Microbiology. 2015;6:1–11. doi: 10.3389/fmicb.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.K., Hernandez-Vargas E.A. Identifiability Challenges in Mathematical Models of Viral Infectious Diseases. IFAC-PapersOnLine. 2015;48(28):2–7. doi: 10.1016/j.ifacol.2015.12.135. [DOI] [Google Scholar]
- Nguyen V.K., Hernandez-Vargas E.A. Windows of opportunity for Ebola virus infection treatment and vaccination. Scientific reports. 2017;7(1):8975. doi: 10.1038/s41598-017-08884-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.K., Klawonn F., Mikolajczyk R., Hernandez-Vargas E.A. Analysis of Practical Identifiability of a Viral Infection Model. PLOS ONE. 2016:e0167568. doi: 10.1371/journal.pone.0167568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.K., Mikolajczyk R., Hernandez-Vargas E.A. High-resolution epidemic simulation using within-host infection and contact data. BMC Public Health. 2018;18(1) doi: 10.1186/s12889-018-5709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.-d., Park W.B., Choe P.G., Choi S.-J., Kim J.-I., Chae J.…Kim N.J. Viral Load Kinetics of MERS Coronavirus Infection. New England Journal of Medicine. 2016;375(13):1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- Parra-Rojas C., Nguyen V.K., Hernandez-Mejia G., Hernandez-Vargas E.A. Neuraminidase inhibitors in influenza treatment and prevention-Is it time to call it a day? Viruses. 2018;10(9):454. doi: 10.3390/v10090454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek K.A., Dor D., Salmeron C., Handel A. Within-host models of high and low pathogenic influenza virus infections: The role of macrophages. PLoS ONE. 2016;11(2):1–16. doi: 10.1371/journal.pone.0150568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek K.A., Huynh G.T., Quinlivan M., Cullinane A., Rong L., Perelson A.S. Modeling Within-Host Dynamics of Influenza Virus Infection Including Immune Responses. PLoS Comput Biol. 2012;8(6):e1002588. doi: 10.1371/journal.pcbi.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M.…HKU/UCH SARS Study Group Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet (London, England) 2003;361(9371):1767–1772. doi: 10.1016/s0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L., Yang, W., Zhang, D., Zhuge, C., & Hong, L. (2020). Epidemic analysis of COVID-19 in China by dynamical modeling,.
- Perelson A.S. Modelling Viral and Immune System Dynamics. Nature Reviews Immunology. 2002;2(1):28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- Perelson A.S., Ribeiro R.M. Modeling the within-host dynamics of HIV infection. BMC Biology. 2013;11(1):96. doi: 10.1186/1741-7007-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkevych M., Kent S.J., Tolstrup M., Lewin S.R., Cooper D.A., Søgaard O.S.…Davenport M.P. Modeling of Experimental Data Supports HIV Reactivation from Latency after Treatment Interruption on Average Once Every 5-8 Days. PLoS Pathogens. 2016;12(8):8–11. doi: 10.1371/journal.ppat.1005740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinky L., Dobrovolny H.M. SARS-CoV-2 coinfections: Could influenza and the common cold be beneficial? Journal of Medical Virology. 2020;(May):1–8. doi: 10.1002/jmv.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather K.A., Wang C.C., Schooley R.T. Reducing transmission of SARS-CoV-2. Science (New York, N.Y.) 2020 doi: 10.1126/science.abc6197. [DOI] [PubMed] [Google Scholar]
- Raue A., Kreutz C., Maiwald T., Bachmann J., Schilling M., Klingmuller U., Timmer J. Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics. 2009;25(15):1923–1929. doi: 10.1093/bioinformatics/btp358. [DOI] [PubMed] [Google Scholar]
- Reluga T.C., Dahari H., Perelson A.S. Analysis if Hepatitis C Virus Infection Models with Hepatocyte Homeostasis. SIAM journal on applied mathematics. 2009;69(4):999–1023. doi: 10.1137/080714579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Azanza C.L., Vargas-Hernandez E.A. The Risk of Lifting COVID-19 Confinement in Mexico. medRxiv. 2020 doi: 10.1101/2020.05.28.20115063. [DOI] [Google Scholar]; p. 2020.05.28.20115063
- Rivadeneira P.S., Caicedo M., Ferramosca A., Gonzalez A.H. 56th ieee conference on decision and control. 2017. Impulsive Zone Model Predictive Control (iZMPC) for Therapeutic Treatments : application to HIV dynamics; pp. 1–6. [Google Scholar]; Melbourne
- Rivadeneira P.S., Moog C.H., Stan G.-B., Brunet C., Raffi F., Ferré V.…Xia X. Mathematical Modeling of HIV Dynamics After Antiretroviral Therapy Initiation: A Review. BioResearch Open Access. 2014;3(5):233–241. doi: 10.1089/biores.2014.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L., Ã A.S.P. Modeling HIV persistence, the latent reservoir, and viral blips. Journal of Theoretical Biology. 2009;260(2):308–331. doi: 10.1016/j.jtbi.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz R.A., Quinlivan M., Elton D., MacRae S., Blunden A.S., Mumford J.A.…Gog J.R. Dynamics of Influenza Virus Infection and Pathology. Journal of Virology. 2010;84(8):3974–3983. doi: 10.1128/JVI.02078-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, C. (2020). Infection with Novel Coronavirus (SARS-CoV- 2) Causes Pneumonia in the Rhesus Macaques Sciences. Preprint at Research Square, (pp. 1–16). [DOI] [PMC free article] [PubMed]
- Sharma-Chawla N., Stegemann-Koniszewski S., Christen H., Boehme J.D., Kershaw O., Schreiber J.…Hernandez-Vargas E.A. In vivo Neutralization of Pro-inflammatory Cytokines During Secondary Streptococcus pneumoniae Infection Post Influenza A Virus Infection. Frontiers in Immunology. 2019;10:1864. doi: 10.3389/fimmu.2019.01864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storn R., Price K. Differential Evolution - A simple and efficient adaptive scheme for global optimization over continuous spaces. Journal of Global Optimization. 1997;11(4):341–359. doi: 10.1023/A:1008202821328. [DOI] [Google Scholar]
- Su K., Ejima K., Ito Y., Iwanami S., Ohashi H. Modelling SARS-CoV-2 Dynamics : Implications for Therapy. medRxiv. 2020 [Google Scholar]
- Thiebaut R., Guedj J., Jacqmin-Gadda H., Chene G., Trimoulet P., Neau D., Commenges D. Estimation of dynamical model parameters taking into account undetectable marker values. BMC medical research methodology. 2006;6(38):1–9. doi: 10.1186/1471-2288-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cerna C.E., Alanis A.Y., Poblete-Castro I., Bermejo-Jambrina M., Hernandez-vargas E.A. A Comparative study of Differential Evolution Algorithms for Parameter Fitting Procedures. IEEE World Congress on Computational Intelligence (WCCI) 2016 doi: 10.1109/CEC.2016.7744385. [DOI] [Google Scholar]; p. In press
- Tyrrell D.A., Myint S.H. University of Texas Medical Branch at Galveston; 1996. Coronaviruses. [PubMed] [Google Scholar]
- Varga Z., Flammer A., Steiger J.P., Haberecker M., Andermatt R., Zinkernagel A.S.…Holger F.R.M. Correspondence Endothelial cell infection and endotheliitis in. The Lancet. 2020;6736(20):19–20. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Heiland R., Craig M., Davis C.L., Ford Versypt A.N., Jenner A.…Macklin P. Rapid community-driven development of a SARS-CoV-2 tissue simulator. bioRxiv. 2020;2 doi: 10.1101/2020.04.02.019075. [DOI] [Google Scholar]; 2020.04.02.019075
- de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T.…Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proceedings of the National Academy of Sciences. 2020:201922083. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A.…Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020:1–10. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xia X. Estimation of HIV/AIDS parameters. Automatica. 2003;39(11):1983–1988. doi: 10.1016/S0005-1098(03)00220-6. [DOI] [Google Scholar]
- Xia X., Moog C. Identifiability of nonlinear systems with application to HIV/AIDS models. IEEE Transactions on Automatic Control. 2003;48(2):330–336. doi: 10.1109/TAC.2002.808494. [DOI] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery. 2020;6(1) doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z.…Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. New England Journal of Medicine. 2020 doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]; p. NEJMc2001737