Abstract
Quorum sensing (QS), a type of chemical communication, allows bacteria to sense and coordinate activities in natural biofilm communities using N-acyl homoserine lactones (AHLs) as one type of signaling molecule. For AHL-based communication to occur, bacteria must produce and recognize the same signals, which activate similar genes in different species. Our current understanding of AHL-QS suggests that signaling between species would arise randomly, which is not probable. We propose that AHL-QS signaling is a mutable and adaptable process, within limits. AHLs are highly-conserved signals, however, their corresponding receptor proteins (LuxR) are highly variable. We suggest that both flexibility and adaptation occur among receptor proteins, allowing for complex signaling networks to develop in biofilms over time.
A Sea of Signals
Quorum sensing (QS) (see Glossary) is a form of chemical communication among bacteria that regulates gene expression and allows microbial communities to respond to environmental changes at the community level ([1,2]; Box 1 and Figure 1). It occurs within and between species of bacteria [3,4] and has also been documented in interkingdom signaling among plants, fungi, and host cells [5,6]. This suggests that a rich chemical language exists between organisms that we largely do not understand.
Box 1. The Genomics of AHL-Mediated QS.
Quorum sensing (QS) is a general term for intercellular signaling systems among microbes that is cell density dependent, and controls gene regulation at a community level. In Vibrio fischeri and many other bacterial species, the LuxI gene produces N-acyl homoserine lactones (AHLs) as the ‘molecule of communication’ or signaling molecule. The LuxI gene is highly conserved (at least 24 known chemical species in nature), which suggests that although diverse, there are a limited number of AHL-type signals that are produced by bacteria. AHL signals are received by the LuxR protein/gene, which is highly variable among species, and is a transcriptional regulator with two domains: one binds to the AHL, and the other, a helix-turn-helix domain, binds to DNA. The LuxR complex binds to a 20 base pair (bp) section of DNA, called the lux box, when it has complexed to a known AHL. This region is in or near the lux promoter region, which is located ~40 bp upstream of the regulated genes. Once the LuxR is bound to the DNA, RNA polymerase is recruited to this promoter region, inducing gene expression (see Figure 2 in main text). In V. fischeri, this process produces bioluminescence, and also increases the production of more LuxI and LuxR proteins.
Figure 1. Quorum Sensing Model in Vibrio fischeri.
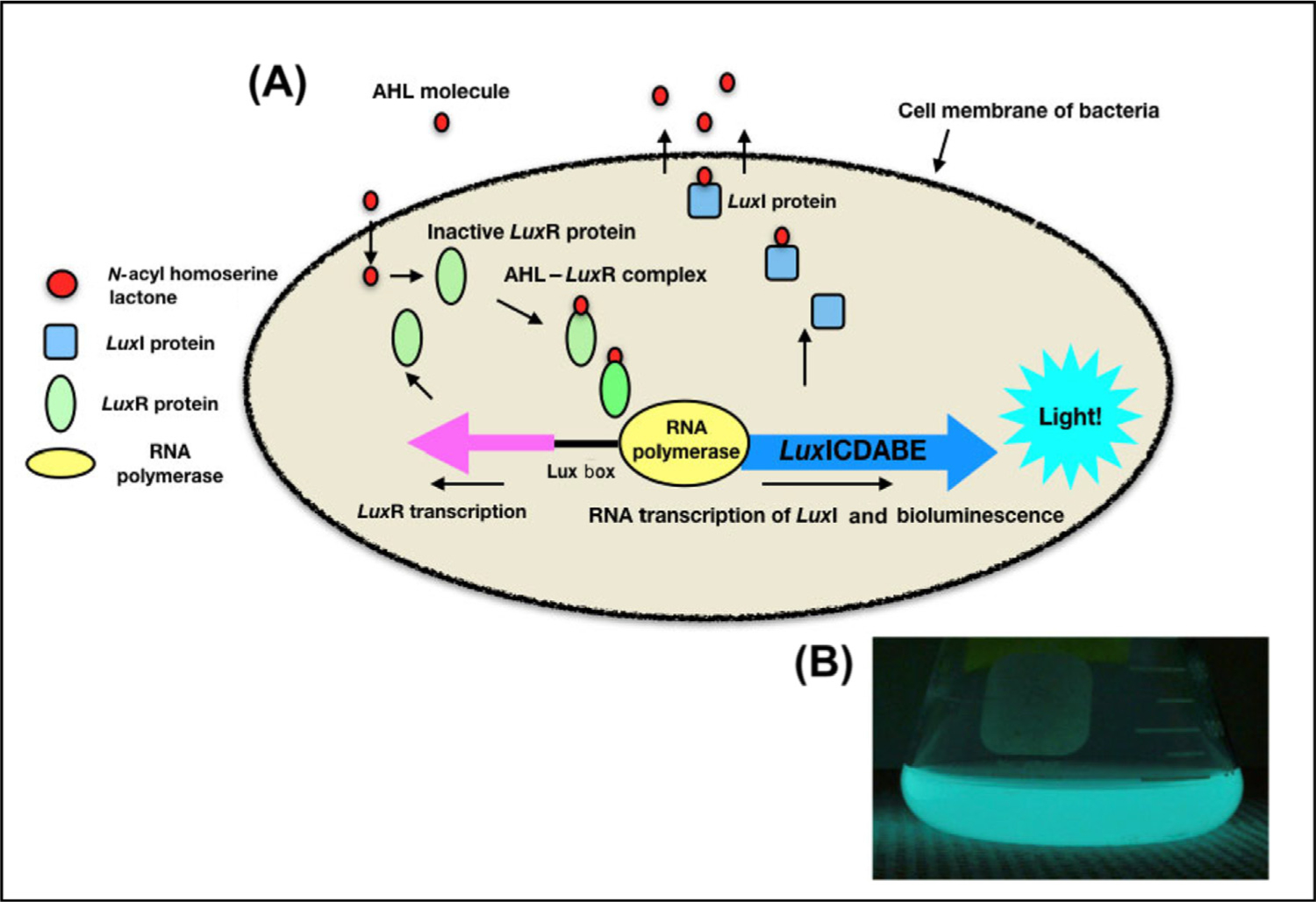
(A) LuxI encodes for the signal of communication, which is N-acyl homoserine lactone synthase or AHL (3-oxo-C6 in this species). This signaling molecule binds to and activates LuxR, a transcriptional regulator protein. The AHL-LuxR protein complex then binds to DNA at an inverted repeat region referred to as the lux box, which is ~40 bp upstream from the transcriptional start site. Working with an RNA polymerase, AHL–LuxR complex stimulates the expression of the downstream genes, luxICDABE, causing bioluminescence (B). At low cell densities, transcription of the luxICDABE operon occurs at basal levels. High cell densities lead to the accumulation of AHLs, which bind and activate the LuxR transcriptional regulator. (B) V. fischeri culture glowing in a beaker due to transcription of bioluminescence luxICDABE operon.
Of the chemical languages that we are aware of, QS that is mediated by N-acyl homoserine lactones (AHLs) as the molecule of communication, are the best studied [7]. AHL-mediated QS (AHL-QS) allows bacteria to synchronize behaviors at high cell densities, particularly within biofilms and microbial mats. It is generally accepted that AHL-QS communicates who and how many neighbors a specific cell has, and information about other cells’ activities and ecological functions [1,8]. Phylogenetic studies of AHL-QS indicate that these systems are ancient and were established early in Earth’s evolutionary history [9,10]. AHL-QS has also been found in a wide variety of microorganisms, including Gram-positive and Gram-negative bacteria as well as Archaea and Cyanobacteria [8,11–14].
Several different molecular forms of AHLs have been documented within microbial mats, as well as within a single species at a given time, creating a sea of signals in nature (Figure 2). For example, AHLs with C4–C18 carbon chains have been found in hypersa-line microbial mats [15,16]. A variety of studies suggest that microorganisms interact using these signals in a variety of ways, including what is called eavesdropping, crosstalk, and self-talk ([2,17–19]; Figure 2). With the high level of social interaction among bacteria, we must ask the question: how do bacteria ‘learn’ to talk to each other in nature?
Figure 2. How Do Bacteria ‘Learn’ to Talk to Each Other?
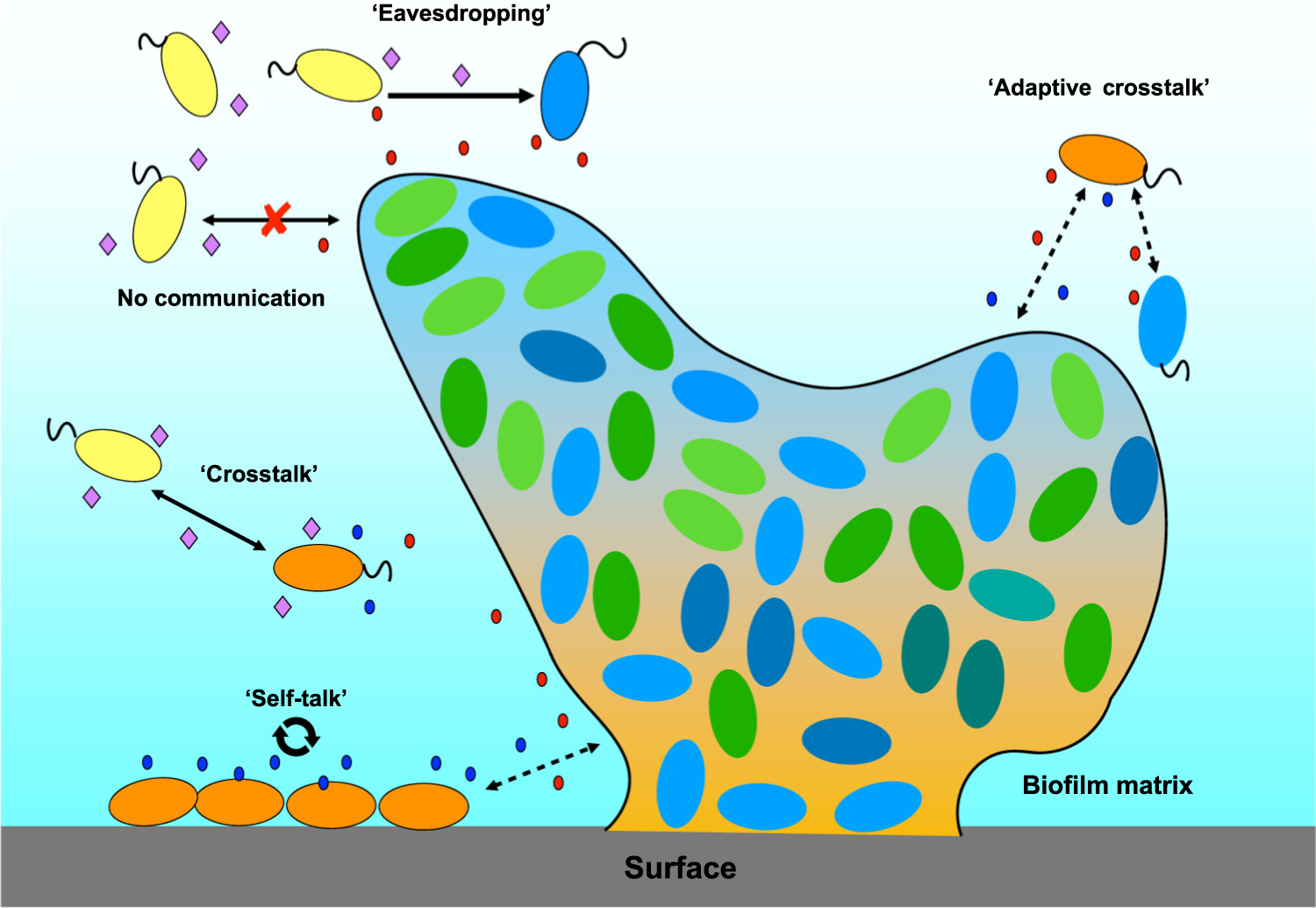
In the figure, a biofilm with blue and green bacterial species is attached to a surface. Cells of other surrounding species produce different signaling molecules [N-acyl homoserine lactone (AHL) types], with a variety of interactions possible. Blue cells can receive signals produced by yellow cells, but yellow cells cannot communicate with blue cells (eavesdropping). Orange and yellow cells can produce and receive some of the same signals (crosstalk), but not all, with orange cells producing one AHL (in blue) that is only perceived by ‘self’ or close kin (self-talk). Over time, orange, blue, and green cells ‘learn’ to perceive the same signals from one another, possibly through exchange of genetic material, promoter movement, or flexibility in the LuxR receptor molecule.
How Do AHL-QS Relationships Develop over Microevolutionary Timescales?
In general, AHL signal molecules are produced by synthase gene LuxI and homologs [1,7], and are perceived by LuxR-type transcriptional regulator protein/gene that detects a specific type of AHL molecule (Box 1 and Figure 1 for more details). LuxI genes are highly conserved [7], suggesting that there are limits to the molecular structure of AHLs. In contrast, LuxR genes are highly variable, with only 18–25% similarity among known LuxR genes [7]. This is supported by a recent bioinformatic study of prokaryotic genomes that found 3550 putative LuxR genes, with 2698 of those being LuxR solos [19]. A second study of only genomes that contained LuxR transcriptional regulators with N-terminal autoinducer-binding domains in the LuxR protein found 4860 LuxR solos [20]. Such high numbers indicate that LuxR receptor genes may be a mutable entity over microevolutionary time frames. Of additional relevance is that 75% of LuxR genes have no known LuxI counterpart within a given genome [19], but many are still able to detect and bind AHL molecules [21–23].
Based on our current understanding of AHL-QS, for bacteria to establish efficient chemical communication with other kinds of bacteria (i.e., crosstalk; Figure 2), two species must: (i) exist in microspatial proximity of each other (~<70 μM [22]), (ii) utilize the same signals, and (iii) activate similar or complementary genes that influence subsequent coordinated activities. If this occurs through stochastic processes, it becomes highly improbable in natural systems. The development of chemical communication between two species in culture has been observed [24] but has not been examined in natural or more complex systems.
We propose that in natural communities, crosstalk through AHL-QS is a process that may evolve among species as the community develops, and is likely a deterministic factor in the development of biofilms over time. This concept was initially suggested by McCusker and colleagues [25] and Holben et al. [26] to understand how biodegradative biofilms improve in efficiency over time. It has also been proposed that as biofilms develop, chemical signaling may attract other cells to a surface to grow [27]. However, the underlying genetic mechanisms were not discussed.
AHL-QS must be an adaptable and flexible (acclimation) process, both at the chemical and genomic levels, for stable ‘crosstalk’ relationships to develop among species. Such communication pathways would provide the capacity to establish new, coordinated interactions as a biofilm community develops and changes over time. Therefore, bacteria must have at least some of the following abilities: (i) detect a wide variety of chemical signals, (ii) incorporate QS circuits from neighbors into their own genomes, (iii) alter which gene sets are activated by QS, and (iv) have some ability to acclimate the conformational changes of the receptor protein (LuxR). The topology of QS gene cassettes, mobilizing genetic elements, and the evolution of LuxR solos may play important roles in this flexible and adaptable system.
Proposed Mechanisms of Adaptability and Flexibility of AHL-QS in Nature
Recent studies have suggested that the bacterial genome is much more flexible than previously thought, with microbes having extremely high and rapid abilities to adapt to wide ranges of environmental variables [28–31]. Understanding microbial flexibility, acclimation, and adaptation is an active area of research, and can be applied to our understanding of AHL-QS. With the current level of knowledge, we list and discuss four mechanisms that may play important roles in the flexibility (or acclimation) and adaptability of AHL-QS in nature, and should be further investigated in QS systems (Figure 3).
Figure 3. Mechanisms of Adaptability and Flexibility of N-Acyl Homoserine Lactone-Mediated Quorum Sensing (AHL-QS) Systems.
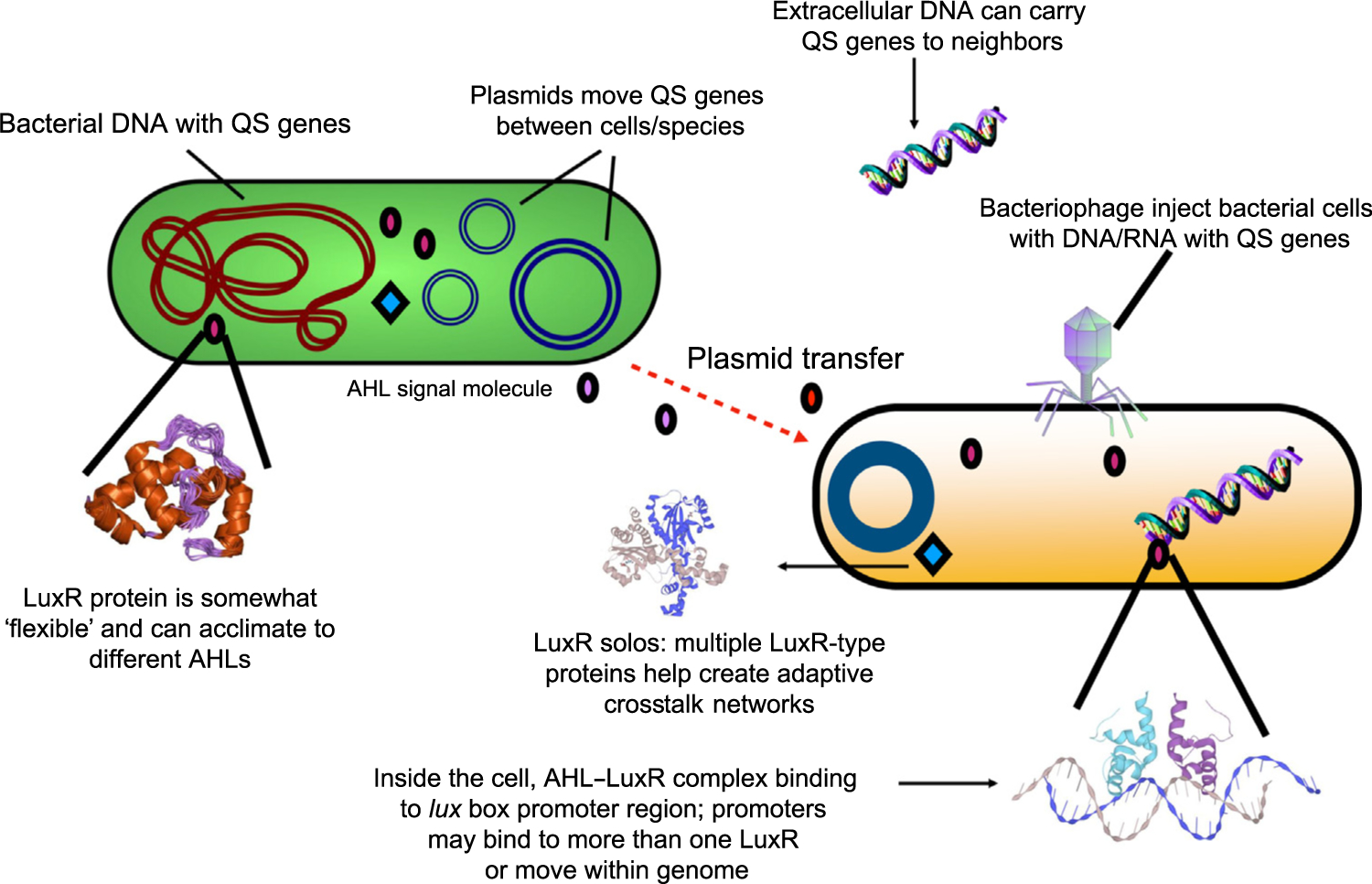
Potential mechanisms of how bacteria ‘learn’ to develop crosstalk. These include mobile genetic elements (GMEs), such as plasmids, extracellular DNA uptake, and bacteriophage infections, among others. LuxR solos are transcriptional regulators without a cognate LuxI gene and may ‘eavesdrop’ on chemical communications of other species. Promoters, where the LuxR–AHL transcriptional regulator complex binds to the DNA to activate transcription, may also play a role in the development of crosstalk networks through the recycling of promoters within a genome (promoter propagation), or promoters that are mobile genetic elements. LuxI/R proteins also have some ability to accept a variety of AHL signals, suggesting some acclimation to new signals may be possible.
Flexibility of LuxI and LuxR Gene/Protein Families
Bacteria typically utilize one or several AHLs, which are produced by the LuxI gene family, to regulate group activities. In addition, many AHL synthases can accept several acyl-ACPs (acyl carrier proteins), which enables bacteria to produce several AHLs from a single AHL synthase. Ortori et al. 2007 [32] found that 24 different AHLs were produced by a single bacterium in response to different culture conditions. However, the LuxI gene family is highly conserved, which limits the structure of AHL protein synthases. A small difference in acyl chain length or a substitution on the chain affects the binding efficiency of an AHL with its cognate LuxR receptor protein, and the ability of the LuxR–AHL complex to activate transcription [33]. It follows that the LuxI gene/protein family is not the most likely source of adaptability in AHL-QS, but it is flexible, allowing for alternations in the AHL type.
Studies have found that the LuxR receptor protein is highly flexible and can bind with multiple AHLs. Lintz and colleagues [34] suggested that signal specificity could be altered by specific residues of LuxR receptor proteins to obtain changes in signal specificity, allowing LuxR proteins to bind with multiple AHLs. Studies have also shown that when AHLs are in receptor protein-binding pockets, their conformation, or overall shape, is not conserved. In addition, LuxR gene/protein families vary slightly in size and can acclimate or adapt to new AHLs over time. Collins et al. [24] illustrated that when laboratory cultures of a bacterium, which normally utilized a 3-oxo-C6-HSL, were exposed over many generations to exogenous C8-AHLs, they developed increased sensitivity and gene activation for the new AHL. Analyses indicated that only small changes in LuxR proteins were needed to allow the bacterium to adapt to the C8-AHL [24], and later a C4-AHL [33]. White and Winans [35] and Weingart et al. [36] revealed that substitutions or deletions of an amino acid at specific locations on TraR, a LuxR homolog, were key to directing changes in the binding of the AHL to TraR and in binding of the TraR–AHL complex to the promoter site on the DNA. Such flexibility and adaptability in the LuxR protein would allow for rapid development of crosstalk networks between species in newly developing biofilms. Hence, it has been suggested that there may be an evolutionary advantage to maintaining plasticity in AHL-QS systems [24].
LuxR proteins are transcriptional regulators, and when the molecule is combined with a given AHL, it attaches to a lux box-type motif within the promoter region, thereby activating transcription of downstream genes. With the high variation in LuxR proteins and its ability to bind with a variety of AHL molecules, LuxR may be able to attach to more than one promotor region or multiple Lux box-type motifs within a genome. This would allow for variation in the genes that are activated by LuxR as well as account for gene expression changes induced by environmentally-altered AHL molecules, or new signals produced by new neighbors.
LuxR Solos
Many genomes have LuxI/R pairs, with the LuxI and LuxR cognates near each other in the genome [1,19,37]. Such AHL-QS circuits are the best studied AHL-QS systems, and may represent true cases of autoinducing, where the signal and the receptor are produced by the same cells and trigger gene expression through ‘self-talk’ (Figure 2). However, additional studies have found that many genomes also contain one or more LuxR solos [19,20,37,38] which are not paired with a nearby LuxI gene. Within a given genome there may be one or multiple LuxR solos with a wide variety of functions [19,20,37].
LuxR solos are involved in gene regulation of virulence, plant growth promotion, nodulation, motility, plasmid transfer, and antibiotic synthesis [19,37]. Some LuxR solos occur in species that do not appear to produce AHLs, but they may detect AHLs with LuxR solos [39,40]. These are a likely source of adaptation and development of ‘crosstalk’ within microbial communities, allowing a cell to ‘eavesdrop’ on new signals (Figure 2). It is also possible that LuxR solos recognize AHLs that are altered by geochemical and photochemical processes during transit between cells. If a signal is altered by the external environment (e.g., oxidation to a 3-oxo form), there is the potential for the signal to have a role as an environmental sensor for the receiving cells [41]. Also, AHLs typically contain an even number of carbon atoms on acyl chains, which result from fatty acid building blocks [32]. Cleavage of the acyl moiety, by acylases, on longer chain (e.g., C12-, C14-) AHLs to shorter chain forms (e.g., C4- to C8-AHL) could, in part, explain the presence of odd-numbered acyl chains (e.g., C7-AHL) that are sometimes abundant in natural samples [15]. Since geochemical and/or enzymatic alterations may be more pronounced in a multispecies biofilm, the altered AHLs, if perceived by cells, could ultimately act as a sensor (e.g., for acylases).
LuxR solos are common in Proteobacteria, but also occur in Archaea and Gram-positive bacteria genomes [38,42]. Subramoni et al. 2015 [20] found that a large number of plant-associated and environmental isolates carried multiple LuxR solos, and that solos were commonly adjacent to transposases and pseudogenes. Hudaiberdiev et al. 2015 [19] found that DNA mobilization elements were common near LuxR solos in bacterial genomes. Such findings suggest that LuxR solos are involved in the establishment of new communication networks, and they may be related to the evolutionary development of long-term communication through cognate LuxI/R pairs seen in mutualistic relationships.
The lux Box: Variability in the Promoter
As stated, the region of DNA where AHL–LuxR complex binds is the lux box-type motif, which consists of an inverted, repeat region within the promoter region. In classical AHL-QS systems of Vibrio spp. lux box-like regions occur upstream of transcriptional start sites (~40 nt) [43–45], and have some similar structure and sequence across species ([7] and references therein). As more AHL-QS circuits are defined however, we learn that AHL-QS regulatory networks vary and are diverse, including the lux box.
The lux box in at least some species are ‘relaxed’ and can bind with more than one LuxR-type regulator, as observed in Pseudomonas aeruginosa [45]. This would provide flexibility and the potential for acclimation to new signals from neighboring species. There are also a variety of AHL-QS gene topologies [46], with promoters flanking both sides of the DNA-binding sites in some genomes. This may allow for a variety of genes to be expressed or repressed, depending on other factors, like secondary transcriptional factors, that may be required to activate AHL-QS circuits.
Another possibility is that of a LuxR-type promoter region representing a ‘generalized promotor.’ In a study of putative mobile promoters, Matus-Garcia et al. [30] found that the promotor regions of eight, nonhomologous coding sequences in bacterial and Archaeal genomes were highly conserved (80% similar), and suggested that bacteria have ‘recyclable’ promoters to avoid evolution of a promoter de novo. Other studies have also concluded that inactive genes (sometimes passed onto neighboring genomes) evolve expression through existing promoters which are copied upstream of the gene where expression is needed [47,48]. This typically occurs via genomic rearrangements or transposable elements that contain active promoters. In this evolutionary concept, it is thought that the promotor and downstream genes would not move together through horizontal gene transfer (HGT), as transcription of a new gene could be lethal for a given cell. However, ‘transcriptional rewiring’ (i.e., making a new gene controlled by a ‘recycled’ promoter), through mobilization of partial or complete promotors by transposable elements, particularly from elsewhere in the genome, would avoid the possibility of a lethal gene being transcribed [30]. Within AHL-QS, such a system would also allow for new communication networks to develop between species. A generalized promoter containing a lux box-like sequence would also allow flexibility and adaptation of different or new LuxR transcriptional factors to bind to multiple promotor sequences. In general, promotors evolve at much faster rates than predicted, and can even quickly evolve de novo from random sequences, suggesting that promoter sequences in bacterial genomes are highly flexible [49].
LuxI/R as Genomic Mobile Elements (GMEs)
There is sufficient evidence to suggest that AHL-QS genes are often associated with genomic mobile elements (GMEs), and that GMEs are involved in the spread of AHL-QS communication networks [5,6,20,31]. This, perhaps more than others, would allow bacteria to ‘learn’ another species’ language through transfer of GMEs. Such features in the genomes of microbes vary widely, and include plasmids, transposons, insertion sequences, and others [31]. These features are the source of HGT and create highly dynamic genomes in microorganisms.
Research of LuxR genes in bacterial genomes has found that QS genes in a given local arrangement (topology or gene cassette) are orthologs with respect to each other, and paralogs with respect to LuxR genes in different topological arrangements. These cassettes are more similar to each other than other QS genes within the same genome [19]. This suggests that QS circuits may move through HGT. Other studies have shown that LuxR solos are mobile elements that occur both on chromosomal DNA and plasmids [20]. In addition, LuxI/R circuits commonly have genes associated with DNA mobilization near them [19], and may suggest that these genes are mobile elements, particularly if DNA mobilization genes are upstream of LuxI/R systems. GMEs may also allow for LuxR genes to be transferred into another species’ plasmid or genome, inserting them in particular locations within the genome to activate specific downstream genes, creating a community-level response through community-level gene expression.
Another potential source of GMEs and HGT of AHL-QS systems is bacteriophage. QS genes have been found in the bacteriophage phiCDHM1, which infects Clostridium difficile, from QS systems that use peptides rather than AHLs [50]. Phage phiCDHM1 has homologs of the accessory gene regulator (agr) QS system, and includes an autoinducing peptide (AIP); it is proposed that these genes were horizontally transferred from the host bacteria [50]. Evolutionary relationships between phage and QS systems may exist but are not well understood, and studies of AHL-QS systems and phage dynamics should be a priority for future research.
Concluding Remarks and Future Directions
Microbial species interact with each other in different manners, from competition to mutualism, and they exhibit social behavior. QS systems are likely involved in all of these interactions. Bacteria must be able to adapt to the chemical language of other species in their proximity and adjust which gene sets are activated. Bacteria, therefore, must have the genetic tools to continuously adjust to new community members and new languages, and we suggest that adaptation and acclimation are both occurring within mixed species microbial communities, regarding AHL-QS gene circuits. These include (but are not limited to): GMEs, promoter propagation, LuxR solos, and flexibility in the LuxI/R proteins themselves. In cases where long-term relationships develop, circuits that become ‘fixed’ may develop, with mutualistic relationships developing between species. However, there are genes present to ‘hear’ new languages, and ‘learn’ to speak those languages within most genomes, helping to adapt to an ever-changing environment.
Future studies of AHL-QS should include the concepts of the pangenome and flexible genomes as well as how acclimation of the LuxI/R proteins themselves may play a role in how bacteria learn new chemical languages. Chemical signaling is, in reality, a way to access the genome (and therefore tools) of other microbial cells in close proximity in order to improve the chances of survival of individual cells. Through genomic and transcriptomic studies of microbial consortia, we can glimpse some of the ways that bacteria have interacted with each other for billions of years, often cooperating to adjust to a changing climate. Such cooperation through chemical signaling may be the key to the success of bacteria throughout Earth’s history (see Outstanding Questions).
Outstanding Questions.
How do signaling relationships develop among disparate species?
Does AHL-QS signaling influence community diversity as biofilms develop, making QS a deterministic factor of community composition? As biofilms develop, early bacterial settlers on surfaces may release AHL-QS signals, attracting closely related species or species with which established AHL-QS crosstalk already exists.
How do a set of genes come under AHL-QS control? This question is central to understanding why and how AHL-QS has developed and also how bacteria can adapt to new environments and enter new communities (e.g., interactions with host tissue in human gut; pathogenic associations with other bacteria).
Do generic LuxR-type promoters exist, and could such promoters be replicated and installed within genomes to elicit expression of cooperative activities? Promoter propagation, which occurs in bacterial genomes, is a potential tool for development of new AHL-QS crosstalk.
What role do small RNAs (sRNAs) play in the evolution of AHL-QS systems between disparate species? Bacterial sRNAs influence how genes are expressed within bacterial cells, including in some AHL-QS systems. What role might they play in the microevolutionary changes in chemical communication?
How is a newly-perceived signal incorporated to then produce beneficial community-level gene expression?
How rapidly can a LuxR mutate in order to recognize and bind to a new signal? Does this process occur in ecological or evolutionary time scales?
What are the relationships between bacteriophage, and possibly other viruses, and AHL-QS systems? Are QS capabilities commonly found in phage genomes, and do viruses selectively infect AHL-QS active cells more or less?
Highlights.
Quorum sensing (QS), one type of social interaction among mixed-species microbial communities, is critical to their gene expression and ability to adjust to changing environmental conditions. Microbial community composition and structure influences QS capabilities and the affect they have on the surrounding environment.
Culturing methods that focus on a single species have limited our understanding of QS. With extensive studies in microbial diversity worldwide, we have learned that single-species communities rarely, if ever, occur in nature.
Microbial diversity on Earth is not static, and may not be quantifiable. Microbes must ‘learn’ to communicate and interact with new species in every environment over time, with adaptable and flexible QS systems. Such systems would allow microbes to utilize and better coordinate the genome capabilities of other nearby species and cells, expanding their abilities to cope with environmental change, and possibly their evolution over long geological periods.
Glossary
- AHL-QS circuit
a gene set consisting of both LuxI and LuxR cognate pair within a genome, producing both the AHL signaling molecule and receiving molecule. This creates ‘self-talk’ or an autoinducing system, causing gene expression changes that are ‘self’ induced.
- Biofilms and microbial mats
surface attached microbial communities in a mucus layer, consisting of Bacteria, Archaea, fungi, and protists in a complex and highly interactive community. Microbial mats differ from biofilms only in that they are thicker than 1 mm and observable to the naked eye.
- Crosstalk
the ability of different microbial species to both produce and receive chemical signals from each other through QS systems.
- Eavesdropping
the ability of species ‘A’ to perceive and possibly utilize signaling molecules from another species (B). However, species ‘B’ cannot perceive signals from species ‘A’.
- LuxI
the name of the gene and protein family that produces AHL signaling molecules in bacteria.
- LuxR
the name of the gene and protein family that binds to AHL signaling molecules, and is a transcriptional regulator in bacteria. LuxR proteins contain two domains: one that binds to a given AHL and one that binds to the DNA at specific promoter sites.
- LuxR solos
transcriptional regulator genes that do not have a paired (cognate) LuxI gene, which produce a specific AHL that can bind to the LuxR solo nearby in the topology of the genome.
- N-acyl homoserine lactone (AHL)
a type of signaling molecule used in QS among microbes, including Bacteria, Cyanobacteria, and Archaea. AHLs are produced within the bacterial cell and released into the environment, with different AHLs having various R group side chains, and chain lengths varying from 4 to 18 carbon atoms as well as a substitution of a carbonyl at the third carbon position.
- Pangenome
is the entire set of genes for all strains within a bacterial clade or group that has a common ancestor. It includes the core genome containing genes present in all strains within the clade, the accessory genome containing dispensable genes present in a subset of the strains, and strain-specific genes. The pangenome is also related to the concept of flexible genomes in microbiology, where often a single species may have strains that have very different genomes.
- Quorum sensing (QS)
a type of intercellular signaling that occurs between and among microbes and controls gene expression at a community level, allowing for group behaviors and responses to the environment. It is density dependent, meaning that the number of microbial cells influences whether genes regulated by QS are expressed or downregulated (repressed).
- Self-talk
when signaling molecules are produced and perceived by the same cells and same species (autoinducing gene regulation).
References
- 1.Waters CM and Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol 21, 319–346 [DOI] [PubMed] [Google Scholar]
- 2.Ng WL and Bassler BL (2009) Bacterial quorum-sensing network architectures. Annu. Rev. Genet 43, 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steidle A et al. (2001) Visualization of N-acylhomoserine lactone-mediated cell–cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol 67, 5761–5770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steidle A et al. (2015) N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147, 3249–3262 [DOI] [PubMed] [Google Scholar]
- 5.Shiner EK et al. (2005) Interkingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol. Rev 29, 935–947 [DOI] [PubMed] [Google Scholar]
- 6.González JF and Venturi V (2013) A novel widespread inter-kingdom signaling circuit. Trends Plant Sci 18, 167–174 [DOI] [PubMed] [Google Scholar]
- 7.Whitehead NA et al. (2001) Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev 25, 365–404 [DOI] [PubMed] [Google Scholar]
- 8.Fuqua C et al. (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet 35, 439–468 [DOI] [PubMed] [Google Scholar]
- 9.Lerat E and Moran NA (2004) The evolutionary history of quorum-sensing systems in bacteria. Mol. Biol. Evol 21, 903–913 [DOI] [PubMed] [Google Scholar]
- 10.Gray KM and Garey JR (2001) The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147, 2379–2387 [DOI] [PubMed] [Google Scholar]
- 11.Tommonaro G et al. (2012) Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb. Ecol 63, 490–495 [DOI] [PubMed] [Google Scholar]
- 12.Biswa P and Doble M (2013) Production of acylated homoserine lactone by Gram-positive bacteria isolated from marine water. FEMS Microbiol. Lett 343, 34–41 [DOI] [PubMed] [Google Scholar]
- 13.Zhang G et al. (2012) Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 6, 1336–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns BP et al. (2013) Quorum sensing in extreme environments. Life 3, 131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decho AW et al. (2009) Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may relate to diel pH. Environ. Microbiol 11, 409–420 [DOI] [PubMed] [Google Scholar]
- 16.Wagner-Dobler I et al. (2005) Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. Chembiochem 6, 2195–2206 [DOI] [PubMed] [Google Scholar]
- 17.Chandler JR et al. (2012) Acyl-homoserine lactone-dependent eavesdropping promotes competition in a laboratory co-culture model. ISME J. 6, 2219–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellington S and Greenberg EP (2019) Quorum sensing signal selectivity and the potential for interspecies cross talk. mBio 10, e00146–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudaiberdiev S et al. (2015) Census of solo LuxR genes in prokaryotic genomes. Front. Cell. Infect. Microbiol 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramoni S et al. (2015) A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria. Front. Cell. Infect. Microbiol 5, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuqua C (2006) The QscR quorum sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol 188, 3169–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gantner S et al. (2006) In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol 56, 188–194 [DOI] [PubMed] [Google Scholar]
- 23.Chugani S and Greenberg EP (2014) An evolving perspective on the Pseudomonas aeruginosa orphan quorum sensing regulator QscR. Front. Cell. Infect. Microbiol 4, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins CH et al. (2005) Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol. Microbiol 55, 712–723 [DOI] [PubMed] [Google Scholar]
- 25.McCusker VW et al. (1988) Biodegradation of carbamothioates in butylate-history soils. Weed Sci. 36, 818–823 [Google Scholar]
- 26.Holben WE et al. (1992) Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol 58, 3941–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall-Stoodley L et al. (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol 2, 95–108 [DOI] [PubMed] [Google Scholar]
- 28.Lozada-Chávez I et al. (2006) Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 34, 3434–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silby MW et al. (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev 35, 652–680 [DOI] [PubMed] [Google Scholar]
- 30.Matus-Garcia M et al. (2012) Promoter propagation in prokaryotes. Nucleic Acids Res. 40, 10032–10040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frost LS et al. (2005) Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol 3, 722–732 [DOI] [PubMed] [Google Scholar]
- 32.Ortori CA et al. (2007) Comprehensive profiling of N-acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole–linear ion trap mass spectrometry. Anal. Bioanal. Chem 387, 497–511 [DOI] [PubMed] [Google Scholar]
- 33.Nasser W and Reverchon S (2007) New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal. Bioanal. Chem 387, 381–390 [DOI] [PubMed] [Google Scholar]
- 34.Lintz MJ et al. (2011) Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc. Natl. Acad. Sci. U. S. A 108, 15763–15768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White CE and Winans SC (2005) Identification of amino acid residues of the Agrobacterium tumefaciens quorum sensing regulator TraR that are critical for positive control of transcription. Mol. Microbiol 55, 1473–1486 [DOI] [PubMed] [Google Scholar]
- 36.Weingart CL et al. (2005) Direct binding of the quorum sensing regulator CepR of Burkolderii cenocepacia to two target promoters in vitro. Mol. Microbiol 57, 452–467 [DOI] [PubMed] [Google Scholar]
- 37.Subramoni S and Venturi V (2009) LuxR-family ‘solos’: bachelor sensors/regulators of signaling molecules. Microbiology 155, 1377–1385 [DOI] [PubMed] [Google Scholar]
- 38.Rajput A and Kumar M (2017) Computational exploration of putative LuxR solos in Archaea and their functional implications in quorum sensing. Front. Microbiol 8, 798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel HK et al. (2014) The kiwifruit emerging pathogen Pseudomonas syringae pv. actinidiae does not produce AHLs but possesses three LuxR solos. PLoS ONE 9, e87862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez P et al. (2015) Stenotrophomonas maltophilia responds to exogenous AHL signals through the LuxR solo SmoR (Smlt1839). Front. Cell. Infect. Microbiol 5, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Decho AW et al. (2011) Chemical challenges to bacterial AHL signaling in the environment. Chem. Rev 111, 86–99 [DOI] [PubMed] [Google Scholar]
- 42.Rajput A and Kumar M (2017) In silico analyses of conservational, functional and phylogenetic distribution of the LuxI and LuxR homologs in Gram-positive bacteria. Sci. Rep 7, 6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egland KA and Greenberg EP (1999) Quorum sensing in Vibrio fischeri: elements of the LuxI promoter. Mol. Microbiol 31, 1197–1204 [DOI] [PubMed] [Google Scholar]
- 44.Stevens AM et al. (1999) Involvement of the RNA polymerase α-subunit C-terminal domain in LuxR-dependent activation of the Vibrio fischeri luminescence genes. J. Bacteriol 181, 4704–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuster M et al. (2004) Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. U. S. A 101, 15833–15839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gelencsér Z et al. (2012) Classifying the topology of AHL-driven quorum sensing circuits in proteobacterial genomes. Sensors 12, 5432–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oren Y et al. (2014) Transfer of noncoding DNA drives regulatory rewiring in bacteria. Proc. Natl. Acad. Sci. U. S. A 111, 16112–16117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somvanshi VS et al. (2012) A single promoter inversion switches Photorhabdus between pathogenic and mutualistic states. Science 337, 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yona AH et al. (2018) Random sequences rapidly evolve into de novo promoters. Nat. Commun 9, 1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hargreaves KR et al. (2014) What does the talking? Quorum sensing signaling genes discovered in a bacteriophage genome. PLoS One 91, e85131. [DOI] [PMC free article] [PubMed] [Google Scholar]


