Abstract
Gtf2ird1 and Gtf2i are two transcription factors (TFs) among the 28 genes deleted in Williams syndrome, and prior mouse models of each TF show behavioral phenotypes. Here we identify their genomic binding sites in the developing brain and test for additive effects of their mutation on transcription and behavior. GTF2IRD1 binding targets were enriched for transcriptional and chromatin regulators and mediators of ubiquitination. GTF2I targets were enriched for signal transduction proteins, including regulators of phosphorylation and WNT. Both TFs are highly enriched at promoters, strongly overlap CTCF binding and topological associating domain boundaries and moderately overlap each other, suggesting epistatic effects. Shared TF targets are enriched for reactive oxygen species-responsive genes, synaptic proteins and transcription regulators such as chromatin modifiers, including a significant number of highly constrained genes and known ASD genes. We next used single and double mutants to test whether mutating both TFs will modify transcriptional and behavioral phenotypes of single Gtf2ird1 mutants, though with the caveat that our Gtf2ird1 mutants, like others previously reported, do produce low levels of a truncated protein product. Despite little difference in DNA binding and transcriptome-wide expression, homozygous Gtf2ird1 mutation caused balance, marble burying and conditioned fear phenotypes. However, mutating Gtf2i in addition to Gtf2ird1 did not further modify transcriptomic or most behavioral phenotypes, suggesting Gtf2ird1 mutation alone was sufficient for the observed phenotypes.
Introduction
The Williams syndrome critical region (WSCR) contains 28 genes that are typically deleted in Williams syndrome (WS) (OMIM#194050). The genes in this region are of interest for their potential to contribute to the unique physical, cognitive and behavioral phenotypes of WS, which include craniofacial dysmorphology, mild to severe intellectual disability, poor visuospatial cognition, balance and coordination problems and a characteristic hypersocial personality (1–3). Single-gene knockout mouse models exist for many of the genes in the region, with differing degrees of face validity for WS phenotypes (4–9). Two genes have been highlighted in the human and mouse literature as playing a large role in the social and cognitive tasks: Gtf2i and Gtf2ird1. While humans with just these single genes mutated have not been described, variants on the remaining chromosome near these genes were among the best associated single nucleotide polymorphisms (SNPs) of the locus (though not genome-wide significant) in quantitative trait loci (QTL) studies of social behavior in WS patients (10). Mouse models of each gene have shown social phenotypes as well as balance and anxiety phenotypes (4,8,9,11–13), though it does not appear that mutations in these two genes alone can reproduce the entirety of the 28-gene deletion phenotype in mice (14). Regardless, since evidence shows that each gene affects similar behaviors, we set out to test the hypothesis that simultaneous knockdown of both genes would cause more severe phenotypes. Investigating the genes together, rather than individually, could provide a more complete understanding of how genes in the same family could combine to contribute to phenotypes.
Gtf2i and Gtf2ird1 are part of the general transcription factor 2i family of genes. A third member, Gtf2ird2, is located in the WSCR that is variably deleted in WS patients with larger deletions (15). This gene family arose from gene duplication events, resulting in high-sequence homology between the genes (16). The defining feature of this gene family is the presence of the helix–loop–helix I repeats, which are involved in DNA and protein binding (17). Gtf2i’s roles include regulating transcriptional activity in the nucleus. However, this multifunctional transcription factor (TF) also exists in the cytoplasm where it conveys messages from extracellular stimuli and regulates calcium entry into the cell (18–20). So far, Gtf2ird1 has only been described in the nucleus of cells and is thought to regulate transcription and associate with chromatin modifiers (21). The DNA binding of these two TFs has only been studied in ES cells and embryonic craniofacial tissues. They recognize similar and disparate genomic loci, suggesting the proteins interact to regulate specific regions of the genome (22,23). However, binding of these TFs has not been studied in the developing brain, which could provide more relevant insight on how the general transcription factor 2i family contributes to cognitive and behavioral phenotypes.
We performed ChIP-seq on GTF2I and GTF2IRD1 in the developing mouse brain to define where these TFs bind and then tested the downstream consequences of disrupting this binding. We used the CRISPR/Cas9 system to make a mouse model with a mutation in Gtf2ird1 alone and a mouse model with mutations in both Gtf2i and Gtf2ird1 to test how adding a Gtf2i mutation modifies the effects of Gtf2ird1 mutation. We showed the mutation in Gtf2ird1 resulted in the production of an N-truncated protein that disrupts the binding of GTF2IRD1 at the Gtf2ird1 promoter and deregulates the transcription of Gtf2ird1, moderately decreasing protein levels so homozygous mutants approximate levels expected from hemizygosity of the WSCR. While there are mild consequences of the mutation on genome-wide transcription, the mutant mouse exhibited clear balance, marble burying and conditioned fear differences. Comparing the single-gene mutant to the double mutant did not reveal more severe transcriptional changes and behavioral phenotypes; however, adding Gtf2i on top of a background of two Gtf2ird1 mutations resulted in abnormal pre-pulse inhibition (PPI) and cued fear conditioning. This suggests Gtf2ird1 drives the majority of the phenotypes observed in current studies, and total protein level, N-terminal truncation or both have functional consequences on DNA binding and behavior.
Results
GTF2I and GTF2IRD1 bind at active promoters and conserved sites
The paralogous TFs, GTF2I and GTF2IRD1, have been implicated in the behavioral phenotypes seen in humans with WS as well as mouse models (4,8,12,13,24,25). However, the underlying mechanisms by which the general transcription factor 2i family acts are not well understood. One approach to begin to identify how these TFs can regulate phenotypes is by identifying where they bind in the genome. This has been done in ES cells and embryonic facial tissues and revealed that both of these TFs bind to genes involved in craniofacial development (22). However, these are not relevant tissues when considering phenotypes related to brain development and subsequent behavior. To address this, we performed ChIP-seq for GTF2IRD1 and GTF2I. We chose to focus on the developing brain at embryonic day 13.5 (E13.5), as this would allow us to identify events that might contribute to changes in brain development and because GTF2IRD1 is not measurably expressed in adult brain (14). At this time point, we showed that the antibodies each pull down their specific target and do not cross-react (Fig. 1A).
Figure 1.
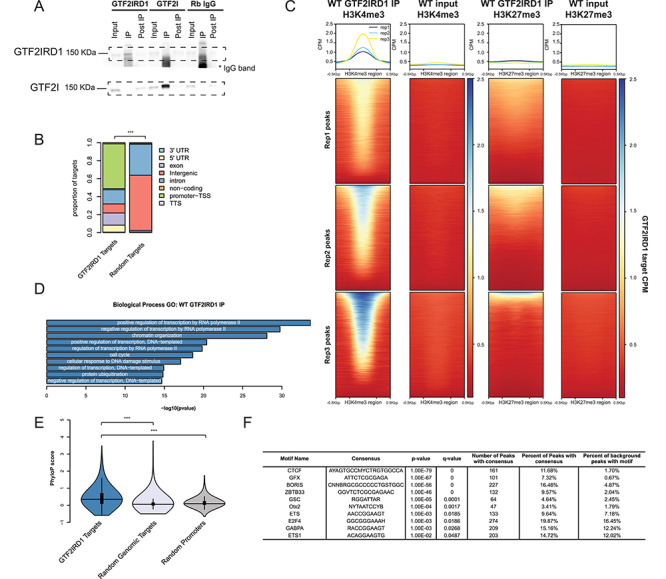
GTF2IRD1 binds preferentially to promoters in conserved, active sites in the genome. (A) GTF2IRD1 and GTF2I IP experiment, followed by immunoblotting for both proteins, shows that each antibody is specific for each protein. (B) GTF2IRD1 binding peaks are annotated primarily in promoters and gene bodies. The distribution of peak annotations is significantly different from random sampling of the genome. (C) GTF2IRD1 peaks were enriched in H3K4me3 sites marking active regions of the genome and to a lesser extent in H3K27me3 sites marking repressed regions. (D) GO analysis of genes that have GTF2IRD1 bound to the promoter. (E) The conservation of sequence in GTF2IRD1-bound peaks is significantly higher than random regions of the genome and a random set of promoter regions. (F) Motifs of transcription factors enriched under GTF2IRD1-bound peaks. *p < 0.05, **p < 0.01, ***p < 0.001.
We identified 1410 peaks that were enriched in the GTF2IRD1 IP samples compared to the input (Supplementary Material, Table S1; Supplementary Material, Fig. S1). The GTF2IRD1-bound regions were enriched in the promoters of genes and along the gene body, more so than would be expected by randomly sampling the genome (Fig. 1B) (χ2 = 1537.8, df = 7, P < 2.2 × 10−16). The bound peaks were found mostly in H3K4me3-bound regions [odds ratio (OR) = 779.5, P < 2.2 × 10−16 Fisher’s exact test (FET), P < 0.001 randomization test (Supplementary Material, Fig. S2A)], suggesting they are in active sites in the genome. Since H3K4me3 sites are enriched for promoter regions as well, we tested this overlap after removing H3K4me3 and GTF2IRD1 peaks annotated at promoter regions, and the overlap is still significant (OR = 234.31, P = 0 FET, P < 0.001 randomization test; Supplementary Material, Fig. S2B). While GTF2IRD1-bound regions were also enriched in repressed regions of the genome as defined by H3K27me3 marks (OR = 4.090, P < 2.2 × 10−16 FET, P < 0.001 randomization test; Supplementary Material, Fig. S2C), only 11% of GTF2IRD1 peaks were found in H3K27me3 regions as opposed to 94% in H3K4me3 regions (Fig. 1C), suggesting GTF2IRD1 may have more of a role in activation than repression.
To understand the common functions of the genes that have GTF2IRD1 bound at the promoter, we performed a GO analysis. The top 10 results were consistent with the functions previously described for GTF2IRD1, specifically regulation of transcription and chromatin organization, but also identified new categories, such as protein ubiquitination (Fig. 1D). To further test these regions for functional consequences, we compared the conservation of GTF2IRD1-bound peaks to a random sample of the genome and to a set of random promoter regions. There was a significant effect overall (F2,3815 = 268.72, P < 2 × 10−16), and post hoc comparison showed the GTF2IRD1 peaks are more conserved than the random genomic targets (P < 1.0 × 10−4) as well as the random promoter regions (P < 1.0 × 10−4) (Fig. 1E). We conducted a motif enrichment analysis using HOMER to identify other factors that share binding sites with GTF2IRD1 (Fig. 1F). The GSC motif, which is similar to the core RGATTR motif for GTF2I and GTF2IRD1, was identified in 4.64% of the targets (26). Interestingly, the CTCF motif was found at 11% of the GTF2IRD1 targets. Similar binding targets were also identified when comparing the gene targets to a background of other promoter regions (Supplementary Material, Fig. S1I). These findings suggest the GTF2IRD1 may modulate chromatin organization both through binding at CTCF sites and through regulation of chromatin modifier genes.
GTF2I ChIP-seq showed similar results to those of GTF2IRD1. We identified 1755 (Supplementary Material, Table S2) wild-type (WT) GTF2I peaks that had significantly higher coverage in the WT IP compared to the KO IP (Supplementary Material, Fig. S3A). These peaks were significantly enriched for promoter regions as well as the gene body when compared to random genomic targets (Fig. 2A) (χ2 = 911.63, df = 7, P < 2.2 × 10−16). Like GTF2IRD1, the majority of the GTF2I peaks (78.7%) overlapped H3K4me3 peaks (OR = 160.98, P < 2.2 × 10−16 FET, P < 0.001 randomization test; Supplementary Material, Fig. S2D), which is still significant when promoter regions are removed from the GTF2I and H3K4me3 sets (OR = 94.92, P = 0 FET, P < 0.001 randomization test; Supplementary Material, Fig. S2E). A smaller subset of peaks (20.7%) overlapped with the H3K27me3 mark (OR = 7.022, P < 2.2 × 10−16 FET, P < 0.001 randomization test; Supplementary Material, Fig. S2F). This suggests that these peaks are located mainly in active regions of the genome (Fig. 2B). Summarizing the common functions of these target genes by GO analysis showed enrichment for biological processes such as intracellular signal transduction and phosphorylation (Fig. 2C). For example, GTF2I binds within the gene body of the Src gene (Fig. 2D), which has been shown to phosphorylate GTF2I in order to activate its transcriptional activity as well as regulate calcium entry into the cell (18,19). The GTF2I binding sites are also significantly more conserved (F2,4816 = 146.51, P < 2 × 10−16) than random sampling of the genome (P < 1.0 × 10−4) and a random set of promoter regions (P < 1.0 × 10−4), further suggesting important functional roles of these regions (Fig. 2E). Motif enrichment of the GTF2I peaks revealed GC-rich binding motifs, such as for the KLF and SP families of TFs, as well as Lhx family members. Finally, we see an enrichment of the CTCF motif, which is fitting as GTF2I has been shown to help target CTCF to specific genomic regions (27) (Fig. 2F). We similarly see enrichment for the Lhx family of TF motifs when we compared the GTF2I-bound regions to a background of promoter sequences; however, the CTCF motif falls lower in the list, but is still significant, when using this background (Supplementary Material, Fig. S3J).
Figure 2.
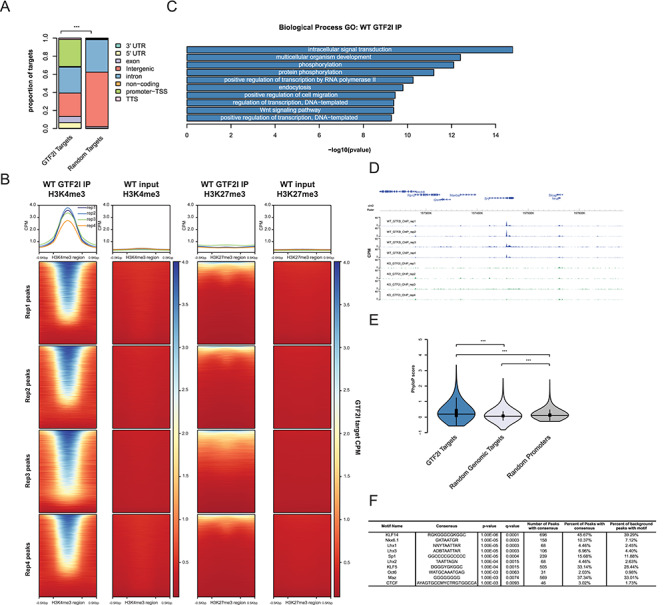
GTF2I binds at promoters in conserved, active sites in the genome. (A) GTF2I binding sites are annotated mostly in gene promoters and the gene body. The distribution of peaks is significantly different than would be expected by chance. (B) GTF2I peaks overlap with H3K4me3 peaks marking active regions, and to a lesser extent GTF2I peaks fall within H3K27me3 peaks marking inactive regions. (C) GO analysis of genes that have GTF2I bound at the promoter. (D) Epigenome browser shot of GTF2I peak bound within the Src gene. (E) Genomic sequence under GTF2I peaks is more conserved than random regions of the genome and a random set of promoter regions. (F) Motifs of transcription factors that are enriched in GTF2I-bound sequences. *p < 0.05, **p < 0.01, ***p < 0.001.
GTF2I and GTF2IRD1 binding sites have distinct features, yet overlap at a subset of promoters
One way in which GTF2I and GTF2IRD1 can interact is by binding the same sites in the genome. We therefore directly compared the regions bound by these proteins. First, we compared the GTF2I and GTF2IRD1 ChIP peaks and found the pattern of their binding sites is significantly different (χ2 = 282.84, df = 7, P < 2.2 × 10−16) (Fig. 3A); while both TFs mainly bind in promoters and the gene body, GTF2IRD1 has a higher proportion of peaks at the promoter compared to GTF2I, whereas GTF2I has more peaks at intergenic regions. Interestingly, when we compared them directly to each other, the GTF2IRD1-bound peaks were significantly more conserved than the GTF2I-bound peaks (t = 7.81, df = 2736.5, P = 8.2 × 10−15) (Fig. 3B). Next, to identify common targets, we identified the genes with both TFs at their promoter and found a significant overlap of 148 genes (OR = 1.4, P = 0.00015 FET) (Fig. 3C). Removing peaks at the promoter for both GTF2I and GTF2IRD1 targets still shows a significant overlap of 77 regions (OR = 167.53, P < 4.97 × 10−137, randomization test P < 0.001; Supplementary Material, Fig. S4A). Motif analysis on the shared peaks showed further enrichment of both CTCF and GSC motifs with very similar results when using other promoter regions as the background set (Fig. 3D; Supplementary Material, Fig. S4B).
Figure 3.

Comparison of GTF2IRD1 and GTF2I binding sites. (A) GTF2I and GTF2IRD1 have different distributions of annotated binding sites. (B) GTF2IRD1-bound sequences are more conserved than GTF2I-bound sequences. (C) The overlap of genes that have GTF2I and GTF2IRD1 bound at their promoters. (D) Motifs of transcription factors that are enriched in regions bound by both GTF2I and GTF2IRD1. (E) GO analysis of genes with both GTF2I and GTF2IRD1 bound at their promoters. (F) Epigenome browser shot of Mapk14 showing peaks for both GTF2I and GTF2IRD1. (G) Enrichment of GTF2IRD1- and GTF2I-bound genes in ASD and conserved gene sets. *p < 0.05, **p < 0.01, ***p < 0.001.
The GO functions of the overlapped genes highlight specific roles in synaptic functioning and signal transduction (Fig. 3E). Mapk14 is an example of a gene involved in signal transduction that has both GTF2I and GTF2IRD1 bound at its promoter (Fig. 3F). Shared targets such as this suggest that there are points of convergence where deleting both genes, such as in WS, might result in synergistic downstream impacts.
We also compared the results of our ChIP-seq experiments in the E13.5 mouse brain for these two TFs with other ChIP studies done for these proteins (22,28) in different systems. Makeyev et al. (22) used ChIP–ChIP to query the binding sites for both GTF2I and GTF2IRD1 in mouse ES cell lines as well as in E10.5 craniofacial tissues. Comparing the genes that had GTF2I peaks at their promoters in our study to results of the Makeyev et al. ES cell line experiment shows an overlap with 207 genes in common (P = 1 FET), and only 12 genes in common between our E13.5 GTF2I ChIP peaks and the peaks they identified at gene promoters in the E10.5 craniofacial tissue (P = 1 FET). In contrast, our GTF2IRD1 peaks at gene promoters did not significantly overlap with their peaks identified in ES cell lines, with only 23 genes in common (P = 1 FET), but they did have a significant overlap with their E10.5 craniofacial peak dataset with 128 genes in common (P = 0.017 FET). For our GTF2I peaks, we also looked at peaks called from ChIP-seq in human iPSCs from Adamo et al. (28). This also showed a significant overlap with 188 genes in common (P = 6.1 × 10−18 FET). These results held even when comparing to the core GTF2I peaks defined by Adamo et al. with 90 genes in common (P = 1.9 × 10−23). While these are each experiments conducted in different cell types and corresponding chromatin environments, these comparisons suggest some commonalities of binding targets across different tissues and species (Supplementary Material, Fig. S5), but also that a substantial amount of binding may depend on existing chromatin context in the cells.
Finally, given the consistent enrichment of CTCF binding sites in both GTF2I- and GTF2IRD1-bound regions, we also compared the targets for each TF to CTCF targets in E14.5 whole brain (29). We found a highly significant overlap between GTF2IRD1 and CTCF peaks, with roughly two-thirds of GTF2IRD1 binding overlapping with CTCF-bound sites (939 shared peaks, OR = 89.60, P < 2.2 × 10−16 FET, P < 0.001 randomization test; Supplementary Material, Fig. S4C). Similarly, GTF2I shared more peaks in common with CTCF (43%) than we would expect by chance (756 shared peaks, OR = 28.16, P < 2.2 × 10−16 FET, P < 0.001 randomization test; Supplementary Material, Fig. S4G). Next, since CTCF has been shown to be present at topological associating domain (TAD) boundaries, we compared the GTF2IRD1 and GTF2I peaks with TAD boundaries determined in E14.5 cortical neurons and found significant overlaps [557 shared peaks, OR = 0.16, P < 2.2 × 10−16 FET, P < 0.001 randomization test and 451 shared peaks, OR = 5.19, P < 2.2 × 10−16 FET, P < 0.001 randomization test, respectively (Supplementary Material, Fig. S4E and I)]. Since CTCF and TAD boundaries are enriched for promoters, we repeated these overlaps after removing promoter regions to test if this was driving the association. There is still a significant enrichment when comparing GTF2IRD1 regions (GTF2IRD1 versus CTCF OR = 40.92, P = 0 FET, P < 0.001 randomization test; GTF2IRD1 versus TAD boundaries OR = 6.94, P = 5.60 × 10−90 FET, P < 0.001 randomization test) and GTF2I regions (GTF2I versus CTCF OR = 15.33, P = 2.06 × 10−266 FET, P < 0.001 randomization test; GTF2I versus TAD boundaries OR = 4.54, P = 6.55 × 10−81 FET, P < 0.001 randomization test; Supplementary Material, Fig. S4D, F, H and J).
Thus, this TF family is enriched at promoters of brain-expressed genes, overlaps substantially at regions binding the chromatin looping protein CTCF and is highly enriched in a subset of TAD boundaries detected in the developing mouse brain. Overall, these TFs are poised to be important regulators of neural development and thus might regulate other genes associated with developmental diseases.
GTF2I and GTF2IRD1 bind promoters of known autism genes
Copy number variations in the WS locus are clearly associated with autism spectrum disorder (30), and recent exome and genome sequencing analyses have identified over 100 more associated loci, mostly containing single genes where loss-of-function mutations can cause ASD (31). Interestingly, these genes are substantially enriched in chromatin modifiers and transcriptional regulators, just like GTF2IRD1 targets. However, it is unclear how mutations in this wide variety of genes all lead to a common cognitive phenotype. One widely proposed possibility is these genes are part of a functional network and mutation of any one gene might disrupt some core facet of transcriptional regulation, though most of the evidence to support this idea comes from analyses of co-expression across development (32), rather than any measure of functional interaction or targeted regulation. Therefore, we next examined whether GTF2IRD1 and GTF2I are positioned to regulate other ASD genes by binding within them or nearby. We found that GTF2IRD1 targets were highly enriched for ASD genes (OR > 3.5, P-value <1.5 × 10−5 FET)—targeting 19 of the 102 genes, including a variety of epigenetic regulators such as DNMT3A, SETD5, NSD1, ADNP and SIN3A (Fig. 3G). GTF2IRD1 was generally bound to its promoters or nearby CpG islands, suggesting direct regulation of these genes. GTF2I was likewise enriched at ASD genes, albeit more moderately (OR = 2.1, P < 0.02 FET), targeting 14 genes. Five genes were targeted by both. Thus, our DNA binding data supports prior co-expression data that suggests a subset of ASD genes may be part of an interrelated module of chromatin-modifying genes and is consistent with prior suggestions that this convergence indicates these genes may have similar downstream pathways leading to disease (33).
It has also been found that genes mutated in ASD tend to be highly constrained in the general human population, with almost no loss-of-function mutations observed across >140 000 exome sequencing samples (summarized as a pLI score of >0.9) (34). Strong constraint, even of heterozygous loss-of-function mutations, suggests that these genes do not tolerate decreased expression and thus require a very precise amount of transcription for normal human survival or reproductive fitness. We find >30% of GTF2IRD1-bound genes meet this criterion, a result highly unlikely to be due to chance (OR = 2.48, P-value<2.2 × 10−16 FET). GTF2I targets also show a significant, but more modest, enrichment (OR = 1.27, P-value <1.1 × 10−7 FET) (Fig. 3G). Together, these results show that these TFs, especially GTF2IRD1, tend to bind genes requiring tight regulation of expression, as indicated by severe phenotypes and intolerance to loss-of-function mutations.
Frameshift mutation in Gtf2ird1 results in truncated protein and affects DNA binding at the Gtf2ird1 promoter
To investigate the functional role of Gtf2ird1 and Gtf2i at these bound sites and understand how these genes interact, we made loss-of-function models of Gtf2ird1 individually and a double mutant with mutations in both Gtf2i and Gtf2ird1. We designed one gRNA for each gene and injected them simultaneously into FVB/NJ mouse embryos to obtain single- and double-gene mutations. We first characterized the consequences of a one base pair adenine insertion in exon 3 of Gtf2ird1. This frameshift mutation introduces a premature stop codon in exon 3, an early constitutively expressed exon, which we expected to trigger non-sense-mediated decay (Fig. 4A). We crossed heterozygous mutant animals to analyze Gtf2i and Gtf2ird1 transcript and protein abundance in heterozygous and homozygous mutants compared to WT littermates (Fig. 4B). The western blots and qPCR were performed using the whole brain at E13.5. As expected, the Gtf2ird1 mutation did not affect Gtf2i transcript or protein levels (Fig. 4C and D). Contrary to our prediction of non-sense-mediated decay, we observed a 1.74-fold increase in the Gtf2ird1 transcript with each copy of the mutation and a 40% reduction of the protein in homozygous mutants compared to WT with no significant difference between the WT and heterozygous mutants (Fig. 4E and F). This suggests the mutation did have an effect on protein abundance and disrupted the normal transcriptional regulation of the gene, with the homozygous mutant modeling protein levels that might be expected in WS.
Figure 4.
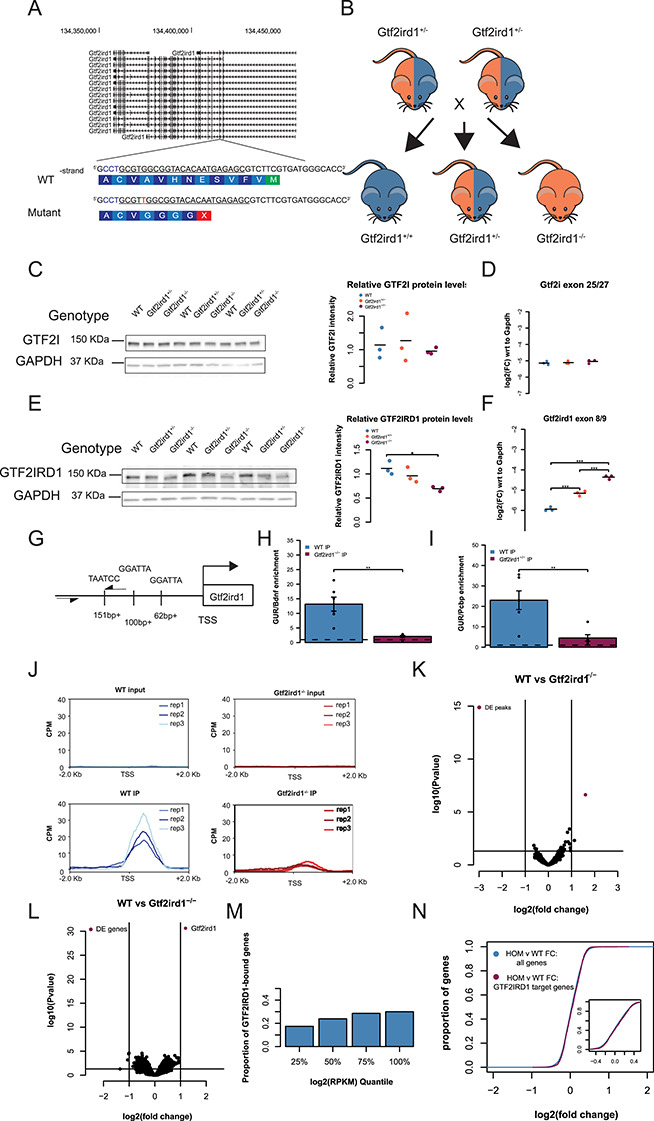
Frameshift mutation in Gtf2ird1 exon 3 results in a decreased amount of an N-truncated protein with diminished binding at Gtf2ird1 promoter and has a little effect on transcription in the brain. (A) The sequence of exon 3 of Gtf2ird1 targeted by the underlined gRNA with the PAM sequence in blue. The mutant allele contains a one base pair insertion of an adenine nucleotide that results in a premature stop codon. (B) Breeding scheme of the intercross of Gtf2ird1+/− to produce genotypes used in the experiments. (C, D) Mutation in Gtf2ird1 does not affect the protein or transcript levels of Gtf2i. (E) Frameshift mutation decreases the amount of protein in Gtf2ird1−/− and causes a slight shift to lower molecular weight. (F) The abundance of Gtf2ird1 transcript increases with increasing dose of the mutation. (G) Schematic of Gtf2ird1 upstream regulatory element (GUR) that shows the three GTF2IRD1 binding motifs. The arrows indicate the location of the primers for amplifying the GUR in the ChIP-qPCR assay. (H, I) WT ChIP of GTF2IRD1 shows enrichment of the GUR over off-target regions. There is more enrichment in the WT genotype compared to the Gtf2ird1−/− genotype. (J) Profile plots of GTF2IRD1 ChIP-seq data confirm diminished binding at the Gtf2ird1 promoter. (K) Differential peak analysis comparing WT and Gtf2ird1−/− ChIP-seq data showed only the peak at Gtf2ird1 is changed between genotypes with an FDR < 0.1. (L) Differential expression analysis in the E13.5 brain comparing WT and Gtf2ird1−/− showed only Gtf2ird1 as changed with FDR < 0.1. (M) The presence of GTF2IRD1 at gene promoters is not evenly distributed across expression levels. (N) The expression of genes bound by GTF2IRD1 is not different compared to all other genes between WT and Gtf2ird1−/− mutants. *p < 0.05, **p < 0.01, ***p < 0.001.
We noticed a slight shift in the homozygous mutant band, which may correspond to loss of the N-terminal end of the protein. Similar results were reported in another mouse model that deleted exon 2 of Gtf2ird1, in which lower levels of an N-terminally truncated protein was caused by a translation re-initiation event at methionine-65 in exon 3 (35). The N-terminal end codes for a conserved leucine zipper, which participates in dimerization as well as DNA binding (35,36). Mutating the leucine zipper was shown to affect binding of the protein to the Gtf2ird1 upstream regulatory (GUR) element located at the promoter of Gtf2ird1. Given the previous findings that GTF2IRD1 negatively autoregulates its own transcription and mutating the leucine zipper affects binding to the GUR, we hypothesized that the frameshift mutation diminished the ability of GTF2IRD1 to bind its promoter resulting in increased transcript abundance. We tested this by performing ChIP-qPCR in the E13.5 brain in WT and Gtf2ird1−/− mutants. In the WT brain, GTF2IRD1 IP enriched for the GUR 13–20 times over off-target sequences, which was significantly higher than the GTF2IRD1 IP in the Gtf2ird1−/− brain (Fig. 4G–I). Taken together, non-sense transcripts of Gtf2ird1 with a stop codon in exon 3 can re-initiate at a lower level to produce a N-truncated protein with a diminished binding capacity at the GUR element.
Truncated GTF2IRD1 does not affect binding genome-wide
Given that the one base pair insertion did not result in a full knockout of the protein but did affect its DNA binding capacity at the GUR of Gtf2ird1, we tested whether the mutant was a loss of function for all DNA binding. We performed ChIP-seq in the E13.5 Gtf2ird1−/− mutants and compared it to WT ChIP-seq data. This comparison confirmed the decrease in binding at the TSS of Gtf2ird1, suggesting the mutation has greatly decreased binding at this locus (Fig. 4J; Supplementary Material, Fig. S6A). Surprisingly, the only peak identified as having differential coverage (FDR < 0.1) between the two genotypes was this peak at the Gtf2ird1 TSS (Fig. 4K). This suggests that the frameshift mutation has a very specific consequence on how GTF2IRD1 binds to its own promoter that does not robustly affect its binding elsewhere in the genome. The Gtf2ird1 promoter has two instances of the R4 core motif in the sense direction and one instance of the motif in the antisense orientation. We searched the sequences under the identified peaks for similar orientations of this binding motif and found three other peaks, none of which showed any difference in binding coverage between genotypes. However, these three other peaks did not match the exact spacing of the R4 motifs found in the Gtf2ird1 promoter. This suggests that the leucine zipper is important for a specific configuration of binding sites that is only present in this one instance in the genome. It has been shown that the three R4 motifs are present in the same orientation and spacing across great evolutionary distances (35).
Gtf2ird1 frameshift mutation shows mild transcriptional differences
The N-truncation of GTF2IRD1 clearly affected its binding at the Gtf2ird1 promoter and affected expression levels. Although we didn’t see genome-wide binding perturbed, it is possible losing the N-terminus, or the decreased protein level, altered the protein’s ability to recruit other transcriptional co-regulators to impact gene expression. Therefore, we tested the effects of this mutation on genome-wide transcription in the E13.5 brain. We compared the whole brain transcriptome of WT littermates to heterozygous and homozygous mutants. Strikingly similar to ChIP-seq data, the only transcript with an FDR < 0.1 was Gtf2ird1, which was affected in the same direction seen in the qPCR results (Fig. 4L; Supplementary Material, Fig. S6B). We leveraged WT ChIP-seq data to test if GTF2IRD1 presence at a promoter correlates with gene expression. Binning the genes according to the expression level showed that the distribution of GTF2IRD1 targets was different than expected by chance (χ2 = 48.83, df = 3, P < 1.42 × 10−10), suggesting highly expressed genes are more likely to have GTF2IRD1 bound at their promoters (Fig. 4M). The majority (1262 peaks) of the GTF2IRD1 peaks at a TSS were at expressed genes, with only 410 peaks next to genes not expressed at detectable levels in the E13.5 brain. To see if there was a more subtle general effect below our sensitivity to detect by analysis of single genes, we tested the bound GTF2IRD1 targets expressed as a population for a shift in expression. We saw a trend toward significance between the bound and unbound genes, but with a small effect size: a mean increase of 0.014-fold change in GTF2IRD1 targets (Kolmogorov–Smirnov test D = 0.038, P = 0.079). In the motif enrichment analysis, motifs similar to the RGATTR core motif, such as GSC, CRX and Lhx, were identified. In case there was a difference between peaks containing this motif and those that do not, we determined which genes had peaks that contained this core motif and specifically looked to see if these genes showed a difference in expression. There is a slight, but significant, downregulation (logFC = −0.037, Kolmogorov–Smirnov test D = 0.133, P = 0.039) of the 112 genes that have a core motif in the GTF2IRD1-bound region (Supplementary Material, Fig. S6C). While small changes are perhaps unsurprising because the frameshift mutation did not disturb binding genome-wide (Fig. 4N), the homozygous mutants do have an overall decrease in protein level of ~50%, which should mimic a WS deletion. Thus, transcriptional consequences of hemizygosity of this gene might be similarly small.
Frameshift mutation is sufficient to affect behavior
Although we observed only small differences in DNA binding and overall brain transcription, another Gtf2ird1 model also reported little to no effects of mutation on transcriptome-wide expression in the brain, yet the model still showed behavioral phenotypes (8,37). Therefore, we tested our mutation for downstream consequences on adult mouse behavior (Table 1). There are many single-gene knockout models of Gtf2ird1, and each shows distinct behavioral differences which are, in some instances, contradictory (8,9,24,38). One consistent phenotype across models is a deficit in motor coordination, which is also affected in individuals with WS. Similarly, we observed a significant effect of genotype (H2 = 16.35, P = 0.0003), on balance. Heterozygous and homozygous animals fell off a ledge sooner than WT littermates (P = 0.0038, P = 0.0007, respectively) (Fig. 5A). Marble burying has not been reported in other Gtf2ird1 models, but in larger WS models that delete the entire syntenic WSCR or the proximal half of the region containing Gtf2ird1 have shown decreased marble burying in mutants (5,39). We observed a similar significant effect of genotype on the number of marbles buried (F2,80 = 6.17, P = 0.0033), with Gtf2ird1−/− mutants burying fewer marbles than WT mice (P = 0.002) (Fig. 5B). Reports of overall activity levels in Gtf2ird1 mouse models have been discrepant (9,24). Here we show there is no main effect of genotype (F2,88 = 1.36, P = 0.263) but a time by genotype interaction (F10,440 = 5.791, P = 3.3 × 10−8) on total distance traveled in a 1-h locomotor task. Though no single time point showed a difference between genotypes when performing post hoc tests, this may reflect an overall difference in habituation to the chamber (Fig. 5C). When taking sex into consideration, there was a main effect of sex (F1,85 = 5.23, P = 0.025), and the genotype by time interaction persists (F10,425 = 5.82, P = 3.06 × 10−8) (Supplementary Material, Fig. S7A and B). Time spent in the center of an open field is used as a measure of anxiety-like behavior in mice. Anxiety-like behaviors in Gtf2ird1 models have also been discrepant in the literature (8). Here we show that there was no main effect of genotype on center variables in this task (F2,88 = 0.88, P = 0.42) (Fig. 5D).
Table 1.
Behaviors and sample sizes for Gtf2ird1+/− × Gtf2ird1+/− cross
| Male | Female | ||||||
|---|---|---|---|---|---|---|---|
| Behavior | Experimenter | WT | Gtf2ird1+/− | Gtf2ird1−/− | WT | Gtf2ird1+/− | Gtf2ird1−/− |
| Cohort | |||||||
| One-hour locomotor activity | Female | 10 | 21 | 12 | 15 | 16 | 17 |
| Ledge | Female | 9 | 21 | 13 | 15 | 17 | 17 |
| Marble burying | Female | 8 | 20 | 10 | 14 | 17 | 14 |
| Pre-pulse inhibition | Female | 8 | 20 | 11 | 14 | 17 | 15 |
| Conditioned fear | Female | 9 | 18 | 10 | 15 | 17 | 17 |
| Shock sensitivity | Female | 9 | 10 | 11 | 15 | 17 | 17 |
Figure 5.
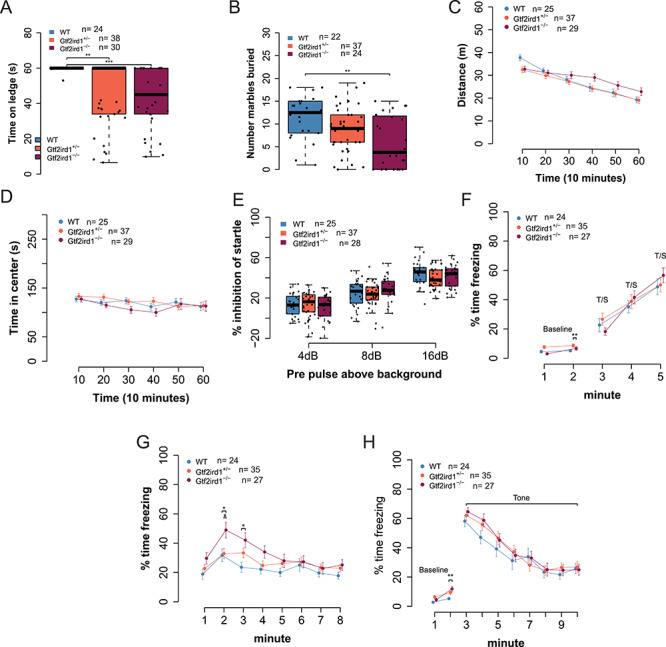
Homozygous frameshift mutation in Gtf2ird1 is sufficient to cause behavioral phenotypes. (A) Homozygous mutants have worse balance than WT littermates in ledge task. (B) Homozygous mutants bury fewer marbles than WT and heterozygous littermates. (C) Overall activity levels are not affected, but a time by genotype interaction shows the mutant animals are slower to habituate to the novel environment. (D) There is no difference in time spent in the center of the apparatus between genotypes. (E) All animals show an increase in startle inhibition when given a pre-pulse of increasing intensity. There is no difference between genotypes. (F) Acquisition phase of fear conditioning paradigm. All animals show the expected increase in freezing to additional foot shocks. (G) Gtf2ird1−/− animals show an early increased contextual fear memory response compared to WT and heterozygous littermates. (H) There were no significant differences between genotypes in cued fear. *p < 0.05, **p < 0.01, ***p < 0.001.
Finally, as individuals with WS also show a high prevalence of phobias, sensitivity to sounds and learning deficits (3,40), we tested sensory motor gating and learning and memory. PPI results from a decreased startle to an auditory stimulus when the startle stimulus is preceded by a smaller stimulus. PPI was reduced in animals with the proximal WSCR region deleted, but mice with the distal deletion or full deletion did not have any abnormal phenotype (6). In our study, there was no main effect of genotype on PPI (F2,87 = 0.24, P = 0.79), but a pre-pulse by genotype interaction (F4,174 = 2.66, P = 0.034), which suggests that for some pre-pulse stimuli, there is a difference between mutants and WT littermates; however, no comparisons survived multiple testing corrections in the post hoc test (Fig. 5E).
In our assessment of learning and memory with the conditioned fear paradigm, there was a main effect of genotype (F2,83 = 4.82, P = 0.010) and minute (F1,83 = 9.75, P = 0.002) on baseline freezing. Post hoc tests on baseline data showed the heterozygous mutants froze more than homozygous mutants during minute one (P = 0.0065). After the baseline, when animals were trained to associate a tone with a foot shock, we observed all mice had increased freezing over time (F2,122 = 26.77, P = 2.28 × 10−10) as expected (Fig. 5F). On the second day, which tested contextual fear memory, all genotypes exhibited a fear memory response as indicated by the significant effect of the context compared to baseline (F1,83 = 173.20, P < 2 × 10−16). Each group froze more during the first 2 min of day 2 than on day 1 (WT, P = 1.98 × 10−8; Gtf2ird1+/−, P = 4.97 × 10−11; Gtf2ird1−/−, P < 2 × 10−16) (Supplementary Material, Fig. S7C). When we analyzed the entire time of the experiment of contextual fear, we similarly saw no main effect of genotype (F2,83 = 2.8946, P = 0.061), but a significant effect of time (F7,581 = 15.05, P < 2 × 10−16) and a time by genotype interaction (F14,581 = 2.01, P = 0.016). Post hoc analysis showed that during minute two, the homozygous animals froze significantly more than the Gtf2ird1+/− mutants (P = 0.026). Similarly, the homozygous mutants froze more than WT littermates during minutes two (P = 0.021) and three (P = 0.012), suggesting an increased contextual fear memory response (Fig. 5G). On day 3 of the experiment, we tested cued fear. During day 3 baseline we saw a difference in freezing between genotypes (F2,83 = 4.13, P = 0.02) as well as a time by genotype interaction (F2,83 = 4.47, P = 0.014), with homozygous mutants freezing more in minute two than WT littermates (P = 0.002). All genotypes had a similar response to the tone (F2,83 = 0.36, P = 0.70) (Fig. 5H). These differences could not be explained by differences in shock sensitivity (flinch, H2 = 2.52, P = 0.28; escape, H2 = 3.13, P = 0.21; vocalization, H2 = 2.20, P = 0.33) (Supplementary Material, Fig. S7D). Thus, mutation of Gtf2ird1 appears to enhance contextual fear learning.
Overall, these behavioral analyses show the N-terminal truncation and/or decreased protein levels of the Gtf2ird1 mutant still result in adult behavioral phenotypes, specifically in the domains of balance, marble burying and fear conditioning. The most severe phenotypes were observed in the homozygous mutants, which may model the haploinsufficiency of WS deletions.
Generation of a Gtf2i and Gtf2ird1 double mutant
The evidence of functional consequences from the one base pair Gtf2ird1 frameshift mutation led us to characterize a double mutant that was generated during the dual gRNA CRISPR/Cas9 injections. This mutant allowed us to test the effects of knocking out Gtf2i combined with a Gtf2ird1 mutation and test different Gtf2ird1 mutations for consistency in phenotypes. The double mutant described here has a two base pair deletion in exon 5 of Gtf2i and a 589 bp deletion that encompasses most of exon 3 of Gtf2ird1 (Fig. 6A). We carried out a heterozygous cross of the double mutants to test the protein and transcript abundance of each gene in the heterozygous and homozygous states. The homozygous double mutant is embryonic lethal due to the lack of Gtf2i, which has been described in other Gtf2i mutants (Fig. 6B) (4,41). We were, however, able to detect homozygous embryos up to E15.5. Thus, we focused our molecular analyses on E13.5 brains. The two base pair deletion in exon 5 of Gtf2i leads to a premature stop codon resulting in a full protein knockout and decreases the transcript abundance consistent with degradation of the mRNA due to non-sense-mediated decay (Fig. 6C and D). The 589 bp deletion in Gtf2ird1 removes all of exon 3 except the first 14 bp. We observed the same increase in transcript abundance that was detected in the one base pair insertion mutation, but this mutation had a larger effect on protein levels across genotypes, with homozygous mutants producing a truncated protein at about 10% of WT levels (Fig. 6E and F).
Figure 6.
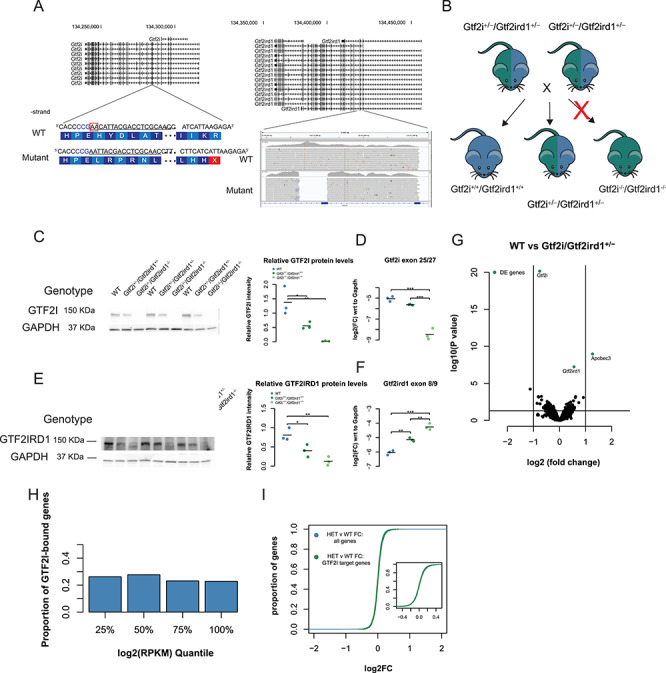
Mutating both Gtf2i and Gtf2ird1 does not result in larger differences in brain transcriptomes. (A) Generation of double mutant. gRNA target is underlined in exon 5 of Gtf2i with the PAM sequence in blue. The two base pair deletion results in a premature stop codon within exon 5. The Gtf2ird1 mutation is a large 589 bp deletion covering most of exon 3 as shown in the IGV browser shot. (B) Heterozygous intercross to generate genotypes for ChIP and RNA-seq experiments. The homozygous double mutants are embryonic lethal but are present up to E15.5. (C) The two base pair deletion in Gtf2i decreases the protein by 50% in heterozygous mutants, and no protein is detected in the homozygous E13.5 brain. (D) The mutation decreases the abundance of Gtf2i transcript consistent with non-sense-mediated decay. (E) The 589 bp deletion in Gtf2ird1 leads to decreased protein levels in heterozygous and homozygous mutants. There is still a small amount of protein made in the homozygous mutant. (F) The 589 bp deletion increases the amount of Gtf2ird1 transcript. (G) Volcano plot comparing the expression in the E13.5 brain of WT and heterozygous double mutants. The highlighted genes represent an FDR < 0.1. (H) The presence of Gtf2i at the promoters does not correlate with the expression of a gene. (I) The fold change of genes between WT and heterozygous double mutants that have GTF2I bound at their promoters were slightly upregulated when compared to the fold change of genes that did not have GTF2I bound. *p < 0.05, **p < 0.01, ***p < 0.001.
Knocking down both Gtf2i and Gtf2ird1 produces mild transcriptomic changes
To test if combined mutation of Gtf2i and Gtf2ird1 had a larger effect on the transcriptome, we performed whole brain RNA-seq analysis on WT E13.5 brains and compared them to Gtf2i+/−/Gtf2ird1+/− littermates. Similar to what was seen in the previous Gtf2ird1−/− mutants, there were only mild differences between the transcriptomes (Fig. 6G). We also compared WT transcriptomes to the homozygous double mutants, which showed greater differences. However, since these mutants have a very severe phenotype, including neural tube closure defects, any direct transcriptional consequences are probably masked by a large number of indirect effects. Indeed, GO term analysis suggested that overall nervous system development and glial cell differentiation are disrupted (Supplementary Material, Fig. S8A and B). We also analyzed GTF2I ChIP-seq data with RNA-seq data. Unlike the enriched binding at highly expressed genes we saw with GTF2IRD1 alone, gene expression levels were not significantly related to GTF2I binding (χ2 = 6.58, df = 3, P = 0.086) (Fig. 6H). This is consistent with a previous report of GTF2I ChIP-seq data. Again, the majority (963 peaks) of the TSS GTF2I peaks were nearby expressed genes, with 458 next to genes not expressed at detectable levels in the E13.5 brain. There is a slight but significant increase in gene expression of genes bound by GTF2I compared to genes that are not (0.0213 logFC, Kolmogorov–Smirnov test D = 0.075, P = 9.50 × 10−5) (Fig. 6I). When we compared the expression of the 200 genes that have the core RGATTR motif in the GTF2I-bound peak, there is a further slight increase in expression (0.031 logFC, Kolmogorov–Smirnov test D = 0.112, P = 0.0154, Supplementary Material, Fig. S8C). Thus, heterozygous Gtf2i and Gtf2ird1 mutation, like Gt2ird1 mutation alone, results in very subtle transcriptional changes.
Double mutants show behavioral consequences similar to single Gtf2ird1 mutants
To test the effects of mutating both Gtf2i and Gtf2ird1 on behavior, we crossed the heterozygous double mutant to the single Gtf2ird1 heterozygous mouse (Fig. 7A). This breeding strategy produced four littermate genotypes, WT, Gtf2ird1+/−, Gtf2i+/−/Gtf2ird1+/− and Gtf2i+/−/Gtf2ird1−/− for direct and well-controlled comparisons. To test for additive effects, the primary comparison was contrasting Gtf2ird1+/− mice to Gtf2i+/−/Gtf2ird1+/− littermates. The remaining genotype, Gtf2i+/−/Gtf2ird1−/−, further tested the effects of heterozygous Gtf2i mutation in the presence of both Gtf2ird1 mutations. To be thorough, we tested protein and transcript abundance of each gene in all four genotypes. As expected, all genotypes with the Gtf2i mutation showed decreased protein and transcript levels. The Gtf2ird1 results reflected what was previously shown for each mutation; however, Gtf2i+/−/Gtf2ird1−/− did not show any further detectable decrease in protein abundance compared to the Gtf2i+/−/Gtf2ird1+/− genotype (Supplementary Material, Fig. S9A–D), with both at about 50% of WT levels.
Figure 7.
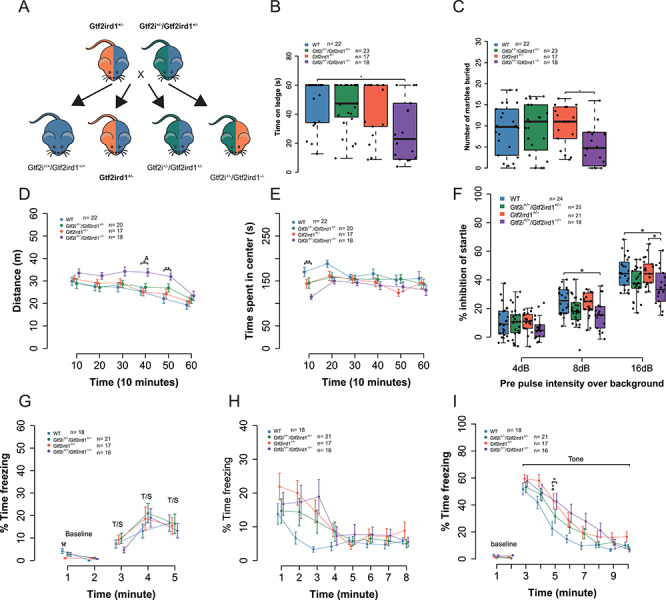
Gtf2i does not modify most of the phenotypes of Gtf2ird1 mutation. (A) Breeding scheme for behavior experiments. (B) The Gtf2i+/−/Gtf2ird1−/− animals fell off ledge sooner than WT littermates. (C) There was a main effect of genotype on marbles buried. Post hoc analysis showed that Gtf2i+/−/Gtf2ird1−/− buried fewer marbles than the Gtf2ird1−/− genotype. (D) Gtf2i+/−/Gtf2ird1−/− had increased overall activity levels in a 1-h activity task. (E) Gtf2i+/−/Gtf2ird1−/− showed a decreased time in the center of the apparatus compared to WT. (F) All animals show an increased startle inhibition when given a pre-pulse of increasing intensity. The Gtf2i+/−/Gtf2ird1−/− mutants show less of an inhibition at higher pre-pulse levels compared to WT and Gtf2ird1+/− animals. (G) All genotypes showed increased freezing with subsequent foot shocks. (H) All genotypes showed a similar contextual fear response. (I) There was a main effect of genotype on cued fear with the Gtf2ird1+/− and Gtf2i+/−/Gtf2ird1−/− genotypes showing an increased fear response compared to WT. *p < 0.05, **p < 0.01, ***p < 0.001.
We repeated the same behaviors performed on the one base pair Gtf2ird1 mutants (Table 2). We saw a similar significant effect of genotype on balance (H3 = 10.68, P = 0.014), with Gtf2i+/−/Gtf2ird1−/− mice falling off sooner compared to WT littermates (P = 0.025) (Fig. 7B). There was no significant difference between the Gtf2ird1+/− and Gtf2i+/−/Gtf2ird1+/− genotypes, suggesting that decreasing the dosage of GTF2I does not strongly modify the Gtf2ird1+/− phenotype. There was a significant effect of genotype on the number of marbles buried (F3,76 = 2.93, P = 0.039). Post hoc analysis showed a significant difference between only Gtf2ird1+/− and Gtf2i+/−/Gtf2ird1−/− littermates (P = 0.050) (Fig. 7C), with a trend in the same direction as was previously seen in the Gtf2ird1−/− mutants. We saw a main effect of genotype on activity levels in the 1-h locomotor task (F3,69 = 3.22, P = 0.028), but we did not see the same main effect of sex (F1,69 = 2.29, P = 0.14), or a sex by genotype interaction (F3,69 = 1.82, P = 0.15); however, we did see a three-way sex by time by genotype interaction (F15,345 = 1.95, P = 0.018). The combined sex data showed Gtf2i+/−/Gtf2ird1−/− mice travel a greater distance than WT and Gtf2ird1+/− mice at time point 40 (Fig. 7D). When we looked at the data by sex, we saw a larger effect in females with Gtf2ird1+/− and Gtf2i+/−/Gtf2ird1+/− genotypes intermediate to Gtf2i+/−/Gtf2ird1−/− (Supplementary Material, Fig. S9E and F). There was also a main effect of genotype on the time spent in the center of the apparatus (F3,69 = 3.60, P = 0.018) that was not seen in the previous Gtf2ird1 cross. Gtf2i+/−/Gtf2ird1−/− mice spent less time in the center during the first 10 min of the task compared to WTs (P = 0.0019) (Fig. 7E).
Table 2.
Behaviors and sample sizes for Gtf2ird1+/− × Gtf2i+/−/Gtf2ird1+/− cross
| Male | Female | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Behavior | Experimenter | WT | Gtf2ird1+/− | Gtf2i+/−/Gtf2ird1+/− | Gtf2i+/−/Gtf2ird1−/− | WT | Gtf2ird1+/− | Gtf2i+/−/Gtf2ird1+/− | Gtf2i+/−/Gtf2ird1−/− |
| Cohort 1 | |||||||||
| One-hour locomotor activity | Male | 8 | 8 | 11 | 7 | 14 | 9 | 9 | 11 |
| Ledge | Male | 8 | 8 | 12 | 7 | 14 | 9 | 11 | 11 |
| Marble burying | Male | 8 | 8 | 12 | 7 | 14 | 9 | 11 | 11 |
| Social approach | Male | 6 | 7 | 10 | 7 | 12 | 6 | 9 | 10 |
| Cohort 2 | |||||||||
| Pre-pulse inhibition | Male | 13 | 6 | 12 | 6 | 11 | 15 | 13 | 12 |
| Conditioned fear | Male | 11 | 4 | 9 | 5 | 7 | 13 | 12 | 11 |
| Shock sensitivity | Male | 12 | 6 | 10 | 6 | 10 | 15 | 13 | 12 |
We also repeated the PPI and conditioned fear memory tasks using this breeding strategy. In contrast to what was observed for PPI in the Gtf2ird1 cross, we saw a significant main effect of genotype (F3,84 = 4.59, P = 0.0051). The Gtf2i+/−/Gtf2ird1−/− mice showed an attenuated PPI response especially at the louder pre-pulse stimuli compared to WT littermates (PPI8, P = 0.02; PPI16, P = 0.018) and Gtf2ird1+/− mice (PPI16, P = 0.02) (Fig. 7F). On day 1 of the conditioned fear task, all genotypes showed increased freezing with subsequent foot shocks as expected. WT animals exhibited higher freezing during minute one of baseline, but this difference diminished during minute two (Fig. 7G). All animals showed a contextual fear memory response when they were re-introduced to the chamber on day 2 (F1,68 = 81.21, P = 3.21 × 10−13) (Supplementary Material, Fig. S9G). While there was no main effect of genotype (F3,68 = 1.61, P = 0.19) (Fig. 7H), the Gtf2ird1+/− and double mutants showed a trend toward increased freezing that was seen in the previous behavior cohort. On day 3, when cued fear was tested, there was a significant effect of genotype on the freezing behavior (F3,68 = 3.17, P = 0.030) and a time by genotype interaction (F21,476 = 1.63, P = 0.040). During minute five of the task, the Gtf2i+/−/Gtf2ird1−/− mutants froze significantly more than WTs (P = 0.030) as did the Gtf2ird1+/− mice (P = 0.024) (Fig. 7I). The cued fear phenotype could not be explained by differences in shock sensitivity (Supplementary Material, Fig. S9H).
Finally, we also tested just these mutants for any enhancement of social behavior, as individuals with WS have increased social motivation. However, at least with the standard social approach task used here, we did not see any difference among the genotypes in time spent investigating a social stimulus (Supplementary Material, Fig. S9I).
By crossing these mutant lines, we tested the hypothesis that the double heterozygous mutant would be more severe than a mutation only affecting Gtf2ird1. Gtf2ird1+/− and Gtf2i+/−/Gtf2ird1+/− genotypes resulted in mild deficits compared to WTs that, in some cases, were intermediate to the Gtf2i+/−/Gtf2ird1−/− phenotype. There were no instances where the Gtf2ird1+/− or Gtf2i+/−/Gtf2ird1+/− genotypes were significantly different from each other, suggesting that in the behaviors we have tested, the Gtf2i mutation does not modify the effects of a Gtf2ird1 mutation. This unique cross also allowed us to characterize a new mouse line Gtf2i+/−/Gtf2ird1−/−, which had the largest impact on behaviors. The phenotypes of Gtf2i+/−/Gtf2ird1−/− were always in the same direction as the phenotypes in the Gtf2ird1−/− mouse model, but we also saw a significant PPI deficit and cued fear difference when the Gtf2i mutation was added. This further supports that the behaviors tested here, such as balance, marble burying and learning and memory, are largely affected by homozygous mutations in Gtf2ird1.
Discussion
We have described the in vivo DNA binding sites of GTF2IRD1 and GTF2I in the developing mouse brain. This is the first description of these two TFs in a tissue that is relevant for the behavioral phenotypes seen in mouse models of WS. GTF2IRD1 showed a preference for active sites and promoter regions. The conservation of GTF2IRD1 targets was higher on average than would be expected by chance, which provides evidence that these are functionally important regions of the genome. The functions of genes bound by GTF2IRD1 include transcriptional regulation, such as chromatin modifiers, as well as posttranslational regulation including protein ubiquitination. A role for GTF2IRD1 in regulating genes involved in protein ubiquitination has not been described before. This supports the role of GTF2IRD1 in regulating chromatin by transcriptionally controlling other chromatin modifiers. These data, along with the localization pattern of GTF2IRD1 in the nucleus and its direct association with other chromatin modifiers such as ZMYM5 (21,42), suggest that GTF2IRD1 can exert its regulation of chromatin at several different levels of biological organization. Motif enrichment analysis of GTF2IRD1 peaks indicated that CTCF cobinds with GTF2IRD1. Consistent with this, GTF2IRD1 is also often present at TAD boundaries where CTCF is known to be enriched. This further suggests GTF2IRD1 may have a role in defining chromatin topology. GTF2I has been shown to interact with and target CTCF to specific sites in the genome (27), so it would be interesting to test if GTF2IRD1 has a similar relationship with CTCF.
Overall, GTF2I showed a similar preference for promoters and active regions, although it had more intergenic targets than GTF2IRD1, and the conservation of GTF2I peaks was significantly lower than GTF2IRD1 peaks. The genes bound by GTF2I were enriched for signal transduction and phosphorylation. Interestingly, GTF2I was bound to the Src gene body. SRC is known to phosphorylate GTF2I to induce its transcriptional activity (18). Phosphorylation of GTF2I by SRC also antagonizes calcium entry into the cell (19). While knocking out Gtf2i did not affect the expression of Src, it would be interesting to understand the functional consequence of GTF2I binding Src, especially since Src knockout mice exhibit similar behaviors as Gtf2i mouse models (43).
The overlap of GTF2I and GTF2IRD1 targets was significant, and the target genes were enriched for synaptic genes, cellular responses to reactive oxygen species and signal transduction. This overlap, particularly with synaptic genes, suggested that these genes could interact via their binding targets to produce cognitive and behavioral phenotypes. To test the difference between combined Gtf2i and Gtf2ird1 mutation and mutation of Gtf2ird1 alone, we characterized two new mouse models. We used the CRISPR/Cas9 system to generate multiple mutations in the two genes individually as well as together from one embryo injection. The ease and combinatorial possibilities of this technology will be amenable to testing many unique combinations of genetic mutations in copy number variant regions, which will be important to fully understand the complex relationships of genes in these disorders.
We found a frameshift mutation expected to trigger non-sense-mediated decay in Gtf2ird1 did not degrade the mRNA but did result in an N-terminal truncation and protein level reduction in the homozygous mutant (14). Even a larger, 589 bp deletion of exon 3 in Gtf2ird1 did not result in mRNA degradation, but did have a larger effect on protein level. This phenomenon of increased Gtf2ird1 RNA levels has been seen in at least three other mouse models of Gtf2ird1 (8,14,35). Two of these were made using a classic homologous recombination removing either exon 2 alone or exon 2 through part of exon 5. In both of these models, Gtf2ird1 transcript was still made, but no in vivo protein analysis was done due to poor-quality antibodies and the undetectably low protein expression in WT mice. The third model also saw the N-terminal truncation. The presence of an aberrant protein that can still bind the genome, such as the mutant described here, could explain the lack of transcriptomic differences in the brain shown here and by others (37). The mutant protein may also still interact with other binding partners and be trafficked to the appropriate genomic loci. This mutation did disrupt the binding of GTF2IRD1 to its own promoter, which resulted in an increase in transcript levels. The property that specifies GTF2IRD1 binding to its own promoter must be unique, as DNA binding genome-wide was not robustly perturbed in the mutant.
In the end, GTF2IRD1 has proven to be a remarkably difficult protein to disrupt in a targeted manner—a finding that may modify the interpretation of prior studies using a variety of mutant lines, including ours (14). Indeed, it took years to establish a sufficiently sensitive immunoblotting protocol for GTF2IRD1, much less a ChIP-seq protocol to study its binding genome-wide. Thus, even in the current study, much of the transcriptional and behavioral characterization was complete prior to discovering that substantial DNA binding remained. Presumably the resiliency of this binding also extends to the mutants we used in our recent test of the sufficiency of Gtf2i family mutants to recapitulate the deletion of the entire locus (14). Therefore, it remains challenging to determine to what extent existing Gtf2ird1 exonic mutants model the loss of this gene in WS, where the whole genic locus is deleted. Interestingly, when the whole locus is deleted in the ‘CD’ complete deletion mice, the elevation of Gtf2ird1 mRNA seen in exonic mutants does not occur (14). However, protein levels appear to stay above 50%, albeit with substantial mouse to mouse variation. Postmortem patient brain samples, if available, may help to resolve the consequences of the human mutation on GTF2IRD1 protein levels. Further, it may be worth revisiting the consequences of Gtf2ird1 mutation following generation of full genic deletions in mice. In the meantime, it may be that some of the homozygous point mutations, though different from the heterozygous mutations of WS, may better model WS protein levels as they can result in a reduction of GTF2IRD1 protein (Fig. 4). Indeed, most studies of Gtf2ird1 mutant behavioral consequences have shown atypical phenotypes in homozygous mutant mice (8,9,24).
It is worth noting that even in those mutants with a 50% reduction in protein, transcriptional changes are very subtle, at least at a steady state. This is similar to findings in other chromatin-modifying knockouts, where final changes in transcription are highly subtle (44–48), even when behavioral consequences can be severe or even lethal, in the case of Mecp2 mutants (49). Thus, a second remaining puzzle about these genes is what their role is in regulating transcription at the majority of their binding sites. One possibility is these genes are essential for regulating the proper dynamics of gene expression, something not captured when assessing a population at a steady state. Another possibility is they affect phenotypes via actions in rare cell types not easily detected in whole brain RNA-seq. Both hypotheses await further experimentation. Finally, it is possible their role might be more in modifying something not well assessed in RNA-seq data, such as DNA methylation, which has shown dysregulation at CTCF sites in blood cells in WS (50). Nonetheless, the strong enrichment of ASD and constrained genes among GTF2I and especially GTF2IRD1 targets suggest that these factors may be key regulators. Such functional interactions suggest common pathways across these chromatin-related forms of ASD and intellectual disability.
Regardless of the resiliency at the protein level, we show heterozygous and homozygous mutations of Gtf2ird1 were sufficient to cause adult behavioral abnormalities. This supports the hypothesis that the N-terminal end of the protein has other important functions beyond DNA binding. Similarly, the N-truncation of GTF2IRD1 did not affect DNA binding genome-wide, but still resulted in behavioral deficits (51). The single Gtf2ird1 homozygous mutant showed balance deficits, which is consistent across many mouse models of WS. We also observed decreased marble burying. This task is thought to be mediated at least in part by hippocampal function, suggesting a possible disruption of the hippocampus caused by this mutation (52). We also observed an increase in contextual fear response, another cognitive task that is thought to be under hippocampal and amygdala regulation. An increase in contextual fear was also seen in another Gtf2ird1 mouse model (53).
Given the prior evidence that these two TFs are both involved in cognitive and behavioral phenotypes of WS (7,54), and the evidence that they shared some binding targets, we tested if having both Gtf2i and Gtf2ird1 mutated could modify the phenotype seen when just Gtf2ird1 was mutated. Contrary to our prediction, we did not see a large effect of adding a Gtf2i mutation to differences in transcriptome-wide expression or behavioral phenotypes. This was also surprising given that we successfully reduced GTF2I protein and it has been described in the literature as regulating transcription (55). Again, whole E13.5 brain analysis could diminish any effects of transcriptional differences in specific cell types. This potential confound could be overcome using single-cell sequencing technologies in the future when those technologies mature and become more reliable for detecting within cell type differences of expression.
It is also worth noting that we did not see any significant changes in the expression of myelination-related genes in heterozygous mutants in the current study or in our prior work where the entire WS locus is deleted (14). This stands in apparent contrast to recent work showing myelination deficits occur downstream of homozygous conditional knockout mutation of Gtf2i only in forebrain neurons (12). However, the most parsimonious explanation for most of the discrepancy is simply the difference between the experimental designs and the conclusions that can be drawn from them—in our case, we are decreasing Gtf2i levels in all cells, more equivalent to levels from a WS mutation, while their study focused primarily on complete removal of Gtf2i just in some neurons. Thus, their study, as a complete loss of function, is more about the role of the Gtf2i gene, rather than the consequences from the ~50% decrease in expression level as seen in WS. As myelination is very well documented to depend on the level of neuronal activity (56), the simplest explanation of the findings across the three papers is that the presence of at least some GTF2I in neurons, potentially in its role as a calcium channel modulator, is required for a normal neuronal activity. Thus, in the complete absence of GTF2I protein in these neurons across development, myelination levels are decreased, and behavior can be disrupted secondary to dysmyelination. This would explain why parallel transcriptomic and behavioral phenotypes did not occur in heterozygous deletions of the whole locus in mice (14), where some GTF2I remains. If the conditional knockout findings are interpreted in that way, then the only remaining discrepancies between the three papers relate to Supplementary Material, Figure S23 of their paper, which shows myelination gene expression changes in a set of non-conditional germline heterozygous Gtf2i mutants. This remaining discrepancy in myelin gene expression is harder to explain. But there are at least four possibilities. First, it could suggest that mutation of Gtf2ird1 and/or other genes in the locus can partially rescue the effect of Gtf2i heterozygosity on myelin gene expression, as we never examined Gtf2i mutation in isolation. Second, it could be regional differences—i.e. the myelination deficits profiled in the cortex (12) do not extend to the hippocampus that was profiled (14). Third, it could be the 50% difference in strain background between the two studies. Fourth, and perhaps the most likely, it could be the difference between the age at which the experiments were conducted: P30 (12) or adult (14). If GTF2I mutation leads to a subtle developmental delay that is resolved by adulthood, it may be the heterozygous mutants were at a slightly more immature (and thus less myelinated) stage when evaluated in (12). Nonetheless, until these possibilities are resolved, it may be premature to test myelination-related therapies in WS.
Regardless, when Gtf2i was knocked down in the presence of two Gtf2ird1 mutations, we saw phenotypes in the same direction as the homozygous one base pair insertion Gtf2ird1 mutant, as well as significant increases in the cued fear memory task. Thus, the behaviors tested in this study seem to be mainly driven by Gtf2ird1 mutant homozygosity, which is consistent across the different mutations. The one exception seemed to be PPI, in which the knockdown of Gtf2i in the presence of two Gtf2ird1 mutations attenuated the effect of the pre-pulse. PPI is thought to be a measure of frontal cortical function and is disrupted in psychiatric diseases such as schizophrenia and ADHD. While PPI has not been tested in patients, our results suggest this would be of interest to examine. There was no effect of homozygous Gtf2ird1 mutation alone on this phenotype, suggesting that Gtf2i and not Gtf2ird1 is playing a larger role. However, the more severe phenotype was seen in the Gtf2i+/−/Gtf2ird1−/− mutant compared to Gtf2i+/−/Gtf2ird1+/−, suggesting a contribution from both genes. This does not exclude the possibility that Gtf2i can modify the phenotype of Gtf2ird1 knockdown in other behavioral domains. To this end, we did also examine whether adding a Gtf2i mutation on top of a Gtf2ird1 mutation altered social behaviors, but did not see any changes in the standard social approach assay. However, on the FVB/AntJ background used here and in F1 FVB/AntJ × C57BL/6J crosses used previously, we have not seen any social behavior disruptions when these genes are mutated or the entire WS locus is deleted (14).
Likewise, the fear conditioning effects of even heterozygous Gtf2ird1 mutation are very clear on the FVB/AntJ background used here, but appear to be masked in F1 FVB × C57BL/6J crosses used in our prior study (14). This indicates that while we did not find evidence for epistasis between Gtf2i and Gft2ird1, there is epistasis between these genes and other loci in the genome. Thus, mouse mapping studies using large cohorts of F2 hybrids might provide an opportunity to leverage this strain difference to find genes that interact with Gtf2ird1 to contribute to these phenotypes. Such studies could help define novel interaction partners for this relatively understudied gene.
Overall, our study has provided the first description of the DNA binding of both GTF2I and GTF2IRD1 in the developing mouse brain and showed they have unique and overlapping targets. These data will be used to inform downstream studies to understand how these TFs interact with the genome. We generated two new mouse models that tested the importance of the N-terminal end of GTF2IRD1 and the effect of mutating both Gtf2i and Gtf2ird1 together. We provided evidence that despite neither gene having much effect on transcription, the Gtf2ird1 mutation affects balance, marble burying, activity levels and fear memory while adding a Gtf2i mutation leads to a larger effect on PPI.
Materials and Methods
Generating genome-edited mice
We generated Gtf2i family mutants as described in (14). To generate unique combinations of gene knockouts, we designed gRNAs targeting early constitutive exons of the mouse Gtf2i and Gtf2ird1 genes. The gRNAs were separately cloned into the pX330 Cas9 expression plasmid (a gift from F. Zhang) and transfected in N2a cells to test for cutting efficiency. DNA was harvested from the cells, and cutting was detected using the T7 endonuclease assay. The gRNAs were transcribed in vitro using the MEGAShortScript kit (Ambion, Austin, TX), and the Cas9 mRNA was in vitro transcribed using the mMessage Machine kit (Ambion). The two gRNAs and Cas9 mRNA were injected into FVB/NJ mouse embryos and implanted into donor females. The resulting offspring were genotyped for mutations with gene-specific primers designed with the Illumina adapter sequences concatenated to their 3′ end to allow for deep sequencing of the amplicons surrounding the expected cut sites. In one line, a large 589 bp deletion in Gtf2ird1 was detected by amplifying 3.5 kb that included exon 2, exon 3 and part of intron 3 and then using a Nextera library prep (Illumina, San Diego, CA) to deep sequence the amplicon. Here we focus on two founder mice obtained from these injections. Founder lines were bred to FVB/AntJ mice to ensure the mutations existed in the germline and, for double mutant founders, on the same chromosome. The mice were further backcrossed until the mutations were on a complete FVB/AntJ background, which differs from the FVB/NJ background at two loci: Tyrc-ch, which gives FVB/AntJ a chinchilla coat color, and the 129P2/OlaHSd WT Pde6b allele, which prevents FVB/AntJ from becoming blind in adulthood. Coat color was identified by eye, and the Pde6b gene was genotyped by PCR. These mouse lines will be available through the MMRRC (66 710, 66 711).
Western blotting
Embryos were harvested on embryonic day 13.5 (E13.5), and the whole brain was dissected in cold PBS and then flash-frozen in liquid nitrogen. The brains were stored at −80°C until they were lysed. The frozen brain was homogenized in 500 μl of 1xRIPA buffer [10 mm Tris–HCl pH 7.5, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% DOC, 0.1% SDS, 10 mm Na3V04, 10 mm NaF, 1× protease inhibitor (Roche, Basil, Switzerland)] along with 1:1000 dilution of RNase inhibitors [RNasin (Promega, Madison, WI) and SUPERaseIn (Thermo Fisher Scientific, Waltham, MA]. The homogenate was incubated on ice for 20 min and then spun at 10 000g for 10 min at 4°C to clear the lysate. The lysate was stored in two aliquots of 100 μl at −80°C for later protein analysis, and 250 μl of the lysate was added to 750 μl of TRIzol LS and stored at −80°C for later RNA extraction and qPCR. The total protein was quantified using the BCA assay, and 25–50 μg of protein in 1× Laemmli buffer with β-mercaptoethanol was loaded onto 4–15% TGX protean gels (Bio-Rad, Hercules, CA). The protein was transferred to a 0.2 μm PVDF membrane by wet transfer. The membrane was blocked with 5% milk in TBST for 1 h at a room temperature. The membrane was cut at the 75 kDa protein marker; the bottom was probed with a GAPDH antibody as an endogenous loading control, while the top was probed with an antibody for either GTF2I or GTF2IRD1. The primary incubation was performed overnight at 4°C. The membrane was washed three times in TBST for 5 min and then incubated with an HRP-conjugated secondary antibody diluted in 5% milk in TBST for 1 h at a room temperature. The blot was washed three times with TBST for 5 min and then incubated with Clarity Western ECL substrate (Bio-Rad) for 5 min. The blot was imaged in a myECL Imager (Thermo Fisher Scientific). Relative protein abundance was quantified using Fiji (NIH) and normalized to GAPDH levels in a reference WT sample. The antibodies used were rabbit anti-GTF2IRD1 (1:500, Novus, NBP1–91973), mouse anti-GTF2I (1:1000 BD Transduction Laboratories, Lexington, KY, BAP-135) and mouse anti-GAPDH (1:10 000, Sigma-Aldrich, St. Louis, MO, G8795), HRP-conjugated goat anti-rabbit IgG (1:2000, Sigma-Aldrich, AP307P) and HRP-conjugated goat anti-mouse IgG (1:2000, Bio-Rad, 1 706 516).
Immunoprecipitation
To test the specificity of the antibodies for GTF2IRD1 and GTF2I, we performed IP with rabbit anti-GTF2IRD1 (4 μg, Novus, NBP1–91973), rabbit anti-GTF2I (2 μg Bethyl Laboratories, Montgomery, TX, A301-330A) and total rabbit IgG (4 μg Jackson ImmunoResearch 011–000-002) on E13.5 brains from WT FVB/ANTJ embryos. Anti-rabbit IgG was coupled to protein G-coated streptavidin magnetic beads and incubated overnight with the brain lysate and primary antibodies. The IP samples were applied to a magnet, and 10 μl of the supernatant was collected for the post-IP sample. The magnetic beads were washed three times with a RIPA buffer.
Western blotting was performed as described above on the input, IP and post-IP sample for each IP condition. Since we did not identify another specific GTF2IRD1 antibody, we blotted with the same antibody that was used for the IP rabbit anti-GTF2IRD1 (1:500, Novus, NBP1–91973). Since the secondary HRP–antibody species is the same as the IP species IgG, we detected a large smear below 150 kDa that is coming from the IgG heavy chain, marked with * on Figure 1. The blot was stripped and then probed with mouse anti-GTF2I (1:1000 BD Transduction Laboratories, Lexington, KY, BAP-135).
Transcript abundance using RT-qPCR
RNA was extracted from TRIzol LS using the Zymo Clean and Concentrator-5 kit with On-Column DNase-I Digestion following the manufacturer’s instructions. The RNA was eluted in 30 μl of RNase-free water and quantified using a NanoDrop 2000 (Thermo Fisher Scientific). One microgram of RNA was transcribed into cDNA using the qScript cDNA synthesis kit (Quanta Biosciences, Beverly, MA). Half a microliter of cDNA was used in a 10 μl PCR reaction with 500 nm of target-specific primers and the PowerUP SYBR green master mix (Applied Biosystems, Foster City, CA). The primers were designed to amplify exons that were constitutively expressed in both Gtf2i (exons 25 and 27) and Gtf2ird1 (exons 8 and 9) and span an intron (14). The RT-qPCR was carried out in a QuantStudio6Flex machine (Applied Biosystems) using the following cycling conditions: (1) 95°C for 20 s, (2) 95°C for 1 s and (3) 60°C for 20 s; then repeat steps 2 and 3 for 40 times. Each target and sample was run in triplicate technical replicates, with three biological replicates for each genotype. The relative transcript abundance was determined using the delta CT method normalizing to Gapdh.
ChIP
Chromatin was prepared as described previously (14). Frozen brains were homogenized in 10 ml of cross-linking buffer [10 mm HEPES pH 7.5, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% formaldehyde (Sigma-Aldrich)]. The homogenate was spun down and resuspended in 5 ml of 1× L1 buffer [50 mm HEPES pH 7.5, 140 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.25% Triton X-100, 0.5% NP40, 10.0% glycerol, 1 mm BGP (Sigma-Aldrich), 1× Na butyrate (Millipore, Burlington, MA), 20 mm NaF, 1× protease inhibitor (Roche)] to release the nuclei. The nuclei were spun down and resuspended in 5 ml of L2 buffer (10 mm Tris–HCl pH 8.0, 200 mm NaCl, 1 mm BGP, 1× Na butyrate, 20 mm NaF, 1× protease inhibitor) and rocked at a room temperature for 5 min. The nuclei were spun down and resuspended in 950 μl of buffer L3 (10mm Tris–HCl pH 8.0, 1 mm EDTA, 1 mm EGTA, 0.3% SDS, 1 mm BGP, 1× Na butyrate, 20 mm NaF, 1× protease inhibitor) and sonicated to a fragment size of 100–500 bp in a Covaris E220 focused-ultrasonicator with 5% duty factor, 140 PIP and 200 cbp. The sonicated chromatin was diluted with 950 μl of L3 buffer and 950 μl of 3× Covaris buffer (20 mm Tris–HCl pH 8.0, 3.0% Triton X-100, 450 mm NaCl, 3 mm EDTA). The diluted chromatin was pre-cleared using 15 μl of protein G-coated streptavidin magnetic beads (Thermo Fisher Scientific) for 2 h at 4°C. For IP, 15 μl of protein G-coated streptavidin beads were conjugated to either 10 μl of GTF2IRD1 antibody (Rb anti-GTF2IRD1, NBP1–91973 LOT:R40410) or 10 μl of GTF2I antibody (Rb anti-GTF2I; Bethyl Laboratories, Montgomery, TX, A301-330A) for 1 h at a room temperature. 80 μl of the pre-cleared lysate was saved for an input sample. 400 μl of the pre-cleared lysate was added to the beads and incubated overnight at 4°C. The IP sample was then washed twice with low-salt wash buffer (10 mm Tris–HCl pH 8.0, 2 mm EDTA, 150 mm NaCl, 1.0% Triton X-100, 0.1% SDS), twice with a high-salt buffer (10 mm Tris–HCl pH 8.0, 2 mm EDTA, 500 mm NaCl, 1.0% Triton X-100, 0.1% SDS), twice with LiCl wash buffer [10 mm Tris–HCl pH 8.0, 1 mm EDTA, 250 mm LiCl (Sigma-Aldrich), 0.5% NaDeoxycholate, 1.0% NP40] and once with TE buffer (10 mm Tris–HCl pH 8.0, 1 mm EDTA). The DNA was eluted from the beads with 200 μl of 1× TE and 1% SDS by incubating at 65°C in an Eppendorf R thermomixer shaking at 1400 rpm. The DNA was de-cross-linked by incubating at 65°C for 15 h in a thermocycler. The RNA was removed by incubating with 10 μg of RNase A (Invitrogen, Carlsbad, CA) at 37°C for 30 min and then treated with 140 μg of Proteinase K (NEB, Ipswich, MA) and incubated at 55°C in a thermomixer mixing at 900 rpm for 2 h. The DNA was extracted with 200 μl of phenol/chloroform/isoamyl alcohol (Ambion) and cleaned up using the Qiagen PCR purification kit and then eluted in 60 μl of an elution buffer. Concentration was assessed using the high sensitivity DNA kit for Qubit quantification (Thermo Fisher Scientific).
ChIP-qPCR
Primers were designed to amplify the upstream regulatory element of Gtf2ird1. Two off-target primers were designed: one 10 kb upstream of the transcription start site of Bdnf and the other 7 kb upstream of the Pcbp3 transcription start site. The input sample was diluted 1:3, 1:30 and 1:300 to create a standard curve for each primer set and sample. Each standard, input and IP sample for each primer set was performed in triplicate in 10 μl reactions using the PowerUP SYBR green master mix (Applied Biosystems) and 250 nm of forward and reverse primers. The reactions were performed in a QuantStudio6Flex machine (Applied Biosystems) with the following cycling conditions: (1) 50°C for 2 min, (2) 95°C for 10 min, (3) 95°C 15 s and (4) 60°C for 1 min; then repeat steps 3–4 for 40 times. The relative concentrations of the input and IP samples were determined from the standard curve for each primer set. Enrichment of the IP samples was determined by dividing the on-target upstream regulatory element relative concentration by the off-target relative concentration.
ChIP-seq
ChIP-seq libraries were prepared using the Swift Accel-NGS 2S plus DNA library prep kits with dual indexing (Swift Biosciences, Ann Arbor, MI). The final libraries were enriched by 13 cycles of PCR. The libraries were sequenced by the Genome Technology Access Center at Washington University School of Medicine on a HiSeq3000 producing 1 × 50 reads.
Raw reads were trimmed of adapter sequences and bases with a quality score below 25 using the Trimmomatic Software (57). The trimmed reads were aligned to the mm10 genome using the default settings of bowtie2 (58). Reads with a mapping quality of less than 10 were removed. Picard tools were used to remove duplicates from the filtered reads (http://broadinstitute.github.io/picard). Macs2 was used to call peaks on the WT IP, Gtf2ird1−/− IP and Gtf2i−/−/Gtf2ird1−/− IP samples with the corresponding sample’s input as the control for each biological replicate (59). Macs2 used an FDR of 0.01 as the threshold to call a significant peak. High confidence peaks were those peaks that had some overlap within each biological replicate for each genotype using bedtools intersect (60). The read coverage for the WT high confidence peaks was determined using bedtools coverage for all genotypes. To identify peaks with differential coverage, we used EdgeR to compare the WT peak coverage files to the corresponding mutant peak coverage; differential peaks were defined as having an FDR < 0.1 (61). To determine GTF2I high confidence peaks, we used peaks that had overlap between all four biological replicates and WT peaks with an FDR < 0.1 and log2FC > 0 when compared to the Gtf2i−/−/Gtf2ird1−/− IP coverage, since this mutation represents a full knockout of the protein.
Annotation of peaks and motif analysis were performed using the HOMER software on the high confidence peaks (62). Peaks were annotated at the transcription start site (TSS) of genes if the peak overlapped the +2.5 kb or −1 kb of the TSS using a custom R script using ensemble version 94 TSS annotations downloaded using BioConductor. The RGATTR motif and reverse complement were searched for in the sequences of the peaks using a custom script. GO analysis on the ChIP target genes was performed using the goseq R package and assuming 22 007 protein coding genes in the genome based on current NCBI annotation. Comparison to ASD genes entailed testing for overlap between ChIP target genes and the genes identified by (31), their table S4, using Fisher’s exact test and assuming 22 007 protein coding genes in the genome based on the current NCBI annotation. Slightly more significant results were obtained when replicating this analysis using the SFARI gene database (63), accessed 8/4/2019, and testing for enrichment of score 1 and 2 ASD genes. We also analyzed pLI scores, downloaded from gnomAD (31) on 9/16/2019, of ChIP peaks. Similar results were obtained using pLI cutoffs of 0.9 and 0.99. For epigenetic overlaps, we used E13.5 H3K4me3 and E13.5 H3K27me3 forebrain narrow bed peak files from the mouse ENCODE project to overlap with our peak datasets (64). deepTools was used to generate bigwig files normalized to the library size for each sample by splitting the genome into 50 bp overlapping bins (65). deepTools was used to visualize the ChIP-seq coverage within the H3K4me3 and H3K27me3 peak regions. The LICR TFBS E14.5 whole brain CTCF peak dataset was downloaded from the UCSC genome browser and lifted over to mm10 coordinates. TAD analysis of E14.5 cortical neuron HiC data (66) was carried out using domains previously called by arrowhead (67,68). Domain boundaries were defined as 10 kb regions centered on the start and end of each domain and lifted over to mm10 coordinates.
Overlaps between GTF2IRD1 and GTF2I peaks and other peak datasets were performed using the bedtools fisher function. The fisher function in bedtools tests the significance of overlap of two sets of regions given a genome size by first identifying the number of overlapping and unique regions for each set. It uses a heuristic based on the size of the regions in the comparison files and the size of the genome provided to estimate the number of possible intervals as the background. We used the mm10 mouse genome size. This method can inflate the P-values of the Fisher test, so we also made a new file with the same number of regions and size of regions randomly sampled from the mouse genome using the bedtools shuffle tool. We compared the random regions with the regions of interest (e.g. CTCF, H3K27me3) and recorded the P-value and OR. We repeated this 1000 times to get a null distribution to which we could compare our experimental ChIP-seq target region P-value (with a minimum of P < 0.0001) and OR. We did this for both the GTF2I and the GTF2IRD1 peak files.
Since these overlaps could be driven mainly by the presence of all these marks at promoter sites, we annotated all comparison files using the HOMER annotatePeaks.pl function. We used grep –v ‘TSS’ to remove all peaks annotated at a promoter-TSS and then repeated Fisher’s exact test using bedtools fisher function and the randomization analysis described above. All results were replicated when excluding promoters in this fashion.
In order to compare our results to the previous studies performed in the literature, we obtained Supplementary Material, Tables S1–S4 from Makeyev et al. (22) which contained lists of genes that had either GTF2I or GTF2IRD1 bound at the promoters in a mouse ES cell line or E10.5 craniofacial tissue. They used two different GTF2IRD1 and GTF2I antibodies that are described here and used a ChIP-CHIP method to determine peaks. We also compared our GTF2I data to the genes identified in human iPSCs from Supplementary Material, Table S6 from Adamo et al. (28). We used the same GTF2I antibody as described in Adamo et al. (28). The significance of overlaps between datasets was assessed using the GeneOverlap package in R (69), which uses Fisher’s exact test and assuming 22 007 protein coding genes in the genome based on the current NCBI annotation as the background.
PhyloP scores for the region underneath the WT ChIP-seq peaks and random genomic regions of the same length were retrieved using the UCSC table browser 60 Vertebrate Conservation PhyloP table. PhyloP scores were obtained from a random subset of promoter regions taken from the HOMER 4.8 promoter region table. The Epigenome browser was used to visualize the ChIP-seq data as tracks (70).
RNA-seq
One microgram of E13.5 whole brain total RNA extracted from TRIzol LS was used as input for rRNA depletion using the NEBNext rRNA Depletion Kit (Human/Mouse/Rat). The rRNA-depleted RNA was used as input for library construction using the NEBNext Ultra II RNA library prep kit for Illumina. The final libraries were indexed and enriched by PCR using the following thermocycler conditions, (1) 98°C for 30 s, (2) 98°C for 10 s, (3) 65°C for 75 s and (4) 65°C for 5 min; (5) hold at 4°C, repeating steps 2–3 six times. The libraries were sequenced by the Genome Technology Access Center at Washington University School of Medicine on a HiSeq3000 producing 1 × 50 reads.
RNA-seq analysis
The raw RNA-seq reads were trimmed of Illumina adapters and bases with quality scores less than 25 using Trimmomatic Software. The trimmed reads were aligned to the mm10 mouse genome using the default parameters of STARv2.6.1b (71). We used HTSeq-count to determine the read counts for features using the Ensembl GRCm38 version 93 gtf file (72). Differential gene expression analysis was done using EdgeR. We compared the expression of genes that are targets of either GTF2IRD1 or GTF2I to non-bound genes by generating a cumulative distribution plot of the average log CPM of the genes between genotypes. GO analysis was performed using the goseq R package.
Data availability
All RNA-seq and ChIP-seq data are available at GEO accession GSE138234.
Behavioral tasks
All animal testing was done with the approval from the Washington University in St. Louis Institutional Animal Care and Use Committee. Mice were group housed in same-sex, mixed-genotype cages with two to five mice per cage in standard mouse cages (dimensions 28.5 × 17.5 × 12 cm) on corn cob bedding. Mice had ad libitum access to food and water and followed a 12-h light-dark cycle (light from 6:00 a.m. to 6:00 p.m.). Animal housing rooms were kept at 20–22°C with a relative humidity of 50%. All mice were maintained on the FVB/AntJ (73) background from Jackson Labs. All behaviors were done in adulthood between ages P58 and P133. A week prior to behavioral testing, mice were handled by the experimenter for habituation. On testing days the mice were moved to habituate to the testing room and the experimenter (when male) for 30 min before testing started.
Ledge
To test balance, we measured how long a mouse could balance on an acrylic ledge with a width of 0.5 cm and a height of 38 cm as described (73). The time when the mouse fell off the ledge was recorded up to 60 s when the trial was stopped. If the mouse fell off within 5 s, the time was restarted and the mouse received another attempt. If the mouse fell off within the first 5 s on a third attempt, that time was recorded. We tested all mice on the ledge twice, with a rest period of at least 20 min between trials. Trial average was used for analysis.
One-hour locomotor activity
We assessed activity levels in a 1-h locomotor task, as previously described (73). Mice were placed in the center of a standard rat-sized cage (dimensions 47.6 × 25.4 × 20.6 cm). The rat-sized cage was located inside a sound-attenuating box with white light at 24 lux. The mice could freely explore the cage for 1 h. An acrylic lid with air holes was placed on top of the cage to prevent mice from jumping out. The position and horizontal movement of the mice were tracked using ANY-maze software (Stoelting Co., Wood Dale, IL: RRID: SCR_014289). The apparatus was divided into two zones: a 33 × 11 cm center zone and an edge zone of 5.5 cm that bordered the cage. The animal was considered in a zone if 80% of the mouse was detected in that zone. ANY-maze recorded the time, distance and number of entries into each zone. After the task, the mouse was returned to its home cage and the apparatus was thoroughly cleaned with 70% ethanol.
Marble burying
Marble burying is a species-specific task that measures the compulsive digging behavior of mice. Normal hippocampal function is thought to be required for normal phenotypes in this task. We tested marble burying as described previously (73). A rat cage was filled with aspen bedding to a depth of 3 cm and placed in a sound-attenuating box with white light set to 24 lux. Twenty marbles were placed on top of the bedding in a 5 × 4 evenly spaced grid. The experimental mouse was placed in the center of the chamber and allowed to freely explore and dig for 30 min. An acrylic lid with air holes was placed on top of the cage to prevent mice from escaping. After 30 min, the animal was returned to its home cage. Two scorers counted the number of marbles not buried (less than two-thirds of the marble was covered with bedding). The number of marbles buried was then determined, and the average of the two scores was used in the analysis. After the marbles were counted, the bedding was disposed of and the cage and marbles were cleaned with 70% ethanol.
Three-chamber social approach
Sociability was tested using the standard three-chamber social approach paradigm (14,73–75). The test apparatus was a plexiglass arena partitioned into three chambers measuring 19.5 × 39 × 22 cm. Rectangular openings measuring 5 × 8 cm allowed the experimental mouse to travel between chambers. The test was made up of three consecutive 10-min trials. In the first trial, the experimental mouse was allowed to habituate in the middle chamber with the openings to the side chambers closed. In the second trial, the experimental mouse could freely explore the whole apparatus. The third trial was the sociability trial. An empty upside-down pencil cup (Galaxy Pencil/Utility Cup, Spectrum Diversified Designs, Inc.) was placed in one side chamber and another pencil cup was placed upside down in the opposite chamber and enclosed a sex- and age-matched WT mouse. A clear plastic drinking cup was placed on top of the pencil cup to prevent the experimental mouse from climbing on top of the pencil cup. The experimental mouse was given 10 min to explore the whole apparatus. The trials were recorded and tracked using the ANY-maze software. ANY-maze calculated the time the experimental mouse spent investigating the social stimulus or empty cup as the time the mouse’s head was within 2 cm of either cup. The first 5 min of the social trial was analyzed using a mixed linear model with genotype and chamber side as fixed effects and the animal as a random effect to account for the repeated measures of the experimental mice. A stimulus animal was only used once per testing session. One day prior to testing, the stimulus mice were habituated to the apparatus and pencil cups for 10 min. The apparatus was cleaned with 2% chlorhexidine (Zoetis), and the pencil cups were cleaned with 70% ethanol between each test.
Pre-pulse inhibition
PPI measures the suppression of the acoustic startle reflex when an animal is presented with a smaller stimulus (pre-pulse) before a more intense stimulus (startle). Acoustic startle response and PPI were measured as previously described (14). Startle Monitor II software was used to run the protocol in a sound-attenuating chamber (Kinder Scientific, LLC, Poway, CA). Mice were secured in a small enclosure and placed on a force-sensitive plate inside the chamber. Animals were acclimated to the test chamber for 5 min before trials began. Startle amplitude was recorded with 1 ms force readings during 65 pseudo-randomized trials that alternated between various stimulus conditions. An auditory stimulus of 120 dB was presented for 40 ms to illicit the startle response. PPI was measured in three different types of trials where the 120 dB stimulus was preceded by pre-pulses of 4, 8 and 16 dB above the background sound level (65 dB). PPI was calculated by using the average startle response (measured in Newtons) of the 120 dB stimulus trials minus the average startle response to the stimulus after each respective pre-pulse level. This was divided by the 120 dB stimulus startle response and multiplied by 100 to get a percent inhibition of startle.
Contextual and cued fear conditioning
Learning and memory were tested using the contextual and cued fear conditioning paradigm as previously described (76). Contextual fear memory is thought to be driven by hippocampal functioning, whereas cued fear is thought to be driven by amygdala functioning. On day 1 of the experiment, animals were placed in an acrylic chamber (26 cm × 18 cm × 18 cm; Med Associates Inc., Fairfax, VT) with a metal grid floor and a peppermint odor from an unobtainable source. The chamber light was on for the duration of the 5-min task. During the first 2 min, the animal freely explored the apparatus to measure baseline freezing. At 100 s, 160 s and 220 s, an 80 dB white noise tone (conditioned stimulus, CS) was played for a duration of 20 s. During the last 2 s of this tone, the mice received a 1.0 mA foot shock (unconditioned stimulus, UCS). The animal’s freezing behavior was monitored by FreezeFrame software (Actimetrics, Evanston, IL) in 0.75 s intervals. Freezing was defined as no movement besides respiration and was used as a measure of the fear response in mice. After the 5-min task, the mice were returned to their home cage. On day 2, we tested contextual fear memory. The mice were placed in the same chamber as day 1 with the peppermint odor, and freezing behavior was measured over 8 min. The first 2 min of day 2 were compared to the first 2 min of day 1 to test for the acquisition of fear memory. The mice were returned to their home cage after the 8-min task. On day 3, to test cued fear, the mice were placed in a new black-and-white chamber that was partitioned into a triangle shape and had a novel coconut scent. The mice were allowed to explore the chamber, and the first 2 min were considered baseline. After minute 2, the 80 dB tone (CS) was played for the remaining 8 min. Freezing behavior was monitored during the entire 10-min task.
Shock sensitivity
We tested the shock sensitivity of the mice to ensure that differences in conditioned fear were not due to differing responses to the shock itself, following a previously established protocol (14). Mice were placed in the apparatus used for day 1 and day 2 of the conditioned fear task. The mice were delivered a 2-s 0.05 mA shock, and their behavior was observed. The shock was increased by 0.05 mA up to 1 mA or until the mice exhibited flinching, escape and vocalization behaviors. Once the mouse had shown each behavior, the test was ended and the final mA used was recorded.
Statistical analysis
All statistical analyses were performed in R v3.4.2 and are reported in Supplementary Material, Table S3. The ANOVA assumption of normality was assessed using the Shapiro–Wilks test and manual inspection of qqPlots, and the assumption of equal variances was assessed with Levene’s test. When appropriate, ANOVA was used to test for main effects and interaction terms. Post hoc analyses were done to compare between genotypes. If the data violated the assumptions of ANOVA, non-parametric tests were performed. If the experiment was performed over time, linear mixed models were used to account for the repeated measures of an animal using the lme4 R package. Post hoc analyses were then conducted to compare between genotypes within time bins. Post hoc analyses were done using the multcomp R package (77). Animals were removed from analysis if they had a value that was 3.29 standard deviations above the mean or had poor video tracking and could not be analyzed.
Supplementary Material
Acknowledgements
We would like to thank the Genome Technology Access Center for their technical support, as well as Dr Beth Kozel for her critical advice on this project. We would also like to thank Dr David Wozniak and the Animal Behavior Core at the Washington University School of Medicine for their support.
Funding
National Institute of Health 1R01MH107515 (J.D.D.); Autism Science Foundation; National Science Foundation Graduate Research Fellowship (DGE-1745038 to N.D.K. and K.R.N.).
Conflict of Interest statement
None of the authors have any conflict of interest that could bias the work presented here.
References
- 1. Merla, G., Brunetti-Pierri, N., Micale, L. and Fusco, C. (2010) Copy number variants at Williams–Beuren syndrome 7q11.23 region. Hum. Genet., 128, 3–26. [DOI] [PubMed] [Google Scholar]
- 2. Meyer-Lindenberg, A., Mervis, C.B. and Faith Berman, K. (2006) Neural mechanisms in Williams syndrome: a unique window to genetic influences on cognition and behaviour. Nat. Rev. Neurosci., 7, 380–393. [DOI] [PubMed] [Google Scholar]
- 3. Korenberg, J.R., Chen, X.-N., Hirota, H., Lai, Z., Bellugi, U., Burian, D., Roe, B. and Matsuoka, R. (2000) VI. Genome structure and cognitive map of Williams syndrome. J. Cogn. Neurosci., 12, 89–107. [DOI] [PubMed] [Google Scholar]
- 4. Sakurai, T., Dorr, N.P., Takahashi, N., McInnes, L.A., Elder, G.A. and Buxbaum, J.D. (2011) Haploinsufficiency of Gtf 2i, a gene deleted in Williams syndrome, leads to increases in social interactions. Autism Res., 4, 28–39. [DOI] [PubMed] [Google Scholar]
- 5. Segura-Puimedon, M., Sahún, I., Velot, E., Dubus, P., Borralleras, C., Rodrigues, A.J., Valero, M.C., Valverde, O., Sousa, N., Herault, Y.et al. (2014) Heterozygous deletion of the Williams–Beuren syndrome critical interval in mice recapitulates most features of the human disorder. Hum. Mol. Genet., 23, 6481–6494. [DOI] [PubMed] [Google Scholar]
- 6. Li, H.H., Roy, M., Kuscuoglu, U., Spencer, C.M., Halm, B., Harrison, K.C., Bayle, J.H., Splendore, A., Ding, F., Meltzer, L.A.et al. (2009) Induced chromosome deletions cause hypersociability and other features of Williams–Beuren syndrome in mice. EMBO Mol. Med., 1, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osborne, L.R. (2010) Animal models of Williams syndrome. Am. J. Med. Genet., 154C, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Young, E.J., Lipina, T., Tam, E., Mandel, A., Clapcote, S.J., Bechard, A.R., Chambers, J., Mount, H.T.J., Fletcher, P.J., Roder, J.C.et al. (2008) Reduced fear and aggression and altered serotonin metabolism in Gtf 2ird1-targeted mice. Genes Brain Behav., 7, 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howard, M.L., Palmer, S.J., Taylor, K.M., Arthurson, G.J., Spitzer, M.W., Du, X., Pang, T.Y.C., Renoir, T., Hardeman, E.C. and Hannan, A.J. (2012) Mutation of Gtf 2ird1 from the Williams–Beuren syndrome critical region results in facial dysplasia, motor dysfunction, and altered vocalisations. Neurobiol. Dis., 45, 913–922. [DOI] [PubMed] [Google Scholar]
- 10. Kopp, N.D., Parrish, P.C.R., Lugo, M., Dougherty, J.D. and Kozel, B.A. (2018) Exome sequencing of 85 Williams–Beuren syndrome cases rules out coding variation as a major contributor to remaining variance in social behavior. Mol. Genet. Genom. Med., 6, 749–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schaefer, M.L., Wong, S.T., Wozniak, D.F., Muglia, L.M., Liauw, J.A., Zhuo, M., Nardi, A., Hartman, R.E., Vogt, S.K., Luedke, C.E.et al. (2000) Altered stress-induced anxiety in adenylyl cyclase type VIII-deficient mice. J. Neurosci., 20, 4809–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barak, B., Zhang, Z., Liu, Y., Nir, A., Trangle, S.S., Ennis, M., Levandowski, K.M., Wang, D., Quast, K., Boulting, G.L.et al. (2019) Neuronal deletion of Gtf 2i, associated with Williams syndrome, causes behavioral and myelin alterations rescuable by a remyelinating drug. Nat. Neurosci., 22, 700. [DOI] [PubMed] [Google Scholar]
- 13. Martin, L.A., Iceberg, E. and Allaf, G. (2018) Consistent hypersocial behavior in mice carrying a deletion of Gtf 2i but no evidence of hyposocial behavior with Gtf 2i duplication: implications for Williams-Beuren syndrome and autism spectrum disorder. Brain Behav., 8, e00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kopp, N., McCullough, K., Maloney, S.E. and Dougherty, J.D. (2019) Gtf 2i and Gtf 2ird1 mutation do not account for the full phenotypic effect of the Williams syndrome critical region in mouse models. Hum. Mol. Genet., 28, 3443–3465. doi: 10.1093/hmg/ddz176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Porter, M.A., Dobson-Stone, C., Kwok, J.B.J., Schofield, P.R., Beckett, W. and Tassabehji, M. (2012) A role for transcription factor GTF2IRD2 in executive function in Williams-Beuren syndrome. PLoS One, 7, e47457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gunbin, K.V. and Ruvinsky, A. (2012) Evolution of general transcription factors. J. Mol. Evol., 76, 28–47. [DOI] [PubMed] [Google Scholar]
- 17. Bayarsaihan, D. and Ruddle, F.H. (2000) Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix–loop–helix domains. PNAS, 97, 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheriyath, V., Desgranges, Z.P. and Roy, A.L. (2002) C-Src-dependent transcriptional activation of TFII-I. J. Biol. Chem., 277, 22798–22805. [DOI] [PubMed] [Google Scholar]
- 19. Caraveo, G., vanRossum, D.B., Patterson, R.L., Snyder, S.H. and Desiderio, S. (2006) Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science, 314, 122–125. [DOI] [PubMed] [Google Scholar]
- 20. Deurloo, M.H.S., Turlova, E., Chen, W.-L., Lin, Y.W., Tam, E., Tassew, N.G., Wu, M., Huang, Y.-C., Crawley, J.N., Monnier, P.P.et al. (2019) Transcription factor 2I regulates neuronal development via TRPC3 in 7q11.23 disorder models. Mol. Neurobiol., 56, 3313–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carmona-Mora, P., Widagdo, J., Tomasetig, F., Canales, C.P., Cha, Y., Lee, W., Alshawaf, A., Dottori, M., Whan, R.M., Hardeman, E.C.et al. (2015) The nuclear localization pattern and interaction partners of GTF2IRD1 demonstrate a role in chromatin regulation. Hum. Genet., 134, 1099–1115. [DOI] [PubMed] [Google Scholar]
- 22. Makeyev, A.V., Enkhmandakh, B., Hong, S.-H., Joshi, P., Shin, D.-G. and Bayarsaihan, D. (2012) Diversity and complexity in chromatin recognition by TFII-I transcription factors in pluripotent embryonic stem cells and embryonic tissues. PLoS One, 7, e44443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bayarsaihan, D., Makeyev, A.V. and Enkhmandakh, B. (2012) Epigenetic modulation by TFII-I during embryonic stem cell differentiation. J. Cell. Biochem., 113, 3056–3060. [DOI] [PubMed] [Google Scholar]
- 24. Schneider, T., Skitt, Z., Liu, Y., Deacon, R.M.J., Flint, J., Karmiloff-Smith, A., Rawlins, J.N.P. and Tassabehji, M. (2012) Anxious, hypoactive phenotype combined with motor deficits in Gtf 2ird1 null mouse model relevant to Williams syndrome. Behav. Brain Res., 233, 458–473. [DOI] [PubMed] [Google Scholar]
- 25. Dai, L., Bellugi, U., Chen, X.-N., Pulst-Korenberg, A.m., Järvinen-Pasley, A., Tirosh-Wagner, T., Eis, P.s., Graham, J., Mills, D., Searcy, Y.et al. (2009) Is it Williams syndrome? GTF2IRD1 implicated in visual–spatial construction and GTF2I in sociability revealed by high resolution arrays. Am. J. Med. Genet., 149A, 302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lazebnik, M.B., Tussie-Luna, M.I. and Roy, A.L. (2008) Determination and functional analysis of the consensus binding site for TFII-I family member BEN, implicated in Williams-Beuren syndrome. J. Biol. Chem., 283, 11078–11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peña-Hernández, R., Marques, M., Hilmi, K., Zhao, T., Saad, A., Alaoui-Jamali, M.A., delRincon, S.V., Ashworth, T., Roy, A.L., Emerson, B.M.et al. (2015) Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. PNAS, 112, E677–E686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adamo, A., Atashpaz, S., Germain, P.-L., Zanella, M., D’Agostino, G., Albertin, V., Chenoweth, J., Micale, L., Fusco, C., Unger, C.et al. (2015) 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat. Genet., 47, 132–141. [DOI] [PubMed] [Google Scholar]
- 29. Yue, F., Cheng, Y., Breschi, A., Vierstra, J., Wu, W., Ryba, T., Sandstrom, R., Ma, Z., Davis, C., Pope, B.D.et al. (2014) A comparative encyclopedia of DNA elements in the mouse genome. Nature, 515, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanders, S.J., Ercan-Sencicek, A.G., Hus, V., Luo, R., Murtha, M.T., Moreno-De-Luca, D., Chu, S.H., Moreau, M.P., Gupta, A.R., Thomson, S.A.et al. (2011) Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron, 70, 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Satterstrom, F.K., Kosmicki, J.A., Wang, J., Breen, M.S., Rubeis, S.D., An, J.-Y., Peng, M., Collins, R.L., Grove, J., Klei, L.et al. (2018) Novel genes for autism implicate both excitatory and inhibitory cell lineages in risk. bioRxiv. doi: 10.1101/484113. [DOI] [Google Scholar]
- 32. Kopp, N., Climer, S. and Dougherty, J.D. (2015) Moving from capstones toward cornerstones: successes and challenges in applying systems biology to identify mechanisms of autism spectrum disorders. Front. Genet., 6, 301–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parikshak, N.N., Luo, R., Zhang, A., Won, H., Lowe, J.K., Chandran, V., Horvath, S. and Geschwind, D.H. (2013) Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell, 155, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karczewski, K.J., Francioli, L.C., Tiao, G., Cummings, B.B., Alföldi, J., Wang, Q., Collins, R.L., Laricchia, K.M., Ganna, A., Birnbaum, D.P.et al. (2019) Variation across 141, 456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. doi: 10.1101/531210. [DOI] [Google Scholar]
- 35. Palmer, S.J., Santucci, N., Widagdo, J., Bontempo, S.J., Taylor, K.M., Tay, E.S.E., Hook, J., Lemckert, F., Gunning, P.W. and Hardeman, E.C. (2010) Negative autoregulation of GTF2IRD1 in Williams-Beuren syndrome via a novel DNA binding mechanism. J. Biol. Chem., 285, 4715–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hinsley, T.A., Cunliffe, P., Tipney, H.J., Brass, A. and Tassabehji, M. (2004) Comparison of TFII-I gene family members deleted in Williams-Beuren syndrome. Protein Sci., 13, 2588–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O’Leary, J. and Osborne, L.R. (2011) Global analysis of gene expression in the developing brain of Gtf 2ird1 knockout mice. PLoS One, 6, e23868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tassabehji, M., Hammond, P., Karmiloff-Smith, A., Thompson, P., Thorgeirsson, S.S., Durkin, M.E., Popescu, N.C., Hutton, T., Metcalfe, K., Rucka, A.et al. (2005) GTF2IRD1 in craniofacial development of humans and mice. Science, 310, 1184–1187. [DOI] [PubMed] [Google Scholar]
- 39. Borralleras, C., Sahun, I., Pérez-Jurado, L.A. and Campuzano, V. (2015) Intracisternal Gtf 2i gene therapy ameliorates deficits in cognition and synaptic plasticity of a mouse model of Williams–Beuren syndrome. Mol. Ther., 23, 1691–1699. doi: 10.1038/mt.2015.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dykens, E.M. (2003) Anxiety, fears, and phobias in persons with Williams syndrome. Dev. Neuropsychol., 23, 291–316. [DOI] [PubMed] [Google Scholar]
- 41. Enkhmandakh, B., Makeyev, A.V., Erdenechimeg, L., Ruddle, F.H., Chimge, N.-O., Tussie-Luna, M.I., Roy, A.L. and Bayarsaihan, D. (2009) Essential functions of the Williams-Beuren syndrome-associated TFII-I genes in embryonic development. PNAS, 106, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Widagdo, J., Taylor, K.M., Gunning, P.W., Hardeman, E.C. and Palmer, S.J. (2012) SUMOylation of GTF2IRD1 regulates protein partner interactions and ubiquitin-mediated degradation. PLoS One, 7, e49283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sinai, L., Ivakine, E.A., Lam, E., Deurloo, M., Dida, J., Zirngibl, R.A., Jung, C., Aubin, J.E., Feng, Z.-P., Yeomans, J.et al. (2015) Disruption of Src is associated with phenotypes related to Williams-Beuren syndrome and altered cellular localization of TFII-I. eNeuro, 2, ENEURO.0016-14. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wade, A.A., Lim, K., Catta-Preta, R. and Nord, A.S. (2019) Common CHD8 genomic targets contrast with model-specific transcriptional impacts of CHD8 haploinsufficiency. Front. Mol. Neurosci., 11, 481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tudor, M., Akbarian, S., Chen, R.Z. and Jaenisch, R. (2002) Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. PNAS, 99, 15536–15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chahrour, M., Jung, S.Y., Shaw, C., Zhou, X., Wong, S.T.C., Qin, J. and Zoghbi, H.Y. (2008) MeCP2, a key contributor to neurological disease, activates and represses transcription. Science, 320, 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stroud, H., Su, S.C., Hrvatin, S., Greben, A.W., Renthal, W., Boxer, L.D., Nagy, M.A., Hochbaum, D.R., Kinde, B., Gabel, H.W.et al. (2017) Early-life gene expression in neurons modulates lasting epigenetic states. Cell, 171, 1151–1164.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fazel Darbandi, S., Robinson Schwartz, S.E., Qi, Q., Catta-Preta, R., Pai, E.L.-L., Mandell, J.D., Everitt, A., Rubin, A., Krasnoff, R.A., Katzman, S.et al. (2018) Neonatal Tbr 1 dosage controls cortical layer 6 connectivity. Neuron, 100, 831–845.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guy, J., Hendrich, B., Holmes, M., Martin, J.E. and Bird, A. (2001) A mouse Mecp 2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet., 27, 322–326. [DOI] [PubMed] [Google Scholar]
- 50. Strong, E., Butcher, D.T., Singhania, R., Mervis, C.B., Morris, C.A., De Carvalho, D., Weksberg, R. and Osborne, L.R. (2015) Symmetrical dose-dependent DNA-methylation profiles in children with deletion or duplication of 7q11.23. Am. J. Hum. Genet., 97, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lucena, J., Pezzi, S., Aso, E., Valero, M.C., Carreiro, C., Dubus, P., Sampaio, A., Segura, M., Barthelemy, I., Zindel, M.Y.et al. (2010) Essential role of the N-terminal region of TFII-I in viability and behavior. BMC Med. Genet., 11, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deacon, R.M.J., Croucher, A. and Rawlins, J.N.P. (2002) Hippocampal cytotoxic lesion effects on species-typical behaviours in mice. Behav. Brain Res., 132, 203–213. [DOI] [PubMed] [Google Scholar]
- 53. van Hagen, J.M., van derGeest, J.N., van derGiessen, R.S., Lagers-van Haselen, G.C., Eussen, H.J.F.M.M., Gille, J.J.P., Govaerts, L.C.P., Wouters, C.H., deCoo, I.F.M., Hoogenraad, C.C.et al. (2007) Contribution of CYLN2 and GTF2IRD1 to neurological and cognitive symptoms in Williams syndrome. Neurobiol. Dis., 26, 112–124. [DOI] [PubMed] [Google Scholar]
- 54. Botta, A., Novelli, G., Mari, A., Novelli, A., Sabani, M., Korenberg, J., Osborne, L.R., Digilio, M.C., Giannotti, A. and Dallapiccola, B. (1999) Detection of an atypical 7q11.23 deletion in Williams syndrome patients which does not include the STX1A and FZD3 genes. J. Med. Genet., 36, 478–480. [PMC free article] [PubMed] [Google Scholar]
- 55. Grueneberg, D.A., Henry, R.W., Brauer, A., Novina, C.D., Cheriyath, V., Roy, A.L. and Gilman, M. (1997) A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev., 11, 2482–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gibson, E.M., Purger, D., Mount, C.W., Goldstein, A.K., Lin, G.L., Wood, L.S., Inema, I., Miller, S.E., Bieri, G., Zuchero, J.B.et al. (2014) Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science, 344, 1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bolger, A.M., Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Langmead, B., Wilks, C., Antonescu, V. and Charles, R. (2019) Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics, 35, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang, Y., Liu, T., Meyer, C.A., Eeckhoute, J., Johnson, D.S., Bernstein, B.E., Nusbaum, C., Myers, R.M., Brown, M., Li, W.et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol., 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Quinlan, A.R. and Hall, I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Robinson, M.D., McCarthy, D.J. and Smyth, G.K. (2010) Edge R: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y.C., Laslo, P., Cheng, J.X., Murre, C., Singh, H. and Glass, C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Abrahams, B.S., Arking, D.E., Campbell, D.B., Mefford, H.C., Morrow, E.M., Weiss, L.A., Menashe, I., Wadkins, T., Banerjee-Basu, S. and Packer, A. (2013) SFARI gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism, 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stamatoyannopoulos, J.A., Snyder, M., Hardison, R., Ren, B., Gingeras, T., Gilbert, D.M., Groudine, M., Bender, M., Kaul, R., Canfield, T.et al. (2012) An encyclopedia of mouse DNA elements (mouse ENCODE). Genome Biol., 13, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ramírez, F., Ryan, D.P., Grüning, B., Bhardwaj, V., Kilpert, F., Richter, A.S., Heyne, S., Dündar, F. and Manke, T. (2016) Deep tools 2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res., 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bonev, B., Cohen, N.M., Szabo, Q., Fritsch, L., Papadopoulos, G.L., Lubling, Y., Xu, X., Lv, X., Hugnot, J.-P., Tanay, A.et al. (2017) Multiscale 3D genome rewiring during mouse neural development. Cell, 171, 557–572.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rao, S.S.P., Huntley, M.H., Durand, N.C., Stamenova, E.K., Bochkov, I.D., Robinson, J.T., Sanborn, A.L., Machol, I., Omer, A.D., Lander, E.S.et al. (2014) A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Clemens, A.W., Wu, D.Y., Moore, J.R., Christian, D.L., Zhao, G. and Gabel, H.W. (2020) MeCP2 Represses Enhancers through Chromosome Topology-Associated DNA Methylation. Molecular Cell, 77, 279–293.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shen, L., Shao, N.-Y., Liu, X., Maze, I., Feng, J. and Nestler, E.J. (2013) Diff reps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li, D., Hsu, S., Purushotham, D., Sears, R.L. and Wang, T. (2019) Wash U Epigenome browser update 2019. Nucleic Acids Res., 47, W158–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dobin, A., Davis, C.A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T.R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Anders, S., Pyl, P.T. and Huber, W. (2015) HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Maloney, S.E., Akula, S., Rieger, M.A., McCullough, K.B., Chandler, K., Corbett, A.M., McGowin, A.E. and Dougherty, J.D. (2018) Examining the reversibility of long-term behavioral disruptions in progeny of maternal SSRI exposure. eNeuro, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nygaard, K.R., Maloney, S.E. and Dougherty, J.D. (2019) Erroneous inference based on a lack of preference within one group: autism, mice, and the social approach task. bioRxiv. doi: 10.1101/530279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Moy, S.S., Nadler, J.J., Young, N.B., Nonneman, R.J., Segall, S.K., Andrade, G.M., Crawley, J.N. and Magnuson, T.R. (2008) Social approach and repetitive behavior in eleven inbred mouse strains. Behav. Brain Res., 191, 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Maloney, S.E., Yuede, C.M., Creeley, C.E., Williams, S.L., Huffman, J.N., Taylor, G.T., Noguchi, K.N. and Wozniak, D.F. (2019) Repeated neonatal isoflurane exposures in the mouse induce apoptotic degenerative changes in the brain and relatively mild long-term behavioral deficits. Sci. Rep., 9, 2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hothorn, T., Bretz, F. and Westfall, P. (2008) Simultaneous inference in general parametric models. Biom. J., 50, 346–363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq and ChIP-seq data are available at GEO accession GSE138234.


