Abstract
Invasive species represent a serious ecological threat for many ecosystems worldwide and provide a unique opportunity to investigate rapid adaptation and evolution. Genetic variation allows populations of organisms to be both robust and adaptable to different environmental conditions over evolutionary timeframes. In contrast, invasive animals can rapidly adapt to new environments, with minimal genetic diversity. Thus, the extent to which environmental effects can trigger epigenetic responses is particularly interesting for understanding the role of epigenetics in rapid adaptation. In this review, we provide a brief overview of the different epigenetic mechanisms that control gene expression, and emphasize the importance of epigenetics for environmental adaptation. We also discuss recent publications that provide important examples for the role of epigenetic mechanisms in environmental adaptation. Furthermore, we present an overview of the current knowledge about epigenetic modulation as an adaptive strategy for invasive species. A particularly interesting example is provided by the marbled crayfish, a novel, monoclonal freshwater crayfish species that has colonized diverse habitats within a few years. Finally, we address important limitations of current approaches and highlight the potential importance of less well-known mechanisms for non-genetic organismal adaptation.
Introduction
It is commonly accepted that differences in the deoxyribonucleic acid (DNA) sequence (i.e., genetic variation) provide organisms with the ability to adapt to different environmental conditions. Natural selection acting on genetic variants explains how organisms can colonize ecological niches over evolutionary timeframes. However, several examples of rapid adaptation and invasion are difficult to explain solely by the selection of genetic variants. As such, epigenetic mechanisms have been increasingly used to explain these phenomena. We delimit epigenetic plasticity from transgenerational epigenetic inheritance, which is controversially discussed and would allow the inheritance of acquired epigenetic traits (Heard and Martienssen 2014). In contrast, epigenetic adaptation is effective within single generations.
Our article focuses on epigenetic adaptation in animals, as several reviews are already available for plants (Richards 2011; Pikaard and Mittelsten Scheid 2014). We begin by defining epigenetics, providing a short overview of the different epigenetic mechanisms, and emphasizing the importance of epigenetics for environmental adaptation. We continue with a detailed description of recent studies that provided important paradigms for the role of epigenetic mechanisms in environmental adaptation. This includes the marbled crayfish, a clonally reproducing animal that has recently colonized various ecosystems worldwide. Finally, we identify important open questions and provide suggestions for future development.
Epigenetic regulation of gene expression
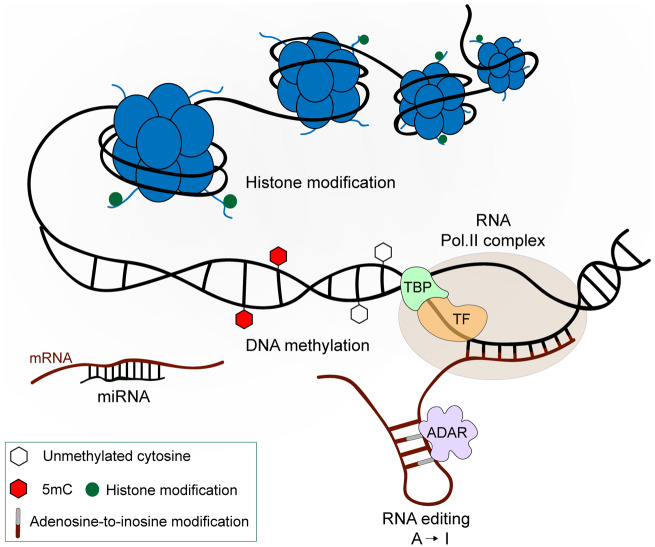
Multicellular organisms are genetically homogeneous, but different cell types and functions arise from the differential expression of genes. It has been shown that differentiating mammalian cells undergo dynamic epigenetic changes, resulting in the establishment of cell-type-specific programs (Gifford et al. 2013; Xie et al. 2013; Roadmap Epigenomics Consortium et al. 2015). Transcription factors primarily control gene expression, and nearly all biological processes are linked to post-synthetic modifications of the three fundamental macromolecules: DNA, ribonucleic acid (RNA), and proteins (Fig. 1). These covalent modifications have been commonly termed as “epigenetic” even though this broad definition has often resulted in misconceptions (Deichmann 2016; Henikoff and Greally 2016).
Fig. 1.
Mechanistic model for epigenetic control of gene expression. Epigenetic mechanisms are important for regulating gene expression and chromatin architecture in eukaryotic cells. The fundamental repeat unit of chromatin is the nucleosome, which is comprised of an octamer of core histone proteins (represented by blue circles). Post-translational modifications (small green circles) of the amino-terminal tails of histone proteins (short blue lines deriving from the histones) affect chromatin structure by fine-tuning the accessibility of the transcription machinery (transcription factors, co-regulators, and the RNA polymerase II complex). DNA methylation (red hexagons) refers to the addition of a methyl group to the five-position of cytosine in the context of CpG dinucleotides. RNA-based mechanisms silence gene expression via complementary base-pairing to mRNA molecules. This evolutionarily conserved mechanism affects gene expression by promoting mRNA degradation or the disruption of protein translation. Adenosine to inosine (A-to-I) RNA editing represents a mechanism to diversify mRNA coding. Because inosine pairs with cytosine, it is a biological mimic for guanosine and can thus alter mRNA coding. The potential of A-to-I editing to diversify the transcriptomic profile represents a possible mechanism to increase phenotypic plasticity and, therefore, aid adaptation to new environments.
Transcription in eukaryotes takes place in the context of chromatin. In general, nucleosomes impede DNA transcription, either by physically obstructing or by compartmentalizing the binding of transcription factors to DNA. An important feature of histones, and particularly of their N-terminal tails, is their ability to carry post-translational modifications, which can affect chromatin structure in different ways. Many of these modifications are dynamically regulated by families of enzymes that write or erase the modifications (Allis and Jenuwein 2016). Besides their role in chromatin remodeling, histone modifications are also responsible for recruiting effector proteins or disrupting their binding to chromatin.
RNA-based mechanisms have also been involved in epigenetic regulation, but are less well understood in animals. Key molecules are noncoding RNAs that belong to several classes. Small interfering RNAs (siRNAs) and microRNAs are derived from longer precursor RNAs by the action of RNAse III-family enzymes, such as Drosha and Dicer, and can inhibit translation or direct mRNA degradation (Kim et al. 2009; Castel and Martienssen 2013). Additionally, siRNAs can regulate gene transcription through transposable element silencing and the interaction with other epigenetic mechanisms, such as DNA methylation and histone modification (Holoch and Moazed 2015). Finally, it should be noted that additional mechanisms that are not traditionally associated with epigenetic gene regulation have the capacity to increase phenotypic plasticity. A prominent example is provided by mRNA editing (Figure 1). The deamination of adenosine to inosine (recognized as guanosine during translation) by the ADAR family of enzymes has the potential to recode codons and diversify the transcriptome by allowing the translation of alternative protein products from a single gene (Eisenberg and Levanon 2018).
DNA methylation represents the most well-studied epigenetic mechanism, and characterizing the biological relevance of this modification and its impact on gene expression has driven research in the field for a long time (Lyko 2018). The dynamic equilibrium between methylation and demethylation modulates gene expression and can be faithfully propagated through many cell generations (Jones 2012). While often associated with transcriptional silencing in mammals, DNA methylation patterns are complex and can affect gene promoters, gene bodies, and repeats differently (Jones 2012; Schubeler 2015).
Mechanistically, the effects of DNA methylation are often explained by their complex association between DNA methylation and transcription factor binding (Schubeler 2015; Yin et al. 2017). An additional model involves the recruitment of methyl-binding proteins (MBPs), which triggers chromatin structural changes and altered gene expression (Klose and Bird 2006). More specifically, MBPs have been shown to bind to methylated DNA, often at gene promoters, resulting in transcriptional repression through the recruitment of histone deacetylases (Klose and Bird 2006). While these models are well-established in vertebrate systems, the role of DNA methylation in invertebrates remains much less understood. Invertebrate methylation patterns can be highly diverse (Bewick et al. 2017) and are often defined by gene body methylation, while promoter methylation is rarely observed. Gene body methylation may reduce spurious RNA polymerase transcription (Neri et al. 2017), which is also known as transcriptional noise (Faure et al. 2017).
From an ecological perspective, epigenetic mechanisms could promote phenotypic plasticity and adaptation to different environments (Verhoeven et al. 2016). Thus, the extent to which environmental effects can trigger epigenetic responses is particularly interesting for understanding the role of epigenetics in animal adaptation.
Key examples for rapid epigenetic adaptation
With the emergence of epigenetics and the development of methods for the detection of epigenetic modifications, many studies have attempted to link adaptive changes to epigenetic changes (Feil and Fraga 2012; Hu and Barrett 2017). More recently, however, several key studies have provided more convincing evidence, based on improved study design and more rigorous methodology. In the following, we provide an overview of several of these particularly interesting examples. For instance, it has been shown that DNA methylation patterns of salmon that were reared in artificial hatcheries were different from salmon that were reared in the wild (Le Luyer et al. 2017). While the phenotypic effects of differentially methylated regions were not investigated, the results were based on sound statistical analysis and included controls for genetic polymorphisms, which represent a major confounding factor in DNA methylation analyses. Overall, the study suggested that adaptive epigenetic changes to the hatchery environment underpin the reduced fitness of hatchery-reared salmon when released in the wild.
Similar observations were also made in terrestrial animals. For example, different methylation patterns were identified in the early stages of a founding population of the brown anole lizard (Anolis sagrei), suggesting a relationship between epigenetic variation and rapid responses to environmental changes (Hu et al. 2019). It was shown that after a 4-days exposure to a new habitat, the lizards had methylation patterns that were distinct from their original habitat. Interestingly, differentially methylated cytosines were detected at genes with functions likely to be relevant to animal plasticity (e.g., signal transduction, immune response, and circadian rhythm). Further integrative studies using whole-genome bisulfite sequencing and RNA sequencing, should improve the understanding of how DNA methylation modulates phenotypic responses to environmental stressors during the colonization of new habitats.
Epigenetic mechanisms have also been implied in the adaptation of the globally distributed scleractinian coral Stylophora pistillata to warmer and more acidic ocean water. Genome-wide DNA methylation analysis revealed pH-dependent differential methylation at genes associated with growth and stress response pathways (Liew et al. 2018). Interestingly, the coral methylome is characterized by gene body methylation, and high levels of gene body methylation were shown to reduce spurious transcription and transcriptional noise (Li et al. 2018). While it appears possible that reduced transcriptional noise facilitates adaptation, variable gene expression has also been suggested as an adaptive mechanism in corals (Kenkel and Matz 2016). Both effects could conceivably be mediated by alterations in gene body methylation.
Another interesting example of an adaptive epigenetic change is provided by certain cavefish, where the environment triggers the degeneration of the eyes. Interestingly, in the Pachón blind morph of the Mexican tetra (Astyanax mexicanus), this process is not accompanied by known genetic mutations in eye developmental genes. Instead, it was shown that promoter hypermethylation could repress eye-specific genes and thus results in defective eye development (Gore et al. 2018). Of note, this study also provides promising results from initial functional experiments, such as a partial rescue of eye phenotypes by injection of a DNA methylation inhibitor (Gore et al. 2018).
Epigenetic adaptation in invasive species
Invasive species represent a serious ecological threat for many ecosystems worldwide and provide a unique opportunity to investigate rapid adaptation and evolution. Invasive animals often show reduced genetic diversity, which is thought to limit the adaptive and evolutionary potential by constraining the availability of new gene variants (Chown et al. 2015). Nevertheless, invasive species are often highly successful in adapting to new and heterogeneous environments, with adaptive plasticity as an essential strategy for rapidly colonizing new habitats and out-competing native species. Commonly neglected or hidden in natural environments, phenotypic plasticity becomes particularly relevant when the invasion of a new or altered environment occurs (Ghalambor et al. 2007; Fox et al. 2019).
Defined as the ability of a genome to express various phenotypes, phenotypic plasticity implies the capacity for multiple adaptive responses to environmental changes, such as environmental stress, rapid growth, and reproduction (Fox et al. 2019). However, the expansion of invasive species represents a genetic paradox, as individuals can adapt rapidly to new, sometimes challenging environments. As discussed before, epigenetic mechanisms can modulate phenotypes without changing genotypes. Through the modulation of gene expression, epigenetic changes can increase phenotypic variation in the absence of genetic diversity, which might facilitate animal adaptation to both biotic and abiotic environmental challenges (Jaenisch and Bird 2003; Duncan et al. 2014).
Epigenetic plasticity can explain organismal adaptation to environmental changes (Hollander et al. 2015). A detailed summary of a significant number of indicative studies that link adaptivity in invasive species to DNA methylation variability has been published recently (Hawes et al. 2018). For example, intrapopulation DNA methylation variability was observed in the invasive house sparrow (Passer domesticus) and proposed as a rapid response mechanism to environmental challenges (Liebl et al. 2013; Sheldon et al. 2018). Additionally, in the pygmy mussel (Xenostrobus secures), global DNA hypomethylation was detected in a recently evolved population and interpreted as a mechanism to promote phenotypic plasticity and thereby facilitate the expansion of invasive populations (Ardura et al. 2017). Finally, in the invasive whitefly (Bemisia tabaci), DNA methyltransferase 1 (DNMT1) knockdown caused reduced thermotolerance, but DNA methylation patterns were not investigated (Dai et al. 2018).
Despite these recent advances, it is worth mentioning that many studies in this field have analyzed DNA methylation using indirect methods and that it will be important to confirm initial findings by more robust and more powerful approaches, such as bisulfite sequencing (Lea et al. 2017). Furthermore, the mechanisms of how the environment shapes the epigenome are mainly unknown. Finally, it will be important to more comprehensively analyze epigenetic gene regulation in invasive animals, in order to better understand and ultimately predict their adaptive and invasive potential. Detailed analyses of select model organisms could be particularly useful for the establishment of more generalizable concepts.
The marbled crayfish as an example for epigenetic adaptation in an invasive species
The marbled crayfish (Procambarus virginalis) represents a novel freshwater crayfish species that emerged from the German aquarium trade about 25 years ago (Scholtz et al. 2003; Lyko 2017). Anthropogenic releases have founded expanding wild populations in several European countries (Chucholl and Pfeiffer 2010; Lipták et al. 2016; Novitsky and Son 2016; Patoka et al. 2016; Pârvulescu et al. 2017; Deidun et al. 2018; Ercoli et al. 2019). Furthermore, marbled crayfish have rapidly invaded ecologically distinct habitats in Madagascar and currently colonize an area that extends over 100.000 km2 (Jones et al. 2009; Kawai et al. 2009; Gutekunst et al. 2018; Andriantsoa et al. 2019).
The marbled crayfish is a parthenogenetic descendant of the sexually reproducing slough crayfish (P. fallax) from Florida (Martin et al. 2010). Its particular mode of reproduction (obligatory apomictic parthenogenesis) results in the generation of a genetically homogeneous, monoclonal population (Gutekunst et al. 2018). Parthenogenetic reproduction is not uncommon in the animal kingdom, but obligatory parthenogenesis is very rare and has been described as an “evolutionary scandal,” as it runs counter to fundamental tenets of evolutionary biology. As genetic polymorphisms are negligible in the marbled crayfish population, the successful adaptation of the animals to different environments cannot be explained by the Darwinian selection of the genetically best-adapted genotype. It is therefore very likely that marbled crayfish use epigenetic mechanisms, such as DNA methylation, to adapt their genome to specific environmental conditions (Figure 2).
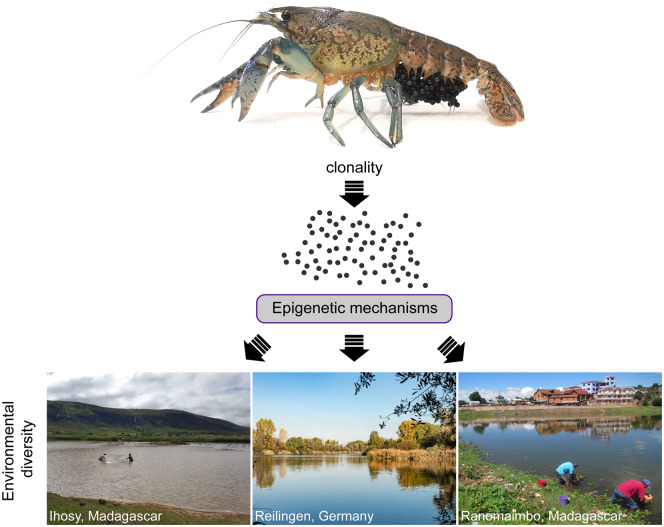
Fig. 2.
Clonal expansion and rapid adaptation of marbled crayfish. The marbled crayfish is a monoclonal, parthenogenetically reproducing species. Individual animals show considerable phenotypic plasticity and adaptivity to various parameters, such as temperature, osmolarity, and pollution. Epigenetic modulation has been implied to marbled crayfish adaptation, but the precise mechanisms remain to be identified. The top panel picture shows an adult marbled crayfish (3 years old) with eggs. The bottom panel displays three ecologically different locations with established marbled crayfish population: Ihosy River (Madagascar), Lake Reilingen (Germany), and Ranomaimbo lake (Madagascar).
The combination of obligatory apomictic parthenogenesis with an extremely young species age (25–30 years) in marbled crayfish creates unique opportunities for elucidating the role of epigenetic mechanisms in invasiveness. Indeed, the genome of the marbled crayfish encodes a conserved and active DNA methylation toolkit (Gatzmann et al. 2018). A detailed analysis of DNA methylation patterns revealed that that the modification is targeted to the gene bodies of housekeeping genes, similar to many other invertebrates (Gatzmann et al. 2018). Interestingly, gene body methylation was found to be inversely correlated with gene expression variability. When compared to the parent species of marbled crayfish (P. fallax), many genes showed reduced levels of gene body methylation and increased levels of gene expression variability (Gatzmann et al. 2018). This might indicate that low levels of gene body methylation promote adaptability (and invasiveness) through increased gene expression variability. This hypothesis will have to be confirmed by a detailed molecular analysis of specific ecotypes and/or by direct experimental approaches that elucidate the functional role of DNA methylation in this organism.
Of note, marbled crayfish possess key prerequisites for a laboratory model, such as suitable size, resistance against handling stress, high fertility, and a relatively short generation time (Vogt 2011). In addition, marbled crayfish combine obligatory apomictic parthenogenesis with an extremely young evolutionary age (25–30 years), thus generating a population with an unparalleled genetic homogeneity. Up to date, this model has been used for research on several biological processes such as development (Seitz et al. 2005), neurobiology (Vilpoux et al. 2006), and epigenetics (Vogt et al. 2008). Furthermore, marbled crayfish has been shown to be very robust against various environmental parameters (Andriantsoa et al. 2019) and represents an excellent model system for modulating environmental conditions in a standardized laboratory setting.
Future directions
Elucidating the role of epigenetic mechanisms in phenotypic plasticity and adaptation will remain an important research topic for many years to come. It will be important to avoid known issues in the design and interpretation of epigenome mapping studies that have plagued the field in the past (Lappalainen and Greally 2017; Lea et al. 2017). In this context, we consider three issues as particularly relevant: (1) Epigenetic patterns can be cell-type specific, but epigenetic profiles are often obtained from whole animals or bulk tissue. For example, two groups from different environments might differ in the cellular composition of a specific tissue, which could introduce a systematic (and potentially large) confounding effect that is unrelated to true epigenetic adaptation. As such, it is important to control the cellular sample composition. (2) Epigenetic effect sizes are often relatively small in ecological studies. The generation of conclusive results, therefore, requires the design of sufficiently powered studies with relatively high sequencing depths and relatively large sample numbers. We recommend that power estimates are included in the study design (Lea et al. 2017). (3) Wild specimens from a single species can have very heterogeneous genetic backgrounds, which can introduce a very strong confounding effect on the analysis. For example, if a cytosine is methylated in one population, but polymorphic in another population, it will be scored as differentially methylated. However, this change is not related to differential epigenetic programming, but rather reflects differences in population genetic structures. Integrated genome/epigenome analysis should be used to resolve this issue.
It will also be critically important to analyze the functional role of epigenetic mechanisms in rapid adaptation and invasiveness. This can be achieved by Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-mediated editing (Rees and Liu 2018) and inactivation of various epigenetic modifier genes, such as DNA methyltransferases, DNA demethylases, and histone modifying enzymes. Knockout animals can then be challenged by changes in environmental parameters that can be easily varied in the laboratory (e.g., temperature, water supplements, and feed components). The increasing availability of complete genome sequences from non-model organisms and the broad application potential of CRISPR-based genome editing tools allow the use of functional approaches in an increasing number of interesting and relevant organisms.
Finally, it is also likely, that additional epigenetic mechanisms will emerge in the context of rapid adaptation and invasiveness. A prominent example is provided by small RNAs that can be loaded into oocytes and sperm and can thus be transmitted from parents to offspring (Chen et al. 2016). If these RNAs have the capacity to modulate genes that are involved in phenotypic plasticity, they can have a profound impact on the adaptive potential of the corresponding organism. Additionally, mRNA editing represents another interesting example of a mechanism that could conceivably play an important role in rapid adaptation (Eisenberg and Levanon 2018). A-to-I mRNA editing is a dynamic process that can diversify the transcriptome, and that has been strongly implicated in adaptive processes (Garrett and Rosenthal 2012). Interestingly, a tradeoff between genome evolution and transcriptome plasticity was suggested in cephalopods, as the genomic regions surrounding the editing sites appeared highly conserved (Liscovitch-Brauer et al. 2017). The high frequency and the conservation of editing sites significantly reduce genetic polymorphisms, and thereby decelerate genome evolution. Thus, RNA editing could provide a paradigm for how animals overcome the lack of genetic diversity and rapidly adapt to new environments by diversifying their transcriptome.
Acknowledgments
We thank Sina Tönges for the pictures of a marbled crayfish and Lake Reilingen. We also thank Ranja Adriantsoa for the pictures of the Ihosy River and the Ranomaimbo lake.
From the symposium “SICB Wide Symposium: Building Bridges from Genome to Phenome: Molecules, Methods and Models” presented at the annual meeting of the Society for Integrative and Comparative Biology January 3–7, 2020 at Austin, Texas.
References
- Allis CD, Jenuwein T.. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17:487–500. [DOI] [PubMed] [Google Scholar]
- Andriantsoa R, Tonges S, Panteleit J, Theissinger K, Carneiro VC, Rasamy J, Lyko F.. 2019. Ecological plasticity and commercial impact of invasive marbled crayfish populations in madagascar. BMC Ecol 19:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardura A, Zaiko A, Moran P, Planes S, Garcia-Vazquez E.. 2017. Epigenetic signatures of invasive status in populations of marine invertebrates. Sci Rep 7:42193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ.. 2017. Evolution of DNA methylation across insects. Mol Biol Evol 34:654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA.. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14:100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan W, Duan E.. 2016. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 17:733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chown SL, Hodgins KA, Griffin PC, Oakeshott JG, Byrne M, Hoffmann AA.. 2015. Biological invasions, climate change and genomics. Evol Appl 8:23–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chucholl C, Pfeiffer M.. 2010. First evidence for an established Marmorkrebs (Decapoda, Astacida, Cambaridae) population in Southwestern Germany, in syntopic occurrence with Orconectes limosus (Rafinesque, 1817). Aquat Invasions 5:405–12. [Google Scholar]
- Dai TM, Lu ZC, Wang YS, Liu WX, Hong XY, Wan FH.. 2018. Molecular characterizations of DNA methyltransferase 3 and its roles in temperature tolerance in the whitefly, Bemisia tabaci Mediterranean. Insect Mol Biol 27:123–32. [DOI] [PubMed] [Google Scholar]
- Deichmann U. 2016. Epigenetics: the origins and evolution of a fashionable topic. Dev Biol 416:249–54. [DOI] [PubMed] [Google Scholar]
- Deidun A, Sciberras A, Formosa J, Zava B, Insacco G, Corsini-Foka M, Crandall KA.. 2018. Invasion by non-indigenous freshwater decapods of Malta and Sicily, central Mediterranean Sea. J Crust Biol 38:748–53. [Google Scholar]
- Duncan EJ, Gluckman PD, Dearden PK.. 2014. Epigenetics, plasticity, and evolution: how do we link epigenetic change to phenotype?. J Exp Zool B Mol Dev Evol 322:208–20. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY.. 2018. A-to-I RNA editing - immune protector and transcriptome diversifier. Nat Rev Genet 19:473–90. [DOI] [PubMed] [Google Scholar]
- Ercoli F, Kaldre K, Paaver T, Gross R.. 2019. First record of an established marbled crayfish Procambarus virginalis (lyko, 2017) population in Estonia. Bioinvasions Rec 8:675–83. [Google Scholar]
- Faure AJ, Schmiedel JM, Lehner B.. 2017. Systematic analysis of the determinants of gene expression noise in embryonic stem cells. Cell Syst 5:471–84.e474. [DOI] [PubMed] [Google Scholar]
- Feil R, Fraga MF.. 2012. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 13:97–109. [DOI] [PubMed] [Google Scholar]
- Fox RJ, Donelson JM, Schunter C, Ravasi T, Gaitan-Espitia JD.. 2019. Beyond buying time: the role of plasticity in phenotypic adaptation to rapid environmental change. Philos Trans R Soc Lond B Biol Sci 374:20180174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S, Rosenthal JJ.. 2012. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science 335:848–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatzmann F, Falckenhayn C, Gutekunst J, Hanna K, Raddatz G, Carneiro VC, Lyko F.. 2018. The methylome of the marbled crayfish links gene body methylation to stable expression of poorly accessible genes. Epigenetics Chromatin 11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalambor CK, McKay JK, Carroll SP, Reznick DN.. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct Ecol 21:394–407. [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. 2013. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153:1149–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore AV, Tomins KA, Iben J, Ma L, Castranova D, Davis AE, Parkhurst A, Jeffery WR, Weinstein BM.. 2018. An epigenetic mechanism for cavefish eye degeneration. Nat Ecol Evol 2:1155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst J, Andriantsoa R, Falckenhayn C, Hanna K, Stein W, Rasamy JR, Lyko F.. 2018. Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat Ecol Evol 2:567–73. [DOI] [PubMed] [Google Scholar]
- Hawes NA, Fidler AE, Tremblay LA, Pochon X, Dunphy BJ, Smith KF.. 2018. Understanding the role of DNA methylation in successful biological invasions: a review. Biol Invasions 20:2285–300. [Google Scholar]
- Heard E, Martienssen RA.. 2014. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Greally JM.. 2016. Epigenetics, cellular memory and gene regulation. Curr Biol 26:R644–8. [DOI] [PubMed] [Google Scholar]
- Hollander J, Snell-Rood E, Foster S.. 2015. New frontiers in phenotypic plasticity and evolution. Heredity (Edinb) 115:273–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D.. 2015. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Askary AM, Thurman TJ, Spiller DA, Palmer TM, Pringle RM, Barrett R.. 2019. The epigenetic signature of colonizing new environments in anolis lizards. Mol Biol Evol 36:2165–70. [DOI] [PubMed] [Google Scholar]
- Hu J, Barrett R.. 2017. Epigenetics in natural animal populations. J Evol Biol 30:1612–32. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A.. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33:245–54. [DOI] [PubMed] [Google Scholar]
- Jones JPG, Rasamy JR, Harvey A, Toon A, Oidtmann B, Randrianarison MH, Raminosoa N, Ravoahangimalala OR.. 2009. The perfect invader: a parthenogenic crayfish poses a new threat to Madagascar’s freshwater biodiversity. Biol Invasions 11:1475–82. [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–92. [DOI] [PubMed] [Google Scholar]
- Kawai T, Scholtz G, Morioka S, Ramanamandimby F, Lukhaup C, Hanamura Y.. 2009. Parthenogenetic alien crayfish (Decapoda: Cambaridae) spreading in Madagascar. J Crust Biol 29:562–7. [Google Scholar]
- Kenkel CD, Matz MV.. 2016. Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat Ecol Evol 1:14. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC.. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10:126–39. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP.. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31:89–97. [DOI] [PubMed] [Google Scholar]
- Lappalainen T, Greally JM.. 2017. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet 18:441–51. [DOI] [PubMed] [Google Scholar]
- Le Luyer J, Laporte M, Beacham TD, Kaukinen KH, Withler RE, Leong JS, Rondeau EB, Koop BF, Bernatchez L.. 2017. Parallel epigenetic modifications induced by hatchery rearing in a pacific salmon. Proc Natl Acad Sci U S A 114:12964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea AJ, Vilgalys TP, Durst PAP, Tung J.. 2017. Maximizing ecological and evolutionary insight in bisulfite sequencing data sets. Nat Ecol Evol 1:1074–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liew YJ, Cui G, Cziesielski MJ, Zahran N, Michell CT, Voolstra CR, Aranda M.. 2018. DNA methylation regulates transcriptional homeostasis of algal endosymbiosis in the coral model aiptasia. Sci Adv 4:eaat2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl AL, Schrey AW, Richards CL, Martin LB.. 2013. Patterns of DNA methylation throughout a range expansion of an introduced songbird. Integr Comp Biol 53:351–8. [DOI] [PubMed] [Google Scholar]
- Liew YJ, Zoccola D, Li Y, Tambutte E, Venn AA, Michell CT, Cui G, Deutekom ES, Kaandorp JA, Voolstra CR, et al. 2018. Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci Adv 4:eaar8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipták B, Mrugała A, Pekárik L, Mutkovič A, Gruľa D, Petrusek A, Kouba A.. 2016. Expansion of the marbled crayfish in Slovakia: beginning of an invasion in the Danube catchment?. J Limnol 75:305–12. [Google Scholar]
- Liscovitch-Brauer N, Alon S, Porath HT, Elstein B, Unger R, Ziv T, Admon A, Levanon EY, Rosenthal JJC, Eisenberg E.. 2017. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 169:191–202 e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F. 2017. The marbled crayfish (Decapoda: Cambaridae) represents an independent new species. Zootaxa 4363:544–52. [DOI] [PubMed] [Google Scholar]
- Lyko F. 2018. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 19:81–92. [DOI] [PubMed] [Google Scholar]
- Martin P, Dorn NJ, Kawai T, van der Heiden C, Scholtz G.. 2010. The enigmatic marmorkrebs (marbled crayfish) is the parthenogenetic form of Procambarus fallax (Hagen, 1870). Contrib Zool 79:107–18. [Google Scholar]
- Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S.. 2017. Intragenic DNA methylation prevents spurious transcription initiation. Nature 543:72–7. [DOI] [PubMed] [Google Scholar]
- Novitsky RA, Son MO.. 2016. The first records of marmorkrebs [Procambarus fallax (Hagen, 1870) f. Virginalis] (Crustacea, Decapoda, Cambaridae) in Ukraine. Ecol Montenegrina 5:44–6. [Google Scholar]
- Pârvulescu L, Togor A, Lele SF, Scheu S, Iinca D, Panteleit J.. 2017. First established population of marbled crayfish Procambarus fallax (Hagen, 1870) f. Virginalis (Decapoda, Cambaridae) in Romania. Bioinvasions Rec 6:357–62. [Google Scholar]
- Patoka J, Buřič M, Kolář V, Bláha M, Petrtýl M, Franta P, Tropek R, Kalous L, Petrusek A, Kouba A.. 2016. Predictions of marbled crayfish establishment in conurbations fulfilled: evidences from the Czech Republic. Biologia 71:1380–5. [Google Scholar]
- Pikaard CS, Mittelsten Scheid O.. 2014. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 6:a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Liu DR.. 2018. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19:770–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. 2011. Natural epigenetic variation in plant species: a view from the field. Curr Opin Plant Biol 14:204–9. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics ConsortiumKundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtz G, Braband A, Tolley L, Reimann A, Mittmann B, Lukhaup C, Steuerwald F, Vogt G.. 2003. Ecology: parthenogenesis in an outsider crayfish. Nature 421:806. [DOI] [PubMed] [Google Scholar]
- Schubeler D. 2015. Function and information content of DNA methylation. Nature 517:321–6. [DOI] [PubMed] [Google Scholar]
- Seitz R, Vilpoux K, Hopp U, Harzsch S, Maier G.. 2005. Ontogeny of the Marmorkrebs (marbled crayfish): a parthenogenetic crayfish with unknown origin and phylogenetic position. J Exp Zool A Comp Exp Biol 303A:393–405. [DOI] [PubMed] [Google Scholar]
- Sheldon EL, Schrey A, Andrew SC, Ragsdale A, Griffith SC.. 2018. Epigenetic and genetic variation among three separate introductions of the house sparrow (Passer domesticus) into Australia. R Soc Open Sci 5:172185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven KJ, vonHoldt BM, Sork VL.. 2016. Epigenetics in ecology and evolution: what we know and what we need to know. Mol Ecol 25:1631–8. [DOI] [PubMed] [Google Scholar]
- Vilpoux K, Sandeman R, Harzsch S.. 2006. Early embryonic development of the central nervous system in the Australian crayfish and the marbled crayfish (Marmorkrebs). Dev Genes Evol 216:209–23. [DOI] [PubMed] [Google Scholar]
- Vogt G. 2011. Marmorkrebs: natural crayfish clone as emerging model for various biological disciplines. J Biosci 36:377–82. [DOI] [PubMed] [Google Scholar]
- Vogt G, Huber M, Thiemann M, van den Boogaart G, Schmitz OJ, Schubart CD.. 2008. Production of different phenotypes from the same genotype in the same environment by developmental variation. J Exp Biol 211:510–523. [DOI] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153:1134–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. 2017. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356:eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]