Resistance-nodulation-division (RND) efflux pumps are important contributors to bacterial antibiotic resistance. In this study, we combined evolutionary sequence analyses, computational structural modeling, and ligand docking to develop a framework that can explain the known antibiotic substrate selectivity differences between two Pseudomonas aeruginosa RND transporters, MexY and MexB. For efficient efflux, antibiotic substrates must possess a “Goldilocks affinity”: binding strong enough to allow interaction with transporter but not so tight as to impede movement through the pump.
KEYWORDS: efflux pumps, antibiotic resistance, Pseudomonas aeruginosa, phylogenetic analysis, computer modeling
ABSTRACT
Resistance-nodulation-division (RND) efflux pumps are important contributors to bacterial antibiotic resistance. In this study, we combined evolutionary sequence analyses, computational structural modeling, and ligand docking to develop a framework that can explain the known antibiotic substrate selectivity differences between two Pseudomonas aeruginosa RND transporters, MexY and MexB. For efficient efflux, antibiotic substrates must possess a “Goldilocks affinity”: binding strong enough to allow interaction with transporter but not so tight as to impede movement through the pump.
INTRODUCTION
The resistance-nodulation-division (RND) efflux pumps are one of five large families of bacterial efflux systems capable of exporting diverse substrates from bacterial cells, including multiple classes of antibiotics (1). Therefore, RND-mediated efflux contributes significantly to clinical multidrug resistance, particularly in Gram-negative pathogens (2, 3). RND pumps are tripartite complexes comprised of homotrimeric inner membrane transporter, homohexameric periplasmic adaptor, and homotrimeric outer membrane components that form a continuous channel for export of molecules from within the cell to the external environment (Fig. 1A). These three pump components are best exemplified by the structurally well-characterized Escherichia coli AcrAB-TolC and Pseudomonas aeruginosa MexAB-OprM RND systems (4, 5).
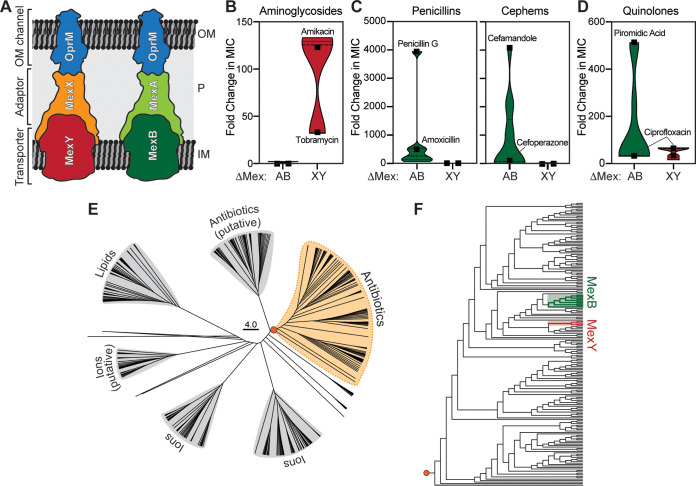
FIG 1.
Substrate selectivity and molecular evolutionary analysis of two P. aeruginosa RND antibiotic efflux systems. (A) Cartoon of the molecular architecture and location of the P. aeruginosa MexXY-OprM and MexAB-OprM RND systems. OM, outer membrane; P, periplasm; IM, inner membrane. (B to D) Violin plots of fold change in antibiotic MIC (following deletion of the indicated RND efflux system compared to the corresponding expressing strain) calculated from published data (10) for aminoglycosides (B), selected β-lactams (shown separately for penicillins and cephems) (C), and quinolones (D). In each group, examples of well-effluxed and poorly effluxed substrates are indicated (full details are shown in Fig. S1). (E) Phylogenetic analysis of the RND superfamily transporter component (i.e., MexY/MexB homologs). (F) Detailed view of the antibiotic efflux RND family exporter clade (the orange circle indicates the branch point used to root the expanded tree). Sequences used to generate conservations within the MexY- and MexB-containing subclades are marked with red and green shading, respectively.
P. aeruginosa possesses 12 RND efflux systems, 4 of which contribute prominently to antibiotic resistance: MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM (6). These four systems have overlapping but distinct substrate preferences and can collectively efflux the majority of clinically relevant antibiotics from the bacterial cell. The regulation and induction of P. aeruginosa efflux pumps are complex and show plasticity under selection pressure in clinical isolates which upregulate MexXY via mutations in regulatory genes (7). Nonetheless, the drug-inducible MexXY-OprM is a well-established and predominant intracellular mechanism of clinical aminoglycoside resistance in P. aeruginosa (8, 9). As a starting point to investigate P. aeruginosa RND substrate selectivity, we reanalyzed available data (10) for laboratory and clinical Pseudomonas strains by calculating the fold change in antibiotic MIC for strains overexpressing specific RND efflux systems compared to their corresponding deletion mutants (ΔMexB or ΔMexY). This analysis allowed simple visualization of the impact on drug sensitivity of the loss of each pump’s activity, with the known strong substrate preference of MexY for aminoglycosides and MexB for β-lactams clearly revealed by the correspondingly greater change in fold MIC in each case (Fig. 1B and C and Fig. S1 in the supplemental material). Interestingly, for carbapenem β-lactams, meropenem is effluxed by MexAB-OprM, while imipenem appears to be a poor substrate for both systems (10). These analyses also confirmed the ability of both pumps to efflux quinolones (Fig. 1D and Fig. S1). Finally, the specific importance of MexXY for aminoglycoside efflux is also supported by studies of its upregulation and activity in cystic fibrosis-associated clinical strains (11, 12). Important differences must therefore exist in the physicochemical properties of the substrate binding regions within MexY and MexB which enable these pumps to bind and efflux their distinct substrates.
We performed phylogenetic and molecular evolution analyses of all known RND transporter proteins (i.e., MexB/MexY homologs). The majority of these homologs (∼128,000 sequences) are present in Gram-negative Proteobacteria due to repetitive gene duplication events that have promoted pump neofunctionalization, i.e., alteration of substrate types and specificity (13). This complete sequence data set was reduced to a collection of ∼700 representative, highly divergent homolog sequences using a 50% sequence identity cutoff (14) and the reduced set was then used for phylogenetic analyses by the evolutionary-trace method (15). The resultant tree divided the RND superfamily into six major clades, including two comprising known or putative antibiotic efflux pumps (Fig. 1E).
We next selected two subgroups of sequences close to either MexB or MexY (shaded regions in Fig. 1F) for detailed analyses using the evolutionary-trace method and subsequent statistical analysis of residue conservation. Among this set, the most distant sequences had ∼30% identity due to the very high sequence divergence in the RND superfamily. Sequences belonging to these groups of close MexB and MexY homologs were pooled and the set was expanded to include all sequences with <90% identity, and these final sets of representative sequences were used to produce the final alignment for functional comparisons (Fig. S2).
To provide a structural framework for interpreting predictions based on these evolutionary analyses, we generated a complete model of MexXY-OprM. First, homology models of the MexX hexamer and MexY trimer were generated in Swiss Model (16) using MexA and MexB (PDB code 6IOL), respectively, as templates. Next, these models and the OprM structure (PDB code 6IOL) were used for alignment-guided docking of the three components in the Schrödinger software (Fig. 2A). The structural features within this model were then compared with the structure of MexAB-OprM (PDB code 6IOL) (Fig. 2B) to identify localized sites of functional divergence between these efflux pumps. The three protomers of RND transporter components can be designated as adopting one of at least three possible states: access, binding, or extrusion (17), corresponding to a stepwise process of initial binding through to transfer to a central channel within the tripartite pump for efflux. These distinct conformational states impact the shape and location of channels and substrate binding pockets (and thus the location of potential discriminatory residues) within each protomer and the assembled trimer. As expected, equivalent conformational states for each protomer are identifiable within the trimeric MexB structure and our MexY model.
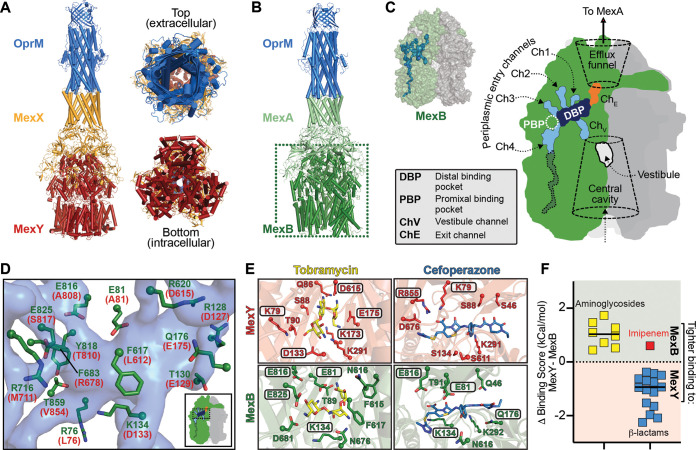
FIG 2.
Substrate selectivity by MexB and MexY is determined by a Goldilocks binding affinity. (A) Three views of the computational model of the P. aeruginosa MexXY-OprM efflux pump. (B) Cryo-electron microscopy (cryo-EM) structure of the P. aeruginosa MexAB-OprM efflux pump (PDB code 6IOL) used to guide modeling of MexXY-OprM. (C) Surface representation of the MexB trimer with a single protomer colored green (other protomers are light and dark gray), with internal channels and pockets determined using Caver shown as a blue surface (this calculation was performed on the protomer in the binding state). A schematic of the same MexB trimer indicating individual channels/pockets, etc., is also shown. (D) Zoomed view of residues lining part of the DBP (semitransparent blue surface) in MexB (green) or the MexY model (red). (E) The aminoglycoside tobramycin and β-lactam cefoperazone can be docked in similar locations within the DBPs of MexY or MexB. Boxed residues making interactions with each drug are those predicted by the evolutionary analyses to be potentially critical for substrate discrimination by these efflux systems. (F) Differences of docking scores for each transporter (MexY-MexB [Table 2]) are plotted for each antibiotic class (aminoglycosides and β-lactams). Note that, consistent with our findings, imipenem (red) is not an efflux substrate of MexAB-OprM.
The MexB binding state protomer was analyzed using Caver (18), and four putative channels for substrate entry as well as the previously identified distal binding pocket (DBP) were located (5) (Fig. 2B and C). The corresponding entry channels and binding pockets were also readily identifiable in the equivalent MexY protomer of our model (data not shown). Two additional regions through which substrates may move, the “vestibule” and “efflux funnel” (19, 20), were also identified in each trimeric transporter assembly (Fig. 2C). However, for subsequent analyses, we focused only on the protein residue networks and physicochemical/structural properties of the DBP in each transporter’s binding state protomer, as this region plays a critical role in substrate interactions (21, 22) and the DBP most completely formed within this protomer state.
Strong enrichment of potential substrate discriminating residues between MexB and MexY were clearly apparent in their DBPs giving each distinct physicochemical properties (Table 1), including amino acid charge distribution, shape, and volume. The proximal region of the MexB DBP (i.e., closer to the entry channels) has a network of negatively charged residues (Glu81, Glu816, and Glu825 [Fig. 2D and Table S1]); in contrast, MexY is more polar or positively charged, with additional enrichment elsewhere in positive charge (Lys79, Lys173, Arg678, and Arg855 [Table S1]). The DBP of MexY is also enriched in small/flexible residues (Ala or Gly) which could provide greater capacity to accommodate larger, more flexible substrates, such as aminoglycosides. Finally, we also note one further observation within the DBPs that at first sight appears counterintuitive given the preference of MexB and MexY for more hydrophobic (e.g., β-lactams) and hydrophilic/positively charged (e.g., aminoglycosides) substrates, respectively: the DBP of MexY is significantly enriched in aliphatic hydrophobic residues compared to MexB.
TABLE 1.
Comparison of the properties of the MexY and MexB DBPs
| Characteristic | % MexY | % MexB |
|---|---|---|
| All polar | 32.4 | 41.2 |
| Positive (R/K) | 14.7 | 17.6 |
| Negative (E/D) | 14.7 | 14.7 |
| Small (A/G) | 8.8 | 5.9 |
| Aliphatic hydrophobic | 17.6 | 2.9 |
| Aromatic/polar (Y/W/H) | 8.8 | 5.9 |
| Aromatic (F) | 2.9 | 11.8 |
We next used Glide SP (23) implemented in the Schrödinger software for flexible docking of aminoglycosides and β-lactams to MexB (PDB code 6IOL) and MexY (our model) to directly explore potential differences in how these molecules might interact with each transporter. Both antibiotic groups are accommodated within the DBP of either MexB or MexY, with similar binding poses corresponding to the best docking score in each case, i.e., tightest predicted binding affinity, as exemplified for the aminoglycoside tobramycin and β-lactam-containing cefoperazone (Fig. 2E). In all cases, the predicted affinities differed considerably, with uniformly higher aminoglycoside affinity for MexB than for MexY and, conversely, higher β-lactam affinity for MexY than for MexB (Table 2 and Fig. 2F). Thus, the predicted binding of nonpreferred substrates compared to preferred substrates is tighter for the DBP of each transporter. Finally, we generated a MexB DBP with more MexY-like properties by in silico substitution of four potentially discriminating residues, resulting in a predicted reduction in affinity of aminoglycosides and an increase in affinity of β-lactams (Table 2). One exception to both observations was for the β-lactam imipenem; however, our finding is also entirely consistent with previous analyses showing that this drug is not a substrate for either MexAB-OprM or MexXY-OprM (10, 24–26).
TABLE 2.
Docking scores for aminoglycoside and β-lactam binding in the MexY and MexB DBPs
| Antibiotic | Flexible docking score (kCal/mol) |
||
|---|---|---|---|
| MexY | MexB | MexBMexY-DBPa | |
| Aminoglycosides | |||
| Amikacin | −7.60 | −8.04 | −5.29 |
| Gentamicin | −5.99 | −7.09 | −5.24 |
| Kanamycin | −5.69 | −7.16 | −5.80 |
| Netilmicin | −5.97 | −6.93 | −6.04 |
| Sisomicin | −6.39 | −6.91 | −5.59 |
| Tobramycin | −5.87 | −7.14 | −5.12 |
| Neomycin | −6.79 | −8.52 | −6.25 |
| Paromomycin | −7.50 | −8.22 | −7.36 |
| β-Lactams | |||
| Carbenicillin | −5.69 | −5.26 | −7.44 |
| Sulbenicillin | −5.18 | −4.80 | −7.87 |
| Flomoxef | −6.13 | −5.18 | −6.91 |
| Moxalactam | −6.90 | −5.25 | −6.76 |
| Imipenemb | −6.73 | −7.36 | −7.38 |
| Meropenem | −6.85 | −5.73 | −6.48 |
| Cefamandole | −5.31 | −4.56 | −6.94 |
| Cefditoren | −4.49 | −2.40 | −7.21 |
| Cefonicid | −6.52 | −5.59 | −7.54 |
| Cefoperazone | −8.50 | −6.25 | −8.98 |
| Cefotetan | −6.45 | −5.82 | −8.95 |
| Ceftaroline fosamil | −7.85 | −5.88 | −8.15 |
| Cefuroxime | −5.22 | −4.73 | −6.95 |
| Cloxacillin | −5.01 | −3.97 | −5.89 |
| Ertapenem | −7.03 | −5.50 | −6.87 |
| Monobactam | −6.33 | −5.63 | −6.51 |
MexBMexY-DBP is the structure of MexB with four in silico amino acid changes at potentially discriminatory residues (E81A, S79K, E816A, and E825S), making the MexB DBP more like MexY.
Imipenem is not a substrate for MexAB-OprM (10).
We propose that substrates must bind the DBP with an affinity that allows them to be both taken up by the transporter and also moved through the pump (effluxed) by the peristaltic movement of the protomer conformational states. In other words, for efficient efflux, substrates should possess a “Goldilocks” affinity: not too weak (resulting in no interaction with the transporter) and not too strong (as tight binding would reduce progress through the pump). These analyses set the scene for further detailed study of substrate selection by the RND systems in P. aeruginosa and other Gram-negative bacteria. Deeper understanding of these mechanisms also holds promise to support design of novel, efflux-resistant antibiotic variants or specific efflux pump inhibitors to counter the challenge of drug resistance conferred by these systems.
Supplementary Material
ACKNOWLEDGMENTS
The work was support in part by Cystic Fibrosis Foundation postdoctoral fellowship DEY18F0 (to D.D.).
We thank Christine M. Dunham and Joanna B. Goldberg for discussions and comments on the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Nikaido H. 2018. RND transporters in the living world. Res Microbiol 169:363–371. doi: 10.1016/j.resmic.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puzari M, Chetia P. 2017. RND efflux pump mediated antibiotic resistance in Gram-negative bacteria Escherichia coli and Pseudomonas aeruginosa: a major issue worldwide. World J Microbiol Biotechnol 33:24. doi: 10.1007/s11274-016-2190-5. [DOI] [PubMed] [Google Scholar]
- 3.Podnecky NL, Rhodes KA, Schweizer HP. 2015. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol 6:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. 2014. Structure of the AcrAB-TolC multidrug efflux pump. Nature 509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsutsumi K, Yonehara R, Ishizaka-Ikeda E, Miyazaki N, Maeda S, Iwasaki K, Nakagawa A, Yamashita E. 2019. Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism. Nat Commun 10:1520. doi: 10.1038/s41467-019-09463-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez L, Hancock RE. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López-Causapé C, Sommer LM, Cabot G, Rubio R, Ocampo-Sosa AA, Johansen HK, Figuerola J, Cantón R, Kidd TJ, Molin S, Oliver A. 2017. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci Rep 7:5555. doi: 10.1038/s41598-017-05621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guenard S, Muller C, Monlezun L, Benas P, Broutin I, Jeannot K, Plesiat P. 2014. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:221–228. doi: 10.1128/AAC.01252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobel ML, McKay GA, Poole K. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 47:3202–3207. doi: 10.1128/aac.47.10.3202-3207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogne C, Aires JR, Bailly C, Hocquet D, Plesiat P. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother 48:1676–1680. doi: 10.1128/aac.48.5.1676-1680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 13.Gorecki K, McEvoy MM. 2020. Phylogenetic analysis reveals an ancient gene duplication as the origin of the MdtABC efflux pump. PLoS One 15:e0228877. doi: 10.1371/journal.pone.0228877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH, UniProt Consortium . 2015. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Innis CA, Shi J, Blundell TL. 2000. Evolutionary trace analysis of TGF-beta and related growth factors: implications for site-directed mutagenesis. Protein Eng 13:839–847. doi: 10.1093/protein/13.12.839. [DOI] [PubMed] [Google Scholar]
- 16.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eicher T, Cha HJ, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grutter MG, Diederichs K, Pos KM. 2012. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci U S A 109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurcik A, Bednar D, Byska J, Marques SM, Furmanova K, Daniel L, Kokkonen P, Brezovsky J, Strnad O, Stourac J, Pavelka A, Manak M, Damborsky J, Kozlikova B. 2018. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 34:3586–3588. doi: 10.1093/bioinformatics/bty386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwama M, Yamasaki S, Nakashima R, Sakurai K, Nishino K, Yamaguchi A. 2018. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat Commun 9:124. doi: 10.1038/s41467-017-02493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 21.Blair JMA, Bavro VN, Ricci V, Modi N, Cacciotto P, Kleinekathӧfer U, Ruggerone P, Vargiu AV, Baylay AJ, Smith HE, Brandon Y, Galloway D, Piddock LJV. 2015. AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc Natl Acad Sci U S A 112:3511–3516. doi: 10.1073/pnas.1419939112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soparkar K, Kinana AD, Weeks JW, Morrison KD, Nikaido H, Misra R. 2015. Reversal of the drug binding pocket defects of the AcrB multidrug efflux pump protein of Escherichia coli. J Bacteriol 197:3255–3264. doi: 10.1128/JB.00547-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. 2004. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 24.Young K, Painter RE, Raghoobar SL, Hairston NN, Racine F, Wisniewski D, Balibar CJ, Villafania A, Zhang R, Sahm DF, Blizzard T, Murgolo N, Hammond ML, Motyl MR. 2019. In vitro studies evaluating the activity of imipenem in combination with relebactam against Pseudomonas aeruginosa. BMC Microbiol 19:150. doi: 10.1186/s12866-019-1522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pournaras S, Maniati M, Spanakis N, Ikonomidis A, Tassios PT, Tsakris A, Legakis NJ, Maniatis AN. 2005. Spread of efflux pump-overexpressing, non-metallo-beta-lactamase-producing, meropenem-resistant but ceftazidime-susceptible Pseudomonas aeruginosa in a region with blaVIM endemicity. J Antimicrob Chemother 56:761–764. doi: 10.1093/jac/dki296. [DOI] [PubMed] [Google Scholar]
- 26.Ziha-Zarifi I, Llanes C, Kohler T, Pechere JC, Plesiat P. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother 43:287–291. doi: 10.1128/AAC.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.