QPX7728 is an investigational ultrabroad-spectrum-beta-lactamase inhibitor (BLI) with potent inhibition of key serine and metallo-beta-lactamases. QPX7728 enhances the potency of many beta-lactams, including carbapenems, in isogenic strains of Gram-negative bacteria producing various beta-lactamases. The potency of meropenem alone and in combination with QPX7728 (tested at fixed concentrations of 1 to 16 μg/ml) was tested against 598 clinical isolates of carbapenem-resistant Enterobacterales (CRE).
KEYWORDS: QPX7728, CRE, metallo-beta-lactamase, serine beta-lactamase
ABSTRACT
QPX7728 is an investigational ultrabroad-spectrum-beta-lactamase inhibitor (BLI) with potent inhibition of key serine and metallo-beta-lactamases. QPX7728 enhances the potency of many beta-lactams, including carbapenems, in isogenic strains of Gram-negative bacteria producing various beta-lactamases. The potency of meropenem alone and in combination with QPX7728 (tested at fixed concentrations of 1 to 16 μg/ml) was tested against 598 clinical isolates of carbapenem-resistant Enterobacterales (CRE). The panel included 363 strains producing serine carbapenemases, 224 strains producing metallo-beta-lactamases (151 NDM, 53 VIM, and 20 IMP), and 50 strains that did not carry any known carbapenemases but were resistant to meropenem (MIC ≥ 4 μg/ml). The panel was also enriched in strains that had various defects in the major porins OmpK35/OmpF and OmpK36/OmpC. Increasing concentrations of QPX7728 restored the potency of meropenem against CRE, with the meropenem MIC90 decreasing from >64 μg/ml to 0.5 μg/ml for QPX7728 (8 μg/ml). QPX7728 significantly increased the potency of meropenem against CRE with multiple resistance mechanisms; the reduction in the meropenem MIC90 with QPX7728 (8 μg/ml) ranged from 32- to >256-fold. Compared with other beta-lactamase inhibitor combinations, meropenem-vaborbactam, ceftazidime-avibactam, and imipenem-relebactam, meropenem with QPX7728 was the most potent beta-lactam–BLI combination tested against all groups of CRE with multiple resistance mechanisms. Defects in OmpK36 in KPC-producing strains markedly decreased the potency of meropenem with vaborbactam (128-fold increase in the MIC90), whereas only an 8- to 16-fold change was observed with QPX7728 plus meropenem. More than 90% of various CRE subsets (including those with reduced permeability) were susceptible to ≤8 μg/ml of meropenem with QPX7728 at 8 μg/ml or lower. The combination of QPX7728 with meropenem against CRE has an attractive microbiological profile in CRE with multiple resistance mechanisms.
INTRODUCTION
Inhibition of beta-lactamase activity with small-molecule inhibitors (beta-lactamase inhibitors [BLIs]) is a broadly recognized strategy to prevent the cleavage of beta-lactams and restore their potency (1, 2). In recent years, the discovery and development of novel inhibitors of beta-lactamases have been some of the most successful directions to address the antimicrobial resistance crisis (3). Since 2015, three new beta-lactamase inhibitors, avibactam, vaborbactam, and relebactam, were introduced into clinical practice (4). All of them have a broader profile of beta-lactamase inhibition than clavulanic acid, tazobactam, and sulbactam in that they inhibit various extended-spectrum beta-lactamases (ESBLs) and carbapenemases, most notably KPC, and class C beta-lactamases. Avibactam and relebactam, both diazabicyclooctanes (DBOs), are structurally and mechanistically related. Vaborbactam is a new chemical class of inhibitor that uses a different pharmacophore based on a cyclic boronate and has a different mechanism of beta-lactamase inhibition than DBOs (5, 6). Unfortunately, none of these new BLIs inhibit members of the class D carbapenemases that are specific to carbapenem-resistant Acinetobacter baumannii, nor do they have any activity against metallo-beta-lactamases (MBLs), whose incidence is increasing geographically and across various species (1, 2). Hence, broadening the spectrum of beta-lactamase inhibition with improved analogs to cover these additional beta-lactamases is an important strategy for the development of new clinically useful therapies.
Increased efflux and reduced permeability are two synergistically acting intrinsic mechanisms of multidrug resistance that are often implicated in the reduced potency of both beta-lactams and beta-lactamase inhibitors. In fact, the potency of all the new BLIs is reduced when these intrinsic mechanisms operate in cells that produce beta-lactamases (7–9). Minimizing the impact of these general intrinsic mechanisms on potency is another rationale for the further optimization of BLIs.
QPX7728 is an investigational BLI based on a boronic acid pharmacophore. Recent biochemical results have demonstrated that QPX7728 is a nanomolar inhibitor of the major representatives of serine and metallo-beta-lactamases from all molecular classes found in bacterial pathogens defined as urgent and serious resistance threats to public health (10). Microbiological experiments with an extensive collection of engineered strains with beta-lactamases showed that QPX7728 inhibits serine class A ESBLs (CTX-M, SHV, TEM, GES, VEB, PER, and OXY) and carbapenemases (KPC-2/3, GES-20, NMC-A, SME-2, VCC-1, SFC-1, and FRI-1) and class C beta-lactamases, both plasmidic (CMY-2, MIR-1, FOX-5, and DHA-1) and chromosomally encoded (AmpC from Pseudomonas aeruginosa, A. baumannii, and Enterobacter cloacae). QPX7728 also inhibits class D beta-lactamases such as the oxacillinase OXA-10 and carbapenemases (OXA-48 and OXA-23/72/58) from Enterobacterales and A. baumannii, respectively. Class B subclass B1 metallo-enzymes, NDM, VIM, some IMPs, GIM-1, and CcrA, are inhibited by QPX7728 (11). Using a panel of isogenic strains of Klebsiella pneumoniae with or without inactivation of the major general porins OmpK35/OmpF and OmpK36/OmpC, the potency of QPX7728 in Enterobacterales was affected much less than the potency of vaborbactam (12).
The potent ultrabroad-spectrum-BLI activity of QPX7728 enhances the potency of multiple beta-lactam antibiotics with varying sensitivity to beta-lactamases and intrinsic resistance mechanisms. This makes it an ideal product to be coadministered in combination with multiple different beta-lactam antibiotics depending on the resistance mechanisms present in the specific pathogen for individualized treatment.
To enable comparisons of the potencies of QPX7728 combinations with multiple antibiotics and across panels of bacteria with different genotypic or phenotypic characteristics, we generated a new metric that reflects the BLI concentration associated with an enhancement of potency. In our discovery program, we determined the concentration of QPX7728 that is required to shift 90% of the tested isolates at or below an existing susceptibility breakpoint for the partner antibiotic tested. This measure of BLI potency against a population of strains (conceptually similar to the MIC90) was termed TPC90 (TPC for target potentiation concentration). This metric can be easily applied to comparisons of the potencies of a beta-lactamase inhibitor combination across bacteria with various genotypic and phenotypic characteristics. The TCP90 is determined using a standard checkerboard technique applied to a population of strains: antibiotic potency against each strain is determined in the presence of increasing concentrations of a BLI (which provides the antibiotic MIC90 at each BLI concentration), and BLI potency is determined in the presence of increasing concentrations of the antibiotic (which provides the TPC90 when the antibiotic concentration corresponds to the susceptibility breakpoint).
As a part of a systematic evaluation of various QPX7728 combinations, we evaluated the impact of QPX7728 on the potency of the carbapenem meropenem and determined the potency of QPX7728 (TCP90) in combination with meropenem in carbapenem-resistant Enterobacterales (CRE) producing serine and metallo-beta-lactamases, including those with permeability defects and including strains resistant to recently approved BLI combinations.
RESULTS
Strains selected for evaluation of QPX7728 in combination with meropenem are enriched with isolates resistant to recently approved BLI-based combinations.
Clinical isolates of carbapenem-resistant Enterobacterales (n = 598) were analyzed for the presence of carbapenemase genes using PCR and sequencing techniques and were shown to contain serine carbapenemases from class A (n = 285; 274 KPC, 8 SME, and 3 NMC-A) and class D (n = 39; 27 OXA-48, 6 OXA-232, 3 OXA-181, and 3 OXA-162), and 224 strains produced metallo-beta-lactamases (151 NDM, 53 VIM, and 20 IMP). The panel also contained 50 strains that did not carry any known carbapenemases but were resistant to meropenem (MIC ≥ 4 μg/ml) and thus were termed non-carbapenemase-producing CRE (non-CP-CRE) (see Table S1 in the supplemental material).
Susceptibility testing demonstrated that carbapenemase-positive, MBL-negative strains in general and KPC-producing strains in particular produced a broad distribution of ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam MICs, particularly enriched in strains from the high end of the distribution of MIC values for these combinations (Fig. S1). For example, the ceftazidime-avibactam susceptibility in carbapenemase-positive, MBL-negative strains in this panel was 90.6%, versus 99.8% reported by the INFORM global surveillance program (13). For KPC-positive/MBL-negative strains, the meropenem-vaborbactam susceptibilities were 85.9% and 90.1% for this panel, compared to 99.2% and 100% for the Clinical and Laboratory Standards Institute (CLSI) and EUCAST breakpoints, respectively, according to the SENTRY global surveillance program (14). The imipenem-relebactam susceptibility was 79.2% in this panel, compared to 98.6% according to the SMART surveillance program (15) (Table 1).
TABLE 1.
MIC distributions for meropenem alone and combined with QPX7728 at various concentrations against 598 carbapenem-resistant strains of Enterobacteralesa
| Treatment | % of strains inhibited at MIC (μg/ml) of: |
MIC50 (μg/ml) | MIC90 (μg/ml) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >64 | |||
| MER alone | 0.5 | 0.5 | 0.5 | 1.2 | 2.7 | 5.5 | 13.2 | 26.3 | 43.5 | 56.5 | 70.7 | 100 | 32 | >64 |
| MER with QPX at concn of: | ||||||||||||||
| 1 μg/ml | 30.9 | 37.8 | 45.7 | 53.3 | 61.5 | 68.6 | 73.2 | 77.8 | 82.3 | 86.8 | 91.0 | 99.8 | 05 | 64 |
| 2 μg/ml | 39.3 | 52.7 | 64.0 | 72.4 | 77.4 | 81.6 | 84.6 | 87.5 | 90.1 | 93.5 | 96.3 | 100 | 0.125 | 16 |
| 4 μg/ml | 51.5 | 66.7 | 76.8 | 83.3 | 86.3 | 89.8 | 93.6 | 96.5 | 97.8 | 98.5 | 99.5 | 100 | ≤0.06 | 4 |
| 8 μg/ml | 66.4 | 76.4 | 86.6 | 92.0 | 95.7 | 98.0 | 99.0 | 99.3 | 99.7 | 100 | 100 | 100 | ≤0.06 | 0.5 |
| 16 μg/ml | 84.3 | 91.1 | 95.0 | 97.8 | 99.5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ≤0.06 | 0.125 |
The panel consisted of 324 strains producing serine carbapenemases from class A (n = 285; 274 KPC, 8 SME, and 3 NMC-A) and class D (n = 39), 224 strains producing metallo-beta-lactamases (151 NDM, 53 VIM, and 20 IMP), and 50 strains that did not carry any known carbapenemases but had a meropenem (MER) MIC of ≥4 μg/ml (non-carbapenemase-producing CRE). QPX, QPX7728.
QPX7728 has weak intrinsic antimicrobial activity.
QPX7728 had some antibacterial activity; 36.5% and 51.8% of MBL-negative and MBL-positive strains, respectively, were inhibited by QPX7728 at 16 μg/ml. A much smaller proportion of strains was inhibited by QPX7728 at 8 μg/ml (4.8% and 7.6% of MBL-negative and MBL-positive strains, respectively). Only one strain in this panel was inhibited by QPX7728 at 4 μg/ml. The higher proportion of MBL-positive strains inhibited by QPX7728 alone might be due to the lower proportion of strains that carry nonfunctional OmpK36/OmpC (Fig. S2), as QPX7728 apparently uses this porin to cross the outer membrane (as inferred from the experiments with K. pneumoniae) (12).
Increasing concentrations of QPX7728 enhance the in vitro potency of meropenem against carbapenem-resistant Enterobacterales.
The MIC of meropenem was evaluated in the presence of five different concentrations of QPX7728 (1 to 16 μg/ml). There was a marked enhancement in the percentage of strains inhibited at each meropenem MIC in the presence of the QPX7728. At MICs of 1 μg/ml and 8 μg/ml, 2.7% and 26.3% of strains were inhibited by meropenem alone, compared to >85% and >95% of strains inhibited at these MICs in the presence of QPX7728 at ≥4 μg/ml, with clear evidence of a QPX7728 dose response (Table 1). For QPX7728 at 16 μg/ml, some of the observed MIC shifts could be due in part to its intrinsic antibacterial activity; however, at concentrations of ≤8 μg/ml, the observed shifts of MIC distributions are attributed mainly to beta-lactamase inhibition.
The pharmacokinetic-pharmacodynamic (PK-PD) breakpoint for meropenem at a dose of 2 g administered every 8 h (q8h) by a 3-h infusion is 8 μg/ml (16, 17); this concentration of meropenem was used to determine the TPC90 of QPX7728 in 598 CRE isolates. The TPC90 of QPX7728 required to shift >90% of CRE isolates to a meropenem MIC of ≤8 μg/ml was 4 μg/ml (Table 1).
QPX7728 enhances the potency of meropenem against all types of CRE, including MBL-positive strains.
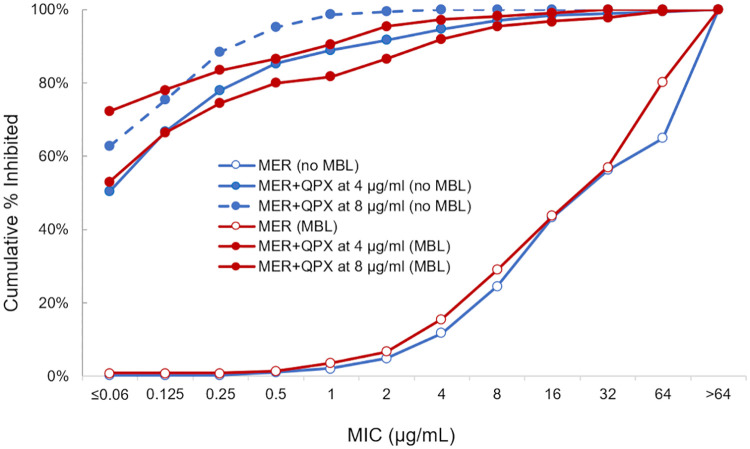
QPX7728 tested at ≥4 μg/ml significantly increased the potency of meropenem against all groups of CRE, including MBL-producing strains. The reduction in meropenem MIC90s ranged from 4- to >64-fold and from 32- to >256-fold in the presence of QPX7728 at 4 μg/ml and 8 μg/ml, respectively (Table 2). A similar shift in the distribution of meropenem MICs was observed in the presence of QPX7728 against strains producing serine and metallo-beta-lactamases (Fig. 1). The mode MICs of meropenem with QPX7728 (4 or 8 μg/ml) for the strains producing all types of carbapenemases (serine and metallo) and non-CP-CRE were ≤0.06 μg/ml and 0.5 μg/ml, respectively (Table 3).
TABLE 2.
In vitro potencies of meropenem alone and combined with QPX7728 at 4 μg/ml and 8 μg/ml and comparator BLI combination agents against carbapenem-resistant strains of Enterobacterales by the carbapenemase present
| Parameter | Value for treatment |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MER | MER + QPX at 1 μg/ml | MER + QPX at 2 μg/ml | MER + QPX at 4 μg/ml | MER + QPX at 8 μg/ml | MER + QPX at 16 μg/ml | MER + VAB at 8 μg/ml | CAZ | CAZ + AVI at 4 μg/ml | IMP | IMP + REL at 4 μg/ml | |
| All (n = 598) | |||||||||||
| MIC50 (μg/ml) | 32 | 0.5 | 0.125 | ≤0.06 | ≤0.06 | ≤0.06 | 4 | >64 | 4 | 32 | 4 |
| MIC90 (μg/ml) | >64 | 64 | 16 | 4 | 0.5 | 0.125 | >64 | >64 | >64 | >64 | 64 |
| % inhibiteda | 26.3 | 77.8 | 87.5 | 96.5 | 99.3 | 100 | 53.7 | 4.2 | 57.4 | 2.7 | 44.5 |
| MBL negative (n = 374) | |||||||||||
| MIC50 (μg/ml) | 32 | 1 | 0.125 | ≤0.06 | ≤0.06 | ≤0.06 | 1 | >64 | 2 | 32 | 0.5 |
| MIC90 (μg/ml) | >64 | >64 | 32 | 2 | 0.5 | 0.125 | 32 | >64 | 8 | >64 | 8 |
| % inhibiteda | 24.6 | 75.9 | 87.4 | 97.1 | 100.0 | 100.0 | 76.7 | 6.4 | 90.6 | 2.9 | 69.5 |
| MBL negative, OXA-48-like negative (n = 325) | |||||||||||
| MIC50 (μg/ml) | 32.0 | 1 | 0.125 | 0.05 | 0.05 | 0.03 | 0.5 | 65 | 2 | 32 | 0.5 |
| MIC90 (μg/ml) | 65.0 | 65 | 32 | 2 | 0.5 | 0.125 | 16 | 65 | 16 | 65 | 4 |
| % inhibiteda | 24.5 | 73.1 | 86.0 | 96.7 | 100.0 | 100.0 | 84.5 | 5.4 | 89.6 | 3.3 | 76.7 |
| KPC (n = 285) | |||||||||||
| MIC50 (μg/ml) | 64 | 0.25 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | 0.25 | >64 | 2 | 64 | 0.5 |
| MIC90 (μg/ml) | >64 | >64 | 64 | 1 | 0.25 | 0.125 | 8 | >64 | 8 | >64 | 4 |
| % inhibiteda | 18.7 | 73.2 | 87.0 | 97.5 | 100.0 | 100.0 | 85.9 | 4.9 | 90.1 | 0.4 | 79.2 |
| K. pneumoniae KPC (n = 230) | |||||||||||
| MIC50 (μg/ml) | >64 | 1 | 0.125 | 0.05 | 0.05 | 0.03 | 0.5 | >64 | 2 | 64 | 0.5 |
| MIC90 (μg/ml) | >64 | >64 | 64 | 1 | 0.25 | 0.125 | 16 | >64 | 16 | >64 | 4 |
| % inhibiteda | 11.7 | 67.4 | 84.3 | 97.0 | 100.0 | 100.0 | 83 | 0.9 | 89.6 | 0.4 | 80.4 |
| OXA-48 (n = 39) | |||||||||||
| MIC50 (μg/ml) | 16 | 0.5 | 0.125 | 0.125 | ≤0.06 | ≤0.06 | 32 | >64 | 2 | 8 | 4 |
| MIC90 (μg/ml) | 64 | 8 | 0.5 | 0.25 | 0.25 | 0.125 | 64 | >64 | 4 | >64 | 64 |
| % inhibiteda | 25.6 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 12.8 | 15.4 | 100.0 | 0.0 | 7.7 |
| Non-CP-CRE (n = 50) | |||||||||||
| MIC50 (μg/ml) | 8 | 4 | 2 | 0.5 | 0.25 | ≤0.06 | 4 | >64 | 2 | 4 | 0.5 |
| MIC90 (μg/ml) | 32 | 16 | 16 | 8 | 1 | 0.5 | 16 | >64 | 16 | 32 | 8 |
| % inhibiteda | 51.2 | 72.0 | 80.0 | 90.7 | 100.0 | 100.0 | 74 | 9.3 | 86.0 | 20.0 | 64.0 |
| MBL (n = 224) | |||||||||||
| MIC50 (μg/ml) | 32 | 0.5 | 0.25 | ≤0.06 | ≤0.06 | ≤0.06 | 32 | >64 | >64 | 32 | 32 |
| MIC90 (μg/ml) | >64 | 64 | 16 | 4 | 1 | 0.125 | >64 | >64 | >64 | >64 | >64 |
| % inhibiteda | 29.0 | 80.8 | 87.5 | 95.5 | 98.2 | 100.0 | 25.9 | 0.4 | 1.8 | 2.2 | 2.7 |
Percentage of strains inhibited at the following concentrations: ≤8 μg/ml/various QPX7728 concentrations for meropenem-QPX7728, ≤4/8 μg/ml (FDA susceptible breakpoint) for meropenem-vaborbactam (VAB), ≤8/4 μg/ml (FDA susceptible breakpoint) for ceftazidime (CAZ)-avibactam (AVI), and ≤1/4 μg/ml (FDA susceptible breakpoint) for imipenem (IMP)-relebactam (REL).
FIG 1.
MIC distributions of meropenem alone or in combination with QPX7728 against carbapenem-resistant Enterobacterales producing either serine beta-lactamases (n = 374) or metallo-beta-lactamases (n = 224). The 374 strains that contain no MBLs include 324 strains producing serine carbapenemases from class A (n = 285; 274 KPC, 8 SME, and 3 NMC-A) and class D (n = 39) and 50 strains that do not carry any known carbapenemases but have meropenem MICs of ≥4 μg/ml (non-carbapenemase-producing CRE [non-CP-CRE]). The 224 strains producing metallo-beta-lactamases include 151 NDM-, 53 VIM-, and 20 IMP-producing strains. Out of the 224 MBL-producing strains, 34 also carry class A or class D serine carbapenemases.
TABLE 3.
In vitro potencies of meropenem alone and combined with QPX7728 at various concentrations against carbapenem-resistant strains of Enterobacterales according to carbapenemase
| CRE (no. of strains) | Agent(s) | No. of strains inhibited at meropenem MIC (μg/ml) of:a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >64 | ||
| KPC (285) | MER | 1 | 0 | 0 | 2 | 3 | 3 | 14 | 30 | 45 | 28 | 29 | 130 |
| MER + QPX at 4 μg/ml | 165 | 43 | 30 | 17 | 9 | 7 | 5 | 2 | 1 | 2 | 2 | 2 | |
| MER + QPX at 8 μg/ml | 203 | 32 | 31 | 11 | 4 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | |
| K. pneumoniae KPC (230) | MER | 0 | 0 | 0 | 1 | 1 | 1 | 6 | 18 | 31 | 21 | 25 | 126 |
| MER + QPX at 4 μg/ml | 116 | 43 | 28 | 15 | 8 | 7 | 5 | 1 | 1 | 2 | 2 | 2 | |
| MER + QPX at 8 μg/ml | 154 | 31 | 29 | 9 | 3 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | |
| Non-K. pneumoniae KPC (53) | MER | 1 | 0 | 0 | 1 | 2 | 2 | 8 | 12 | 12 | 7 | 4 | 4 |
| MER + QPX at 4 μg/ml | 47 | 0 | 2 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| MER + QPX at 8 μg/ml | 47 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| OXA-48 (39) | MER | 0 | 0 | 0 | 1 | 0 | 2 | 2 | 5 | 13 | 11 | 4 | 1 |
| MER + QPX at 4 μg/ml | 19 | 14 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| MER + QPX at 8 μg/ml | 23 | 11 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Non-CP-CRE (50) | MER | 0 | 0 | 0 | 0 | 1 | 5 | 10 | 13 | 12 | 9 | 0 | 0 |
| MER + QPX 4 μg/ml | 5 | 4 | 7 | 10 | 4 | 3 | 6 | 7 | 4 | 0 | 0 | 0 | |
| MER + QPX at 8 μg/ml | 9 | 4 | 13 | 14 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| MBL (224) | MER | 2 | 0 | 0 | 1 | 5 | 7 | 20 | 30 | 33 | 30 | 52 | 44 |
| MER + QPX at 4 μg/ml | 119 | 30 | 18 | 12 | 4 | 11 | 12 | 8 | 3 | 2 | 4 | 1 | |
| MER + QPX at 8 μg/ml | 162 | 13 | 12 | 7 | 9 | 11 | 4 | 2 | 2 | 2 | 0 | 0 | |
| NDM (151) | MER | 2 | 0 | 0 | 0 | 4 | 5 | 13 | 12 | 19 | 20 | 42 | 34 |
| MER + QPX at 4 μg/ml | 79 | 25 | 14 | 7 | 1 | 2 | 10 | 5 | 3 | 2 | 2 | 1 | |
| MER + QPX at 8 μg/ml | 113 | 8 | 4 | 2 | 7 | 10 | 2 | 2 | 2 | 1 | 0 | 0 | |
| VIM (53) | MER | 0 | 0 | 0 | 1 | 1 | 2 | 3 | 14 | 6 | 9 | 7 | 10 |
| MER + QPX at 4 μg/ml | 32 | 3 | 4 | 3 | 2 | 6 | 1 | 1 | 0 | 0 | 1 | 0 | |
| MER + QPX at 8 μg/ml | 36 | 4 | 6 | 4 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
| IMP (20) | MER | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 | 8 | 1 | 3 | 0 |
| MER + QPX at 4 μg/ml | 8 | 2 | 0 | 2 | 1 | 3 | 1 | 2 | 0 | 0 | 1 | 0 | |
| MER + QPX at 8 μg/ml | 13 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
Mode MIS is in bold.
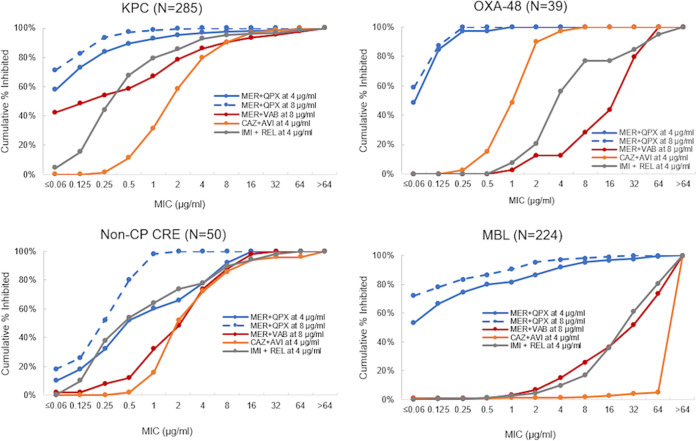
Based on the MIC50/MIC90, meropenem with QPX7728 was more potent than ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam against all groups of CRE, including MBL-negative strains, e.g., KPC producers, OXA-48-like producers, and non-CP-CRE (Table 2 and Fig. 2). The TPC90 of QPX7728 for each group of CRE was 4 μg/ml (Table 2).
FIG 2.
MIC distributions of various beta-lactam–BLI combinations against carbapenem-resistant Enterobacterales according to the type of CRE.
QPX7728 in combination with meropenem has excellent potency against CRE with permeability defects due to porin mutations.
We have recently shown that the potency of QPX7728 was reduced (albeit to a much lesser degree than vaborbactam) in strains that carried nonfunctional or partially functional porin OmpK36 (12). To extend this work, we assessed the reduction in meropenem MICs with various concentrations of QPX7728 in a subset of isolates that carried various defects in OmpK36/OmpC and OmpK35/OmpF to determine the QPX7728 TPC90 against this panel of strains.
The functional status of OmpK36/OmpC (inferred from sequence and expression analyses) was characterized in 501 isolates. In addition, ompK35 was sequenced in 488 of these isolates. Various OmpK36/OmpC defects were detected in 244 out of 501 isolates (48.7%), with 202 (62.3%) for MBL-negative and 42 (23.7%) for MBL-positive strains. These defects included complete inactivation (n = 67) due to frameshift and nonsense mutations and partial inactivation (n = 163) mainly due to the duplication of two amino acids, Gly134 and Asp135 (GD repeat), located within the L3 internal loop, resulting in the constriction of the channel (18), and reduced gene expression (n = 14) mainly due to various insertions in the promoter region was also observed (19) (Table S3). Nonfunctional OmpK35/OmpF (due to frameshift, nonsense, and insertion sequence mutations) was observed in 320 strains (65.6%), and 192 strains (39.3%) had defects in both major porins OmpK35/OmpF and OmpK36/OmpC.
In agreement with our data from laboratory-derived strains (12), the inactivation of OmpK35 did not affect the potency of QPX7728; the meropenem MIC distributions in the presence of QPX7728 at 4 μg/ml or 8 μg/ml remained very similar for isolates with either functional or nonfunctional OmpK35/OmpF (the meropenem MIC90 with 4 or 8 μg/ml of QPX7728 was 4 μg/ml or 0.5 μg/ml, respectively, compared to >64 μg/ml for meropenem alone) (Table S4a). Defects in OmpK36/OmpC were associated with changes in meropenem-QPX7728 MIC distributions; the MIC90 of meropenem in the presence of QPX7728 was increased 16- to 32-fold (Table 4 and Table S4b) from 0.25 μg/ml to 8 μg/ml and from ≤0.06 μg/ml to 1 μg/ml for QPX7728 at 4 μg/ml and 8 μg/ml, respectively. The inactivation of OmpK35 in strains that had various defects in OmpK36 did not result in any additional shift of meropenem-QPX7728 MICs, indicating no additional loss of QPX7728 potency (Table S4c).
TABLE 4.
In vitro activities of meropenem alone and combined with QPX7728 at 4 μg/ml and 8 μg/ml and comparator BLI combination agents against carbapenem-resistant strains of Enterobacterales according to the functional status of OmpK35/OmpF and OmpK36/OmpC and the carbapenemase present
| Parameter | Value |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Defective OmpK36b |
Functional OmpK36 |
|||||||||
| MER + QPX at 4 μg/ml | MER + QPX at 8 μg/ml | MER + VAB at 8 μg/ml | CAZ + AVI at 4 μg/ml | IMI + REL at 4 μg/ml | MER + QPX at 4 μg/ml | MER + QPX at 8 μg/ml | MER + VAB at 8 μg/ml | CAZ + AVI at 4 μg/ml | IMP + REL at 4 μg/ml | |
| All (n = 501) | n = 244 | n = 257 | ||||||||
| MIC50 (μg/ml) | 0.25 | 0.125 | 4 | 4 | 2 | ≤0.06 | ≤0.06 | 4 | >64 | 4 |
| MIC90 (μg/ml) | 8 | 1 | >64 | >64 | >64 | 0.25 | ≤0.06 | 64 | >64 | 64 |
| % inhibiteda | 92.6 | 98.8 | 51.6 | 73.8 | 49.2 | 99.6 | 99.6 | 55.3 | 44.4 | 44.7 |
| MBL negative (n = 324) | n = 202 | n = 122 | ||||||||
| MIC50 (μg/ml) | 0.25 | 0.125 | 4 | 2 | 1 | ≤0.06 | ≤0.06 | ≤0.06 | 2 | 0.25 |
| MIC90 (μg/ml) | 4 | 0.5 | 32 | 16 | 8 | 0.125 | ≤0.06 | 1 | 8 | 1 |
| % inhibiteda | 95.0 | 100.0 | 62.4 | 89.1 | 59.4 | 100.0 | 100.0 | 94.3 | 91.0 | 90.2 |
| K. pneumoniae KPC (n = 230) | n = 135 | n = 95 | ||||||||
| MIC50 (μg/ml) | 0.25 | 0.125 | 2 | 4 | 0.5 | ≤0.06 | ≤0.06 | ≤0.06 | 2 | 0.25 |
| MIC90 (μg/ml) | 2 | 0.5 | 32 | 16 | 4 | 0.125 | ≤0.06 | 0.25 | 8 | 1 |
| % inhibiteda | 94.8 | 100.0 | 72.6 | 88.1 | 70.4 | 100.0 | 100.0 | 97.9 | 91.6 | 94.7 |
| MBL (n = 177) | n = 42 | n = 135 | ||||||||
| MIC50 (μg/ml) | 4 | 1 | >64 | >64 | >64 | ≤0.06 | ≤0.06 | 32 | >64 | 32 |
| MIC90 (μg/ml) | 32 | 4 | >64 | >64 | >64 | 0.5 | 0.125 | >64 | >64 | 64 |
| % inhibiteda | 81.0 | 92.9 | 0.0 | 0.0 | 0.0 | 99.3 | 99.3 | 20.0 | 2.2 | 3.7 |
Percentage of strains inhibited at the following concentrations: ≤8 μg/ml/various QPX7728 concentrations for meropenem-QPX7728, ≤4/8 μg/ml (FDA susceptible breakpoint) for meropenem-vaborbactam, ≤8/4 μg/ml (FDA susceptible breakpoint) for ceftazidime-avibactam, and ≤1/4 μg/ml (FDA susceptible breakpoint) for imipenem-relebactam.
OmpK36 was considered defective if frameshift and nonsense mutations; large insertions; a duplication of two amino acids, Gly134 and Asp135 (GD repeat), located within the L3 internal loop, resulting in the constriction of the channel; or reduced expression due to insertions in the promoter region have been identified. Otherwise, it was assumed to be functional.
Despite the negative impact of various defects in OmpK36/OmpC on QPX7728 potency, 4 μg/ml of QPX7728 was sufficient to shift the meropenem MICs to ≤8 μg/ml for 95% of non-MBL-producing CRE. In the MBL-positive strains with OmpK36/OmpC-deficient isolates, 81% of the meropenem MICs were shifted to ≤8 μg/ml when tested with QPX7728 at 4 μg/ml, and with 8 μg/ml of QPX7728, 92.9% of MBL-positive OmpK36-deficient isolates had meropenem MICs of ≤8 μg/ml (Table 4); thus, the QPX7728 TPC90 values remained in the 4- to 8-μg/ml range for this subset of isolates with MBL or KPC beta-lactamases combined with intrinsic resistance mechanisms that are not addressed by the existing beta-lactamase inhibitor combinations ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam (Table 4 and Fig. 3).
FIG 3.
MIC distributions of various beta-lactam–BLI combinations against KPC-producing strains of K. pneumoniae with defective (DEF) (n = 135) or functional (FL) (n = 95) OmpK36. OmpK36 was considered defective if frameshift and nonsense mutations, large insertions, a duplication of two amino acids (Gly134 and Asp135 [GD repeat], located within the L3 internal loop, resulting in the constriction of the channel), or reduced expression due to insertions in the promoter region has been identified. Otherwise, it was assumed to be functional.
DISCUSSION
QPX7728 is an ultrabroad-spectrum-beta-lactamase inhibitor that shows a significant enhancement of the potencies of various beta-lactam antibiotics against Gram-negative bacteria producing diverse beta-lactamases (10–12). In this study, we demonstrated that QPX7728 significantly enhanced the potency of meropenem against carbapenem-resistant Enterobacterales producing serine and metallo-beta-lactamases. The mode MIC of meropenem with QPX7728 (at 4 or 8 μg/ml) for all groups of carbapenemase-producing CRE, including MBL-producing strains, was ≤0.06 μg/ml. This activity was observed in a panel that contained a high number of MBL-producing isolates (37%) as well as serine carbapenemase-producing strains from the high end of the distribution of MIC values for the recently approved beta-lactamase–BLI combinations ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam. Not surprisingly, meropenem with QPX7728 was the most potent beta-lactamase–BLI combination against MBL-producing strains. But it was also a more potent combination than ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam against strains producing the serine carbapenemases KPC and OXA and against non-carbapenemase-producing CRE.
Mutations in drug permeability and efflux have been increasingly reported in clinical isolates of Enterobacterales (9, 19–21). Using the panel of KPC-producing strains, we demonstrated that OmpK36/OmpC deficiency appeared to affect QPX7728 to a significantly lesser degree than vaborbactam. Meropenem with QPX7728 had excellent potency against strains with permeability defects due to various ompK36-ompC mutations; while the MIC90 of meropenem plus QPX7728 was increased 16- to 32-fold in the strains with permeability defects, it remained below its PK-PD breakpoint (8 μg/ml) with QPX7728 concentrations of 4 to 8 μg/ml. This makes meropenem with QPX7728 (at either 4 μg/ml or 8 μg/ml) a more potent combination than ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam against OmpK36-deficient isolates, including KPC- and OXA-producing producing strains.
A key objective of this study was to determine the concentration of QPX7728 that is required to shift the meropenem MICs for >90% of isolates to below the PK-PD breakpoint for meropenem (≤8 μg/ml when administered as 2 g every 8 h as a 3-h infusion). This QPX7728 concentration, or TPC90 (TPC for target potentiation concentration), was 8 μg/ml or lower.
As we develop dosage regimens for QPX7728 for use with different partner beta-lactams, the TPC90 will be an important metric for the translation of BLI potency into exposures that are expected to be associated with BLI activity in vivo against target pathogens, including MBL-producing strains with various defects in the major porin OmpK36/OmpC. Studies are under way to evaluate QPX7728 in combination with meropenem against other target pathogens, such as carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa, as well as in combinations with other potential partner beta-lactam antibiotics.
MATERIALS AND METHODS
Bacterial strains.
A total of 598 clinical isolates of carbapenem-resistant Enterobacterales were from the Qpex collection of strains. The majority of strains originated from various global survey programs and were acquired from JMI (North Liberty, IA) and IHMA (Schaumburg, IL). The panel also included 89 strains from the CDC and FDA Antibiotic Resistance (AR) Isolate Bank (https://wwwn.cdc.gov/ARIsolateBank/). The strains were from multiple countries (244 from the United States, 193 from Europe, 43 from Latin America, 8 from the Middle East, 101 from Asia-Pacific and South Pacific countries, and 9 from Africa) and were isolated from 2001 to 2017. The majority of isolates were cultured from 2014 to 2017 (408 isolates), 142 isolates were from 2010 to 2013, and 48 isolates were from 2001 to 2009. The panel included 432 Klebsiella pneumoniae isolates, 59 Escherichia coli isolates, 55 Enterobacter cloacae complex isolates, and 52 isolates from other species of Enterobacterales (see Table S1 in the supplemental material).
Detection of carbapenemase genes.
Quantitative PCR (qPCR) was used to detect the presence of carbapenemase genes.
A single colony was grown in cation-adjusted Mueller-Hinton broth (CA-MHB) with shaking at 37°C until the optical density at 600 nm (OD600) reached ∼0.5. A total of 0.3 ml of the culture was centrifuged for 1 min, and the cells were resuspended in 0.1 ml of water. The cell suspension was heated at 95°C for 10 min and stored at −20°C for future use as the template for qPCR. qPCR was performed on an ABIPrism 7000 sequencing instrument (Applied Biosystems) using SYBR select master mix (catalog number 4472919; Thermo Fisher). The template preparations were diluted 10-fold with water, and 9 μl of the dilution was used in a qPCR mixture that also contained 10 μl of SYBR select master mix (2×) and 1 μl of qPCR primer pair mix to make the final concentration of forward and reverse primers 0.5 μM. The primer sequences for qPCR are listed in Table S5. Primers representing a conserved region of the bacterial 16S rRNA gene, Univ-5-qF and Univ-5-qR, were used as internal controls. The qPCR was run in duplicate with the following thermal cycling conditions: 50°C for 2 min and 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 70°C for 45 s.
The qPCR threshold cycle (CT) value of a beta-lactamase gene to be tested was normalized to the internal control by subtracting the CT of the test gene from the CT of Univ-5 (16S rRNA gene) from the same strain. To compare the signals of a test gene between strains, a commonly used strain known to carry the test gene was designated the standard strain for that gene, and its copy number is designated 1. The normalized CT of the standard strain was subtracted from that of a test strain. The difference (ΔCT) was used as a logarithmic power (base 2) to calculate the relative copy number of the test gene in the test strain. For the detection of a test gene, the gene is considered present in a strain if the relative copy number of the test gene is >0.01 and the thermodissociation curve is similar to that of the standard strain. Otherwise, the gene is considered absent in the strain.
Sequence analysis of the ompK35-ompF and ompK36-ompC genes.
The ompK35-ompF and ompK36-ompC genes were amplified by PCR using the primer pairs shown in Table S5. The PCR products cover the coding sequences and about 70 to 100 bp of flanking regions, allowing sequencing of the entire coding sequence of the corresponding genes. PCR was run using DreamTaq green PCR master mix (catalog number K1082; Thermo Fisher) with the following thermal cycling conditions: 95°C for 5 min; 35 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 90 s; and, finally, 72°C for 10 min. The PCR products were sequenced at Eton Biosciences (San Diego, CA).
Determination of expression of the ompK36-ompC genes.
Single colonies grown overnight from a plate were inoculated into CA-MHB and grown at 37°C until an OD600 of 0.7 was obtained. Cell pellets were collected by centrifugation, and total RNA was isolated using an Ambion RiboPure-Bacteria RNA isolation kit (Thermo Fisher, San Diego, CA). Residual DNA in the RNA samples was removed by treatment with DNase I, according to the manufacturer’s instructions. Reverse transcription (RT) was performed using a TaqMan reverse transcriptase reagent kit (Thermo Fisher, San Diego, CA). A mixture of reverse primers for ompK36-ompC and the internal control housekeeping gene rpoB (Table S5), each at a final concentration of 0.5 μM, was used as the RT primers. Two microliters of RNA samples were used in a total RT reaction volume of 10 μl. The RT reaction mixture was diluted 10-fold and used for quantitative real-time PCR performed on an ABIPrism 7000 sequencer (Applied Biosystems) using SYBR select master mix (Thermo Fisher). For these reactions, 9 μl of the diluted RT reaction mixture was used as the template and mixed with 10 μl of SYBR select master mix (2×) and 1 μl of a qPCR primer pair mixture (Table S5) to make the final concentration of forward and reverse primers 0.5 μM. The qPCR was run with the following thermal cycling conditions: 55°C for 2 min and 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. qPCR was carried out in duplicate. The qPCR results (CT values) were normalized with the housekeeping gene rpoB by subtracting the CT value of a porin gene from the CT value of the rpoB gene of the same RT reaction mixture. To compare the mRNA levels of a porin gene in a test strain and the control strain, the normalized CT value of the control strain was subtracted from that of the test strain, and the difference (ΔCT) was used as a logarithmic power (base 2) to calculate the relative normalized level of mRNA. The averages and standard deviations of relative normalized mRNA levels in duplicated qPCRs from the same strain were calculated.
Antimicrobial susceptibility testing.
Bacterial isolates were subjected to broth microdilution susceptibility testing, performed according to CLSI methods (22), using panels prepared in-house. A checkerboard assay conforming to the Moody procedures in the Clinical Microbiology Procedures Handbook (23) was used to evaluate the effect of various concentrations of QPX7728 on the meropenem MIC. Meropenem was purchased from Carbosynth, and all other antibiotics were obtained from Sigma-Aldrich. All beta-lactamase inhibitors were synthesized at Qpex Biopharma, San Diego, CA.
Supplementary Material
ACKNOWLEDGMENTS
This project has been funded in whole or in part with federal funds from the Department of Health and Human Services, Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority (BARDA), under OTA number HHSO100201600026C.
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for the manuscript, take responsibility for the integrity of the work as a whole, and have given final approval to the version to be published.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bush K, Bradford PA. 2019. Interplay between beta-lactamases and new beta-lactamase inhibitors. Nat Rev Microbiol 17:295–306. doi: 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- 2.Papp-Wallace KM. 2019. The latest advances in beta-lactam/beta-lactamase inhibitor combinations for the treatment of Gram-negative bacterial infections. Expert Opin Pharmacother 20:2169–2184. doi: 10.1080/14656566.2019.1660772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lasko MJ, Nicolau DP. 2020. Carbapenem-resistant Enterobacterales: considerations for treatment in the era of new antimicrobials and evolving enzymology. Curr Infect Dis Rep 22:6. doi: 10.1007/s11908-020-0716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pogue JM, Bonomo RA, Kaye KS. 2019. Ceftazidime/avibactam, meropenem/vaborbactam, or both? Clinical and formulary considerations. Clin Infect Dis 68:519–524. doi: 10.1093/cid/ciy576. [DOI] [PubMed] [Google Scholar]
- 5.Tsivkovski R, Lomovskaya O. 2020. Biochemical activity of vaborbactam. Antimicrob Agents Chemother 64:e01935-19. doi: 10.1128/AAC.01935-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsivkovski R, Lomovskaya O. 2020. Potency of vaborbactam is less affected than avibactam in strains producing KPC-2 mutations that confer resistance to ceftazidime-avibactam. Antimicrob Agents Chemother 64:e01936-19. doi: 10.1128/AAC.01936-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomez-Simmonds A, Stump S, Giddins MJ, Annavajhala MK, Uhlemann AC. 2018. Clonal background, resistance gene profile, and porin gene mutations modulate in vitro susceptibility to imipenem-relebactam in diverse Enterobacteriaceae. Antimicrob Agents Chemother 62:e00573-18. doi: 10.1128/AAC.00573-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomovskaya O, Sun D, Rubio-Aparicio D, Nelson K, Tsivkovski R, Griffith DC, Dudley MN. 2017. Vaborbactam: spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob Agents Chemother 61:e01443-17. doi: 10.1128/AAC.01443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson K, Hemarajata P, Sun D, Rubio-Aparicio D, Tsivkovski R, Yang S, Sebra R, Kasarskis A, Nguyen H, Hanson BM, Leopold S, Weinstock G, Lomovskaya O, Humphries RM. 2017. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother 61:e00989-17. doi: 10.1128/AAC.00989-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsivkovski R, Totrov M, Lomovskaya O. 2020. Biochemical characterization of QPX7728, a new ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-beta-lactamases. Antimicrob Agents Chemother 64:e00130-20. doi: 10.1128/AAC.00130-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomovskaya O, Tsivkovski R, Nelson K, Rubio-Aparicio D, Sun D, Totrov M, Dudley MN. 2020. Spectrum of beta-lactamase inhibition by the cyclic boronate QPX7728, an ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-beta-lactamases: enhancement of activity of multiple antibiotics against isogenic strains expressing single beta-lactamases. Antimicrob Agents Chemother 64:e00212-20. doi: 10.1128/AAC.00212-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomovskaya O, Nelson K, Rubio-Aparicio D, Tsivkovski R, Sun D, Dudley MN. 2020. Impact of intrinsic resistance mechanisms on potency of QPX7728, a new ultrabroad-spectrum beta-lactamase inhibitor of serine and metallo-beta-lactamases in Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Antimicrob Agents Chemother 64:e00552-20. doi: 10.1128/AAC.00552-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiliopoulou I, Kazmierczak K, Stone GG. 2020. In vitro activity of ceftazidime/avibactam against isolates of carbapenem-non-susceptible Enterobacteriaceae collected during the INFORM global surveillance programme (2015–17). J Antimicrob Chemother 75:384–391. doi: 10.1093/jac/dkz456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castanheira M, Doyle TB, Kantro V, Mendes RE, Shortridge D. 2020. Meropenem-vaborbactam activity against carbapenem-resistant Enterobacterales isolates collected in U.S. hospitals during 2016 to 2018. Antimicrob Agents Chemother 64:e01951-19. doi: 10.1128/AAC.01951-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lob SH, Karlowsky JA, Young K, Motyl MR, Hawser S, Kothari ND, Sahm DF. 2020. In vitro activity of imipenem-relebactam against resistant phenotypes of Enterobacteriaceae and Pseudomonas aeruginosa isolated from intraabdominal and urinary tract infection samples—SMART Surveillance Europe 2015–2017. J Glob Antimicrob Resist 69:207–217. doi: 10.1099/jmm.0.001142. [DOI] [PubMed] [Google Scholar]
- 16.Kuti JL, Dandekar PK, Nightingale CH, Nicolau DP. 2003. Use of Monte Carlo simulation to design an optimized pharmacodynamic dosing strategy for meropenem. J Clin Pharmacol 43:1116–1123. doi: 10.1177/0091270003257225. [DOI] [PubMed] [Google Scholar]
- 17.Lee LS, Kinzig-Schippers M, Nafziger AN, Ma L, Sorgel F, Jones RN, Drusano GL, Bertino JS Jr. 2010. Comparison of 30-min and 3-h infusion regimens for imipenem/cilastatin and for meropenem evaluated by Monte Carlo simulation. Diagn Microbiol Infect Dis 68:251–258. doi: 10.1016/j.diagmicrobio.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shields RK, Clancy CJ, Hao B, Chen L, Press EG, Iovine NM, Kreiswirth BN, Nguyen MH. 2015. Effects of Klebsiella pneumoniae carbapenemase subtypes, extended-spectrum beta-lactamases, and porin mutations on the in vitro activity of ceftazidime-avibactam against carbapenem-resistant K. pneumoniae. Antimicrob Agents Chemother 59:5793–5797. doi: 10.1128/AAC.00548-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordmann P, Poirel L. 2019. Epidemiology and diagnostics of carbapenem resistance in Gram-negative bacteria. Clin Infect Dis 69:S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shields RK, McCreary EK, Marini RV, Kline EG, Jones CE, Hao B, Chen L, Kreiswirth BN, Doi Y, Clancy CJ, Nguyen MH. 18 November 2019. Early experience with meropenem-vaborbactam for treatment of carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis doi: 10.1093/cid/ciz1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed, M100-Ed30 CLSI, Wayne, PA. [Google Scholar]
- 23.Moody J. 2007. Synergism testing: broth microdilution checkerboard, Chapter 5.12 In Garcia LS. (ed), Clinical microbiology procedures handbook, 3rd ed ASM Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.