Abstract
The objective of this study was to explore the relationships between ruminal microbial populations from Angus steers that were divergent in carcass traits related to adipose accumulation. Twenty-four feedlot-finished Angus steers (age: 538 ± 21 d; body weight following lairage: 593.9 ± 43.7 kg) were slaughtered, and ruminal contents and carcass data were collected. Ruminal microbial deoxyribonucleic acid (DNA) extraction and 16S ribosomal ribonucleic acid (rRNA) gene sequencing were performed to determine microbial relative abundances, to estimate microbial diversity, and to predict microbial metabolic pathways. A variety of correlation analyses and one-way ANOVA were performed to investigate the relationships between the rumen microbiome and carcass traits. Marbling score (P = 0.001) and longissimus lipid content (P = 0.009) were positively correlated to Chao1 Richness Index, suggesting that increased intramuscular fat was associated with increased numbers of ruminal microbial species. The phyla Tenericutes and TM7 were negatively correlated (P ≤ 0.05) to marbling score and longissimus lipid content, indicating that lower abundances of these phyla may be associated with improvements in intramuscular fat content. Greater abundance of the bacterial family S24-7 was positively correlated (P = 0.002) to marbling score. Analysis by marbling classification revealed further linkages to microbial richness (P ≤ 0.063), diversity (P = 0.044), and S24-7 (P < 0.001) populations. Computational prediction of the microbial metabolic pathways revealed no differences (P ≥ 0.05) in metabolic pathway expression in rumen microbes between steers in the high- and low-marbling classes. Several phyla, families, and genera were positively correlated (P ≤ 0.05) to both rib fat thickness and yield grade. Collectively, our results suggest that microbial composition is associated to differing performance in carcass adipose traits. Overall, most of the bacterial taxa correlated to the intramuscular and subcutaneous fat depots did not overlap, suggesting the microbial population end products likely impacted adipose accumulation largely via separate adipogenic pathways of the host animal.
Keywords: carcass quality, lipid, marbling, microbiome, rumen, S24-7
Introduction
The rumen of cattle is capable of fermenting a wide variety of feedstuffs because it contains a diverse and competitive consortium of bacteria, protozoa, archaea, and fungi that are collectively recognized as the rumen microbiome (Hungate, 1966; Sharp et al., 1998; Mackie et al., 2000). The ruminal microbial population produces fermentation products such as volatile fatty acids (VFA) and microbial crude protein that contribute to meeting the host’s requirements for energy and protein, respectively (Hungate, 1966; Russell and Wallace, 1997). Nonetheless, the rumen microbiota are divergent in their functional group(s), or ecological niche(s) (e.g., amylolytic, proteolytic, and cellulolytic), as well as in their metabolic pathways, which ultimately dictates the production of fermentation end products and their impact on the host animal (Hungate, 1966). Thus, understanding the microbial community composition and the impact of population changes on end product contributions is key to understanding how the microbial population impacts host production of meat, milk, and fiber (e.g., wool and hair) through the nutrients produced by the microbes, which are then absorbed and metabolized by the host.
In recent years, several studies have explored the interactions between the ruminal microbial populations and ruminant production traits, such as feed efficiency (Myer et al., 2015; Shabat et al., 2016; Ellison et al., 2017; Schären et al., 2018; Li et al., 2019), methane emission (Kamke et al., 2016; Wallace et al., 2019), and milk fatty acid composition (Buitenhuis et al., 2019). Throughout these studies, trends in microbial relative abundances were linked to production trait differences, suggesting that the rumen microbiome impacted animal performance. Although these relationships are not necessarily a new discovery, the new methodologies that are now available continually allow us to visualize the host–microbial relationship in greater detail. Nonetheless, while improvements in production efficiency are clearly of great economic value to the beef industry (Samarajeewa et al., 2012), an area of similar value that has received little correlative investigation to date is the relationship between the rumen microbiome and beef carcass merit. The inherent value in a beef carcass is determined by consumer perception, which dictates the economic return to the producer. Thus, producing a beef carcass that is both high quality and high quantity supports the relationship that the industry has with consumers and increases income directly to the producer.
Expectations set by the consumers for overall beef palatability include a tender, juicy, and flavorful beef product (Smith and Carpenter, 1974; O’Quinn et al., 2018). Marbling (intramuscular fat) is a large influencer of palatability (O’Quinn et al., 2018); hence, marbling is critical to the value of a beef carcass. For producers, profitability is influenced not only by beef quality through improved marbling, but also by the quantity of beef produced. Thus, yield grades (which can be significantly affected by seam fat, determined via kidney, pelvic, heart fat percentage) represent the amount of boneless retail product that is generated and, therefore, can also impact producer profitability (Trenkle, 2001; Forristall et al., 2002).
The objective of this study was to conduct a preliminary investigation into the relationships between the rumen microbial populations and both carcass quality and yield traits in Angus steers. We hypothesized that steers with differing carcass merit in terms of marbling, longissimus lipid content, adjusted 12th rib fat thickness, and yield grades would have different ruminal microbiota compositions, which would result in different expressions of the microbial metabolic pathways.
Materials and Methods
All animals used in this study were humanely managed under the University of Georgia (UGA) Animal Care and Use Committee guidelines (AUP #A2012 11-006-R1).
Experimental animals
The 24 Angus steers utilized in this study were a subsample selected from a group of 60 commercial Angus steers produced for an ongoing study on selection for feed efficiency and carcass quality. Information about the selection, management, and diets of the steers were reported by Detweiler et al. (2019) in a summary of the study’s first 3 yr. Briefly, the 24 Angus steers were part of the fifth generation of offspring that had been divergently selected for marbling and feed efficiency. Overall, there were eight sires represented in the subsample that were divergent in terms of residual average daily gain (low or high) and marbling (high or average) expected progeny differences (EPD), but that were similar in terms of other EPD. While the selection of the subsample was based on the investigation of the digestive tract microbiome in relation to feed efficiency, as detailed by Welch et al. (2020), additional data collection on the carcasses allowed for the present study to explore the relationships between the rumen microbiome and carcass traits.
The steers were produced from the cow herd at the Northwest Georgia Research and Education Center in Calhoun, GA (34°30′N, 84°57′W). The calves were born around January 2017 (calving season length: 47 d), weaned in September, and transported to Brasstown, NC (35°10′N, 83°23′W), shortly after weaning. The steers were backgrounded on pasture until they entered the feedlot at approximately 13 to 14 mo of age. During the approximately 4 mo that they spent in the feedlot, the steers completed a 70-d feed intake trial using a GrowSafe (GrowSafe Systems Ltd., Airdrie, Alberta, Canada) bunk system where average daily gain (0.98 ± 0.16 kg/d), average daily dry matter intake (11.65 ± 2.02 kg/d), residual feed intake (−0.04 ± 1.83 kg/d), and dry matter gain to feed ratios (0.09 ± 0.02) were measured. All steers were nutritionally managed as a contemporary group with the nutrient composition of the finishing diet (dry matter basis) being: net energy maintenance 95.17 mcal/cwt, net energy gain 65.00 mcal/cwt, crude protein 14.5%, roughage 12.4%, rough neutral detergent fiber 6.9%, fat 5.3%, calcium 0.7%, phosphorus 0.5%, potassium 0.7%, magnesium 0.2%, sulfur 0.3%, and added salt 0.2%. No growth enhancement technologies such as hormone implants, beta-adrenergic agonists (e.g., ractopamine), or ionophores (e.g., monensin) were utilized during any phase of production. Upon exiting the feedlot, the steers were evaluated for residual feed intake, an indicator of feed efficiency. The 12 most-efficient and 12 least-efficient steers were selected and humanely slaughtered (age 538 ± 21 d; body weight following lairage 593.9 ± 43.7 kg) at the UGA Meat Science Technology Center, a federally inspected meat plant in Athens, GA (33°57′N, 83°22′W).
Sample and carcass data collection
The 24 steers were fasted and held overnight with access to water and were harvested the following morning under USDA inspection. Ruminal contents were aseptically collected into 50-mL conical tubes following the evisceration process and were frozen at −20 °C until further analysis. The rumen contents were collected from both the ventral caudal and ventral cranial pouches, in order to get a representative sample of the fluid-associated microbial populations in the rumen, and represent the microbial populations of steers that were fasted (approx. 24 h) prior to slaughter.
Carcasses were chilled at 2 °C for 48 h before a trained evaluator collected USDA yield and quality measures, including adjusted 12th rib fat thickness, hot carcass weight, kidney pelvic heart fat, ribeye area, overall carcass maturity, and marbling scores (recorded to a tenth of a marbling score; 500 = Modest00, 600 = Moderate00, 700 = Slightly Abundant00, and 800 = Moderately Abundant00). A 1.27-cm longissimus dorsi steak from the anterior end of the loin was removed, trimmed of all subcutaneous and intermuscular fat, vacuum-packaged, and immediately frozen at −20 °C for proximate analysis.
Proximate analysis
For proximate analysis, the 1.27-cm steaks were thawed overnight at 6 °C, powder homogenized in liquid nitrogen, and then refrozen for subsequent analysis. To determine the lipid and moisture content, duplicate samples of approximately 2.0 g were weighed and sealed into separate ANKOM bags (Ankom Technology, Fairport, NY). Samples were placed in a drying oven (model no.: 1350 FM, Sheldon Manufacturing Co., Cornelius, OR) at 100 °C overnight, cooled in a desiccator for 10 min, and weighed to determine the amount of moisture lost from the sample. Dried samples were placed in the Crude Fat Extractor (model no.: ANKOM XT15, Ankom Technology, Fairport, NY). To remove residual ether after extraction, samples were placed back in the 100 °C drying oven for 15 min, cooled for 10 min in a desiccator, and weighed to determine their crude fat content. Samples were reanalyzed if the coefficient of variation was greater than 10% between duplicates.
DNA extraction and 16S rRNA gene sequencing
Ruminal samples were thawed and processed following the deoxyribonucleic acid (DNA) extraction procedure described by Rothrock et al. (2014). Briefly, this procedure requires 0.33 g of sample (as-is basis) to be weighed out into a 2-mL Lysing Matrix E tube (MP Biomedicals LLC, Irvine, CA) prior to mechanical and enzymatic steps to achieve DNA extraction. Cells were homogenized and disrupted via a FastPrep 24 Instrument (MP Biomedicals LLC, Irvine, CA) and InhibitEX Tablets (QIAGEN, Venlo, Limburg, The Netherlands) were added for enzymatic inhibition. Elution and purification of DNA were completed using an automated robotic workstation (QIAcube; QIAGEN, Venlo, Limburg, The Netherlands). Extracted DNA purity and concentration were evaluated using a Synergy H4 Hybrid Multi-Mode Microplate Reader along with the Take3 Micro-Volume Plate (BioTek Instruments Inc., Winooski, VT).
The extracted DNA samples were prepared for 16S ribosomal ribonucleic acid (rRNA) gene sequencing by the Georgia Genomics and Bioinformatics Core (https://dna.uga.edu). The DNA amplification was performed using the forward: S-D-Bact-0341-b-S-17 (5′-CCTACGGGNGGCWGCAG-3′) and reverse: S-D-Bact-0785-a-A-21 (5′-GACTACHVGGGTATCTAATCC-3′) primer pair (Klindworth et al., 2013). Amplified DNA was sequenced using the Illumina MiSeq platform and a V3 reagent kit (Illumina Inc., San Diego, CA).
16S rRNA gene sequencing data analysis
The DNA sequence data were first demultiplexed, according to the barcodes applied during the amplification process, before being converted into FASTQ files. Next, using BBMerge Paired Read Merger v37.64, the high-quality pair-end reads were merged. Once merged, the data files were analyzed using the QIIME pipeline v1.9.1 (Caporaso et al., 2010). The data were then quality filtered and combined into one single FASTA file. Following, the nucleotide sequences were clustered according to the Greengenes database (gg_13_8_otus) into operational taxonomic units (OTU), which were defined at the threshold of 97% similarity. For further analysis, singleton OTU were removed, and the sequencing depth was set at 17,542 sequences per sample.
Alpha-diversity
Alpha-diversity of a microbial ecosystem is a measure of within-sample variation (Fisher et al., 1943; Whittaker, 1960). For the present study, we computed five alpha-diversity measures: Chao1 Richness Index (Chao1), Pielou’s Evenness Index (Evenness), Faith’s Phylogenetic Diversity Index (Pdive), the total number of OTU, and Shannon Index. Of these five measures, two represent species diversity (Shannon Index and Pdive), two represent richness (Chao1 and OTU), and one measures species evenness (Evenness). Briefly, species evenness represents how equally abundant the species are in the sample, while richness can be described as the number of different species within a sample (Tuomisto, 2012). Both richness and evenness are accounted for when estimating microbial diversity (Lloyd and Ghelardi, 1964; Pielou, 1966).
Prediction of metabolic pathways
Prediction of biochemical pathways expressed by the ruminal microbial communities were obtained using v1.1.4 of the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) methodology, which utilizes the 16S rRNA gene as a marker, coupled with a database of reference genomes, to provide the predicted functional composition of microbiotas (Langille et al., 2013). The Greengenes database (gg_13_5_otus) and QIIME were used to generate a closed-reference OTU table, which was normalized using 16S rRNA gene copy numbers. Rumen metagenomes were then predicted and categorized into functions based on Kyoto Encyclopedia of Genes and Genomes (KEGG) level 3 pathways (Kanehisa et al., 2014).
Statistical analysis
Data analysis was performed using Minitab v19.2 (Minitab LLC, State College, PA). Means and SD for carcass traits were analyzed using a two-sample t-test. Pearson correlations were performed between microbial traits including alpha-diversity indices and microbial taxa (at the phylum, family, and genus levels of classification) in relation to carcass traits (i.e., marbling score, longissimus lipid content, adjusted 12th rib fat thickness, and yield grade). A correlation coefficient greater than or equal to the absolute value of 0.40 (i.e., r ≥ ±0.40) was used as the threshold for reporting since correlation coefficients larger than ±0.40 are considered of moderate or higher strength. Moreover, a correlation coefficient greater than or equal to ±0.40 consistently corresponded to a P-value ≤ 0.05 for our data. Linear regression plots between microbial and carcass traits were depicted using a fitted line plot containing all the observations (n = 24). Quadratic effects were not significant for these comparisons.
The steers with the 10 highest and 10 lowest numeric marbling scores were grouped into the high-marbling and low-marbling classes, respectively. With respect to yield grade, a lower number indicates a greater percentage of boneless retail product and was considered a more desirable outcome. The steers with the 10 lowest and 10 highest yield grades were grouped into the best- and worst-yield grade classes, respectively. One-way ANOVA was performed for marbling classes (n = 10 steers per class) and yield grade classes (n = 10 steers per class) in relation to alpha-diversity traits and microbial families. Only microbial families with a relative abundance greater than or equal to 0.45% were considered in these analyses. Analyses of the microbial metabolic pathways (n = 10 steers per class) were performed using a one-way ANOVA with the marbling classifications (low- or high-marbling) as fixed factors. Significance was set at P ≤ 0.05 with tendencies declared at 0.05 < P ≤ 0.10 for all statistical tests.
Results and Discussion
Carcass quality measures
Descriptive statistics
The mean values for carcass quality-related traits in the 24 steers were 155 for overall maturity, 670 for marbling score, USDA High Choice for quality grade, and 9.4% for longissimus lipid content (Table 1). In comparison, the National Beef Quality Audit of 2016 reported mean values for fed steers to be 159 for overall maturity, 467 for marbling score, and USDA Low Choice for quality grade (Boykin et al., 2017).
Table 1.
Means and SD1 for carcass traits in Angus steers within the various populations analyzed in this study
| Populations | |||||
|---|---|---|---|---|---|
| Marbling class | Yield grade class | ||||
| Carcass trait | Experimental (n = 24) | High (n = 10) | Low (n = 10) | Best (n = 10) | Worst (n = 10) |
| Overall maturity2 | 155 (11.4) |
156 (13.5) | 156 (10.7) | 153 (14.9) | 157 (8.23) |
| Marbling score3 | 670 (94.8) |
760a (56.0) | 579b (40.4) | 670 (96.4) |
678 (106.4) |
| USDA quality grade4 | Choice+ | Prime– | Choice° | Choice+ | Choice+ |
| Longissimus lipid content, % | 9.4 (2.60) |
10.9a (2.11) | 7.7b (2.22) | 8.8 (2.29) |
10.0 (3.17) |
| Hot carcass weight, kg | 375.9 (27.48) |
377.1 (27.55) | 370.5 (26.40) | 367.7 (32.61) | 384.9 (24.48) |
| Adjusted 12th rib fat thickness, cm | 1.3 (0.28) |
1.4 (0.33) | 1.3 (0.25) | 1.1a (0.12) | 1.6b (0.20) |
| Ribeye area, cm2 | 79.4 (6.92) |
80.1 (8.38) | 79.7 (6.40) | 83.6a (7.68) | 75.2b (4.56) |
| Kidney, pelvic, heart fat, % | 2.2 (0.36) |
2.1 (0.21) | 2.2 (0.48) | 2.2 (0.41) |
2.3 (0.35) |
| Yield grade | 3.5 (0.59) |
3.5 (0.73) | 3.4 (0.41) | 2.9a (0.30) | 4.0b (0.39) |
| Dressing percentage, % | 61.7 (1.32) |
61.8 (1.64) | 61.7 (1.22) | 61.8 (1.32) | 61.8 (1.23) |
1Means are followed by the SD in parentheses.
2Overall maturity is the average of bone and lean maturity where A00 = 100 and B00 = 200.
3Marbling score was converted to a numeric scale where 500 = Modest00, 600 = Moderate00, 700 = Slightly Abundant00, and 800 = Moderately Abundant00.
4Choice° = USDA Average Choice, Choice+ = USDA High Choice, and Prime– = USDA Low Prime
a,bDenotes carcass trait means that differ (P ≤ 0.050) between classes.
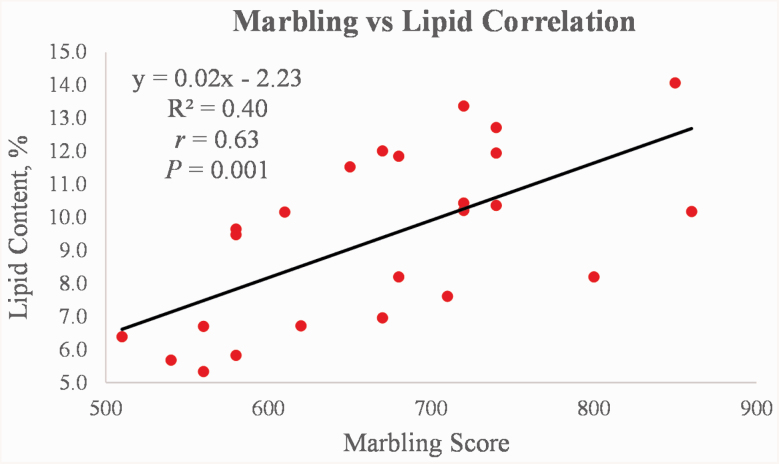
Marbling scores are an estimate of intramuscular fat in the longissimus muscle of cattle and are relatively representative of the chemical (true) lipid content (Dryden, 1967). The correlation between marbling scores and the percentage of lipid found within the longissimus muscle in the present study was r = 0.63 (P = 0.001; Figure 1), whereas other studies have reported correlations ranging from 0.79 to 0.91 (Wellington and Stouffer, 1959; Armbruster et al., 1983; Cameron et al., 1994).
Figure 1.
Correlation between marbling scores and chemical lipid content in the longissimus muscle of Angus steers (n = 24). The regression equation, R2 value, correlation coefficient, and P-value are indicated for the given relationship.
Alpha-diversity of the rumen
Only significant correlation coefficients (r ≥ ±0.40; P ≤ 0.050) between marbling scores and longissimus lipid content with ruminal microbial traits such as diversity, richness, and different microbial taxa were reported (Table 2). Chao1, an estimator of species richness, was positively correlated (P ≤ 0.009) with both marbling score (r = 0.62) and lipid content (r = 0.52), suggesting that a greater number of ruminal bacterial species may be associated with higher marbling scores and greater longissimus lipid content. This result was further supported by the positive correlations (P ≤ 0.043) between marbling score (r = 0.48) and longissimus lipid content (r = 0.42) with the number of observed OTU, another measure of microbial species richness. Similarly, Pdive, which indicates species diversity, was positively correlated (P ≤ 0.030) with marbling score (r = 0.49) and longissimus lipid content (r = 0.44). While few studies exist in cattle that relate ruminal microbial diversity and richness to intramuscular fat content, it has been shown in humans that obesity is related to reduced microbial diversity (Turnbaugh et al., 2009; Le Chatelier et al., 2013); however, other studies have failed to consistently support these findings (Bondia-Pons et al., 2014; Walters et al., 2014).
Table 2.
Correlation between marbling and longissimus lipid content with different microbial traits1 assessed in the rumen of Angus steers (n = 24)
| Trait | Pearson correlation | P-value |
|---|---|---|
| Marbling | ||
| Chao1 | 0.615 | 0.001 |
| OTU | 0.484 | 0.017 |
| Pdive | 0.491 | 0.015 |
| Tenericutes | −0.409 | 0.047 |
| TM7 | −0.561 | 0.004 |
| Verrucomicrobia | 0.428 | 0.037 |
| Erysipelotrichaceae | −0.422 | 0.040 |
| F16 | −0.562 | 0.004 |
| Rikenellaceae | 0.441 | 0.031 |
| S24-7 | 0.610 | 0.002 |
| Verrucomicrobiaceae | 0.409 | 0.047 |
| Akkermansia | 0.409 | 0.047 |
| Blautia | 0.409 | 0.047 |
| Klebsiella | 0.426 | 0.038 |
| Moryella | 0.456 | 0.025 |
| Peptostreptococcus | 0.426 | 0.038 |
| Pseudomonas | −0.474 | 0.019 |
| RFN20 | −0.473 | 0.020 |
| Selenomonas | 0.434 | 0.034 |
| Lipid content | ||
| Chao1 | 0.519 | 0.009 |
| OTU | 0.417 | 0.043 |
| Pdive | 0.444 | 0.030 |
| Actinobacteria | 0.430 | 0.036 |
| Cyanobacteria | −0.532 | 0.007 |
| Proteobacteria | −0.508 | 0.011 |
| Tenericutes | −0.406 | 0.049 |
| TM7 | −0.413 | 0.045 |
| Verrucomicrobia | 0.433 | 0.034 |
| Coriobacteriaceae | 0.405 | 0.049 |
| F16 | −0.413 | 0.045 |
| Succinivibrionaceae | −0.413 | 0.045 |
| Dorea | 0.438 | 0.032 |
| Moryella | 0.489 | 0.015 |
| Pseudomonas | −0.420 | 0.041 |
| RFN20 | −0.440 | 0.031 |
| Succinivibrio | −0.476 | 0.019 |
1Only microbial traits that had a significant correlation (r ≥ ±0.40; P ≤ 0.050) with marbling and lipid content are shown.
Bacterial taxa of the rumen
Many ruminal bacterial phyla have been identified, yet the ecological roles in the complex rumen ecosystem of many of these phyla remain unclear. The phylum Verrucomicrobia was positively correlated (P ≤ 0.037) to both marbling score (r = 0.43) and longissimus lipid content (r = 0.43), whereas Actinobacteria (r = 0.43) was also positively correlated (P = 0.036) to lipid content (Table 2). In general, Verrucomicrobia makes up less than 1% of the ruminal microbial population and is only found in the solid-associated fraction of the rumen contents (Cunha et al., 2011; Jewell et al., 2015). Nevertheless, members of this phylum have been found to contribute considerably to diverse polysaccharide degradation, despite their low prevalence in the ruminal population (Martinez-Garcia et al., 2012). Genome analysis has further revealed that Verrucomicrobia, in comparison with other bacterial genomes, has a diverse selection of carbohydrate-degrading glycoside hydrolases (Martinez-Garcia et al., 2012), which could partially explain its association with the solid fraction of the rumen contents, where it is able to degrade complex sugars in feedstuffs such as starch, hemicellulose, and cellulose. Actinobacteria makes up a slightly larger portion of the rumen microbial population at 2% to 3% (Pandya et al., 2010), and while the phylum is comprised of a diverse population of bacteria, all Actinobacteria that have been cultured from the ruminant digestive tract use pectin, cellulose, or xylan as their sole carbon source (Tan et al., 2014).
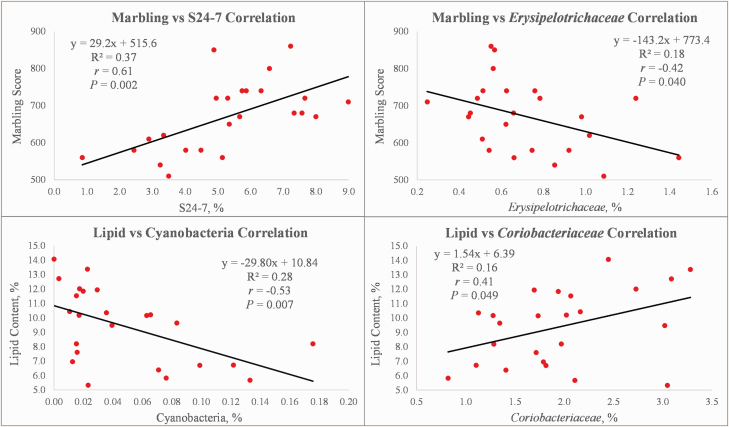
The bacterial phyla Tenericutes (r = −0.41) and TM7 (r = −0.56) were negatively correlated (P ≤ 0.047) with marbling score, while the phyla Cyanobacteria (r = −0.53; Figure 2), Proteobacteria (r = −0.51), Tenericutes (r = −0.41), and TM7 (r = −0.41) were negatively correlated (P ≤ 0.049) with longissimus lipid content (Table 2), suggesting that lower abundances of these phyla may indirectly contribute to improved marbling scores and greater longissimus lipid content of carcasses. The phylum Tenericutes consists of a single class known as Mollicutes. Mollicutes is further comprised of four bacterial orders (Mycoplasmatales, Entomoplasmatales, Acholeplasmatales, and Anaeroplasmatales), of which several members have been found to be animal pathogens and parasites (Ludwig et al., 2010; Zhan et al., 2017), which could partially explain why greater abundances of the phylum Tenericutes were associated with reduced carcass quality in terms of intramuscular fat deposition.
Figure 2.
Fitted line plots depicting the relationships between marbling scores and longissimus lipid content with selected bacterial taxa within the rumen of Angus steers (n = 24). The regression equation, R2 value, correlation coefficient, and P-value are indicated for the given relationship.
The bacterial family S24-7 (also referred to as Muribaculaceae) was positively correlated (r = 0.61; P = 0.002) with marbling score (Figure 2). Similarly, Fang et al. (2017) found that S24-7 members (two OTU in the cecum and three fecal OTU) were associated with increased intramuscular fat in pigs. Early research suggests that the S24-7 family is comprised of three trophic guilds consisting of α-glucan, host glycan, and plant glycan-based (hemicellulose and pectin) carbohydrate utilizers (Ormerod et al., 2016). All known S24-7 members harbor genes for α-amylases indicating that while members may be able to operate in separate niches using specialized carbohydrate sources, starch utilization is conserved across the family (Ormerod et al., 2016; Lagkouvardos et al., 2019). Further research suggests that the capacity to produce propionate is widespread throughout this bacterial family with increased abundances being linked to increased propionate production (Ormerod et al., 2016; Smith et al., 2019, 2020, preprint). Since propionate is the preferred precursor for intramuscular fat via its connection to glucose (Smith and Crouse, 1984; Smith et al., 2018; Wandita et al., 2018), it is possible that the association between increased abundances of this bacterial family and increased marbling scores could be due in part to an increase in the production of the precursor propionate; however, further research is needed along with metabolic data such as VFA concentrations in order to better understand this relationship.
The family F16 is a part of the phylum TM7 and was negatively correlated with both marbling score (r = −0.56; P = 0.004) and longissimus lipid content (r = −0.41; P = 0.045), as was the TM7 phylum as a whole (Table 2). There were a wide variety of other bacterial families correlated with marbling score, including Erysipelotrichaceae (r = −0.42; P = 0.040; Figure 2), Rikenellaceae (r = 0.44; P = 0.031), and Verrucomicrobiaceae (r = 0.41; P = 0.047). Other bacterial families were also correlated with longissimus lipid content: Coriobacteriaceae (r = 0.41; P = 0.049; Figure 2) was positively correlated, whereas Succinivibrionaceae (r = −0.41; P = 0.045) was negatively correlated.
Several genera were positively correlated (P ≤ 0.047) with marbling scores, such as Akkermansia (r = 0.41), Blautia (r = 0.41), Klebsiella (r = 0.43), Peptostreptococcus (r = 0.43), and Selenomonas (r = 0.43) (Table 2). While Blautia is typically found at relatively low abundances throughout the mammalian gut (Eren et al., 2015), some members of the genus are capable of utilizing H2 to produce acetate (Rey et al., 2010), which could result in a shuttling of energy from methane production toward acetate production that would ultimately result in less energy waste (Li et al., 2019). A similar shifting of H2 disposal from methane to acetate production via acetogenesis has been noted by members of the genus Peptostreptococcus (Nollet et al., 1997). Overall, reducing the dietary energy lost and increasing VFA concentrations through increased abundances of these genera could potentially lead to more energy available to the host for fat deposition; however, more metabolic data are needed to support this theory. Meanwhile, well-known members of the Selenomonas genus such as Selenomonas ruminantium are known for producing ruminal propionate from lactate via the succinate–propionate pathway (Paynter and Elsden, 1970; Scheifinger and Wolin, 1973; Wallace, 1978). Therefore, increased populations of Selenomonas could result in greater propionate production, and since propionate is a gluconeogenic precursor, the increased propionate could be linked to the increased intramuscular fat via glucose being the main lipogenic precursor of intramuscular adipose tissue (Smith and Crouse, 1984; Smith et al., 2018; Wandita et al., 2018).
In addition, other genera were correlated (P ≤ 0.041) to both marbling score and longissimus lipid content, respectively, including Moryella (r = 0.46, r = 0.49), Pseudomonas (r = −0.48, r = −0.42), and RFN20 (r = −0.47, r = −0.44). Genera that were only correlated (P ≤ 0.032) to longissimus lipid content included Dorea (r = 0.44) and Succinivibrio (r = −0.48).
Carcass yield measures
Descriptive statistics
Carcass descriptive statistics (n = 24) relating to yield traits in the present study revealed mean values of 375.9 kg for hot carcass weight, 1.3 cm for adjusted 12th rib fat thickness, 79.4 cm2 for ribeye area, 3.5 for yield grade, 61.7% for dressing percentage, and 2.2% for kidney, pelvic, heart fat (Table 1). Similarly, the National Beef Quality Audit of 2016 reported mean yield characteristics for fed steers to be 398.2 kg for hot carcass weight, 1.3 cm for adjusted fat thickness, 88.9 cm2 for longissimus muscle area, 3.1 for yield grade, and 2.0% for kidney, pelvic, heart fat (Boykin et al., 2017).
Bacterial taxa of the rumen
No microbial diversity, richness, or evenness traits were correlated (P ≥ 0.050) to yield grade or rib fat thickness (data not shown). The bacterial phyla, families, and genera with significant correlations (P ≤ 0.050; r ≥ ±0.40) were largely the same for both yield grade and rib fat thickness (Table 3), which is logical given the incorporation of rib fat thickness into the yield grade calculations (USDA, 1965).
Table 3.
Correlation between yield grade and adjusted 12th rib fat thickness with different microbial taxa1 (phyla, families, and genera) assessed in the rumen of Angus steers (n = 24)
| Trait | Pearson correlation | P-value |
|---|---|---|
| Yield grade | ||
| Actinobacteria | 0.517 | 0.010 |
| Bifidobacteriaceae | 0.491 | 0.015 |
| Coriobacteriaceae | 0.450 | 0.027 |
| Peptococcaceae | 0.472 | 0.020 |
| Bifidobacterium | 0.488 | 0.015 |
| Bulleidia | 0.479 | 0.018 |
| Dorea | 0.410 | 0.047 |
| Peptococcus | 0.500 | 0.013 |
| Rib fat thickness | ||
| Actinobacteria | 0.613 | 0.001 |
| Bifidobacteriaceae | 0.439 | 0.032 |
| Coriobacteriaceae | 0.589 | 0.002 |
| Erysipelotrichaceae | 0.413 | 0.045 |
| Peptococcaceae | 0.560 | 0.004 |
| Bifidobacterium | 0.436 | 0.033 |
| Bulleidia | 0.554 | 0.005 |
| Butyrivibrio | 0.405 | 0.049 |
| Peptococcus | 0.582 | 0.003 |
1Only microbial taxa that had a significant correlation (r ≥ ±0.40; P ≤ 0.050) with yield grade and rib fat thickness are shown.
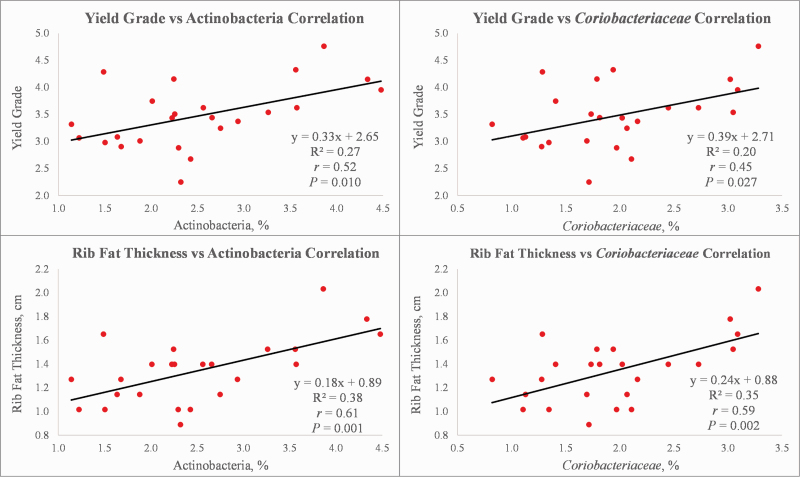
The phylum Actinobacteria was positively correlated (P ≤ 0.010) to yield grade (r = 0.52) and rib fat thickness (r = 0.62) (Figure 3). The families Bifidobacteriaceae (r = 0.49, r = 0.44), Coriobacteriaceae (r = 0.45, r = 0.59; Figure 3), and Peptococcaceae (r = 0.47, r = 0.56) were positively correlated to yield grade (P ≤ 0.027) and rib fat thickness (P ≤ 0.032), respectively (Table 3). In addition, the family Erysipelotrichaceae was positively correlated to rib fat thickness (r = 0.41; P = 0.045). Erysipelotrichaceae has been correlated with host lipid metabolism (Martínez et al., 2009; Zhang et al., 2009; Spencer et al., 2011); however, in the present study, we observed a negative correlation between the family Erysipelotrichaceae and marbling score, suggesting that members of the family may be more closely associated with subcutaneous rather than intramuscular fat accumulation. In other studies, Coriobacteriaceae populations were associated with host lipid metabolism via conditions, such as hypercholesterolemia, hepatic metabolism, and obesity (Martínez et al., 2009, 2013; Zhang et al., 2009; Claus et al., 2011). In the present study, Coriobacteriaceae was positively correlated with longissimus lipid content as well as rib fat thickness and yield grade, suggesting that this family may contribute to adipose accumulation via a more general role in lipid metabolism.
Figure 3.
Fitted line plots depicting the relationships between adjusted 12th rib fat thickness and yield grade with selected bacterial taxa within the rumen of Angus steers (n = 24). The regression equation, R2 value, correlation coefficient, and P-value are indicated for the given relationship.
The genus Bifidobacterium, which is a well-known gastrointestinal bacterium that produces primarily acetate and lactate (Falony et al., 2006), was positively correlated (P ≤ 0.033) with yield grade (r = 0.49) and rib fat thickness (r = 0.44) (Table 3). Bulleidia, in the family Erysipelotrichaceae, also produces acetate (Downes et al., 2000) and was positively correlated (P ≤ 0.018) with both yield grade (r = 0.48) and rib fat thickness (r = 0.55). Previous research has shown that both Bifidobacterium and Bulleidia were positively correlated with milk fat yield in first lactation cows (Jami et al., 2014) likely due in part to their contribution of acetate, and the use of acetate as the primary substrate for milk fat synthesis (Urrutia and Harvatine, 2017). Further research is needed to investigate the relationships between these bacterial genera and their potential end product contributions to lipid metabolism. The genus Peptococcus was also positively correlated (P ≤ 0.013) with yield grade (r = 0.50) and rib fat thickness (r = 0.58). Lastly, yield grade was uniquely correlated to Dorea (r = 0.41; P = 0.047), whereas the Butyrivibrio (r = 0.41; P = 0.049) correlation was unique to rib fat thickness.
Marbling classes
Descriptive statistics
Average marbling scores (P < 0.001) for the high- and low-marbling classes (n = 10 steers per class) were 760 and 579, respectively, translating to Low Prime (Slightly Abundant60) and Average Choice (Modest79) USDA quality grades (Table 1). Aside from longissimus lipid content, no other carcass traits differed (P ≥ 0.180) between the high- and low-marbling classes.
Alpha-diversity of the rumen
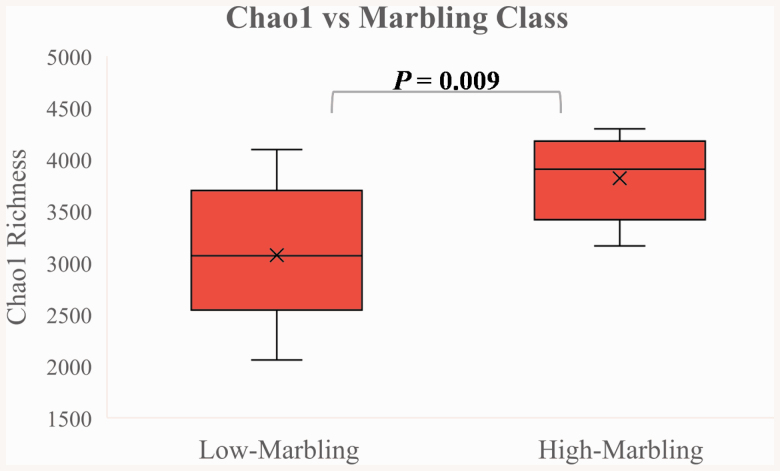
Chao1 richness was greater (P = 0.009) in the rumen of the high- compared to the low-marbling steers, further suggesting that the high-marbling steers had a greater number of microbial species present in their rumen (Figure 4). These results were supported by the trend in the number of unique ruminal OTU (P = 0.063) observed, with high-marbling steers tending to have more OTU compared to low-marbling steers (Table 4). Pdive is a measure of species diversity that accounts for both microbial richness and evenness, and it was greater (P = 0.044) in the rumen of the high-marbling compared to the low-marbling steers.
Figure 4.
Chao1 index (an estimator of microbial richness) measured in the rumen of Angus steers with distinct marbling scores (n = 10 steers per class). A one-way ANOVA revealed differences (P = 0.009) between the two marbling groups. Whisker bars indicate the minimum and maximum values, while the “x” represents the mean value.
Table 4.
Analysis of variance for alpha-diversity traits in the rumen of Angus steers from different marbling classifications (n = 10 steers per class)
| Marbling class | ||||
|---|---|---|---|---|
| Diversity trait | Low | High | Standard error | P-value |
| Chao1 | 3,075 | 3,819 | 180.2 | 0.009* |
| Evenness | 0.788 | 0.794 | 0.0081 | 0.603 |
| Pdive | 120.3 | 132.3 | 3.90 | 0.044* |
| OTU | 1,736 | 1,967 | 82.4 | 0.063 |
| Shannon Index | 8.47 | 8.68 | 0.129 | 0.254 |
*Denotes a diversity trait with a significant difference (P ≤ 0.050) in representation between the two classes of steers.
Bacterial families of the rumen
Analysis of bacterial families with relative abundances of ≥0.45% in the rumen of the marbling class steers revealed that the only family to differ was the family S24-7 (P < 0.001) such that the high-marbling steers had greater abundances (6.35%) than did the low-marbling steers (3.53%) (Table 5). These results are similar to previously reported correlations between the abundance of bacterium in the S24-7 family and increased intramuscular fat content in pigs (Fang et al., 2017).
Table 5.
Analysis of variance for microbial families1 found in the rumen of Angus steers from different marbling classifications (n = 10 steers per class)
| Marbling class | ||||
|---|---|---|---|---|
| Family | Low | High | Standard error | P-value |
| Christensenellaceae | 0.47 | 0.51 | 0.123 | 0.826 |
| Clostridiaceae | 1.38 | 1.35 | 0.206 | 0.902 |
| Coriobacteriaceae | 1.85 | 2.01 | 0.236 | 0.630 |
| Erysipelotrichaceae | 0.84 | 0.63 | 0.087 | 0.109 |
| F16 | 0.69 | 0.32 | 0.155 | 0.115 |
| Lachnospiraceae | 15.06 | 15.29 | 1.254 | 0.898 |
| Methanobacteriaceae | 1.28 | 1.20 | 0.245 | 0.815 |
| Mogibacteriaceae | 1.61 | 2.02 | 0.221 | 0.206 |
| Order_Bacteroidales2 | 6.83 | 7.88 | 0.621 | 0.248 |
| Order_Clostridiales2 | 12.75 | 13.15 | 0.735 | 0.701 |
| Paraprevotellaceae | 2.21 | 1.89 | 0.316 | 0.481 |
| Pirellulaceae | 0.53 | 0.72 | 0.132 | 0.319 |
| Prevotellaceae | 20.28 | 18.63 | 1.664 | 0.490 |
| Ruminococcaceae | 18.75 | 17.25 | 1.319 | 0.432 |
| S24-7 | 3.53 | 6.35 | 0.416 | <0.001* |
| Spirochaetaceae | 2.02 | 1.34 | 0.315 | 0.146 |
| Succinivibrionaceae | 0.93 | 0.45 | 0.418 | 0.426 |
| Veillonellaceae | 3.61 | 3.96 | 0.673 | 0.720 |
1Only microbial families with an abundance of ≥ 0.45% are shown.
2Denotes an unidentified family within the listed taxonomical Order.
*Denotes a family with a significant difference (P ≤ 0.050) in relative abundances between the two classes of steers.
Prediction of metabolic pathways in the rumen
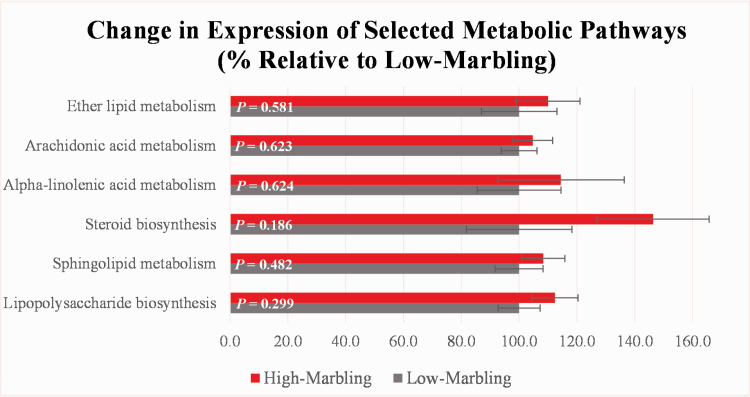
Analysis using the PICRUSt methodology to predict microbial metabolic pathways revealed no significant differences (P ≥ 0.050) in metabolic function between the high- and low-marbling classes (Figure 5); however, in spite of the small sample size (n = 10 steers per class), some metabolic pathways relating to lipid metabolism did differ numerically between marbling classes. In general, ruminal microbes from the high-marbling steers had greater expression of the lipid metabolism pathways as compared to those microbes found within the rumens of the low-marbling steers. For example, the steroid biosynthesis pathway was numerically greater (P = 0.190) in the high-marbling steers (146%) ruminal microbial populations compared to low-marbling steers (100%), suggesting that bacteria within the rumen of the high-marbling steers were producing a greater amount of steroid than those in the rumen of the low-marbling steers.
Figure 5.
Results using the PICRUSt methodology to predict the expression of microbial metabolic pathways (level 3 KEGG pathways shown) within the rumen of Angus steers from different marbling classifications (n = 10 steers per class). The expression of the pathways was set at 100% for the low-marbling steers.
Overall, the ability of the PICRUSt methodology to predict microbial metabolic pathways is largely dependent on the genomes available in the reference database (Langille et al., 2013; Zeng et al., 2015). Furthermore, the PICRUSt database is optimized to predict the functional pathways of microbes in the human gut, and, therefore, caution must be used when applying it to microbial populations from ecosystems that are not as well-characterized (Langille et al., 2013). Predicted metabolic pathway analysis is a useful strategy that can offer relatively inexpensive, yet valuable insights into unclear ruminal metabolic activities. However, metabolomics analysis that measures the activity of intermediate microbial metabolism is needed to fully elucidate interactions between the members of the microbial ecosystem and their ruminant host.
Yield grade classes
Descriptive statistics
Two carcass measures differed (P ≤ 0.011) between the yield grade classes (n = 10 steers per class): adjusted 12th rib fat thickness (1.1 vs. 1.6 cm) and ribeye area (83.6 vs. 75.2 cm2), which resulted in divergent yield grades (2.9 vs. 4.0) for the best- and worst-yield grade classes, respectively (Table 1). No other carcass traits differed (P ≥ 0.200) between classes.
Alpha-diversity of the rumen
There were no significant differences in ruminal microbial species diversity, richness, or evenness across yield grade classes (Table 6); however, there was a tendency (P = 0.067) for Evenness to be closer to 1 in the best-yield grade class steers compared to the worst-yield grade class, suggesting that ruminal microbial species abundances were more equally distributed in the best-yield grade class steers.
Table 6.
Analysis of variance for alpha-diversity traits in the rumen of Angus steers from different yield grade classifications (n = 10 steers per class)
| Yield grade class | ||||
|---|---|---|---|---|
| Diversity trait | Worst | Best | Standard error | P-value |
| Chao1 | 3,369 | 3,432 | 205.1 | 0.829 |
| Evenness | 0.779 | 0.799 | 0.0072 | 0.067 |
| Pdive | 123.1 | 126.4 | 4.00 | 0.568 |
| OTU | 1,767 | 1,870 | 80.9 | 0.383 |
| Shannon Index | 8.40 | 8.67 | 0.118 | 0.112 |
Bacterial families of the rumen
Bacterial families with ruminal abundances ≥ 0.45% were analyzed between the yield grade classes (Table 7). The family Coriobacteriaceae (P = 0.005) had lower ruminal abundances in the best-yield grade (1.52%) compared to the worst-yield grade (2.40%) class steers. Similarly, there was a tendency for the best-yield grade class steers (0.62%) to have lower abundances of the family Erysipelotrichaceae (P = 0.058) compared to the worst-yield grade class steers (0.87%). The family Christensenellaceae (P = 0.045), while having relatively low overall abundance in the rumen, was greater in the best-yield grade class steers (0.56%) compared to the worst-yield grade class steers (0.27%).
Table 7.
Analysis of variance for microbial families1 found in the rumen of Angus steers from different yield grade classifications (n = 10 steers per class)
| Yield grade class | ||||
|---|---|---|---|---|
| Family | Worst | Best | Standard error | P-value |
| Christensenellaceae | 0.27 | 0.56 | 0.094 | 0.045* |
| Clostridiaceae | 1.19 | 1.37 | 0.194 | 0.519 |
| Coriobacteriaceae | 2.40 | 1.52 | 0.194 | 0.005* |
| Erysipelotrichaceae | 0.87 | 0.62 | 0.087 | 0.058 |
| F16 | 0.47 | 0.46 | 0.169 | 0.966 |
| Lachnospiraceae | 15.76 | 13.39 | 1.319 | 0.220 |
| Methanobacteriaceae | 1.26 | 1.22 | 0.242 | 0.895 |
| Mogibacteriaceae | 1.65 | 1.64 | 0.210 | 0.979 |
| Order_Bacteroidales2 | 7.41 | 7.37 | 0.673 | 0.968 |
| Order_Clostridiales2 | 12.44 | 12.44 | 0.693 | 0.998 |
| Paraprevotellaceae | 2.12 | 2.42 | 0.301 | 0.498 |
| Pirellulaceae | 0.47 | 0.61 | 0.105 | 0.366 |
| Prevotellaceae | 20.72 | 21.34 | 1.602 | 0.788 |
| Ruminococcaceae | 16.40 | 17.85 | 1.373 | 0.466 |
| S24-7 | 5.06 | 5.60 | 0.656 | 0.569 |
| Spirochaetaceae | 2.17 | 1.69 | 0.372 | 0.379 |
| Succinivibrionaceae | 0.84 | 0.61 | 0.421 | 0.703 |
| Veillonellaceae | 3.62 | 3.55 | 0.681 | 0.948 |
1Only microbial families with an abundance of ≥ 0.45% are shown.
2Denotes an unidentified family within the listed taxonomical Order.
*Denotes a family with a significant difference (P ≤ 0.050) in relative abundances between the two classes of steers.
Conclusions
Overall, our findings indicate that improvements in marbling score as well as increased longissimus lipid content of carcasses were highly correlated with increased microbial richness, suggesting that a greater number of bacterial species in the rumen may be associated with increased fat content in the longissimus muscle. Several bacterial phyla, families, and genera were correlated to important carcass traits, such as marbling score, longissimus lipid content, adjusted 12th rib fat thickness, and calculated yield grade. Of particular interest was the bacterial family S24-7 and its correlation with increased marbling scores, which was initially noted in pigs, and is further supported by our findings. Surprisingly, few bacterial taxa overlapped in their correlations to the two studied fat depots, subcutaneous (rib fat and yield grade) and intramuscular (marbling and longissimus lipid content) fat. Thus, our results suggest that while a number of bacterial taxa appear to play some role in adipose tissue accumulation in each depot, their influence may be largely accomplished via separate host metabolic pathways for each fat depot. The separation between bacteria linked to each fat depot could be due in part to the different regulatory pathways that control de novo fatty acid synthesis for each depot, as reported by Smith and Crouse (1984); however, metabolic data regarding the microbes’ contribution to lipid metabolism are needed to better understand the potential relationships between the rumen microbiome and carcass performance. Collectively, these findings serve as a basis for continued exploration into the relationships between the ruminal microbial populations and beef carcass merit, which can potentially be a method to improve carcass quality and producer profitability.
Acknowledgments
We would like to express our appreciation to the staff at the Northwest Georgia Research and Education Center and Ridgefield Farms, LLC, for the management of the cattle in the project. We would also like to thank Brasstown Beef, LLC, the Georgia Agricultural Commodity Commission for Beef, and the Angus Foundation for financial support of this project.
Glossary
Abbreviations
- Chao1
Chao1 Richness Index
- DNA
Deoxyribonucleic Acid
- EPD
expected progeny difference
- Evenness
Pielou’s Evenness Index
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OTU
operational taxonomic units
- Pdive
Faith’s Phylogenetic Diversity Index
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- rRNA
Ribosomal Ribonucleic Acid
- VFA
volatile fatty acid
Conflict of interest statement
The authors declare no conflicts of interest regarding the publication of this manuscript.
Literature Cited
- Armbruster G, Nour A, Thonney M, and Stouffer J. . 1983. Changes in cooking losses and sensory attributes of Angus and Holstein beef with increasing carcass weight, marbling score or longissimus ether extract. J. Food Sci. 48:835–840. doi: 10.1111/j.1365-2621.1983.tb14911.x [DOI] [Google Scholar]
- Bondia-Pons I, Maukonen J, Mattila I, Rissanen A, Saarela M, Kaprio J, Hakkarainen A, Lundbom J, Lundbom N, Hyötyläinen T, . et al. 2014. Metabolome and fecal microbiota in monozygotic twin pairs discordant for weight: a Big Mac challenge. FASEB J. 28:4169–4179. doi: 10.1096/fj.14-250167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boykin C A, Eastwood L C, Harris M K, Hale D S, Kerth C R, Griffin D B, Arnold A N, Hasty J D, Belk K E, Woerner D R, . et al. 2017. National Beef Quality Audit-2016: In-plant survey of carcass characteristics related to quality, quantity, and value of fed steers and heifers. J. Anim. Sci. 95:2993–3002. doi: 10.2527/jas.2017.1543 [DOI] [PubMed] [Google Scholar]
- Buitenhuis B, Lassen J, Noel S J, Plichta D R, Sørensen P, Difford G F, and Poulsen N A. . 2019. Impact of the rumen microbiome on milk fatty acid composition of Holstein cattle. Genet. Sel. Evol. 51:23. doi: 10.1186/s12711-019-0464-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron P J, Zembayashi M, Lunt D K, Mitsuhashi T, Mitsumoto M, Ozawa S, and Smith S B. . 1994. Relationship between Japanese beef marbling standard and intramuscular lipid in the M. longissimus thoracis of Japanese Black and American Wagyu Cattle. Meat Sci. 38:361–364. doi: 10.1016/0309-1740(94)90125-2 [DOI] [PubMed] [Google Scholar]
- Caporaso J G, Kuczynski J, Stombaugh J, Bittinger K, Bushman F D, Costello E K, Fierer N, Peña A G, Goodrich J K, Gordon J I, . et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus S P, Ellero S L, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want E J, de Waziers I, Cloarec O, . et al. 2011. Colonization-induced host-gut microbial metabolic interaction. mBio. 2:e00271–e00210. doi: 10.1128/mBio.00271-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha I S, Barreto C C, Costa O Y, Bomfim M A, Castro A P, Kruger R H, and Quirino B F. . 2011. Bacteria and Archaea community structure in the rumen microbiome of goats (Capra hircus) from the semiarid region of Brazil. Anaerobe 17:118–124. doi: 10.1016/j.anaerobe.2011.04.018 [DOI] [PubMed] [Google Scholar]
- Detweiler R A, Pringle T D, Rekaya R, Wells J B, and Segers J R. . 2019. The impact of selection using residual average daily gain and marbling EPDs on growth, performance, and carcass traits in Angus steers1. J. Anim. Sci. 97:2450–2459. doi: 10.1093/jas/skz124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes J, Olsvik B, Hiom S J, Spratt D A, Cheeseman S L, Olsen I, Weightman A J, and Wade W G. . 2000. Bulleidia extructa gen. nov., sp. nov., isolated from the oral cavity. Int. J. Syst. Evol. Microbiol. 50(Pt 3):979–983. doi: 10.1099/00207713-50-3-979 [DOI] [PubMed] [Google Scholar]
- Dryden F D. 1967. The relationship of certain chemical constituents of beef muscle to its quality [master thesis]. Tucson:University of Arizona. [Google Scholar]
- Ellison M, Conant G, Lamberson W, Cockrum R, Austin K, Rule D, and Cammack K. . 2017. Diet and feed efficiency status affect rumen microbial profiles of sheep. Small Rumin. Res. 156:12–19. doi: 10.1016/j.smallrumres.2017.08.009 [DOI] [Google Scholar]
- Eren A M, Sogin M L, Morrison H G, Vineis J H, Fisher J C, Newton R J, and McLellan S L. . 2015. A single genus in the gut microbiome reflects host preference and specificity. ISME J. 9:90–100. doi: 10.1038/ismej.2014.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G, Vlachou A, Verbrugghe K, and De Vuyst L. . 2006. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 72:7835–7841. doi: 10.1128/AEM.01296-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Xiong X, Su Y, Huang L, and Chen C. . 2017. 16S rRNA gene-based association study identified microbial taxa associated with pork intramuscular fat content in feces and cecum lumen. BMC Microbiol. 17:162. doi: 10.1186/s12866-017-1055-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R A, Corbet A S, and Williams C B. . 1943. The relation between the number of species and the number of individuals in a random sample of an animal population. J. Anim. Ecol. 12:42–58. doi: 10.2307/1411 [DOI] [Google Scholar]
- Forristall C, May G J, and Lawrence J D. . 2002. Assessing the cost of beef quality. Proceedings of the NCR-134 Conference on Applied Commodity Price Analysis, Forecasting, and Market Risk Management; April 22–23, 2002.St Louis (MO). doi: 10.22004/ag.econ.19060 [DOI] [Google Scholar]
- Hungate R E. 1966. The rumen and its microbes. 1st ed. New York (NY):Academic Press Inc. [Google Scholar]
- Jami E, White B A, and Mizrahi I. . 2014. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One. 9:e85423. doi: 10.1371/journal.pone.0085423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell K A, McCormick C A, Odt C L, Weimer P J, and Suen G. . 2015. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl. Environ. Microbiol. 81:4697–4710. doi: 10.1128/AEM.00720-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamke J, Kittelmann S, Soni P, Li Y, Tavendale M, Ganesh S, Janssen P H, Shi W, Froula J, Rubin E M, . et al. 2016. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 4:56. doi: 10.1186/s40168-016-0201-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, and Tanabe M. . 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42(Database issue):D199–D205. doi: 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, and Glöckner F O. . 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Lesker T R, Hitch T C A, Gálvez E J C, Smit N, Neuhaus K, Wang J, Baines J F, Abt B, Stecher B, . et al. 2019. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 7:28. doi: 10.1186/s40168-019-0637-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M G, Zaneveld J, Caporaso J G, McDonald D, Knights D, Reyes J A, Clemente J C, Burkepile D E, Vega Thurber R L, Knight R, . et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31:814–821. doi: 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J M, Kennedy S, . et al. ; MetaHIT consortium. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500:541–546. doi: 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- Li F, Hitch T C A, Chen Y, Creevey C J, and Guan L L. . 2019. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 7:6. doi: 10.1186/s40168-019-0618-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd M, and Ghelardi R J. . 1964. A table for calculating the equitability’component of species diversity. J. Anim. Ecol. 33:217–225. doi: 10.2307/2628 [DOI] [Google Scholar]
- Ludwig W, Euzeby J, and Whitman W B. . 2010. Road map of the phyla Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes. In: Krieg, N.R., W. Ludwig, W. B. Whitman, B. P. Hedlund, B. J. Paster, J. T. Staley, N. Ward, and D. Brown, editors. Bergey’s manual of systematic bacteriology. New York, NY: Springer; p. 1–19. [Google Scholar]
- Mackie R, Aminov R, White B, and McSweeney C. . 2000. Molecular ecology and diversity in gut microbial ecosystems. In: Cronjé, P.B., editor. Ruminant physiology: digestion, metabolism, growth and reproduction. New York (NY):CABI Publishing; p. 61–77. [Google Scholar]
- Martínez I, Perdicaro D J, Brown A W, Hammons S, Carden T J, Carr T P, Eskridge K M, and Walter J. . 2013. Diet-induced alterations of host cholesterol metabolism are likely to affect the gut microbiota composition in hamsters. Appl. Environ. Microbiol. 79:516–524. doi: 10.1128/AEM.03046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Wallace G, Zhang C, Legge R, Benson A K, Carr T P, Moriyama E N, and Walter J. . 2009. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl. Environ. Microbiol. 75:4175–4184. doi: 10.1128/AEM.00380-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia M, Brazel D M, Swan B K, Arnosti C, Chain P S, Reitenga K G, Xie G, Poulton N J, Lluesma Gomez M, Masland D E, . et al. 2012. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One. 7:e35314. doi: 10.1371/journal.pone.0035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer P R, Smith T P, Wells J E, Kuehn L A, and Freetly H C. . 2015. Rumen microbiome from steers differing in feed efficiency. PLoS One. 10:e0129174. doi: 10.1371/journal.pone.0129174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet L, Vande Velde I, and Verstraete W. . 1997. Effect of the addition of Peptostreptococcus productus ATCC35244 on the gastro-intestinal microbiota and its activity, as simulated in an in vitro simulator of the human gastro-intestinal tract. Appl. Microbiol. Biotechnol. 48:99–104. doi: 10.1007/s002530051022 [DOI] [PubMed] [Google Scholar]
- O’Quinn T G, Legako J F, Brooks J C, and Miller M F. . 2018. Evaluation of the contribution of tenderness, juiciness, and flavor to the overall consumer beef eating experience. Transl. Anim. Sci. 2:26–36. doi: 10.1093/tas/txx008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormerod K L, Wood D L, Lachner N, Gellatly S L, Daly J N, Parsons J D, Dal’Molin C G, Palfreyman R W, Nielsen L K, Cooper M A, . et al. 2016. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 4:36. doi: 10.1186/s40168-016-0181-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya P R, Singh K M, Parnerkar S, Tripathi A K, Mehta H H, Rank D N, Kothari R K, and Joshi C G. . 2010. Bacterial diversity in the rumen of Indian Surti buffalo (Bubalus bubalis), assessed by 16S rDNA analysis. J. Appl. Genet. 51:395–402. doi: 10.1007/BF03208869 [DOI] [PubMed] [Google Scholar]
- Paynter M J, and Elsden S R. . 1970. Mechanism of propionate formation by Selenomonas ruminantium, a rumen micro-organism. J. Gen. Microbiol. 61:1–7. doi: 10.1099/00221287-61-1-1 [DOI] [PubMed] [Google Scholar]
- Pielou E C. 1966. Species-diversity and pattern-diversity in the study of ecological succession. J. Theor. Biol. 10:370–383. doi: 10.1016/0022-5193(66)90133-0 [DOI] [PubMed] [Google Scholar]
- Rey F E, Faith J J, Bain J, Muehlbauer M J, Stevens R D, Newgard C B, and Gordon J I. . 2010. Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 285:22082–22090. doi: 10.1074/jbc.M110.117713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock M J Jr, Hiett K L, Gamble J, Caudill A C, Cicconi-Hogan K M, and Caporaso J G. . 2014. A hybrid DNA extraction method for the qualitative and quantitative assessment of bacterial communities from poultry production samples. J. Vis. Exp. 94:e52161. doi: 10.3791/52161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J, and Wallace R. . 1997. Energy-yielding and energy-consuming reactions. In: Hobson P N., and Stewart C S, editors. The rumen microbial ecosystem. Dordrecht:Springer; p. 246–282. [Google Scholar]
- Samarajeewa S, Hailu G, Jeffrey S, and Bredahl M. . 2012. Analysis of production efficiency of beef cow/calf farms in Alberta. Appl. Econ. 44:313–322. doi: 10.1080/00036846.2010.507173 [DOI] [Google Scholar]
- Schären M, Frahm J, Kersten S, Meyer U, Hummel J, Breves G, and Dänicke S. . 2018. Interrelations between the rumen microbiota and production, behavioral, rumen fermentation, metabolic, and immunological attributes of dairy cows. J. Dairy Sci. 101:4615–4637. doi: 10.3168/jds.2017-13736 [DOI] [PubMed] [Google Scholar]
- Scheifinger C C, and Wolin M J. . 1973. Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 26:789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabat S K, Sasson G, Doron-Faigenboim A, Durman T, Yaacoby S, Berg Miller M E, White B A, Shterzer N, and Mizrahi I. . 2016. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10:2958–2972. doi: 10.1038/ismej.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp R, Ziemer C J, Stern M D, and Stahl D A. . 1998. Taxon-specific associations between protozoal and methanogen populations in the rumen and a model rumen system. FEMS Microbiol. Ecol. 26:71–78. doi: 10.1111/j.1574-6941.1998.tb01563.x [DOI] [Google Scholar]
- Smith S B, Blackmon T L, Sawyer J E, Miller R K, Baber J R, Morrill J C, Cabral A R, and Wickersham T A. . 2018. Glucose and acetate metabolism in bovine intramuscular and subcutaneous adipose tissues from steers infused with glucose, propionate, or acetate. J. Anim. Sci. 96:921–929. doi: 10.1093/jas/sky017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G C, and Carpenter Z L. . 1974. Eating quality of animal products and their fat content. Changing the fat content and composition of animal products. Washington (DC):National Academy Press; p. 124–137. [Google Scholar]
- Smith S B, and Crouse J D. . 1984. Relative contributions of acetate, lactate and glucose to lipogenesis in bovine intramuscular and subcutaneous adipose tissue. J. Nutr. 114:792–800. doi: 10.1093/jn/114.4.792 [DOI] [PubMed] [Google Scholar]
- Smith B J, Miller R A, Ericsson A C, Harrison D C, Strong R, and Schmidt T M. . 2019. Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol. 19:130. doi: 10.1186/s12866-019-1494-7, preprint: not peer reviewed [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B J, Miller R A, and Schmidt T M. . 2020. Muribaculaceae genomes assembled from metagenomes suggest genetic drivers of differential response to acarbose treatment in mice. biorXiv. doi: 10.1101/2020.07.01.183202, preprint: July 2020 not peer reviewed [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M D, Hamp T J, Reid R W, Fischer L M, Zeisel S H, and Fodor A A. . 2011. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140:976–986. doi: 10.1053/j.gastro.2010.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Deng Q, and Cao L. . 2014. Ruminant feces harbor diverse uncultured symbiotic actinobacteria. World J. Microbiol. Biotechnol. 30:1093–1100. doi: 10.1007/s11274-013-1529-4 [DOI] [PubMed] [Google Scholar]
- Trenkle A. 2001. Strategies for optimizing value of finished cattle in value-based marketing grids. In: Beef Research Report, 2000. Ames (IA):Iowa State University. [Google Scholar]
- Tuomisto H. 2012. An updated consumer’s guide to evenness and related indices. Oikos. 121:1203–1218. doi: 10.1111/j.1600-0706.2011.19897.x [DOI] [Google Scholar]
- Turnbaugh P J, Hamady M, Yatsunenko T, Cantarel B L, Duncan A, Ley R E, Sogin M L, Jones W J, Roe B A, Affourtit J P, . et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia N L, and Harvatine K J. . 2017. Acetate dose-dependently stimulates milk fat synthesis in lactating dairy cows. J. Nutr. 147:763–769. doi: 10.3945/jn.116.245001 [DOI] [PubMed] [Google Scholar]
- USDA 1965. Official United States standards for grades of carcass beef. Washington (DC):USDA. [Google Scholar]
- Wallace R J. 1978. Control of lactate production by Selenomonas ruminantium: homotropic activation of lactate dehydrogenase by pyruvate. J. Gen. Microbiol. 107:45–52. doi: 10.1099/00221287-107-1-45 [DOI] [PubMed] [Google Scholar]
- Wallace R J, Sasson G, Garnsworthy P C, Tapio I, Gregson E, Bani P, Huhtanen P, Bayat A R, Strozzi F, Biscarini F, . et al. 2019. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 5:eaav8391. doi: 10.1126/sciadv.aav8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters W A, Xu Z, and Knight R. . 2014. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588:4223–4233. doi: 10.1016/j.febslet.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandita T G, Joshi N, Kim H H, An S J, and Hwang S G. . 2018. Pre-adipocyte determination and adipocyte differentiation of stromal vascular cells isolated from intramuscular tissue of Hanwoo beef cattle treated by acetate and propionate. Trop. Anim. Sci. J. 41:207–214. doi: 10.5398/tasj.2018.41.3.207 [DOI] [Google Scholar]
- Welch C B, Lourenco J M, Davis D B, Krause T R, Carmichael M N, Rothrock M J, Pringle T D, and Callaway T R. . 2020. The impact of feed efficiency selection on the ruminal, cecal, and fecal microbiomes of Angus steers from a commercial feedlot. J. Anim. Sci. 98:1–10. doi: 10.1093/jas/skaa230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington G H, and Stouffer J R. . 1959. Beef marbling: its estimation and influence on tenderness and juiciness. Ithaca, New York: Cornell University Agricultural Experiment Station. [Google Scholar]
- Whittaker R H. 1960. Vegetation of the Siskiyou mountains, Oregon and California. Ecol. Monogr. 30:279–338. doi: 10.2307/1943563 [DOI] [Google Scholar]
- Zeng B, Han S, Wang P, Wen B, Jian W, Guo W, Yu Z, Du D, Fu X, Kong F, . et al. 2015. The bacterial communities associated with fecal types and body weight of rex rabbits. Sci. Rep. 5:9342. doi: 10.1038/srep09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, Liu M, Wu C, Su X, Zhan K, and qi Zhao G. . 2017. Effects of alfalfa flavonoids extract on the microbial flora of dairy cow rumen. Asian-Australas. J. Anim. Sci. 30:1261–1269. doi: 10.5713/ajas.16.0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, DiBaise J K, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell M D, Wing R, Rittmann B E, . et al. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365–2370. doi: 10.1073/pnas.0812600106 [DOI] [PMC free article] [PubMed] [Google Scholar]