Abstract
The nicotine metabolite ratio (NMR; 3-hydroxycotinine/cotinine) is an index of CYP2A6 activity. CYP2A6 is responsible for nicotine’s metabolic inactivation and variation in the NMR/CYP2A6 is associated with several smoking behaviors. Our aim was to integrate established alleles and novel genome-wide association studies (GWAS) signals to create a weighted genetic risk score (wGRS) for the CYP2A6 gene for European-ancestry populations. The wGRS was compared with a previous CYP2A6 gene scoring approach designed for an alternative phenotype (C2/N2; cotinine-d2/(nicotine-d2 + cotinine-d2)). CYP2A6 genotypes and the NMR were assessed in European-ancestry participants. The wGRS training set included N = 933 smokers recruited to the Pharmacogenetics of Nicotine Addiction and Treatment clinical trial [NCT01314001]. The replication cohort included N = 196 smokers recruited to the Quit 2 Live clinical trial [NCT01836276]. Comparisons between the two CYP2A6 phenotypes and with fractional clearance were made in a laboratory-based pharmacokinetic study (N = 92 participants). In both the training and replication sets, the wGRS, which included seven CYP2A6 variants, explained 33.8% (P < 0.001) of the variance in NMR, providing improved predictive power to the NMR phenotype when compared with other CYP2A6 gene scoring approaches. NMR and C2/N2 were strongly correlated to nicotine clearance (ρ = 0.70 and ρ = 0.79, respectively; P < 0.001), and to one another (ρ = 0.82; P < 0.001); however reduced function genotypes occurred in slow NMR but throughout C2/N2. The wGRS was able to predict smoking quantity and nicotine intake, to discriminate between NMR slow and normal metabolizers (AUC = 0.79; P < 0.001), and to replicate previous NMR-stratified cessation outcomes showing unique treatment outcomes between metabolizer groups.
Clinical Trial Registrations
NCT01314001 and NCT01836276.
Keywords: CYP2A6, European-ancestry, genetic risk score, nicotine metabolism, pharmacogenetics
1 |. INTRODUCTION
Nicotine is the primary psychoactive compound in cigarettes, responsible for tobacco’s addictive properties.1 Nicotine undergoes CYP2A6-mediated metabolism to cotinine (COT)2 and COT is further metabolized to 3-hydroxycotinine (3HC) exclusively by CYP2A6.3,4 The 3HC/COT ratio, known as the nicotine metabolite ratio (NMR), is a well-established index of CYP2A6 activity.3 The NMR is strongly associated with CYP2A6 genotype, highly correlated with total nicotine clearance,3 and is associated with smoking behaviors including acquisition,5 cigarettes/day,6–8 smoking topography,9 nicotine dependence,7,8,10 and cessation outcomes.11–15
Heritability estimates for NMR derived from European-ancestry twins range from 60% to 80%.16,17 Among European-ancestry individuals, common loss- (*2, *4) and decrease- (*9, *12) of-function * alleles [pharmvar.org] explain approximately 20% of total NMR variation.17 Since these * alleles are primarily associated with haplotypes that lead to reduced activity, they are less effective in stratifying faster metabolizers, resulting in a wide range of variability among normal metabolizers.
Several genome-wide association studies (GWAS) of the NMR have shown most (more than 98%) of the genome-wide significant variants are concentrated in or around CYP2A6.16,18–20 Among European-ancestry cohorts, the top replicated hit is rs56113850, a CYP2A6 intronic SNP, explaining approximately 14% to 22% of NMR variation.16 Putative independent signals (via conditional analysis) included rs56113850, rs113288603, and esv2663194 (ie, CYP2A6*12) among Finnish smokers,16 and rs56113850, rs2316204, and rs1801272 (ie, CYP2A6*2) among European-ancestry smokers recruited to the Pharmacogenetics of Nicotine Addiction and Treatment (PNAT2) trial (unpublished observations). In addition, novel CYP2A6 diplotypes constructed from rs28399453, rs150298687, rs7260629, and rs57837628 have been identified through next generation sequencing.21
Measuring ad libitum NMR requires that individuals are smoking at regular intervals so that cotinine remains in steady state. A CYP2A6 genetic risk score would improve assessment in instances where ad libitum NMR is unavailable, including among nonsmokers, intermittent smokers, and former smokers, as well as in studies in which DNA is available, but other biological matrices are not. Genetic risk scores have been described for multiple complex diseases and behaviors, by aggregating multiple genetic variants into a single predictive measure.22,23 CYP2A6 is involved in the pathogenesis of multiple diseases and metabolism of several clinical substrates. For example, CYP2A6 activates tobacco-specific nitrosamines; greater CYP2A6 activity is associated with an increased risk for lung cancer8,24 and several tobacco-related illnesses (eg, COPD and type-2 diabetes).25,26 Among CYP2A6 substrates also include tegafur, letrozole, metronidazole, and efavirenz.27,28 A recent set of polygenic risk scores (PRSs) developed by Chen and colleagues demonstrated an ability to capture 9.2% to 16% of the variation in nicotine metabolism markers, but these PRSs were unable to predict either smoking quantity or cessation.29 Improved estimation of the impact of different CYP2A6 genotypes on enzyme activity will enhance the clinical utility of available genotype data. Specifically, improving interpretation of the effect of CYP2A6 gene variants on CYP2A6 enzyme activity will facilitate the ability to use genetics-based approaches to study the influence of CYP2A6 on disease risk and/or drug metabolism in the absence of a measured phenotype.
A CYP2A6 multiplicative model, also referred to as the CYP2A6 metric,29 was constructed by Bloom and colleagues on an alternative CYP2A6 phenotype (COT-d2/(NIC-d2 + COT-d2); abbreviated C2/N2, also referred to as the metabolism proportion29), where COT-d2 represents dideutero-cotinine and NIC-d2 represents dideutero-nicotine.30 The model used seven CYP2A6 polymorphisms (*1A(51A), *1D-Y351H, *2, *4, *9, *12, and *14) and explained approximately 70% of the variance in the C2/N2 measure. Because of nicotine’s short half-life (1–2 h) and cotinine’s long half-life (16–19 h),1 C2/N2 is usually quantified in a laboratory setting where deuterated compounds are consumed orally. In contrast, because of the long half-life of cotinine and formation dependence of 3-hydroxycotinine on cotinine,1,3 NMR can be reliably derived from nicotine derived from ad-libitum smoking31–33 and is minimally affected by other enzyme pathways.34–36 We are unaware of studies directly associating the laboratory phenotype C2/N2 with smoking behaviors, including response to smoking cessation therapies. It is also unclear how C2/N2 correlates with nicotine clearance or NMR, or whether the multiplicative metric, which was primarily designed to fit the C2/N2 measure, would be effective in predicting NMR.
This study seeks to (1) assess how well a previous CYP2A6 multiplicative gene scoring approach based on the C2/N2 phenotype predicts NMR; (2) develop an improved genotype model, a weighted genetic risk score (wGRS), specifically designed to predict NMR among those of European-ancestry through the integration of NMR GWAS signals with established * alleles and evaluate this wGRS in an independent cohort to validate model generalizability; (3) compare C2/N2 and NMR directly with each other and with nicotine clearance; (4) evaluate the wGRS’s relationship to smoking quantity and nicotine intake; (5) examine the ability of the wGRS to discriminate between slow and normal metabolizers based on NMR cut-points implicated in clinical outcomes; and lastly, (6) compare the wGRS with the NMR in predicting smoking cessation outcomes.
2 |. MATERIALS AND METHODS
2.1 |. Study populations
Each study was approved by institutional review boards at all participating sites and at the University of Toronto. Participants providing written informed consent for DNA sample collection and release of de-identified information to investigators underwent genotyping.
2.1.1 |. Training set
A total of N = 933 treatment-seeking European-ancestry smokers were recruited to the PNAT2 clinical trial [NCT01314001], where ancestry was determined from GWAS data using principal components analysis19; 96.8% of European-ancestry smokers and 98.5% of African-ancestry smokers in the PNAT2 trial had genetic ancestries concordant with self-reported ancestry.19 Study details are described elsewhere.12 The NMR was measured from whole blood32 collected at intake. Smoking quantity in PNAT2 was assessed at intake using self-reported cigarettes per day (CPD), assessed both as a continuous variable, and by a 4-level ordered grouping strategy (CPD ≤ 10; 11 ≤ CPD ≤ 20; 21 ≤ CPD ≤ 30; and CPD ≥ 31) as previously described.29 Nicotine intake at this time point was assessed by the sum of cotinine and 3-hydroxycotinine (COT+3HC), a superior biomarker to cotinine alone6,37 as cotinine can overestimate smoking quantity in CYP2A6 slow metabolizers because of reduced COT metabolism to 3HC.38
2.1.2 |. Replication cohort
A total of N = 196 treatment-seeking European-ancestry smokers were recruited to the Quit-2-Live (Q2L) clinical trial [NCT01836276], where ancestry was self-reported. Study details are described elsewhere.39 The NMR was measured from whole blood collected at intake.
2.1.3 |. Laboratory-based pharmacokinetic study
A total of N = 92 European-ancestry participants (N = 44 smokers and N = 48 nonsmokers) were recruited to the Pharmacogenetic Study of Nicotine Metabolism, where ancestry was self-reported. Study details are described elsewhere.3 Fractional clearance to cotinine, C2/N2, and the NMR were derived from single-dose oral administration of deuterium-labeled nicotine (nicotine-d2). We refer to this version of the NMR as NMR-d2, which has also been referred to as experimentally ingested NMR (eNMR) by others.29 Metabolite concentrations for C2/N2 and the NMR were determined from plasma samples collected at 30 minutes and 6 hours following oral administration of nicotine-d2, respectively, as previously described.3,30
2.2 |. Genotyping
Copy number variants were determined through TaqMan copy number assays (ThermoFisher Scientific, Waltham, Massachusetts, USA) according to the manufacturer’s protocol and previous studies.40 A gene conversion variant in the CYP2A6 3′ flanking region (CYP2A6*1B) was determined by a two-step PCR assay as described previously.41 For the training set, single nucleotide variants were genotyped using an Illumina HumanOmniExpressExome-8 version 1.2 array with a custom add-on containing more than 2500 additional variants; details on genotyping, quality control procedures, and imputation are found elsewhere.19 For the replication cohort and the laboratory-based pharmacokinetic study, single nucleotide variants were genotyped by quantitative polymerase chain reaction (qPCR) approaches including TaqMan (rs56113850, rs113288603, rs1801272 (CYP2A6*2)) and two-step PCR (rs2316204, rs1137115 (CYP2A6*1A), rs28399433 (CYP2A6*9), rs28399435 (CYP2A6*14)) assays (ThermoFisher Scientific) as described.41 Genotype frequencies for each of the SNPs were in Hardy-Weinberg equilibrium (P > 0.05) in each of the cohorts. Genotyping methods were cross-validated in a subset of training set samples (N = 141) to confirm validity of comparisons between SNPs that were imputed in the training set (ie, rs113288603, rs56113850, and rs2316204) and qPCR results. rs113288603 and rs2316204 yielded 100% concordance between genotyping methods; rs56113850 yielded a 94% concordance rate, with mismatches found predominantly in samples with a CYP2A6 deletion (CYP2A6*4).
2.3 |. Gene scoring models
2.3.1 |. Multiplicative model
The multiplicative model was employed as described previously using described variants and parameters.42 Briefly, model parameters were fit to the C2/N2 phenotype and included genotypes for six CYP2A6 * alleles (*1A(51A) (rs1137115), *2 (rs1801272), *4 (gene deletion), *9 (rs28399433), *12 (CYP2A6/2A7 hybrid), and *14 (rs28399435)), with final scores ranging from 0.44 to 0.90. CYP2A6*1D-Y351H is a rare allele that was not identified in our training set.
2.3.2 |. Weighted genetic risk score model
An additive wGRS was constructed to the variation in the NMR in the training set, where the selection of variants came from four variant sets. Set 1 were independent signals identified by conditional analysis in the training set. Details on GWAS methodology and conditional analysis for the training set (variant set 1) were identical to methods used previously for the African-ancestry participants recruited to the PNAT2 clinical trial.19 Set 2 were independent signals identified from another large-scale European-ancestry GWAS of the NMR.16 Set 3 were CYP2A6 * alleles not identified in sets 1 and 2, but common in European-ancestry populations, and set 4 were SNPs identified in a recent CYP2A6 sequencing study.
Using the combination of independent signals (variant sets 1 & 2) as the initial baseline set of variants for the wGRS model, additional established CYP2A6 gene variants that are frequently studied in candidate gene analyses were tested. These were from set 3 CYP2A6 * alleles (*4 (gene deletion), *9 (rs28399433), *1A(51A) (rs1137115), *1B (58 base-pair gene conversion in the 3′ UTR of CYP2A6), and *14 (rs28399435)), as well as set 4 SNPs identified in a recent CYP2A6 sequencing study (rs28399453, rs150298687, rs7260629, and rs57837628; Tanner et al., 2017). These additional variant sets (3 & 4) were investigated by evaluating the variance (R2) captured (in combination with the independent signals from variant sets 1 & 2). Only the variants which contributed an additional source of variance to the NMR phenotype (ie, increased the R2) were added into the wGRS model. Scores were created by summing the number of risk alleles weighted by their unstandardized effect sizes. The use of unstandardized betas is to retain the unit of measurement from the GWAS analysis, where standardized variables invite bias because of sampling error. Betas were estimated from frequentist additive linear regression models (using SNPTEST, version 2.5.2)43 of the NMR phenotype in the training set, adjusted for principal components 1 and 2, and unstandardized through multiplying betas by the standard deviation (SD) of the NMR in the training set (SD = 0.205). To evaluate an individual’s wGRS, the number of risk alleles were summed using their assigned weights (Table 1); this summed value was then adjusted by the addition of 2.0 to create a final score with a positive range of values, assigning individuals with the CYP2A6 diplotype *1/*1, ie, those without the included wGRS decrease or increase of function variants, a score of 2.0 to resemble other CYP gene activity scores, eg, CYP2D644 or CYP2C1945 and as outlined by the Clinical Pharmacogenetics Implementation Consortium (CPIC) to assign phenotypes based on CYP genotypes.46 This will enhance the clinical implementation of CYP2A6 genetic data by facilitating the conversion of the wGRS to an analogous CYP activity score. For example, an individual with the risk allelesT/C for rs56113850, T/T for rs2316204, A/T for CYP2A6*2, and reference alleles for the remaining genotypes would be as assigned an activity score of 2.046 (ie, [{0.135 + 0.16 − 0.25} + 2.0]).
TABLE 1.
The seven variants included in the weighted genetic risk score (wGRS)
| Variant | Reference Allelec | Risk Allele | Location with Respect to CYP2A6 Gene | Beta per Risk Alleled | Weight per Risk Allelee |
|---|---|---|---|---|---|
| rs56113850a,b | T | C | Intron 4 | +0.657 | +0.135 |
| rs2316204a | C | T | 5 kb 3′ | +0.388 | +0.080 |
| rs113288603b | C | T | 6 kb 5′ | −0.122 | −0.025 |
| *2 (rs1801272)a | A | T | Exon 3 (L160H) | −1.221 | −0.250 |
| *9 (rs28399433) | A | C | Promoter (TATA box) | −0.778 | −0.160 |
| *4 (CYP2A6 deletion) | - | Deletion | Deletion of exons 1–9 | −1.707 | −0.350 |
| *12 (CYP2A6/2A7 hybrid)b | - | Hybrid | Translocation of exons 1–2 | −1.329 | −0.272 |
| *1 (no wGRS variants) | - | - | - | - | 0.000 |
Independent signals identified from conditional analyses in the nicotine metabolite ratio (NMR) genome-wide association studies (GWAS) in Pharmacogenetics of Nicotine Addiction and Treatment (PNAT2) European-ancestry smokers (training set).
Independent signals identified from conditional analyses in the meta-GWAS of the NMR in Finnish European-ancestry smokers.16
Reference alleles are in relation to the positive strand of the genome orientation.
Betas were estimated from frequentist additive linear regression models (using SNPTEST, version 2.5.2)43 of the NMR phenotype in the training set, principal components 1 and 2 were included as covariates.
The change in NMR, and thus the “weight per risk allele” was estimated by multiplying the standard deviation (SD) of NMR (SD = 0.205 in training set sample) by the effect size (ie, beta) of the risk allele.
2.4 |. Statistical analysis
All statistical analyses were completed using SPSS version 20 (IBM Corporation) and MedCalc version 17.4 (MedCalc Software). The Shapiro-Wilk test was used to test for normality in dependent variables. Variables which were not normally distributed were log-transformed.33 Linear regression was used to assess NMR variation accounted for by the gene scoring models, and the overall contribution to NMR variation was assessed after controlling for known NMR covariates (sex, age, and body mass index [BMI])47 reflecting factors that influence the NMR in addition to CYP2A6 genetic variants. Correlations between variables in the laboratory-based pharmacokinetic study were assessed using Spearman rank correlation coefficients. Linear regression and Spearman rank correlations were used to evaluate the relationship between the wGRS and measures of smoking quantity and nicotine intake. Receiver operating characteristic (ROC) curve analyses were conducted with two NMR definitions of normal metabolizers (NMR ≥ 0.26 and ≥ 0.31) coding slow and normal dichotomously as the outcome variable, with the various gene scoring models included as continuous predictors. The Youden’s J index was used to determine the criterion for the optimal cut-point in the wGRS. Logistic regression was used to evaluate end-of-treatment quit rates (nicotine patch vs. varenicline) within slow and normal metabolizers defined by NMR or the wGRS. An interaction between treatment and metabolizer group was evaluated as the ratio of odds ratios (ORRs).12
3 |. RESULTS
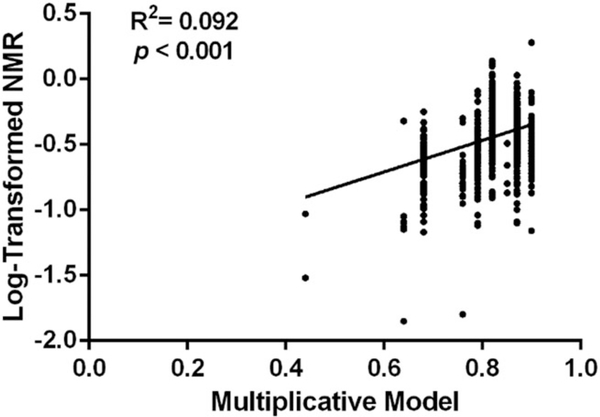
The multiplicative model, originally developed to predict C2/N2 following oral nicotine administration,30 explained 9.2% of the variance (R2) in log-transformed NMR (log-NMR) in the training set (Figure 1). Including non-CYP2A6 covariates (sex, age, and BMI) known to influence NMR47 in the model modestly increased the amount of log-NMR variability captured (R2 = 12.4%).
FIGURE 1.
Linear regression analysis of the relationship between the multiplicative model score as described previously42 and log-transformed nicotine metabolite ratio (NMR) in the training set (N = 933). The multiplicative model explained 9.2% of the variance in log-NMR. When non-CYP2A6 covariates (sex, age, and BMI)47 were additionally included, the model explained 12.4% of the variance in log-NMR
The final wGRS model included seven variants (Table 1), derived from the four variant sets comprising five putative independent signals identified using conditional analyses in two exclusively European-ancestry NMR GWASs, as described in variant sets 1 and 2, and two additional functionally relevant CYP2A6 * alleles common (minor allele frequency > 1%) in European-ancestry populations [pharmvar.org], as described in variant set 3. Versions of the model including other common CYP2A6 * alleles (*1A(51A), *1B, and *14; variant set 3), and novel diplotypes (constructed from rs28399453, rs150298687, rs7260629, and rs57837628; variant set 4)21 yielded poorer fit to the NMR phenotype and were consequently excluded, suggesting the impact of these excluded variants is captured by the independent signals. The wGRS model was based on an additive genetic model (see individual variant weights, Table 1), which reflects the observed influence of CYP2A6 variant alleles on the NMR, as illustrated for rs56113850 with incrementing allele dosages (Figure S1).
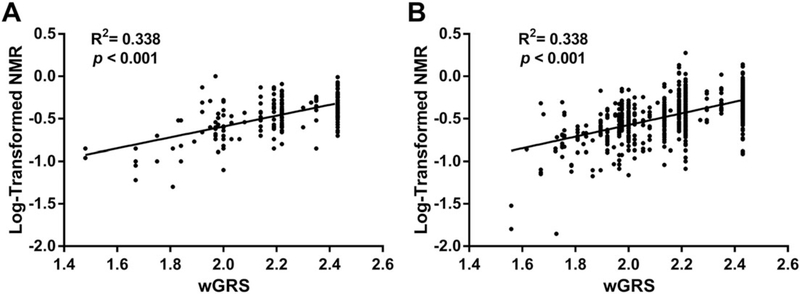
The wGRS was assessed in an independent cohort of treatment-seeking smokers (replication cohort) to validate the model’s generalizability. The developed wGRS model explained 33.8% of the log-NMR variance in the replication cohort. When sex, age, and BMI were included as covariates, the overall model explained 41.1% of the log-NMR variance (Figure 2A). In the training set, the variance in log-NMR explained exclusively by the wGRS model was similar (R2 = 33.8%). After including known NMR covariates (sex, age, and BMI), the overall model explained 37.6% of the log-NMR variance in the training set (Figure 2B).
FIGURE 2.
Linear regression analysis of the relationship between the 7-variant weighted genetic risk score (wGRS) and log-transformed NMR in the A, replication cohort (N = 196) and B, training set (N = 933). The wGRS explained 33.8% of the variance in log-NMR in both cohorts. When non-CYP2A6 covariates (sex, age, and BMI)47 were additionally included, the model explained 41.1% and 37.6% of the variance in the replication cohort and training set, respectively
Variations of the optimal 7-variant wGRS were then tested in the replication cohort and training set using a subset of the seven variants (Table 2). A 4-variant wGRS including only the common and functionally-relevant CYP2A6 * alleles (*2, *4, *9, and *12) was tested, representing variants commonly used to genotype CYP2A6 and excluding for the three additional variants in the 7-variant wGRS to come from the recent GWASs (rs56113850, rs2316204, rs13288603). The 4-variant wGRS explained 23.1% of the log-NMR variance (31.3% including covariates of sex, age, and BMI) in the replication cohort and 29.6% (33.2% including covariates) in the training set. A 6-variant version was also tested; the 6-variant model may be applicable in instances where copy-number genotyping for *4 (CYP2A6 gene deletion) or *12 (CYP2A6/CYP2A7 hybrid allele) is not feasible. This 6-variant model excluded *4, and used rs28399442 as a proposed surrogate marker for *12.48 This version of the model was marginally inferior to the full 7-variant wGRS, explaining 29.3% of the log-NMR variance (33.0% including covariates of sex, age, and BMI) in the training set; rs28399442 was not genotyped in the replication cohort. In evaluating the performance of rs28399442 at capturing the *12 hybrid allele, the rs28399442 surrogate marker correctly identified 34/41 (82.9%) of the *12 alleles previously identified using qPCR copy number variation analysis in the training set, with no false positives. Furthermore, there were negligible differences in the log-NMR variance explained between a 6-variant model that included the *12 surrogate SNP (ie, rs28399442) and a 6-variant model that included the *12 allele determined through qPCR copy number variation analysis: R2 = 29.3% versus 28.9%, respectively (excluding covariates), and R2 = 33.0% versus 32.7%, respectively (including covariates).
TABLE 2.
Linear regression analysis of the relationship between alternative weighted genetic risk score (wGRS) models and log-transformed nicotine metabolite ratio (NMR) in the replication cohort (N = 196) and training set (N = 933)
|
R2; P value |
R2; P value |
|||
|---|---|---|---|---|
| (without covariates) |
(with covariates)e |
|||
| Model | Replication | Training | Replication | Training |
| 7-variant wGRSa | 33.8%; P < 0.001 | 33.8%; P < 0.001 | 41.1%; P < 0.001 | 37.6%; P < 0.001 |
| 4-variant wGRSb | 23.1%; P < 0.001 | 29.6%; P < 0.001 | 31.3%; P < 0.001 | 33.2%; P < 0.001 |
| 6-variant wGRSc | N/A | 29.3%; P < 0.001 | N/A | 33.0%; P < 0.001 |
| 6-variant wGRSd | 33.3; P < 0.001 | 28.9%; P < 0.001 | 40.4%; P < 0.001 | 32.7%; P < 0.001 |
Abbreviation: N/A (Not Applicable)—rs28399442 (*12 Surrogate) not genotyped in replication cohort
*2, *4, *9, *12, rs56113850, rs2316204, rs113288603.
*2, *4, *9, *12.
*2, *9, rs28399442 (*12 Surrogate), rs56113850, rs2316204, rs113288603.
*2, *9, *12, rs56113850, rs2316204, rs113288603.
Covariates included: sex, age, and BMI.47
In contrast to previously used methods to classify individuals by CYP2A6 * alleles into slow, intermediate, or normal metabolizers (Figure 3A), the semi-continuous range of values from the wGRS can be used either to replicate similar broad categorizations as demonstrated by splitting the wGRS scale by tertiles (Figure 3B), or more refined divisions using, for example, quintiles (Figure 3C).
FIGURE 3.
Tukey box-and-whisker plots of nicotine metabolite ratio (NMR) distributions. Data is from the training set (N = 933) split as a function of A, * allele groupings; SM, slow metabolizers; IM, intermediate metabolizers; NM, normal metabolizers. V/V, two variant * alleles B, wGRS scale (1.300–2.430) split into tertiles (T, tertile) and C, weighted genetic risk score (wGRS) scale (1.300–2.430) split into quintiles (Q, quintile)
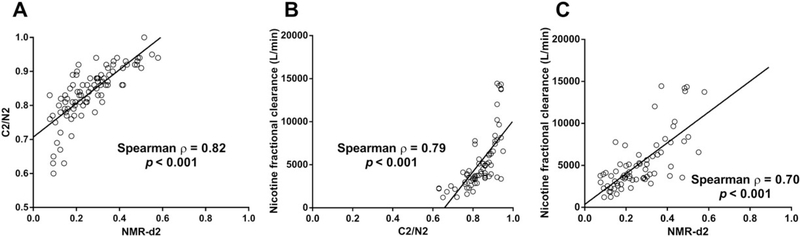
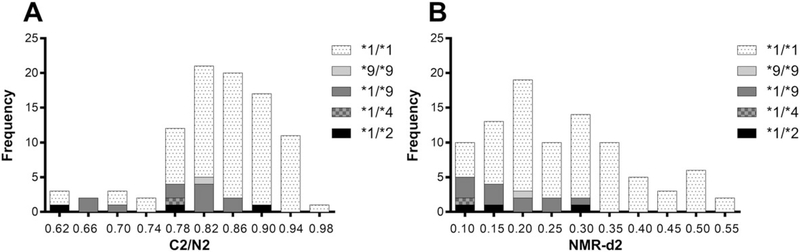
The C2/N2 and NMR-d2 ratios were compared following oral nicotine administration in the laboratory-based pharmacokinetic study. Following oral nicotine-d2, the C2/N2 and NMR-d2 ratios were significantly and strongly correlated with each other (ρ = 0.82) (Figure 4A). Furthermore, both the C2/N2 and NMR-d2 ratios were significantly and strongly correlated with nicotine fractional clearance to cotinine (ρ = 0.79 and ρ = 0.70, respectively) (Figures 4B, 4C). Decrease/loss of function CYP2A6 * alleles (*2, *4, and *9) were distributed throughout the range of C2/N2 values (Figure 5A), in contrast to the NMR where they were concentrated exclusively in the slower half of the NMR range (Figure 5B). The lack of association of these alleles with lower C2/N2 suggests C2/N2 does not represent genetic variation in CYP2A6, which may contribute to the relatively poor ability of the multiplicative model, based on C2/N2, to predict NMR (Figure 1).
FIGURE 4.
Correlations between nicotine kinetic parameters in the laboratory-based pharmacokinetic study (N = 92). Correlations between the CYP2A6 phenotype ratios C2/N2 and NMR-d2 measured at 30 and 360 minutes, respectively are shown in A. Correlations between nicotine fractional clearance (L/min) and C2/N2, and between nicotine fractional clearance (L/min) and NMR-d2 are shown in B, and C, respectively
FIGURE 5.
The distribution of A, C2/N2 and B, NMR-d2 in the laboratory-based pharmacokinetic study (N = 92) as a frequency histogram color-coded by genotype class. Decrease/loss of function CYP2A6 * alleles (*2, *4, and *9) were distributed throughout the range of C2/N2 values A, these CYP2A6 * alleles (*2, *4, and *9) were concentrated exclusively in the slower half of the NMR-d2 range B
The wGRS was significantly associated with smoking quantity, as defined by a 4-level grouping order of CPD (R2 = 0.8%, P = 0.006), as well as by Spearman correlation when entering CPD as a continuous variable (ρ = 0.085, P = 0.009). Likewise, the wGRS was significantly associated with nicotine intake, as defined by a log-transformed sum of COT + 3HC (R2 = 2.5%, P < 0.001), as well by Spearman correlation to COT + 3HC as an untransformed variable (ρ = 0.132, P < 0.001).
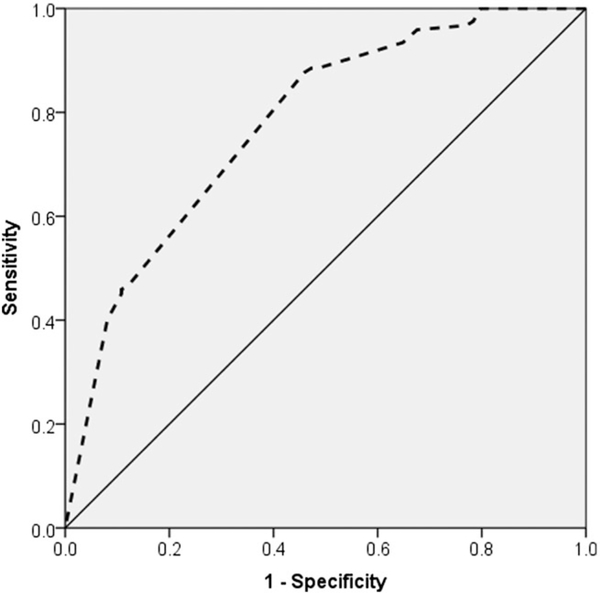
ROC curve analyses were performed to assess the ability of the wGRS to discriminate between slow and normal metabolizers in the replication cohort, using the NMR cut-point of 0.31 (Figure 6) used in the original PNAT2 smoking cessation clinical trial to prospectively stratify slow and normal metabolizers for treatment randomization.12 The wGRS model, excluding covariates, showed fair to good diagnostic ability to discriminate between slow and normal metabolizers at the 0.31 cut-point, yielding a significant area under the curve (AUC) of 0.78 (95% confidence interval (CI), 0.71–0.85) in the replication cohort. The Youden index J statistic indicated an optimal cut-point wGRS-2.14 to best identify normal metabolizers based on the NMR ≥ 0.31 definition. The wGRS yielded a similar AUC and Youden index J statistic in the training set when dichotomizing NMR metabolism groups based on the 0.31 cut-point: 0.79 (95% CI, 0.76–0.82) and 2.19, respectively (Figure S2). The diagnostic ability of the wGRS appeared superior to the multiplicative model in the training set, which in comparison yielded an AUC of 0.60 (95% CI, 0.55–0.64) for the NMR 0.31 cut-point (Figure S2). Moreover, the wGRS showed consistent diagnostic validity at another cited NMR cut-point used to evaluate smoking cessation treatment outcomes (NMR: 0.2613), yielding an AUC of 0.78 (95% CI, 0.71–0.86) and 0.81 (95% CI, 0.78–0.85) in the replication cohort and training set, respectively, compared with the multiplicative model which yielded an AUC of 0.65 (95% CI, 0.60–0.71) in the training set.
FIGURE 6.
Receiver-operating-characteristic (ROC) curve of the weighted genetic risk score (wGRS) (dashed line) in discriminating CYP2A6 slow and fast metabolizer groups using an NMR cut-point of 0.31 in the replication cohort. The wGRS for an nicotine metabolite ratio (NMR) cut-point of 0.31 (slow: NMR < 0.31, normal: NMR ≥ 0.31)12 yielded an area under the curve (AUC) estimate of 0.78; P < 0.001 (95% confidence interval (CI), 0.71–0.85). The wGRS for an NMR cut-point of 0.26 (slow: NMR < 0.26, normal: NMR ≥ 0.26)13 yielded an AUC estimate of 0.78; P < 0.001 (95% CI, 0.71–0.86) (not shown)
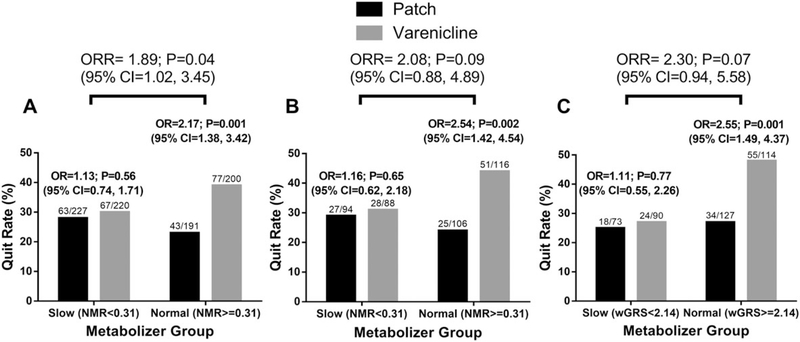
In the placebo-controlled PNAT2 clinical trial, focusing on the active treatment arms as the main hypothesis,12 N = 838 multiracial smokers were randomized to varenicline or nicotine patch based on pretreatment NMR. Normal metabolizers (NMR ≥ 0.31) experienced significantly higher end-of-treatment quit rates on varenicline compared with the nicotine patch, while slow metabolizers (NMR < 0.31) had similar quit rates on varenicline and the nicotine patch resulting in a significant NMR-by-treatment interaction (ratio of odds ratio, ORR = 1.89; 95% CI, 1.02–3.45; Figure 7A). Compared with the complete multiracial cohort receiving varenicline or nicotine patch (N = 838), in the genetically-determined European-ancestry subset (N = 404 training set smokers of the 838 that were randomized to the varenicline or nicotine patch treatment arms), a similar NMR metabolism group-by-treatment interaction (ORR = 2.08; 95% CI, 0.88–4.89) on quitting was observed (Figure 7B). Moreover, substituting NMR with our wGRS (Figure 7C) (normal metabolizers defined as wGRS ≥ 2.14, as described above) produced a similar ORR of 2.30 (95% CI, 0.94–5.58).
FIGURE 7.
End-of-treatment quit rates by treatment group and metabolizer group. Odds ratios (OR) with 95% confidence intervals (CI) comparing the efficacy of varenicline versus the nicotine patch. Metabolizer-by-treatment interaction effects on end-of-treatment quit rates evaluated by the ratio of odds ratios (ORR) with 95% CI. A, NMR stratification (slow: NMR < 0.31, normal: NMR ≥ 0.31) in the complete varenicline and nicotine patch treatment arms from intent-to-treat dataset (N = 838).12 B, NMR stratification (slow: NMR < 0.31, normal: NMR ≥ 0.31) in the genetically determined European-ancestry subset of the varenicline and nicotine patch treatment arms (N = 404). (C) wGRS stratification (slow: wGRS < 2.14, normal: wGRS ≥ 2.14) in the genetically determined European-ancestry subset of the varenicline and nicotine patch treatment arms (N = 404)
Likewise, the relative treatment effects within metabolizer group in the N = 404 subset were comparable to the observations from the N = 838 dataset (Figure 7A) when stratified by the NMR (Figure 7B), and the wGRS (Figure 7C), where normal metabolizers showed significantly higher quit rates on varenicline versus nicotine patch (OR = 2.54, P = 0.002 by the NMR or OR = 2.55, P = 0.001 by the wGRS compared with OR = 2.17, P = 0.001 by the NMR in the N = 838 dataset), while slow metabolizers demonstrated similar quit rates between treatments (OR = 1.16, P = 0.65 by the NMR or OR = 1.11, P = 0.77 by the wGRS compared with OR = 1.13, P = 0.56 by the NMR in the N = 838 dataset) (Figures 7A–C). Using the Youden J Statistic identified in the training set (wGRS≥2.19) versus the replication cohort (wGRS≥2.14), a similar treatment-by-group effect was observed (Figure S3).
4 |. DISCUSSION
We present a simple 7-variant approach to translate CYP2A6 genotypes into a semi-continuous CYP2A6 genetic measure for use in European-ancestry populations. The model improves on previous approaches, such as traditional broad categorization of composite CYP2A6 genotypes based on CYP2A6 * alleles into slow, intermediate, and normal metabolizers, by translating CYP2A6 genetic information to a semi-continuous metric predictive of the NMR. Moreover, our findings replicated in an independent cohort of treatment-seeking smokers, as demonstrated by a similar proportion of log-NMR variance explained by the wGRS. This successful replication suggests that parameter estimates for the wGRS are precise and may be extended to further studies of European-ancestry populations seeking to classify CYP2A6 genotypes.
Our wGRS model is superior to the multiplicative metric, originally modeled on C2/N2, for predicting NMR variation. This finding is intriguing since, following oral nicotine, both C2/N2 and NMR-d2 were significantly correlated with each other (Figure 4A) and to nicotine fractional clearance to cotinine (Figures 4B and 4C). The discrepancy in NMR prediction may be due, at least in part, to the observation that well-characterized decrease/loss of function CYP2A6 * alleles (*2, *4, and *9) are found across the C2/N2 distribution, in contrast to the NMR where these variant alleles are concentrated exclusively in the slower half of the NMR range (Figures 5A and 5B). This finding suggests that C2/N2, unlike the NMR, does not exclusively reflect CYP2A6 activity, and may be less suitable for phenotyping CYP2A6 and for fitting CYP2A6 genotypes for genetic scoring. Furthermore, the wGRS model, using the same variants and weights as described to predict the NMR, revealed comparable predictive power (12%) to the multiplicative model (11%) in predicting C2/N2 (Figure S4), substantially lower than previously reported for the multiplicative model (70%).30
After including known NMR covariates, the wGRS model explained approximately 35% of NMR variation suggesting, based on NMR heritability estimates of 60% to 80%, that additional genetic variation is yet to be characterized in CYP2A6, and perhaps in additional regulatory or pharmacokinetic genes. Despite the noted linkage disequilibrium between the reduce-of-function CYP2A6 * alleles and the independent signals identified through conditional analysis in GWASs,16 the inclusion of these independent GWAS signals explained more of the variation in the NMR than the CYP2A6 * alleles alone (Table 2), suggesting that some of the contribution of these GWAS hits to NMR variability is independent of their linkage to CYP2A6 * alleles. Conditional analysis, while informative for identifying independent signals in GWASs, is a relatively conservative approach; many additional CYP2A6 genetic variants not included here likely contribute to the variation in the NMR. In all, 719 genome-wide significant variants were identified in a meta-GWAS of the NMR in Finnish smokers16; assessment of these variants using approaches beyond traditional univariate analyses may explain a larger portion of the variation in the NMR. Emerging computational approaches, such as those involving more sophisticated predictive modeling (eg, regularized regression)49 may help account for the complex linkage disequilibrium and haplotype structures between associated variants, aiding in the prediction of the NMR. One study limitation was the lack of principal components-based ancestry determination in the replication cohort. However, the high concordance between PC-based and self-reported ancestry in the training set (96.8%), as observed in other studies (eg, 96.9% in Sucheston et al50), and the high similarity in wGRS fit between the training set and replication cohort (Figure 2) suggests this is a minimal limitation. However, this may contribute to a poorer fit of the wGRS in other studies, especially when the degree of genetic admixture among study participants is high.
The wGRS significantly associated to measures of smoking quantity and nicotine intake as assessed by the relationships to CPD and COT+3HC, respectively. Through ROC analyses, we demonstrated that the wGRS was favorable to the multiplicative approach in distinguishing slow from normal NMR metabolizers based on NMR cut-points that have previously reflected distinct smoking cessation outcomes (Figure 6 and Figure S2).12,13 The wGRS was able to replicate the within-metabolizer cessation outcomes observed by baseline pretreatment NMR, where normal (NMR ≥ 0.31 or wGRS ≥ 2.14) but not slow metabolizers show significant differences in quit outcomes between varenicline and nicotine patch treatments (Figure 7). Similar interaction effect sizes (ORRs) were noted between NMR and the wGRS approaches, but we were underpowered in the European-ancestry subset (N = 404 of 838) to observe statistical significance; similar and significant effect sizes (ORs) were noted between treatments within normal metabolizer groups, where no differences between treatments were observed among slow metabolizers (Figure 7A–C).
Here we have focused on developing a CYP2A6 gene-scoring approach in European-ancestry populations. Because of the unique differences in SNP rankings based on association strength (P values) and linkage disequilibrium patterns between different ancestral populations,18 developing unique gene-scoring approaches according to ancestry is likely necessary. Of note, in a meta-GWAS for the NMR in a cohort of exclusively African-ancestry smokers,19 a distinct list of independent signals was yielded compared with those identified in European-ancestry smokers, and only approximately 40% of the overall significant hits overlapped those identified in the GWAS of Finnish smokers.16 Bayesian fine-mapping approaches will likely aid in the identification of the causal SNPs that these independent signals are tagging and may give rise to a unified approach in translating CYP2A6 gene-scoring approaches across multiple ancestries, and individuals of mixed ancestry. However, we demonstrated that our current approach of combining independent GWAS signals with functional CYP2A6 * alleles provides a good and immediately available method of translating CYP2A6 genetic variants and warrants extension to other ancestries.
In summary, we have developed an original genomics approach to translate a small subset of seven CYP2A6 genetic variants into a single semi-continuous genetic score. This model replicated in an external cohort, indicating generalizability, associated with measures of smoking quantity and nicotine intake, and showed the ability to replicate NMR-based clinical outcomes for slow and normal metabolizers. Our wGRS approach represents a practical approach for diverse studies seeking to understand the contribution of CYP2A6 genetic variation to tobacco dependence, as well as tobacco-related diseases, and potentially to the metabolism of other clinical substrates.
Supplementary Material
ACKNOWLEDGEMENTS
Computations were performed on the Centre for Addiction and Mental Health (CAMH) Specialized Computing Cluster (SCC), funded by the Canada Foundation for Innovation Research Hospital Fund. We also acknowledge these sources of funding: Canada Research Chair in Pharmacogenomics (R.F.T.); the National Institutes of Health (NIH) grants PGRN DA020830 (R.F.T. and C.L.) and R01-DA031815 (N.L.N.); Canadian Institutes of Health Research (CIHR) grant FDN-154294 (R.F.T.); the Campbell Family Mental Health Research Institute of the Centre for Addiction and Mental Health (CAMH); the CAMH Foundation; the Canada Foundation for Innovation (#20289 and #16014); and the Ontario Ministry of Research and Innovation. We acknowledge additional members of the PGRN-PNAT Research Group including Frank Leone, Henry Glick, Angela Pinto, Paul Sanborn, Peter Gariti, Richard Landis (University of Pennsylvania); Maria Novalen, Bin Zhao, Ewa Hoffmann, Qian Zhou, Adel Aziziyeh (CAMH/University of Toronto); Martin Mahoney (Roswell Cancer Center, University of Buffalo); Maher Karam-Hage (The University of Texas M.D. Anderson Cancer Center); David Conti (University of Southern California); and Andrew Bergen (SRI International). This publication was made possible by the Pharmacogenomics Research Network-RIKEN Global Alliance (PGRN-RIKEN), which is supported by the RIKEN Center for Integrative Medical Science and the NIH Pharmacogenomics Research Network (GM115370).
Funding information
Ontario Ministry of Research and Innovation; Canada Foundation for Innovation, Grant/ Award Numbers: 16014 and 20289; CAMH Foundation; Campbell Family Mental Health Research Institute of the Centre for Addiction and Mental Health; Canadian Institutes of Health Research, Grant/Award Number: FDN-154294; National Institutes of Health (NIH), Grant/Award Numbers: R01-DA031815 and PGRN DA020830; Canada Research Chair in Pharmacogenomics
Footnotes
DISCLOSURE/CONFLICT OF INTEREST
R.F.T. has consulted for Quinn Emmanual and Apotex on unrelated topics. N.L.B. has consulted with pharmaceutical companies that market or are developing smoking cessation therapies and has been a paid expert witness in litigation against tobacco companies.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Benowitz NL, Hukkanen J, Jacob P. Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol. 2009;192:29–60. 10.1007/978-3-540-69248-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakajima M, Yamamoto T, Nunoya K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996a;24:1212–1217. [PubMed] [Google Scholar]
- 3.Dempsey D, Tutka P, Jacob P, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76(1):64–72. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima M, Yamamoto T, Nunoya K, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996b;277:1010–1015. [PubMed] [Google Scholar]
- 5.O’Loughlin J, Paradis G, Kim W, et al. Genetically decreased CYP2A6 and the risk of tobacco dependence: a prospective study of novice smokers. Tob Control. 2004;13(4):422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11(4):400–409. [DOI] [PubMed] [Google Scholar]
- 7.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14(9):615–626. [DOI] [PubMed] [Google Scholar]
- 8.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103(17):1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strasser AA, Benowitz NL, Pinto AG, et al. Nicotine metabolite ratio predicts smoking topography and carcinogen biomarker level. Cancer Epidemiol Biomarkers Prev. 2011;20(2):234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sofuoglu M, Herman AI, Nadim H, Jatlow P. Rapid nicotine clearance is associated with greater reward and heart rate increases from intravenous nicotine. Neuropsychopharmacology. 2012;37(6):1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerman C, Jepson C, Wileyto EP, et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clin Pharmacol Ther. 2010;87(5):553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerman C, Schnoll RA, Hawk LW Jr, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respir Med. 2015;3(2):131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerman C, Tyndale R, Patterson F, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79(6):600–608. [DOI] [PubMed] [Google Scholar]
- 14.Patterson F, Schnoll RA, Wileyto EP, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther. 2008;84(3):320–325. [DOI] [PubMed] [Google Scholar]
- 15.Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav. 2009;92(1):6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loukola A, Buchwald J, Gupta R, et al. A genome-wide association study of a biomarker of nicotine metabolism. PLoS Genet. 2015;11(9): e1005498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL. Genetic and environmental influences on the ratio of 3’hydroxycotinine to cotinine in plasma and urine. Pharmacogenet Genomics. 2009;19(5):388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baurley JW, Edlund CK, Pardamean CI, et al. Genome-wide association of the laboratory-based nicotine metabolite ratio in three ancestries. Nicotine Tob Res. 2016;18(9):1837–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chenoweth MJ, Ware JJ, Zhu AZX, et al. Genome-wide association study of a nicotine metabolism biomarker in African American smokers: impact of chromosome 19 genetic influences. Addiction. 2018;113(3):509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel YM, Park SL, Han Y, et al. Novel association of genetic markers affecting CYP2A6 activity and lung cancer risk. Cancer Res.2016;76(19):5768–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner JA, Zhu AZ, Claw KG, et al. Novel CYP2A6 diplotypes identified through next-generation sequencing are associated with invitro and in-vivo nicotine metabolism. Pharmacogenet Genomics. 2018; 28(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Song Y. Building genetic scores to predict risk of complex diseases in humans: is it possible? Diabetes. 2010;59(11):2729–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor AE, Davey Smith G, Munafo MR. Associations of coffee genetic risk scores with consumption of coffee, tea and other beverages in the UK biobank. Addiction. 2018;113(1):148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKay JD, Hung RJ, Han Y, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017; 49(7):1126–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho MH, Castaldi PJ, Wan ES, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21(4):947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T, Chen WQ, David SP, et al. Interaction between heavy smoking and CYP2A6 genotypes on type 2 diabetes and its possible pathways. Eur J Endocrinol. 2011;165(6):961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce RE, Cohen-Wolkowiez M, Sampson MR, Kearns GL. The role of human cytochrome P450 enzymes in the formation of 2-Hydroxymetronidazole: CYP2A6 is the high affinity (low K(m)) catalyst. Drug Metab Dispos: Bethesda, MD. 2013;41(9):1686–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanner JA, Tyndale RF. Variation in CYP2A6 activity and personalized medicine. J Pers Med. 2017;7(4):18 10.3390/jpm7040018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen LS, Hartz SM, Baker TB, Ma Y, N LS, Bierut LJ. Use of polygenic risk scores of nicotine metabolism in predicting smoking behaviors. Pharmacogenomics. 2018;19(18):1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloom J, Hinrichs AL, Wang JC, et al. The contribution of common CYP2A6 alleles to variation in nicotine metabolism among European-Americans. Pharmacogenet Genomics. 2011;21(7):403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mooney ME, Li ZZ, Murphy SE, Pentel PR, Le C, Hatsukami DK. Stability of the nicotine metabolite ratio in ad libitum and reducing smokers. Cancer Epidemiol Biomarkers Prev. 2008;17(6):1396–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St.Helen G, Novalen M, Heitjan DF, et al. Reproducibility of the nicotine metabolite ratio in cigarette smokers. Cancer Epidemiol Biomarkers Prev. 2012;21:1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanner JA, Novalen M, Jatlow P, et al. Nicotine metabolite ratio (3-hydroxycotinine/cotinine) in plasma and urine by different analytical methods and laboratories: implications for clinical implementation. Cancer Epidemiol Biomarkers Prev. 2015;24(8):1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chenoweth MJ, Zhu AZ, Sanderson Cox L, Ahluwalia JS, Benowitz NL,Tyndale RF. Variation in P450 oxidoreductase (POR) A503V and flavincontaining monooxygenase (FMO)-3 E158K is associated with minor alterations in nicotine metabolism, but does not alter cigarette consumption. Pharmacogenet Genomics. 2014a;24(3):172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taghavi T, St Helen G, Benowitz NL, Tyndale RF. Effect of UGT2B10, UGT2B17, FMO3, and OCT2 genetic variation on nicotine and cotinine pharmacokinetics and smoking in African Americans. Pharmacogenet Genomics. 2017;27(4):143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu AZ, Zhou Q, Cox LS, Ahluwalia JS, Benowitz NL, Tyndale RF. Variation in trans-3′-hydroxycotinine glucuronidation does not alter the nicotine metabolite ratio or nicotine intake. PLoS ONE. 2013b;8(8):e70938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benowitz NL, Dains KM, Dempsey D, Yu L, Jacob P 3rd. Estimation of nicotine dose after low-level exposure using plasma and urine nicotine metabolites. Cancer Epidemiol Biomarkers Prev. 2010;19(5):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu AZ, Renner CC, Hatsukami DK, et al. The ability of plasma cotinine to predict nicotine and carcinogen exposure is altered by differences in CYP2A6: the influence of genetics, race, and sex. Cancer Epidemiol Biomarkers Prev. 2013a;22(4):708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nollen NL, Cox LS, Yu Q, et al. A clinical trial to examine disparities in quitting between African American and white adult smokers: design, accrual, and baseline characteristics. Contemp Clin Trials. 2016;47:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu M, Koyama T, Kishimoto I, Yamazaki H. Dataset for genotyping validation of cytochrome P450 2A6 whole-gene deletion (CYP2A6*4) by real-time polymerase chain reaction platforms. Data Brief. 2015;5:642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wassenaar CA, Zhou Q, Tyndale RF. CYP2A6 genotyping methods and strategies using real-time and endpoint PCR platforms. Pharmacogenomics. 2016;17:147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olfson E, Bloom J, Bertelsen S, et al. CYP2A6 metabolism in the development of smoking behaviors in young adults. Addict Biol. 2018;23(1):437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. [DOI] [PubMed] [Google Scholar]
- 44.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83(2):234–242. [DOI] [PubMed] [Google Scholar]
- 45.Mrazek DA, Biernacka JM, O’Kane DJ, et al. CYP2C19 variation and citalopram response. Pharmacogenet Genomics. 2011;21(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical pharmacogenetics implementation consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014;95(4):376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chenoweth MJ, Novalen M, Hawk LW Jr, et al. Known and novel sources of variability in the nicotine metabolite ratio in a large sample of treatment-seeking smokers. Cancer Epidemiol Biomarkers Prev. 2014b;23(9):1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloom AJ, Harari O, Martinez M, et al. Use of a predictive model derived from in vivo endophenotype measurements to demonstrate associations with a complex locus, CYP2A6. Hum Mol Genet. 2012;21(13):3050–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baurley JW, McMahan CS, Ervin CM, Pardamean B, Bergen AW. Biosignature discovery for substance use disorders using statistical learning. Trends Mol Med. 2018;24(2):221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sucheston LE, Bensen JT, Xu Z, et al. Genetic ancestry, self-reported race and ethnicity in African Americans and European Americans in the PCaP cohort. PLoS ONE. 2012;7(3):e30950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.