By a combination of nuclear magnetic resonance (NMR) and mass spectroscopy of cross-linked peptides, we show that in contrast to human immunodeficiency virus type 1 (HIV-1), the C-terminal residues of the unstructured cytoplasmic tail of Mason-Pfizer monkey virus (M-PMV) Env interact with the matrix domain (MA). Based on biochemical data and molecular modeling, we propose that individual cytoplasmic tail (CT) monomers of a trimeric complex bind MA molecules belonging to different neighboring trimers, which may stabilize the MA orientation at the membrane by the formation of a membrane-bound net of interlinked Gag and CT trimers. This also corresponds with the concept that the membrane-bound MA of Gag recruits Env through interaction with the full-length CT, while CT truncation during maturation attenuates the interaction to facilitate uncoating. We propose a model suggesting different arrangements of MA-CT complexes between a D-type and C-type retroviruses with short and long CTs, respectively.
KEYWORDS: Env, M-PMV, Retrovirus, cytoplasmic tail, protein interactions
ABSTRACT
Retroviral envelope glycoprotein (Env) is essential for the specific recognition of the host cell and the initial phase of infection. As reported for human immunodeficiency virus (HIV), the recruitment of Env into a retroviral membrane envelope is mediated through its interaction with a Gag polyprotein precursor of structural proteins. This interaction, occurring between the matrix domain (MA) of Gag and the cytoplasmic tail (CT) of the transmembrane domain of Env, takes place at the host cell plasma membrane. To determine whether the MA of Mason-Pfizer monkey virus (M-PMV) also interacts directly with the CT of Env, we mimicked the in vivo conditions in an in vitro experiment by using a CT in its physiological trimeric conformation mediated by the trimerization motif of the GCN4 yeast transcription factor. The MA protein was used at the concentration shifting the equilibrium to its trimeric form. The direct interaction between MA and CT was confirmed by a pulldown assay. Through the combination of nuclear magnetic resonance (NMR) spectroscopy and protein cross-linking followed by mass spectrometry analysis, the residues involved in mutual interactions were determined. NMR has shown that the C terminus of the CT is bound to the C-terminal part of MA. In addition, protein cross-linking confirmed the close proximity of the N-terminal part of CT and the N terminus of MA, which is enabled in vivo by their location at the membrane. These results are in agreement with the previously determined orientation of MA on the membrane and support the already observed mechanisms of M-PMV virus-like particle transport and budding.
IMPORTANCE By a combination of nuclear magnetic resonance (NMR) and mass spectroscopy of cross-linked peptides, we show that in contrast to human immunodeficiency virus type 1 (HIV-1), the C-terminal residues of the unstructured cytoplasmic tail of Mason-Pfizer monkey virus (M-PMV) Env interact with the matrix domain (MA). Based on biochemical data and molecular modeling, we propose that individual cytoplasmic tail (CT) monomers of a trimeric complex bind MA molecules belonging to different neighboring trimers, which may stabilize the MA orientation at the membrane by the formation of a membrane-bound net of interlinked Gag and CT trimers. This also corresponds with the concept that the membrane-bound MA of Gag recruits Env through interaction with the full-length CT, while CT truncation during maturation attenuates the interaction to facilitate uncoating. We propose a model suggesting different arrangements of MA-CT complexes between a D-type and C-type retroviruses with short and long CTs, respectively.
INTRODUCTION
Mason-Pfizer monkey virus (M-PMV) is a D-type retrovirus that has been considered a favorable model for the study of immature particle assembly and trafficking because it preassembles in the cytoplasm of infected cells (1, 2). Due to both temporally and spatially separated processes of assembly and budding, M-PMV has been utilized to study individual steps of the retroviral life cycle. It has been repeatedly shown that M-PMV experiments have a great potential for generalization to complex retroviruses (3). This mainly applies to the assembly and intracellular targeting of viral proteins and particles. By using a set of Rab GTPase-based endosomal markers, we have recently shown that M-PMV Env localizes at intracellular vesicles. Based on biochemical and live imaging data, we have proposed a model of Env-mediated intracellular trafficking of M-PMV Gag immature particles (4). However, little is known about the interaction between Env and retroviral structural proteins. It was previously shown that the cytoplasmic tail (CT) of Env plays an important role in the trafficking of immature M-PMV particles to the site of retroviral budding (2, 5), and it was well documented that a tyrosine motif in the CT controls the incorporation of M-PMV Env into virions (6). It has also been suggested that Env interacts directly with Gag, which is crucial for the specific incorporation of Env into the virus envelope in human immunodeficiency virus (HIV) (7) and simian immunodeficiency virus (SIV) (8).
Even for HIV-1, the most-studied retrovirus, the site of the primary contact of Env with Gag in the host cell and a detailed mechanism of Env incorporation into the virus envelope have not been satisfactorily explained. In contrast to cytoplasmically synthesized Gag, the Env precursor is formed at the endoplasmic reticulum (ER), which is the site of the removal of signal peptide, folding, initial glycosylation, and trimerization (9). Upon translocation into the Golgi apparatus, the Env precursor is fully glycosylated and processed by furin-like protease into surface (SU) and transmembrane (TM) moieties, which remain membrane associated in the form of a trimer composed of SU-TM heterodimers. It has recently been shown that the cleavage of HIV-1 Env (gp160) is required for the proper functioning of HIV-1 Env complex and makes HIV-1 resistant to neutralizing antibodies (10). In contrast, the uncleaved HIV-1 gp160 is degraded in lysosomes (9). In the mature virion, the Env trimeric complex mediates specific attachment of the virus to the surface of the targeted host cell and virus entry by promoting TM-induced fusion of viral and cell membranes (11, 12).
As mentioned above, there is evidence that in D-type retroviruses, the matrix domain (MA) of Gag interacts with Env at the membranes of intracytoplasmic vesicles, and this interaction regulates the transport of preassembled immature M-PMV particles to the plasma membrane (2, 4, 5). The Env-mediated transport of M-PMV Gag is apparently connected with endocytic trafficking of Env (2, 4). In addition, it has recently been well documented that endosomal recycling of HIV-1 Env is essential for its interaction with Gag at distinct microdomains of the plasma membrane and recruitment into the budding particles (13).
HIV-1 CT is apparently the part of Env responsible for the interaction with the MA ensuring the recruitment of the Env complex into the virus membrane (14, 15). It has been clearly demonstrated that the mutation of HIV-1 CT prevents Env incorporation, but it may be compensated for by some mutations in the MA (16). Other mutations in the HIV-1 MA have also been reported to block the incorporation of native Env (17, 18). Furthermore, there is evidence that the MA trimers in lentiviruses effectively interact with the trimeric CT. Investigation of HIV-1 MA mutants with impaired trimerization showed that the MA trimeric interface is required for efficient incorporation of Env into the virus envelope (19, 20). Interestingly, the truncation of the HIV-1 CT restores the recruitment of Env into the envelope of the virus with impaired MA trimerization, which also leads to the restoration of the infectivity of such mutated virus (20). This indicates that the central cavity within the trimeric lattice of HIV-1 MA is essential for efficient incorporation of extremely long CTs (>100 amino acids) shared among lentiviruses, except for feline immunodeficiency virus (13, 20). However, most retroviruses, including M-PMV, have a relatively short CT, ranging from 25 to 38 amino acids (21). The question remains whether MA trimerization is required also for the accommodation of these short CTs.
In some retroviruses, the C-terminal part of a short CT is further processed by virus-encoded protease, as shown for murine leukemia virus (MLV) and M-PMV (22, 23). This event may modulate a mutual affinity of the SU and TM Env subunits, as shown in MLV (24). Analysis of MLV and HIV-1 mutants indicated that these viruses contain virus-specific sequences in the CT that are responsible for different Env-mediated membrane fusogenicity. The different membrane fusogenicity is thus attributable to the processing of the CT and possibly the different compositions of the HIV-1 and MLV membranes (25).
Crystal structures of the fusion domains of M-PMV and Xenotropic murine leukemia virus-related virus suggest the presence of electrostatic interactions within extramembrane regions of the TMs that stabilize the Env conformation to overcome the energy barrier required for fusion (26). Only recently, the structural organization of HIV-1 TM has been published, and the results indicate that in contrast to the unstructured N-terminal extramembrane part, the C-terminal moiety consists of three membrane-bound amphipathic α-helices (27).
RESULTS
Protein preparation.
We employed several techniques and different forms of M-PMV MA or CT to investigate the interaction between the MA and CT.
We expressed both nonmyristoylated and myristoylated versions of M-PMV MA proteins in Escherichia coli. We also produced in E. coli a nonmyristoylated version of MA with a C-terminal extension with 18 residues from phosphoprotein (PP; which naturally follows the MA in Gag) and a His tag (MAPPHis) or glutathione transferase (GST) (MAPPGST). The myristoylated MA was also produced in E. coli by using a two-plasmid system ensuring the production of both an MA and yeast myristoyltransferase. Both MA proteins were purified using a hexahistidine anchor. The presence of PP allowed us to use M-PMV protease to produce nonmyristoylated MA without the affinity tag and to separate the nonmyristoylated from myristoylated MA, since the myristoylated MAPPHis is not cleaved by M-PMV protease as described by Prchal et al. (28). We did not treat MAPPGST with the protease, because we used the affinity tag for the pulldown.
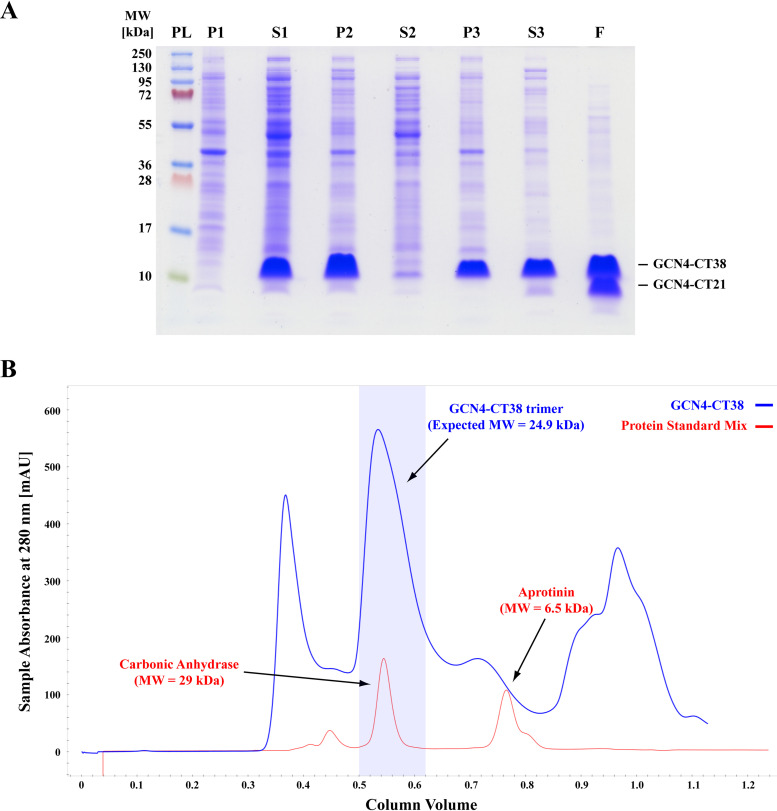
It is well known that retroviral Env glycoproteins occur as trimers where the trimerization is mediated by membrane-spanning TM domains. We induced membrane-independent trimerization in vitro by flanking M-PMV TM with the trimerization leucine-zipper motif of the GCN4 yeast transcription factor. As mentioned above, M-PMV protease truncates the cytotail consisting of 38 amino acids (CT38) during maturation. This cleavage occurs between residues H21 and Y22 (6), corresponding to residues H54 an Y55 in GCN4-CT38. To test whether this truncation has an influence on the interaction with MA, we prepared the truncated CT construct consisting of 21 amino acids in fusion with GCN4 (GCN4-CT21). Since the M-PMV GCN4-CT38 lacks any affinity tag, it was purified by ammonium sulfate precipitation followed by size exclusion chromatography. SDS-PAGE, reporting the whole purification process (see Fig. 1A), shows sufficient purity of the final GCN4-CT38 sample obtained by size exclusion chromatography. Additionally, the size exclusion chromatography confirmed the presence of a major product of molecular size corresponding to the expected size of a trimer (see Fig. 1B). However, despite the presence of protease inhibitors, part of GCN4-CT38 was shortened to GCN4-CT21 during the purification process, and the signals of both forms are visible in the final sample of GCN4-CT. To perform the pulldown assay with GCN4-CT, we also prepared both naturally occurring versions of M-PMV CT (CT38 and CT21) with N-terminal fusions with a hemagglutinin (HA) tag and GCN4 (HA-GCN4-CT38 and HA-GCN4-CT21, respectively).
FIG 1.
Purification of GCN4-CT38. (A) Electrophoreogram illustrating the purification of GCN4-CT38. (B) Chromatogram illustrating the separation of the protein standard mix (Sigma-Aldrich; red) and the GCN4-CT38 sample (the same as in lane S3 panel A; blue). PL, Thermo Scientific PageRuler Plus prestained protein ladder; P1 and S1, the pellet and supernatant, respectively, of the centrifuged lysate of E. coli BL21(DE3) producing GCN4-CT38; P2 and S2, the pellet and supernatant, respectively, obtained after the precipitation by 33% ammonium sulfate; P3 and S3, the pellet and supernatant, respectively, obtained after the overnight resuspension of GCN4-CT precipitated in P2; F, the final GCN4-CT purified from S3 by size exclusion chromatography and concentrated using microfiltration.
The interaction between nonmyristoylated MAPPGST and HA-GCN4-CT38 was examined by the pulldown assay, where also HA-GCN4-CT21 and GST were used as controls. In addition, we used nuclear magnetic resonance (NMR) for the determination of the interaction interface between the nonmyristoylated MA and GCN4-CT38. The differences between the interactions of nonmyristoylated and myristoylated versions of MA with GCN4-CT38 and GCN4-CT21 were also studied. Finally, we used protein cross-linking for the further description of the MA-CT complex, to detect the residues occurring in close proximity of nonmyristoylated MAPPHis and GCN4-CT38 or GCN4-CT21.
Pulldown assay showing interaction of GCN4-CT38 and MA.
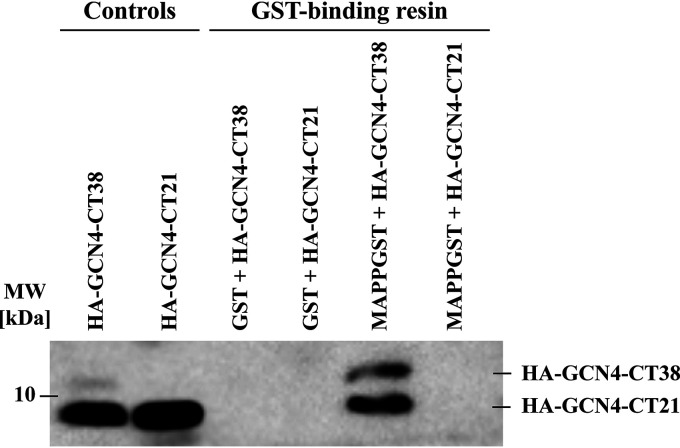
To determine the occurrence of a specific interaction of the MA with the CT, we used HA-GCN4-CT38 and MAPPGST. The proteins were incubated together for 5 days and then loaded onto GST-binding resin (Merck). After the unbound proteins were washed out, the bound proteins were visualized by immunoblot with anti-HA antibody. GST and HA-GCN4-CT21 were used as negative controls. The results of the pulldown assay (Fig. 2) clearly show that the CT has an affinity to the MA, because HA-GCN4-CT38 was pulled down on GST-binding resin only after its incubation with MAPPGST. Furthermore, HA-GCN4-CT21 did not interact with the MA, proving that neither the HA, the GCN4, nor the CT21 sequence has an affinity to MAPPGST. Nonspecific interactions were not observed between HA-GCN4-CT38 or HA-GCN4-CT21 and GST either. Unfortunately, HA-GCN4-CT38 was already cleaved to HA-GCN4-CT21 during the protein preparation (as shown in Fig. 1A) and was further processed during the incubation of the samples, despite the presence of protease inhibitors. Approximately 90% of HA-GCN4-CT38 was processed to HA-GCN4-CT21 during the incubation before the results of the pulldown assays were visualized. The protein was cleaved between residues V53 and H54, which was confirmed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (data not shown). The presence of HA-GCN4-CT21 bound to the GST-binding resin through MAPPGST along with the full-length HA-GCN4-CT38 can be explained by the gradual formation of the heterologous protein complexes of HA-GCN4-CT38 with HA-GCN4-CT21 in the incubated sample of HA-GCN4-CT38. Therefore, most of HA-GCN4-CT38 was cleaved to HA-GCN4-CT21, and the initial HA-GCN4-CT38 trimers likely contained one or two shortened HA-GCN4-CT21 chains. In addition, HA-GCN4-CT21 was thus detected in the fraction of HA-GCN4-CT38 interacting with MA. Importantly, the HA-GCN4-CT38 was significantly enriched upon binding to the MA protein (Fig. 2, cf. lanes 1 and 5, i.e., HA-GCN4-CT38 and MAPPGST+HA-GCN4-CT38, respectively).
FIG 2.
Pulldown assay showing the interaction between MA and CT. For the visualization of the pulldown by immunoblot, the anti-HA monoclonal antibody was used. The controls, HA-GCN4-CT38 and HA-GCN4-CT21, show the positions of purified proteins in the gel. Despite the fact that HA-GCN4-CT38 was truncated during the incubation with HA-GCN4-CT21, the remaining portion of intact full-length HA-GCN4-CT38 within the resulting heterotrimers was sufficient for the interaction with MAPPGST. GST-binding resin lines show the affinity of the HA-GCN4-CT38 to immobilized MAPPGST.
NMR spectroscopic analysis of M-PMV GCN4-CT38 and MA interaction.
To study the interaction between GCN4-CT38 and MA using NMR spectroscopy, we prepared GCN4-CT38 uniformly labeled with carbon (13C) and nitrogen (15N). Although the signals of the protein backbone were assigned, the signals of the atoms from the GCN4 and N-terminal part of the CT were not visible, probably due to the fast relaxation of the nuclei caused by the size and rigidity of the GCN4 domain (see Fig. 3A for detailed information). Since we detected signals of fewer than one-half of the residues and the data from the visible ones implied that the peptide was intrinsically disordered, we did not attempt to determine the structure of GCN4-CT38.
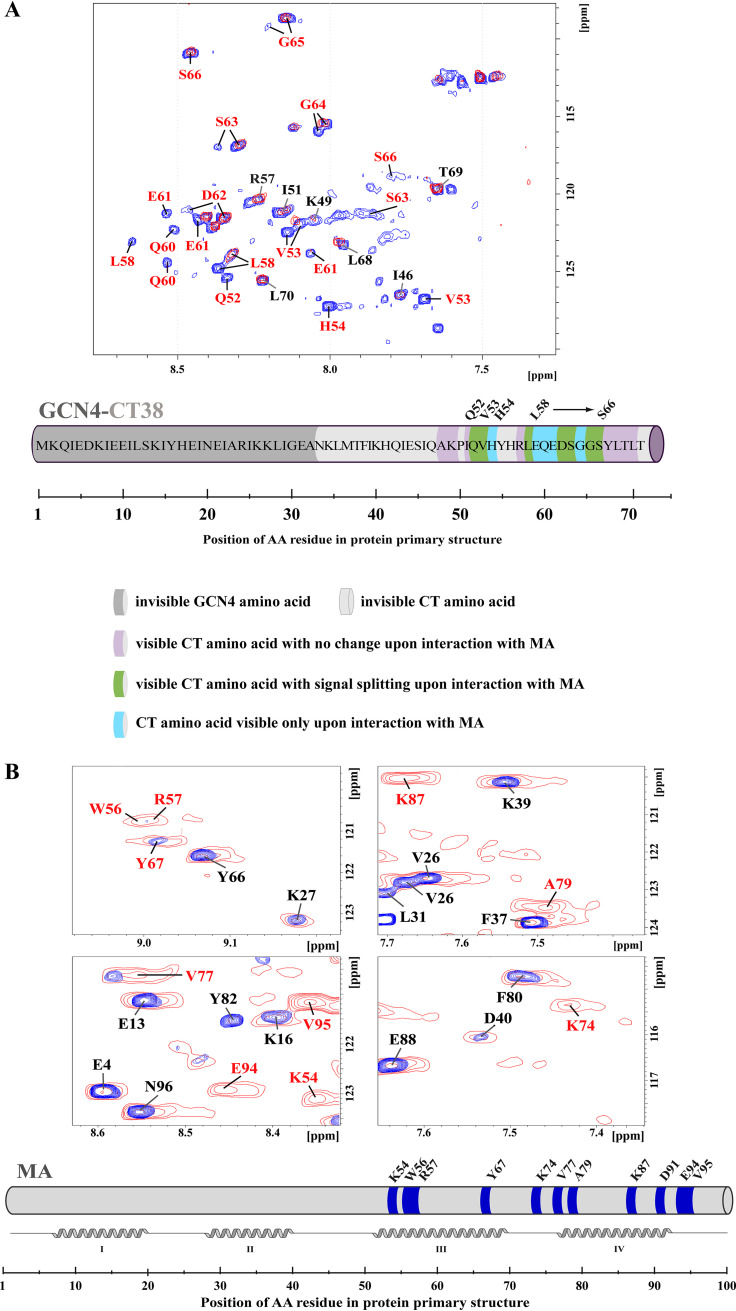
FIG 3.
NMR analysis of the interaction interface between GCN4-CT38 and MA. (A) Overlaid HN-HSQC spectra of free 15N-labeled GCN4-CT38 (red) and 15N-labeled GCN4-CT38 with 10-fold molar excess of the nonmyristoylated MA 3 days after mixing (blue), (B) Overlaid HN-HSQC spectra of the free 15N-labeled nonmyristoylated MA (red) and the 15N-labeled nonmyristoylated MA with 10-fold molar excess of GCN4-CT38 3 days after mixing (blue). The NMR signals that changed upon the interaction with MA (labeled in red) indicate the residues of CT38 or MA, which are involved in the interaction. The NMR signals that remained unchanged upon the interaction are labeled in black. In the schematic view of the MA sequence, the noninteracting residues of the MA are colored in gray, and the residues interacting with the CT38 are colored in blue.
When the excess of nonlabeled nonmyristoylated MA was added to labeled GCN4-CT38 and the protein mixture was equilibrated for at least 3 days, we observed signals splitting of several residues (Fig. 3A). Most of the CT38 signals in the HN spectrum split into two or three peaks, indicating that the trimer had lost its symmetry due to the interaction. We also found new signals of several CT38 residues that were not observed in the absence of the MA. This was likely caused by their interaction-mediated shifts from positions overlapping with signals of other residues. GCN4-CT38 interacted with both myristoylated and nonmyristoylated MAs, but the interaction with the nonmyristoylated one was stronger. The interaction was slow on the NMR timescale, i.e., the complex remained stable for more than 1 s. During the experiment with labeled GCN4-CT38, the formation of its stable complex with the MA took several days. This can be explained by slow MA oligomerization, which is in agreement with our previous data showing that it takes a similar time to achieve the MA monomer-dimer-trimer equilibrium.
To determine the CT38 interaction interface on the MA, we prepared both myristoylated and nonmyristoylated MA uniformly labeled with nitrogen (15N). The results were similar for both proteins: the interaction with GCN4-CT38 was slow on the NMR timescale, although the nonmyristoylated MA showed stronger interaction than the myristoylated one. We observed intensive signal broadening of backbone HN group signals of residues K54, W56, R57, Y67, K74, V77, A79, K87, D91, E94, and V95. The interacting residues are located in the patch formed by helices III and IV (Fig. 3B). To determine whether the M-PMV CT cleavage during the maturation affects the affinity of Gag to Env in the mature virus, we also tested the interaction of the MA with this truncated CT21 N-terminally extended with GCN4. We observed no interaction with the CT21 (Fig. 4).
FIG 4.
NMR analysis of the interaction interface between the GCN4-CT21 and the MA. The overlaid HN-HSQC spectra of the 15N-labeled GCN4-CT21 either free (red) or incubated for 3 days with 10-fold molar excess of the nonmyristoylated MA (blue). No significant changes in signal positions or intensities were observed. The individual signals of GCN4-CT21 were not assigned, because no interaction was observed.
Interacting interface of GCN4-CT38 and MA revealed by protein cross-linking.
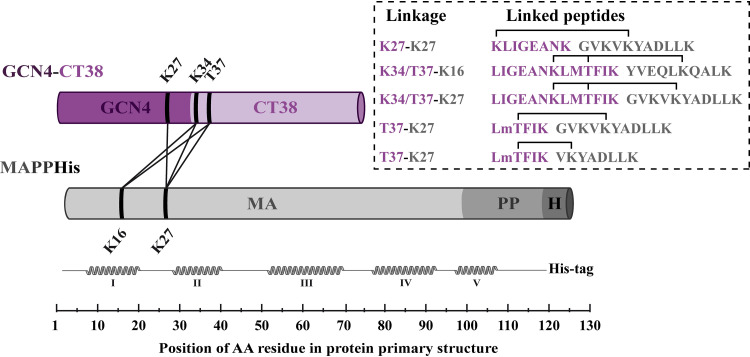
We also used a cross-linking mass spectrometry (XL-MS) approach as an independent method enabling the mapping of the interaction interfaces in protein complexes. We used homobifunctional bis(sulfosuccinimidyl)suberate (BS3) to cross-link mainly the residues containing primary amines (lysines or N-terminal residues), which are in close proximity (29). Through XL-MS analysis of the complex of nonmyristoylated MAPPHis with GCN4-CT38, we identified five cross-links between MA and GCN4-CT38 (Fig. 5), suggesting that the cross-linked residues of both proteins are in close proximity. This provided additional data of the MA-CT complex topology, especially for the N-terminal part of the CT, which had not been visible in the NMR experiment. All the identified cross-links (Table 1) joined the residues of the N-terminal part of the MA sequence (K16 and K27) with the residues located in the N-terminal part of the CT in the GCN4-CT38 (K34 and T37) and one residue located in the C-terminal part of GCN4 domain (K27). Since the analysis was performed in three repetitions and no other cross-linked residues of different parts of the proteins were found, the results indicate the close proximity of the N-terminal parts of the CT and the MA in the MA-CT complex. In the complex of the MA with GCN4-CT38, also the GCN4 residue K27 was linked with the MA residue K27, but this result is simply based on the fact that this residue is also in close proximity to the N-terminal part of the CT sequence.
FIG 5.
Linkages identified between the MA and GCN4-CT38 by XL-MS. The schemes of the proteins are displayed in violet (GCN4-CT38) or gray (MA) cylinders. The GCN4 part of the GCN4-CT38 is colored a dark shade of violet, and the CT part is a light shade of violet. The MA, PP, and His tag parts of MAPPHis are colored different shades of gray. The linkages identified in three repetitions of the analysis are depicted as lines between cylinders. The linearized structure of the MA is presented under the MA with schematically displayed helices in the MA structure. The sequences of identified cross-linked peptides are shown in the rectangle on the right, where the sequences and residues of GCN4-CT38 are colored violet and the sequences and residues of the MA are colored gray. The obtained spectra of the cross-links K34/T37-K16 and K34/T37-K27 did not allow determination of whether the residues K16 and K27 of MAPPHis interacted with the residue K34 or T37 of GCN4-CT38.
TABLE 1.
Cross-links between GCN4-CT38 and MAPPHis identified by XL-MSa
| m/z measured | Calculated mol wt (Da) | Charge state | Mass deviation (ppm) | GCN4-CT38 peptide | MAPPHis peptide | Linked residues (GCN4-CT38 to MAPPHis) |
|---|---|---|---|---|---|---|
| 561.589 | 2,243.337 | 4 | −0.67 | KLIGEANK | GVKVKYADLLK | K27-K27 |
| 709.409 | 2,834.610 | 4 | 1.90 | LIGEANKLMTFIK | YVEQLKQALK | K34/T37-K16 |
| 712.924 | 2,848.662 | 4 | 4.36 | LIGEANKLMTFIK | GVKVKYADLLK | K34/T37-K27 |
| 715.097 | 2,143.275 | 3 | 0.63 | LMTFIK | GVKVKYADLLK | T37-K27 |
| 620.367 | 1,859.090 | 3 | −1.43 | LMTFIK | VKYADLLK | T37-K27 |
For all the cross-links, the table shows the measured m/z value, the calculated molecular weight in daltons, the charge state, mass deviation, GCN4-CT38 and MAPPHis peptides, and their residues linked by BS3. The linked residues are marked in bold in the peptide sequences.
Labeled tandem mass spectrometry (MS/MS) spectra of the identified cross-links between the MA and GCN4-CT38 and their individual tables of all identified fragment ions are shown in Fig. S1 to S7 and Tables S1 to S5, respectively, in the supplemental material.
We have also used XL-MS to verify the NMR results showing lack of interaction of nonmyristoylated MAPPHis with truncated GCN4-CT21. Indeed, we have not identified any cross-links between these two proteins which would meet the parameters of reliably identified cross-links, and we have detected only a few nonspecific linkages mediated by the protein’s N-terminal amino group (data not shown).
MA-CT38 complex model.
Based on the interacting residues identified by NMR, we calculated the structure of the complex consisting of a trimeric MA (30) and a CT38 peptide without the GCN4 domain. We used aa trimeric M-PMV MA structure, because the N terminus of HIV-1 Gag forms trimers upon the interaction with the plasma membrane (14, 31, 32). In the interaction interface (Fig. 6A), the C terminus of the CT38 interacts with the C-terminal part of the MA, where helices III and IV form the CT38 binding site. The CT38 chain binds in parallel with these helices. Its C terminus is located close to the MA trimerization site, whereas its N-terminal part is oriented to the other side of the MA molecule. Additionally, the calculated structure of the CT38 complex with the monomeric MA provided the same results. A comparison of our proposed MA and CT38 complex with the membrane-bound MA (33) indicated that the MA-interacting site for CT38 is not obstructed by the interaction with the membrane.
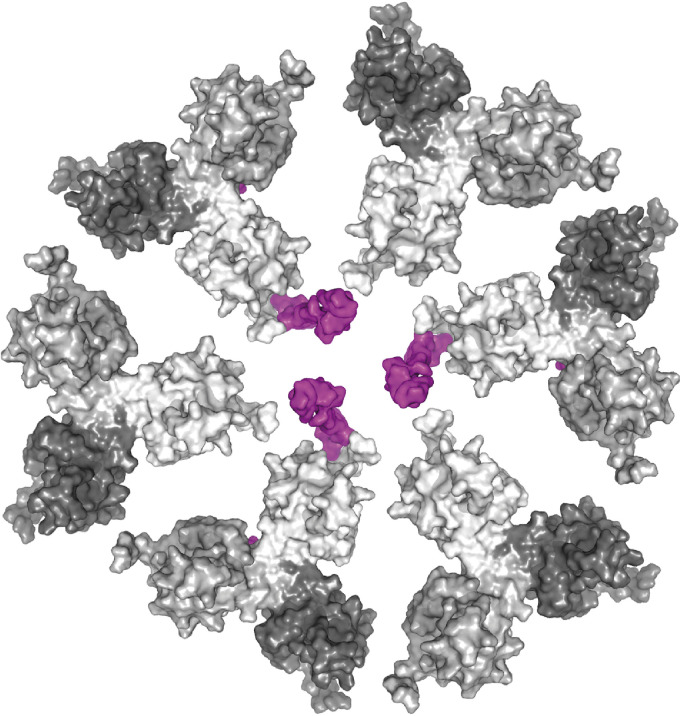
FIG 6.
Schematic model of the organization of MA and CT trimers at the membrane (a view from the inner side of the membrane). MA trimers and CT38 molecules are shown in surface representation. Single MA monomers are colored in different shades of gray, and CT38 is colored violet. Six MA molecules of individual MA trimers form a hexameric structure, with three MA trimers in complex with three individual CT38 domains of the M-PMV TM Env trimer anchored in the plasma membrane.
Based on the cross-linking data, we constructed a model of the MA-CT38 complex by adjusting backbone dihedral angles of the N-terminal part of the CT38 in the MA-CT38 complex calculated by HADDOCK. It is evident that the CT38 can be oriented in a position suitable for the interaction with the MA and where its N terminus can reach the membrane (Fig. 6B).
DISCUSSION
The contact between Gag and Env polyproteins is essential for the infectivity of retroviruses (34). Mutation studies of HIV-1 identified several residues of the MA that are responsible for Env recruitment (16). However, the MA and CT interactions can significantly differ between M-PMV and HIV-1, with short and extremely long CTs, respectively (35). The different morphogeneses of the D- and C-type retroviruses is another reason for the possible differences in Env recruitment, which may initiate at different cellular membranes.
The pulldown experiment confirmed that in contrast to M-PMV CT21, the CT38 directly interacted with the MA. This proves that only the immature-like M-PMV CT (i.e., CT38) interacts with the MA of Gag. During the experiment, the full-length CT38 was partially truncated to its mature form despite the presence of protease inhibitors. We believe that MA binds to the portion of CT38 molecules within the CT38 and CT21 heterotrimers bound to MAPPGST. This conclusion is supported by differential binding of MA to the CT38 and CT21 confirmed by NMR and XL-MS.
This correlates well with existing data showing that upon its incorporation, the M-PMV CT38 is proteolytically shortened by 16 C-terminal residues during virus maturation (22). It was shown that the truncation of the CT is essential for membrane fusion during the early phase of the M-PMV life cycle (36). Nevertheless, the newly formed particles are defective in the recruitment of Env if the truncation occurs prematurely before Env reaches the membrane. Although the truncation of the last four C-terminal residues does not affect M-PMV Env incorporation into virions, the truncation of 11 residues (i.e., the truncation after E26) leads to a significant decrease of Env incorporation, and Env with an even shorter CT is not incorporated at all (36). The importance of MA-membrane dissociation was suggested for HIV-1, where the proteolytically released MA becomes a part of the preintegration complex (37).
The interaction interface of the CT38 with the MA comprises residues up to S33, which is in good agreement with the fact that the truncation after Y34 had no effect on the incorporation of the CT into virions. Moreover, Song et al. found that the replacement of residues 18 to 25 by alanines leads to a decrease of Env incorporation (6, 38). We identified the same residues by NMR as those interacting with the MA. Therefore, we conclude that residues 20 to 33 of the CT38 are critical for the formation of a stable MA-CT complex.
Brody et al. reported that the M-PMV MA mutations A79V and T41I/T78I block the cleavage of the CT during virus maturation (22). We showed that these residues located in helices III and IV belong among those that interact with the CT. None of the MA residues interacting with the CT are involved in the interaction of the MA with the plasma membrane (33, 39) and all are thus available for interaction with the CT. Changes in NMR spectra showed that some residues participating in MA oligomerization (30) interact also with the CT38, suggesting the effect of the MA-CT interaction on MA oligomerization. Additionally, GCN4-CT38 exhibited a stronger interaction with the nonmyristoylated MA than the myristoylated one. We assume that upon interaction with the membrane, the myristoyl is exposed from the MA hydrophobic pocket and the MA molecule adopts a conformation similar to that of the nonmyristoylated MA. Correspondingly, the nonmyristoylated M-PMV MA favors the trimeric arrangement more than the myristoylated one (30, 39). This supports the theory that the MA with exposed myristoyl tends to form trimers on the membrane. It also explains the stronger interaction of the CT38 with the nonmyristoylated MA and may indicate that a virus-like particle recruits the CT after interaction with the membrane. This is in agreement with the data showing that the CT trimers are recruited into assembling the HIV-1 Gag lattice by binding to MA trimers (40, 41).
In addition to NMR, we used mass spectroscopy of peptides cross-linked by BS3 covalently joining lysine residues in a distance of up to 26 to 30 Å (29). Also, tyrosine, threonine, or serine residues may form stable cross-linking products (42–45). The identified cross-links proved the close proximity of the N-terminal parts of both MA and GCN4-CT38. This interaction interface does not interfere with the M-PMV MA trimerization region occurring on the other side of the MA molecule (30). However, the cross-linked K27 at the N terminus of M-PMV MA mediates the interaction with the plasma membrane (33), suggesting that also the N-terminal part of CT38 points toward the membrane. This is in good agreement with the fact that the GCN4 domain in our construct substitutes for the transmembrane domain of Env.
In our XL-MS experiment, we did not detect the same region as observed by NMR. This can be due to the absence of lysines in the C-terminal part of the CT38. Although several serines, tyrosines, and threonines (enabling also BS3 cross-linking) are located in this region, their reactivity with BS3 is significantly lower. Using NMR, we identified several lysine residues (K54, K74, K87, K92, and K93) in the region of the MA interacting with the CT38, but probably the steric hindrance at the tight interaction interface prevented BS3 from cross-linking the interacting residues.
In summary, our approach provides initial information about the MA-CT38 complex at the plasma membrane, where the MA as the N-terminal domain of Gag first binds to the membrane and then uses the CT to recruit Env into the viral particle. Then, CT truncation during the maturation step disturbs the interaction and enables the matured viral particle to infect a new host cell and facilitates the release of the capsid into the cytoplasm of the host cell (36, 46, 47). We propose a model where individual CT molecules in a trimeric complex bind MA monomers belonging to different neighboring trimers, which may stabilize the MA orientation at the membrane through the formation of the membrane-bound net of interlinked Gag and CT trimers (Fig. 7). The model differs from that proposed for HIV-1, where each CT trimer is positioned above a single MA trimer protruding to different hexamer holes (41). This seems to be another difference between HIV-1 and M-PMV, with extremely long and rather short CTs, respectively. The HIV-1 CT interacts with the MA through its N-terminal α-helical domains within LLP-3 and LLP-1 (41). In contrast, our data along with previous mutagenesis studies (36) suggest that the short M-PMV CT is unstructured and its C-terminal moiety is responsible for the interaction with the MA. Both models share the principle where the trimerization of the MA and the CT supports mutual interactions of these proteins, but because the M-PMV CT is much shorter and the M-PMV MA trimer is organized differently from the HIV-1 MA trimer (30), the organization of the complex must be different.
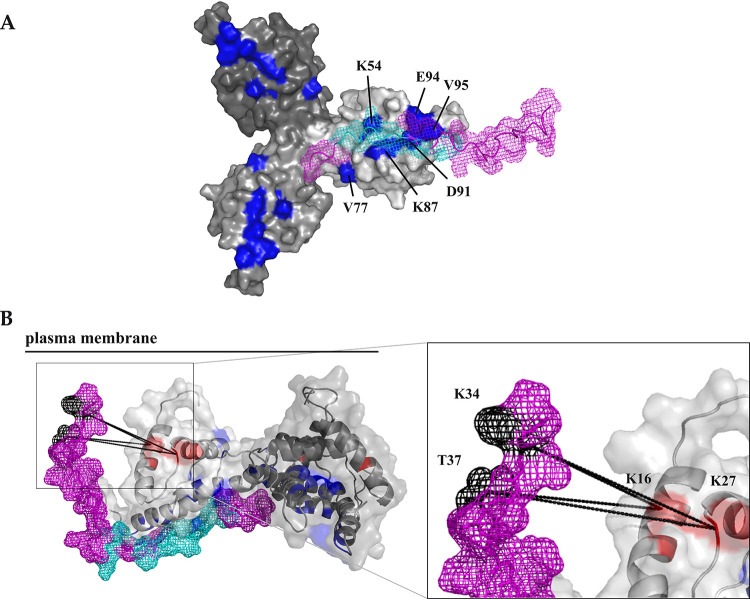
FIG 7.
Model of the CT complex with a trimeric MA. (A) MA-CT38 complex calculated in HADDOCK based on NMR results. (B) Model of the MA-CT complex created based on HADDOCK calculation (the C-terminal part of the CT38) and the results of XL-MS (the N-terminal part of the CT), where the N-terminal part of the CT sequence was twisted around the MA to meet the restrains determined by identified cross-links. Trimeric MA (30) is shown in the surface representation (A) or in the cartoon representation with a transparent surface (B), and single MA monomers are colored in different shades of gray. The MA residues identified by NMR spectroscopy as interacting with the CT38 are colored blue, and the MA residues forming cross-links with the CT38 residues are colored red. CT38 is shown in ribbon with mesh surface representation colored violet. The CT38 residues identified by NMR as interacting with the MA are colored, cyan and the CT38 residues forming cross-links with MA residues are colored black. The cross-links are displayed as dashed lines. The plasma membrane is depicted as a full line to express the orientation of the MA-CT38 complex on the membrane.
MATERIALS AND METHODS
Protein production and isolation.
Myristoylated and nonmyristoylated M-PMV MAs and MAPPHis (a version of the MA extended with 18 amino acid residues of the polyprotein Gag structure on the C terminus of the MA and hexahistidine tag) were prepared as previously described (28). A similar procedure was used to prepare MAPPGST. The only differences were the use of GST-binding agarose (Sigma) instead of Ni-nitrilotriacetic acid (NTA) agarose, the elution using 20 mM glutathione, and the absence of M-PMV protease in the purification procedure.
For the expression of M-PMV CT, plasmid encoding CT38 N-terminally extended with a gene encoding a leucine-zipper motif of GCN4 yeast transcription factor (GCN4-CT38) was used for the transformation of E. coli BL21(DE3). Cells were disrupted using One-Shot cell disrupter (Constant Systems). The protein was precipitated by 33% ammonium sulfate. After solubilization, the protein was further purified using size exclusion chromatography on a HiLoad 26/60 Superdex 75PG column (Amersham) and concentrated using microfiltration. The interaction studies were performed in a buffer containing 100 mM phosphate (pH 6), 100 mM NaCl, and 0.02% β-mercaptoethanol.
GCN4-CT21 was expressed and purified similarly to GCN4-CT38, with the only difference being the use of a supernatant from 50% ammonium sulfate precipitation.
For the pulldown assay, a full-length version of GCN4-CT38 was prepared with the HA tag sequence on its N terminus (HA-GCN4-CT38). The shortened version (HA-GCN4-CT21) was prepared by cleaving the full-length version with M-PMV protease.
For the NMR experiments, the proteins were uniformly labeled with nitrogen (15N) or were double labeled with carbon (13C) and nitrogen (15N).
Pulldown assay.
HA-GCN4-CT21 or HA-GCN4-CT38 at 0.18 mM concentration was mixed in 3.6-fold molar excess with GST or MAPPGST, and the final mixture was incubated at room temperature for 5 days. Afterward, the mixtures were alkalized by 5 M NaOH to pH 8 to ensure the conditions for the binding of GST to GST-binding resin. Then, 200 μl of GST-binding resin suspension (Novagen) was added to each mixture, and the suspensions were incubated at room temperature for 1 h. The GST-binding resin was then separated from the solution and washed with 500 μl of pulldown buffer (100 mM NaCl, 100 mM phosphate, 0.02% β-mercaptoethanol [vol/vol], pH 8). The washing step was repeated three times, and a sample of the GST-binding resin was collected after each wash. The samples of the GST-binding resin were analyzed by SDS-PAGE with subsequent immunoblot detection by the primary mouse monoclonal antibody against the HA tag (Sigma-Aldrich).
NMR spectroscopy.
All NMR data were measured on a Bruker Avance III 600‐MHz NMR spectrometer equipped with a cryoprobe. The spectra were processed using Topspin 3.5 software (Bruker BioSpin, GmbH) and further analyzed by Sparky (48) and CcpNmr analysis software (49). MA backbone atom assignments were taken from the BMRB database entry 34029 (39). The backbone atoms of CT38 were assigned using the standard test of triple-resonance experiments. The interactions were observed by monitoring chemical-shift changes in 1H/15N heteronuclear single quantum coherence (HN-HSQC) spectra. For these experiments, only one of the interaction partners was uniformly labeled by nitrogen (15N). The proteins were mixed in the ratios of 1:1 to 1:5 for 3 days before the measurement.
Protein cross-linking and digestion.
The 1 mM MAPPHis and GCN4-CT38 or GCN4-CT21 protein solutions were mixed in equimolar ratios, and the mixture was incubated at room temperature for 3 days to stabilize the interactions prior to protein cross-linking. The protein mixture was then treated by 5-fold molar excess of BS3 (Thermo Fisher Scientific) for 30 min. The reaction was quenched by the addition of 1 M Tris-HCl to such a volume as to achieve 50-fold molar excess of Tris-HCl to the concentration of BS3. The cross-linked proteins were treated with 0.1 M dithiothreitol at 56°C for 45 min and subsequently with 0.5 M iodoacetamide at room temperature in the dark for 30 min. Afterwards, the cross-linked proteins were digested at 37°C for 2 h with Pierce MS-grade trypsin (Thermo Scientific), using the trypsin-to-substrate ratio of 1:20 (wt/wt). The digestion was terminated by the addition of trifluoroacetic acid (TFA) to a final concentration of 0.5% (vol/vol). The obtained peptide mixture was then desalted by ZipTip C18 microtips (Millipore) according to the manufacturer’s instructions.
Nano-HPLC/Nano-ESI-Orbitrap mass spectrometry.
The analysis of the cross-linked peptide mixtures obtained was carried out on an Ultimate 3000 rapid-separation liquid chromatography (RSLC) nano-high pressure liquid chromatography (HPLC) system (Dionex) coupled to the EASY-Spray NG nano-electrospray ionization (ESI) source of an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). The peptides were dissolved in 0.1% formic acid (FA) and separated using reversed-phase C18 columns (μ-precolumn, Acclaim PepMap 100 C18, 300 μm by 5 mm, 5 μm; separation column, Acclaim PepMap RSLC C18, 75 μm by 250 mm, 2 μm [Thermo Fisher Scientific]). The peptides were eluted and separated using gradients from 5% to 12% B (varied between 0 and 3 min), 12% to 35% (varied between 3 and 52 min), 35% to 90% B (1 min), 90% B (53 to 57 min), 90% to 5% B (1 min), and 5% B (58 to 65 min) with a constant flow rate of 300 nl/min, where solvent A was 0.1% FA and solvent B was 100% acetonitrile containing 0.1% FA.
MS data were acquired in the data-dependent MS/MS mode with the high-energy collisional dissociation (HCD) fragmentation technique. Each high-resolution full scan (m/z 350 to 2,000 Da, R = 60,000) in the Orbitrap was followed by product ion scans (R = 30,000) also in the Orbitrap, starting with the most intense signal in the full-scan mass spectrum (an isolation window of 1.6 Th). The automatic gain control (AGC) value was set to 400,000 (MS) and 50,000 (MS/MS), while the maximum accumulation time was set to 50 ms (MS and MS/MS). Precursor ions with charge states from 2+ to 6+ were selected for the fragmentation. Dynamic exclusion (exclusion duration, 10 s; exclusion window, 10 ppm) was enabled.
Identification of cross-linked products.
The measured spectra were analyzed with the StavroX (version 3.6.6) (50) search engine for the identification of cross-linked peptides. The input data Mascot generic format (MGF) obtained from raw data by MSConvert (ProteoWizard) was used, and the search parameters were set as follows: three potential missed cleavage sites on the C terminus of K and R; the cleavage site on the C terminus of K blocked when it forms a cross-link; carbamidomethylation of cysteines as static modification; oxidation of methionines as variable modification (maximum, three); the minimum peptide length, 2; disuccinimidyl suberate (DSS)/BS3 as cross-linker with the K and the N-terminal amine group as the first linkable site; the N-terminal amine group, K, S, T, or Y as the second linkable site; 5 ppm as MS mass tolerance; 10 ppm as MS/MS mass tolerance; the lower mass limit of 200 Da; the upper mass limit of 6,000 Da; the signal-to-noise (S/N) ratio of 2.0; the possible ion types a, b, and y, with the maximum of three neutral losses per cross-link; and the application of the prescore with a value of >10% intensity as the minimum fraction of signal intensity identified compared to the total ion current. The results were filtered with the false-discovery rate cutoff of 5%, only taking into account the candidate scans providing ions of cross-linked fragmented peptides as well as the minimum cross-link sequence coverage of 40%.
Calculation of the MA-CT complex model.
The structure of the MA-CT complex was calculated by the HADDOCK 2.2 webserver (51). As an input for docking, we used the structure of nonmyristoylated MA trimer (30) and an extended structure of CT peptide without the GCN4 domain generated in the X-PLOR NIH 2.2 program (52). The residues determined by NMR spectroscopy were used as active. Passive residues were determined automatically. Docking was performed according to the standard protocol provided by the webserver. The resulting structures were clustered by structural similarity based on root mean square deviation (RSMD), and the cluster with the lowest overall energy was selected. The structure was visualized using PyMOL 0.99 software (https://pymol.org/2/).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Czech Science Foundation (GACR) grant no. GA17-24281S and by the European Regional Development Fund project ChemBioDrug (no. CZ.02.1.01/0.0/0.0/16_019/0000729).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Rhee SS, Hunter E. 1990. A single amino-acid substitution within the matrix protein of a type-D retrovirus converts its morphogenesis to that of a type-C retrovirus. Cell 63:77–86. doi: 10.1016/0092-8674(90)90289-Q. [DOI] [PubMed] [Google Scholar]
- 2.Sfakianos JN, Hunter E. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671–680. doi: 10.1034/j.1600-0854.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- 3.Schur FKM, Hagen WJH, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JAG. 2015. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature 517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 4.Proksova PG, Lipov J, Zelenka J, Hunter E, Langerova H, Rumlova M, Ruml T. 2018. Mason-Pfizer monkey virus envelope glycoprotein cycling and its vesicular co-transport with immature particles. Viruses 10:575. doi: 10.3390/v10100575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira LE, Clark J, Grznarova P, Wen X, LaCasse R, Ruml T, Spearman P, Hunter E. 2014. Direct evidence for intracellular anterograde co-transport of M-PMV Gag and Env on microtubules. Virology 449:109–119. doi: 10.1016/j.virol.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song C, Dubay SR, Hunter E. 2003. A tyrosine motif in the cytoplasmic domain of Mason-Pfizer monkey virus is essential for the incorporation of glycoprotein into virions. J Virol 77:5192–5200. doi: 10.1128/jvi.77.9.5192-5200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosson P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J 15:5783–5788. doi: 10.1002/j.1460-2075.1996.tb00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vincent MJ, Melsen LR, Martin AS, Compans RW. 1999. Intracellular interaction of simian immunodeficiency virus Gag and Env proteins. J Virol 73:8138–8144. doi: 10.1128/JVI.73.10.8138-8144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willey RL, Bonifacino JS, Potts BJ, Martin MA, Klausner RD. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci U S A 85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castillo-Menendez LR, Witt K, Espy N, Princiotto A, Madani N, Pacheco B, Finzi A, Sodroski J. 2018. Comparison of uncleaved and mature human immunodeficiency virus membrane envelope glycoprotein trimers. J Virol 92:e00277-18. doi: 10.1128/JVI.00277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. 2005. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J Virol 79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol 67:2747–2755. doi: 10.1128/JVI.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschman J, Qi M, Ding L, Hammonds J, Dienger-Stambaugh K, Wang JJ, Lapierre LA, Goldenring JR, Spearman P. 2017. HIV-1 envelope glycoprotein trafficking through the endosomal recycling compartment is required for particle incorporation. J Virol 92:e01893-17. doi: 10.1128/JVI.01893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci U S A 93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol 244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 16.Murakami T, Freed EO. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol 74:3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freed EO, Martin MA. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol 69:1984–1989. doi: 10.1128/JVI.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger HG. 1995. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol 69:3824–3830. doi: 10.1128/JVI.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tedbury PR, Ablan SD, Freed EO. 2013. Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure. PLoS Pathog 9:e1003739. doi: 10.1371/journal.ppat.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tedbury PR, Novikova M, Ablan SD, Freed EO. 2016. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc Natl Acad Sci U S A 113:e182–e190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tedbury PR, Freed EO. 2015. The cytoplasmic tail of retroviral envelope glycoproteins. Prog Mol Biol Transl Sci 129:253–284. doi: 10.1016/bs.pmbts.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brody BA, Rhee SS, Sommerfelt MA, Hunter E. 1992. A viral protease-mediated cleavage of the transmembrane glycoprotein of Mason-Pfizer monkey virus can be suppressed by mutations within the matrix protein. Proc Natl Acad Sci U S A 89:3443–3447. doi: 10.1073/pnas.89.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green N, Shinnick TM, Witte O, Ponticelli A, Sutcliffe JG, Lerner RA. 1981. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci U S A 78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguilar HC, Anderson WF, Cannon PM. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R peptide. J Virol 77:1281–1291. doi: 10.1128/jvi.77.2.1281-1291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song YE, Olinger GY, Janaka SK, Johnson MC. 2019. Sequence determinants in gammaretroviral Env cytoplasmic tails dictate virus-specific pseudotyping compatibility. J Virol 93:e02172-18. doi: 10.1128/JVI.02172-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aydin H, Cook JD, Lee JE. 2014. Crystal structures of beta- and gammaretrovirus fusion proteins reveal a role for electrostatic stapling in viral entry. J Virol 88:143–153. doi: 10.1128/JVI.02023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy RE, Samal AB, Vlach J, Saad JS. 2017. Solution structure and membrane interaction of the cytoplasmic tail of HIV-1 gp41 protein. Structure 25:1708.e5–1718.e5. doi: 10.1016/j.str.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prchal J, Junkova P, Strmiskova M, Lipov J, Hynek R, Ruml T, Hrabal R. 2011. Expression and purification of myristoylated matrix protein of Mason-Pfizer monkey virus for NMR and MS measurements. Protein Expr Purif 79:122–127. doi: 10.1016/j.pep.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merkley ED, Rysavy S, Kahraman A, Hafen RP, Daggett V, Adkins JN. 2014. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci 23:747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vlach J, Srb P, Prchal J, Grocky M, Lang J, Ruml T, Hrabal R. 2009. Nonmyristoylated matrix protein from the Mason-Pfizer monkey virus forms oligomers. J Mol Biol 390:967–980. doi: 10.1016/j.jmb.2009.05.063. [DOI] [PubMed] [Google Scholar]
- 31.Morikawa Y, Zhang WH, Hockley DJ, Nermut MV, Jones IM. 1998. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J Virol 72:7659–7663. doi: 10.1128/JVI.72.9.7659-7663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci U S A 101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junkova P, Prchal J, Spiwok V, Pleskot R, Kadlec J, Krasny L, Hynek R, Hrabal R, Ruml T. 2016. Molecular aspects of the interaction between Mason-Pfizer monkey virus matrix protein and artificial phospholipid membrane. Proteins 84:1717–1727. doi: 10.1002/prot.25156. [DOI] [PubMed] [Google Scholar]
- 34.Murakami T. 2008. Roles of the interactions between Env and Gag proteins in the HIV-1 replication cycle. Microbiol Immunol 52:287–295. doi: 10.1111/j.1348-0421.2008.00008.x. [DOI] [PubMed] [Google Scholar]
- 35.Steckbeck JD, Craigo JK, Barnes CO, Montelaro RC. 2011. Highly conserved structural properties of the C-terminal tail of HIV-1 gp41 protein despite substantial sequence variation among diverse clades: implications for functions in viral replication. J Biol Chem 286:27156–27166. doi: 10.1074/jbc.M111.258855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brody BA, Rhee SS, Hunter E. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J Virol 68:4620–4627. doi: 10.1128/JVI.68.7.4620-4627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermida-Matsumoto L, Resh MD. 1999. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55(gag) and p17MA. J Virol 73:1902–1908. doi: 10.1128/JVI.73.3.1902-1908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song C, Micoli K, Bauerova H, Pichova I, Hunter E. 2005. Amino acid residues in the cytoplasmic domain of the Mason-Pfizer monkey virus glycoprotein critical for its incorporation into virions. J Virol 79:11559–11568. doi: 10.1128/JVI.79.18.11559-11568.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prchal J, Srb P, Hunter E, Ruml T, Hrabal R. 2012. The structure of myristoylated Mason-Pfizer monkey virus matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in its membrane binding. J Mol Biol 423:427–438. doi: 10.1016/j.jmb.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alfadhli A, Mack A, Ritchie C, Cylinder I, Harper L, Tedbury PR, Freed EO, Barklis E. 2016. Trimer enhancement mutation effects on HIV-1 matrix protein binding activities. J Virol 90:5657–5664. doi: 10.1128/JVI.00509-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alfadhli A, Staubus AO, Tedbury PR, Novikova M, Freed EO, Barklis E. 2019. Analysis of HIV-1 matrix-envelope cytoplasmic tail interactions. J Virol 93:e01079-19. doi: 10.1128/JVI.01079-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leavell MD, Novak P, Behrens CR, Schoeniger JS, Kruppa GH. 2004. Strategy for selective chemical cross-linking of tyrosine and lysine residues. J Am Soc Mass Spectrom 15:1604–1611. doi: 10.1016/j.jasms.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Swaim CL, Smith JB, Smith DL. 2004. Unexpected products from the reaction of the synthetic cross-linker 3,3′-dithiobis(sulfosuccinimidyl propionate), DTSSP with peptides. J Am Soc Mass Spectrom 15:736–749. doi: 10.1016/j.jasms.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 44.Kalkhof S, Sinz A. 2008. Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal Bioanal Chem 392:305–312. doi: 10.1007/s00216-008-2231-5. [DOI] [PubMed] [Google Scholar]
- 45.Madler S, Bich C, Touboul D, Zenobi R. 2009. Chemical cross-linking with NHS esters: a systematic study on amino acid reactivities. J Mass Spectrom 44:694–706. doi: 10.1002/jms.1544. [DOI] [PubMed] [Google Scholar]
- 46.Hunter E, Swanstrom R. 1990. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol 157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 47.Rawat SS, Viard M, Gallo SA, Rein A, Blumenthal R, Puri A. 2003. Modulation of entry of enveloped viruses by cholesterol and sphingolipids (review). Mol Membr Biol 20:243–254. doi: 10.1080/0968768031000104944. [DOI] [PubMed] [Google Scholar]
- 48.Goddard TD, Kneller DG. 2002. SPARKY 3. University of California, San Francisco, San Francisco, CA: https://iop.vast.ac.vn/theor/conferences/smp/1st/kaminuma/UCSFComputerGraphicsLab/index-16.htm. [Google Scholar]
- 49.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 50.Gotze M, Pettelkau J, Schaks S, Bosse K, Ihling CH, Krauth F, Fritzsche R, Kuhn U, Sinz A. 2012. StavroX–a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom 23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 51.van Zundert GCP, Rodrigues J, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, van Dijk M, de Vries SJ, Bonvin A. 2016. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol 428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Schwieters CD, Kuszewski JJ, Tjandra N, Marius Clore G. 2003. The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160:65–73. doi: 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.