Host cells use a number of factors to defend against viral infection. Viruses are, however, in an arms race with their host cells to overcome these defense mechanisms. The tumor suppressor protein p53 is an important sensor of cell stress induced by oncogenic insults or viral infections, which upon activation induces various pathways to ensure the integrity of cells. Viruses have to counteract many functions of p53, but complex DNA viruses such as cytomegaloviruses may also utilize some p53 functions for their own benefit. In this study, we discovered that the M25 proteins of mouse cytomegalovirus interact with p53 and mediate its accumulation during infection. Interaction with the M25 proteins sequesters p53 molecules in nuclear dot-like structures, limiting their availability for activation of a subset of p53-regulated target genes. Understanding the interaction between viral proteins and p53 may allow to develop new therapeutic strategies against cytomegalovirus and other viruses.
KEYWORDS: M25 proteins, cytomegalovirus, p53, virus host cell interaction
ABSTRACT
To ensure productive infection, herpesviruses utilize tegument proteins and nonstructural regulatory proteins to counteract cellular defense mechanisms and to reprogram cellular pathways. The M25 proteins of mouse cytomegalovirus (MCMV) belong to the betaherpesvirus UL25 gene family that encodes viral proteins implicated with regulatory functions. Through affinity purification and mass spectrometric analysis, we discovered the tumor suppressor protein p53 as a host factor interacting with the M25 proteins. M25-p53 interaction in infected and transfected cells was confirmed by coimmunoprecipitation. Moreover, the proteins colocalized in nuclear dot-like structures upon both infection and inducible expression of the two M25 isoforms. p53 accumulated in wild-type MCMV-infected cells, while this did not occur upon infection with a mutant lacking the M25 gene. Both M25 proteins were able to mediate the effect, identifying them as the first CMV proteins responsible for p53 accumulation during infection. Interaction with M25 proteins led to substantial prolongation of the half-life of p53. In contrast to the higher abundance of the p53 protein in wild-type MCMV-infected cells, the transcript levels of the prominent p53 target genes Cdkn1a and Mdm2 were diminished compared to cells infected with the ΔM25 mutant, and this was associated with reduced binding of p53 to responsive elements within the respective promoters. Notably, the productivity of the M25 deletion mutant was partially rescued on p53-negative fibroblasts. We propose that the MCMV M25 proteins sequester p53 molecules in the nucleus of infected cells, reducing their availability for activating a subset of p53-regulated genes, thereby dampening the antiviral role of p53.
IMPORTANCE Host cells use a number of factors to defend against viral infection. Viruses are, however, in an arms race with their host cells to overcome these defense mechanisms. The tumor suppressor protein p53 is an important sensor of cell stress induced by oncogenic insults or viral infections, which upon activation induces various pathways to ensure the integrity of cells. Viruses have to counteract many functions of p53, but complex DNA viruses such as cytomegaloviruses may also utilize some p53 functions for their own benefit. In this study, we discovered that the M25 proteins of mouse cytomegalovirus interact with p53 and mediate its accumulation during infection. Interaction with the M25 proteins sequesters p53 molecules in nuclear dot-like structures, limiting their availability for activation of a subset of p53-regulated target genes. Understanding the interaction between viral proteins and p53 may allow to develop new therapeutic strategies against cytomegalovirus and other viruses.
INTRODUCTION
Cytomegaloviruses (CMV) colonize various mammals and underwent millions of years of coevolution with their respective host species. Besides the prototypic human cytomegalovirus (HCMV), which is a highly prevalent human pathogen that can lead to serious disease, particularly in immunocompromised patients and in congenitally infected children, the CMV species infecting mice, rats, guinea pigs, and rhesus macaques have become useful models to study CMV pathogenesis (1–4). The different CMVs are so well adapted that they replicate exclusively in their respective host. For successful replication viruses have to overcome various host defense mechanisms and stress responses acting at the cellular and organismal level. CMVs have developed impressively large genomes and devote a substantial part of their genomic information to gene products counteracting these defense mechanisms. Some viral proteins with such functions are incorporated into the virus particles, typically within the tegument protein layer of virions located between the nucleocapsid and the viral lipid membrane, and can therefore be delivered into cells immediately at the onset of the infection process, allowing to disarm intrinsic cellular defense pathways early on (5). Other regulatory viral proteins are expressed during the immediate early and early infection phases, thereby securing the successful productive infection cycle. Although CMVs target essentially the same cellular pathways, they often evolved different gene products that perform this task, providing us with useful tools to dissect and investigate host defense mechanisms.
The injection of viral DNA genomes into the cell nucleus at the onset of infection and particularly their amplification during viral DNA replication remains not undetected by the cell and induces a genotoxic stress response. This and several other stresses associated with viral infection, such as metabolic stress and RNA stress, lead almost inevitably to activation of the tumor suppressor protein p53. Investigation of p53 over more than 40 years revealed its central role in preventing oncogenesis; notably, however, this protein was originally identified as a binding partner of the regulatory large T protein of the simian virus 40 polyomavirus (reviewed in reference 6), and it has been speculated that p53 evolved to block invading viruses (7–9). p53 acts primarily as a transcription factor, which upon activation regulates the expression of a plethora of cellular genes (10). Some of the immediate consequences of p53 activation are cell cycle arrest at the G1/S-phase transition and induction of DNA repair mechanisms to restore the integrity of the cellular genome. Upon prolonged DNA damage p53 initiates apoptosis. Cell death is detrimental for the propagation of viruses, thus forcing them to interfere with this process. Moreover, many DNA viruses influence the host cell cycle in order to create conditions favorable for viral DNA replication (11), often requiring interference with p53-regulated checkpoint control. HCMV drives quiescent cells into G1 phase but then stops further progression at a check point between the G1 and S phases, although a series of S-phase proteins are expressed in the resulting state (reviewed in references 12 and 13). Almost all DNA viruses have developed strategies to interfere with the activation of p53. A strategy used by papillomaviruses is to promote the proteasomal degradation of p53, whereas complex DNA viruses such as herpes- and adenoviruses use more refined mechanisms to prevent the transcriptional activation of cellular pathways by p53, which are unfavorable for their replication (14, 15). Human CMV sequesters p53 in the cell nucleus, leading to its accumulation over the course of infection (16–19). Although it is likely that HCMV blocks some activities of p53, studies with p53-deficient cells suggested that the virus may also utilize p53 for its own benefit (20, 21).
The members of the UL25 gene family of betaherpesviruses encode structural components of the viral tegument, as well as nonstructural proteins (22–25), and several of them have been implicated in countering antiviral defense pathways (26–29). In previous work, we and others have characterized the two proteins encoded by the mouse CMV (MCMV) M25 gene, an abundant tegument protein and a nonstructural early protein (30–32). Interestingly, an MCMV mutant lacking the M25 gene did not lead to the typical cytopathic effect of infected cells, and rearrangement of the actin cytoskeleton as observed after wild-type (WT) MCMV infection did not occur (32). Other phenotypes of the ΔM25 mutant were growth to lower titers and diminished cell-to-cell spread. We did not detect differences between the ΔM25 mutant and WT MCMV in the kinetics of viral protein expression or replication of the viral DNA genome, pointing to a deficit of the ΔM25 mutant in release of infectious particles (32). In a first attempt to unravel the functions of the M25 proteins, we aimed to identify cellular proteins interacting with them. One of the discovered interaction partners is p53, and we show that the M25 proteins are able and sufficient to induce nuclear accumulation of p53 in infected cells and also when expressed independent of viral infection. Our data suggest that the M25 proteins sequester p53 in the nuclei of MCMV-infected cells, reducing the availability of p53 molecules to induce p53-regulated pathways, and thus modulate the consequences of p53 activation.
RESULTS
The MCMV M25 proteins interact with the host factor p53.
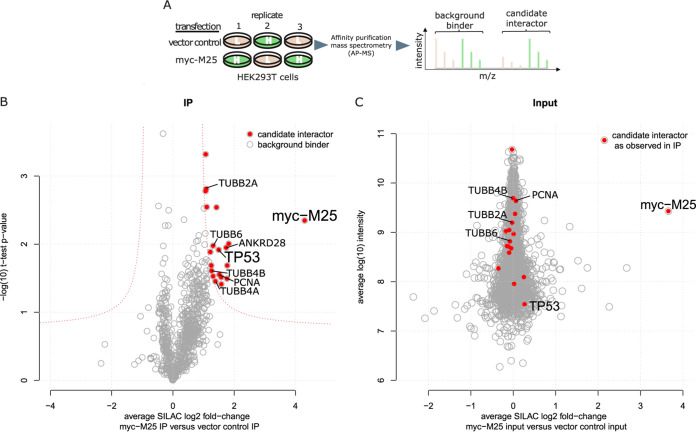
Herpesvirus tegument proteins are introduced into cells immediately upon viral entry, and many of them exert a regulatory function in modulating cellular pathways, a role they share with various nonstructural viral proteins expressed early in infection (1, 5). The MCMV M25 gene encodes both a tegument protein of 130 kDa (here called pM25l) and a nonstructural isoform of 105 kDa (pM25s) (30–32). To check which cellular pathways are affected by the M25 proteins, we sought to identify interacting host factors via quantitative affinity purification-mass spectrometry (AP-MS). The principle is based on comparing the abundance of proteins copurifying with M25 to an internal control.
To this end, we transfected SILAC (stable isotope labeling of amino acids in cell culture) light- and heavy-isotope-labeled HEK293T cells with an expression plasmid coding for a myc-tagged version of the M25l protein or with an empty plasmid vector. HEK293T cells were chosen because they can be transfected with high efficiency and the SILAC technique for these cells was already established in our laboratory. Three independent experiments, including a label-swap, were performed. At 48 h posttransfection, pM25l and interacting proteins were enriched by anti-myc affinity purification, digested, and analyzed by liquid chromatography coupled to mass spectrometry (Fig. 1A). After proteomic analysis of all three samples, we depicted the average SILAC fold change for all quantified proteins over their t test P values in a volcano plot (Fig. 1B). We accepted 19 proteins as candidate interactors based on a combination of both values (see Table S1 in the supplemental material). Importantly, the abundance of these candidates in the lysate of HEK293T cells was not affected by transfection of the M25 plasmid (Fig. 1C). This indicated that their enrichment is not due to changes in abundance in the input but instead due to specific coprecipitation with pM251. The list of interactors included components of the cytoskeleton (TUBB2A, TUBB4A/B, and TUBB6 [beta-2A, -4A, -4B, and -6 chains of the tubulin family]), a regulatory subunit of protein phosphatase 6 (ANKRD28 [ankyrin repeat domain-containing protein 28]), and PCNA (proliferating cell nuclear antigen), a component of the cellular DNA replication machinery, as well as the tumor suppressor protein p53.
FIG 1.
Identification of cellular proteins interacting with myc-tagged pM25l using quantitative AP-MS. (A) Workflow. (Left) SILAC light (L)- and heavy (H)-labeled HEK293T cells were transfected in triplicates (n = 3) with either constructs encoding pM25l (myc-M25) or vector control. (Middle) Bait and prey proteins were enriched by affinity purification before being subjected to LC-MS/MS. (Right) Schematic depiction of a MS1 spectrum. Proteins from control and IP samples were discriminated based on the mass shift of the corresponding SILAC-labeled peptides (L, beige; H, green). Differences in signal intensities between differentially SILAC-labeled peptides were used to calculate enrichment ratios. (B) Candidate interactors were discriminated from background binders based on a combination of the effect size (the average SILAC log2-fold change of all replicates) and statistical significance (–log10 [t test P value] of the replicate IPs). Candidate interactors and the bait are highlighted red. (C) Proteomic assessment of input samples indicates that candidate interactors (red) are equally abundant in both transfection conditions. Protein abundance (log10 intensity) is plotted over SILAC fold changes.
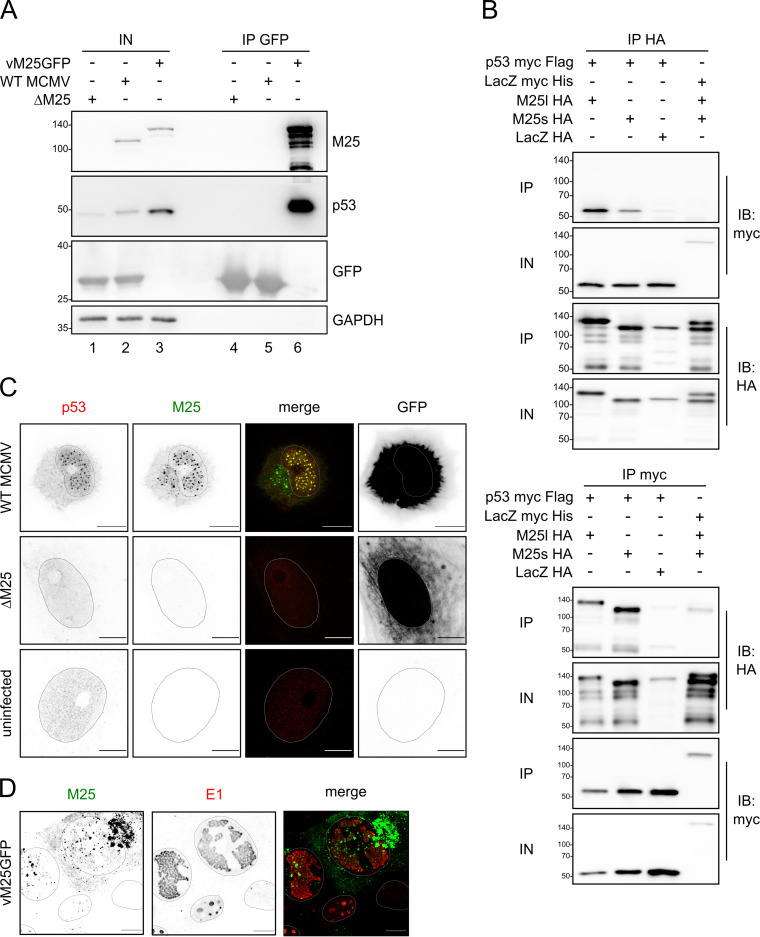
We chose to focus on p53 because it is an important sensor of cellular stress and a known antiviral factor. Moreover, previous work by other groups revealed that CMV modulates p53 abundance during infection (16–19), and yet the viral proteins involved in this process remained unknown. First, we verified the interaction between M25 and p53 in MCMV-infected murine NIH 3T3 cells, not least because the initial findings were obtained for human cells, and used a tagged virus (vM25GFP) expressing M25 protein versions with a C-terminal fusion of green fluorescent protein (GFP) for this purpose. Pulldown of the M25-GFP fusion proteins from lysates of infected cells by use of GFP nanobodies led to the coprecipitation of p53 (Fig. 2A, second panel, lane 6). No p53 was detectable when lysates of cells infected with WT MCMV, expressing the untagged, native M25 proteins, or the mutant ΔM25 lacking the M25 open reading frame (ORF) were used, despite strong GFP expression by these viruses and GFP enrichment upon pulldown with GFP nanobodies (Fig. 2A, third row, lanes 4 and 5). This indicated that binding of p53 is mediated by the M25 moiety of the M25-GFP fusion proteins.
FIG 2.
M25 proteins interact with p53. (A) NIH 3T3 cells were infected with the indicated viruses (at MOI 1) and harvested 36 h p.i. GFP and GFP-tagged M25 proteins were precipitated with GFP nanobodies, and immunoblotting was performed with antibodies for the specified proteins. (B) p53-deficient HCT116 cells were transfected with plasmids coding for the indicated proteins, and immunoprecipitation was performed with an HA (top panel) or a myc antibody (bottom panel) using cell lysates prepared 24 h posttransfection. Antibodies used for immunoblotting are indicated in the right margin. IP, precipitated proteins; IN, input lysates. (C) MEF were infected with WT MCMV or the ΔM25 mutant at an MOI of 1 or were left uninfected, and at 36 h p.i. the cells were labeled with the indicated antibodies and analyzed by confocal microscopy. Images to the left are depicted in inverse mode, with positive staining appearing in black and gray. Infected cells were identified based on GFP expression (right panels). Please note that the apparently different GFP distribution is due to the different effects of WT MCMV and the ΔM25 mutant on cell morphology. While WT MCMV-infected cells become spherical, leaving a small cytoplasmic rim around the cell nucleus, ΔM25-infected cells retain an elongated shape. (D) NIH 3T3 cells were infected with the tagged virus vM25GFP for 24 h, followed by immunolabeling with an antibody against the E1 protein. The M25-GFP fusion protein was detected via GFP fluorescence. Shapes of the nuclei were determined based on Hoechst staining. Images are representative of at least 20 cells analyzed per condition. Scale bars, 10 μm.
Next, we tested the interaction with p53 upon expression of the M25 proteins in the absence of other viral proteins. p53-deficient HCT116 cells were transfected with plasmids coding for pM25l or pM25s tagged with a hemagglutinin (HA) epitope and with an expression vector encoding myc-tagged murine p53 (Fig. 2B). Control plasmids encoding myc-tagged LacZ and HA-tagged LacZ protein were included. These cells were chosen because they could easily be transfected and lacked p53 and the candidate proteins were efficiently expressed upon transfection. After immunoprecipitation (IP) with an HA antibody, the HA-tagged M25 proteins and the HA-tagged LacZ were enriched (Fig. 2B, top, third panel), and p53 coprecipitated with the M25 proteins (Fig. 2B top, lanes 1 and 2), but not with the HA-tagged LacZ protein, and there was also no coprecipitation of the M25 proteins with myc-tagged LacZ. Upon reciprocal IP with a myc antibody, p53 and the myc-tagged LacZ were enriched, and both pM25l and pM25s were found to coprecipitate with p53 (Fig. 2B, bottom, first panel), whereas there was only a weak signal for the control protein or when LacZ was used as bait. Taken together, these data indicate that the M25 proteins bind to p53 both in both infected and transfected cells.
The observed interaction prompted us to examine the localization of M25 and p53 in infected NIH 3T3 cells and for comparison also in a primary cell type, murine embryonic fibroblasts (MEF). The findings were identical for these cells and are reported here for MEF (Fig. 2C). In uninfected cells p53 was present at low level, sometimes near to the detection limit of the immunofluorescence microscopy technique (IF) (Fig. 2C, third row). In WT MCMV-infected cells, p53 was enriched in dot-like structures in the nucleus, colocalizing with the M25 proteins. The p53 pattern becomes particularly detectable during the late phase of the infection cycle as shown in Fig. 2C (top row) for cells 36 h postinfection (p.i.). In agreement with our previous results (32), at this time point the M25 proteins were also present in a structure in the cytoplasm adjacent to the nucleus, representing the cytoplasmic virus assembly compartment, where secondary envelopment of the viral capsids takes place (33, 34). Notably, no p53 association with this M25-containing structure in infected cells was observed (Fig. 2C, top row). In cells infected with the ΔM25 deletion mutant, a comparable change of the p53 distribution as in WT MCMV-infected cells was not detectable (Fig. 2C, second row). Next, we analyzed the distribution of the M25-containing dots in the cell nucleus. NIH 3T3 cells were infected with the tagged virus vM25GFP and labeled with antibodies against the early 1 (E1) proteins encoded by the MCMV ORF M112/113. The E1 proteins are known to localize to prereplication centers and later in infection to replication centers (35). The image in Fig. 2D depicts cells in different phases of the infection cycle. The cell on the upper right side, for instance, is in the late infection phase characterized by large E1-positive replication centers. The M25 dots in the nucleus were located not only within the replication centers but also outside them. There was also a faint M25 labeling, which overlapped with the E1 signals (Fig. 2D, compare first and second images). The cell at the bottom of the image is in an early infection stage, with E1 labeling of structures that probably represent prereplication centers. In this cell, the M25 dots were small, and little overlap between the M25-GFP and E1 signals was detected. Accordingly, the M25 proteins do not primarily localize to prereplication centers, and late in infection M25 dots are distributed throughout the nucleus, including the replication centers. Moreover, a subpopulation of the M25 proteins (faint signal in the nucleus) colocalizes with the E1 proteins within the replication centers.
Accumulation of p53 in WT MCMV-infected cells but not in cells infected with M25 deletion mutants.
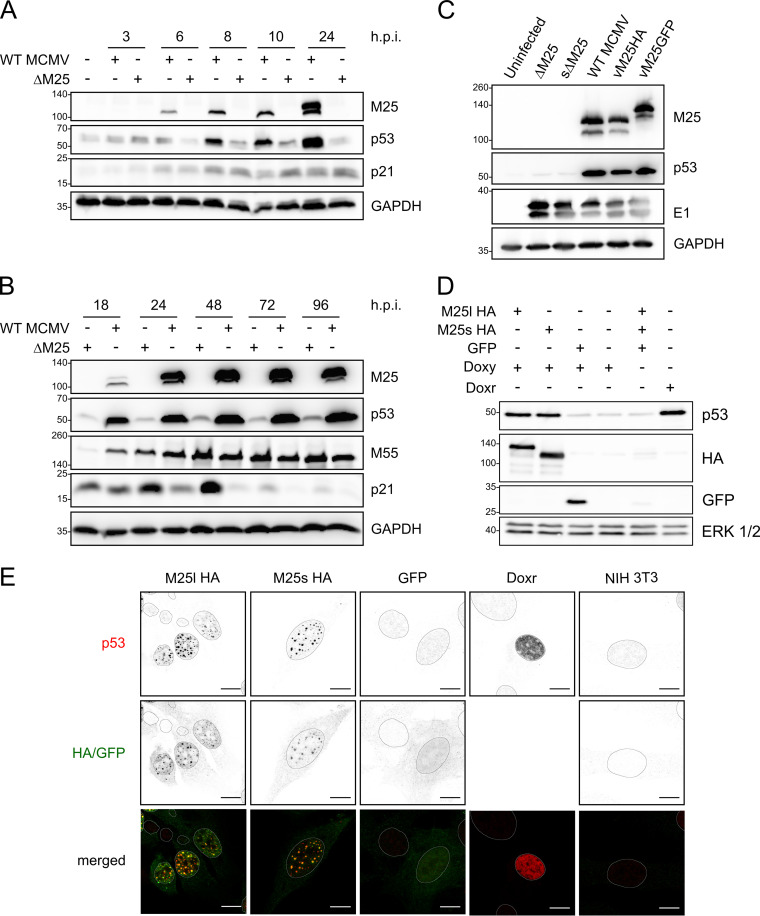
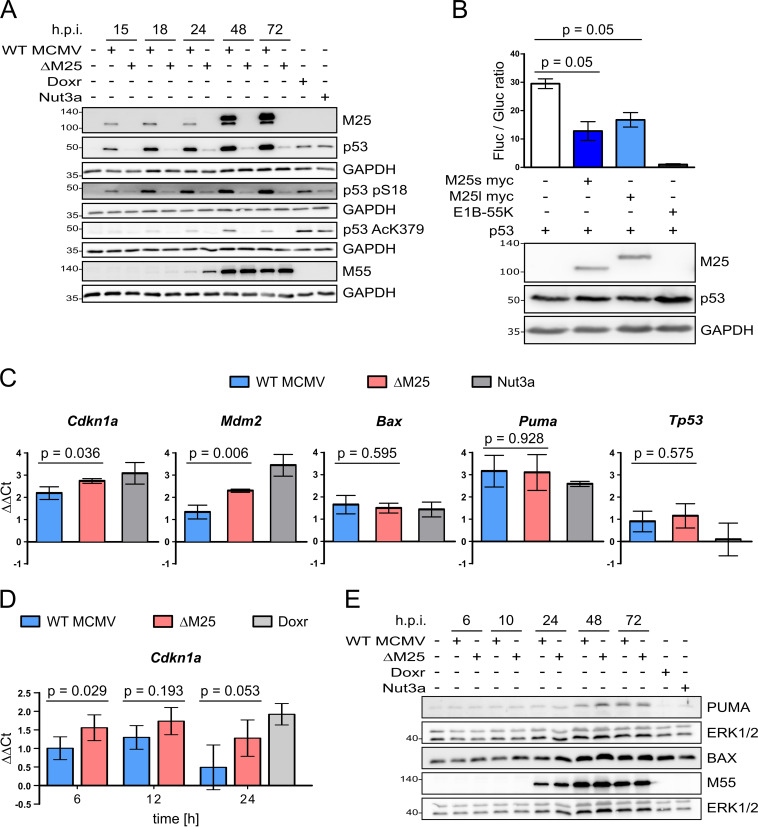
The presence of p53 in dot-like structures in the nuclei of MCMV-infected cells raised the question whether this resulted from altered p53 expression. Therefore, the p53 levels were assessed in WT MCMV- or ΔM25-infected NIH 3T3 cells at different time points of the lytic infection cycle. As early as 8 h p.i., higher levels of p53 were detected in WT MCMV-infected cells compared to ΔM25-infected cells (Fig. 3A, second row), and there was a further substantial increase of p53 levels in the late phase of infection (24 to 48 h) (Fig. 3B, second row). Notably, the p53 accumulation did go in parallel with an increase in M25 protein amounts. In cells infected with the M25 deletion mutant p53 levels were slightly higher than in uninfected cells (Fig. 3A, second row, first and each subsequent second lane), and yet—in marked contrast to WT MCMV-infected cells—a comparable further increase of p53 amounts was not observed at late time points of infection (Fig. 3B). The described effects occurred consistently in NIH 3T3 cells and primary MEF and for different M25-expressing viruses and M25 deletion mutants (Fig. 3C). To test whether the larger amounts go along with increased transcriptional activity of p53, we examined the expression of the cyclin-dependent kinase inhibitor 1 (encoded by the Cdkn1a gene and here called p21 protein), an important protein induced after p53 activation, which is involved in cell cycle regulation (36). Already early in infection p21 expression was rising compared to uninfected cells, although initially there was no difference between WT MCMV- and ΔM25-infected cells (Fig. 3A). Subsequently, particularly visible at 24 and 48 h, p21 amounts were further enhanced in ΔM25-infected cells, whereas this was not the case in WT MCMV-infected cells (Fig. 3B). This occurred in marked contrast to the smaller amounts of p53 in ΔM25-infected cells. At very late times p21 levels were diminished in cells infected with either virus, an effect which has been similarly described for HCMV-infected cells (37). Nevertheless, p21 levels remained slightly higher in ΔM25 than in WT MCMV infected cells. Thus, in contrast to the expectation, the increased amounts of p53 were not associated with a higher expression of p21, one of the prominent p53 target genes, suggesting that the M25 proteins may negatively regulate transcriptional activation by p53.
FIG 3.
p53 accumulates in WT MCMV-infected cells and in M25-expressing cell lines. (A and B) NIH 3T3 cells were infected with WT MCMV or the ΔM25 mutant (MOI of 3) for the indicated time periods, and lysates were analyzed by immunoblotting with the indicated antibodies. GAPDH served as a loading control, and the viral M25 and M55 proteins were used as infection marker. (C) Using the indicated viruses NIH 3T3 cells were infected as in panels A and B, and cell lysates prepared at 36 h p.i. were analyzed by immunoblotting with antibodies specific for the indicated proteins. The viral E1 protein served as an infection marker. (D) NIH 3T3-derived cell lines encoding pM25l-HA, pM25s-HA, or GFP and parental NIH 3T3 cells were treated with doxycycline (Doxy) for 24 h or left untreated as indicated. Doxorubicin (Doxr) treatment of NIH 3T3 cells (for 24 h) was used for inducing genotoxic stress (positive control). Cell lysates were analyzed by immunoblotting using the indicated antibodies. The HA antibody detected the M25 proteins, and ERK1/2 (extracellular signal-regulated kinase 1/2) served as a loading control. (E) The cell lines expressing the proteins indicated at the top and NIH 3T3 cells (panels at the right) were treated with doxycycline for 24 h, followed by immunolabeling with the indicated antibodies. GFP was detected using its autofluorescence, and doxorubicin (Doxr) treatment of NIH 3T3 cells was used as a positive control to induce p53 expression (second last panels on the right). Scale bars, 10 μm.
The M25 proteins are sufficient for accumulation of p53.
To substantiate the results, we made use of murine fibroblast cell lines that express the M25 proteins in a doxycycline-inducible manner. In brief, NIH 3T3 cells were transduced with lentiviral vectors encoding HA-tagged M25l or M25s isoforms or GFP, a reverse tetracycline-controlled transactivator (rtTA) and a puromycin selection marker. Transduced cells were selected by propagation in the presence of puromycin. After doxycycline treatment for 24 h, the M25l and M25s proteins, as well as GFP, were present in substantial amounts in cells transduced with the respective vectors, whereas this was not the case in the absence of doxycycline treatment (Fig. 3D, second and third rows). Most importantly, p53 levels were increased in cells expressing the M25 proteins, almost as strong as in parental NIH 3T3 cells treated with doxorubicin for 24 h (Fig. 3D, first row). Doxorubicin induces genotoxic stress, which leads to activation and stabilization of p53. IF experiments with the cell lines induced with doxycycline revealed the presence of p53 in dot-like structures in the nuclei of those lines that express the M25 proteins, and p53 colocalized with the M25 proteins (Fig. 3E, panels in first and second columns). A similar effect was neither detected in the GFP-expressing cell line nor detected in parental NIH 3T3 cells (Fig. 3E, third and fifth columns). Notably, in NIH 3T3 cells treated with doxorubicin (for control) the distribution of p53 differed in that it was dispersed throughout the nucleus (Fig. 3E, fourth column). These experiments indicated that both the M25l and the M25s protein alone are able to mediate p53 accumulation and p53 enrichment in dot-like structures, indicating that no other viral proteins are required for the effects seen in WT MCMV-infected cells.
The M25 proteins prolong the half-life of p53.
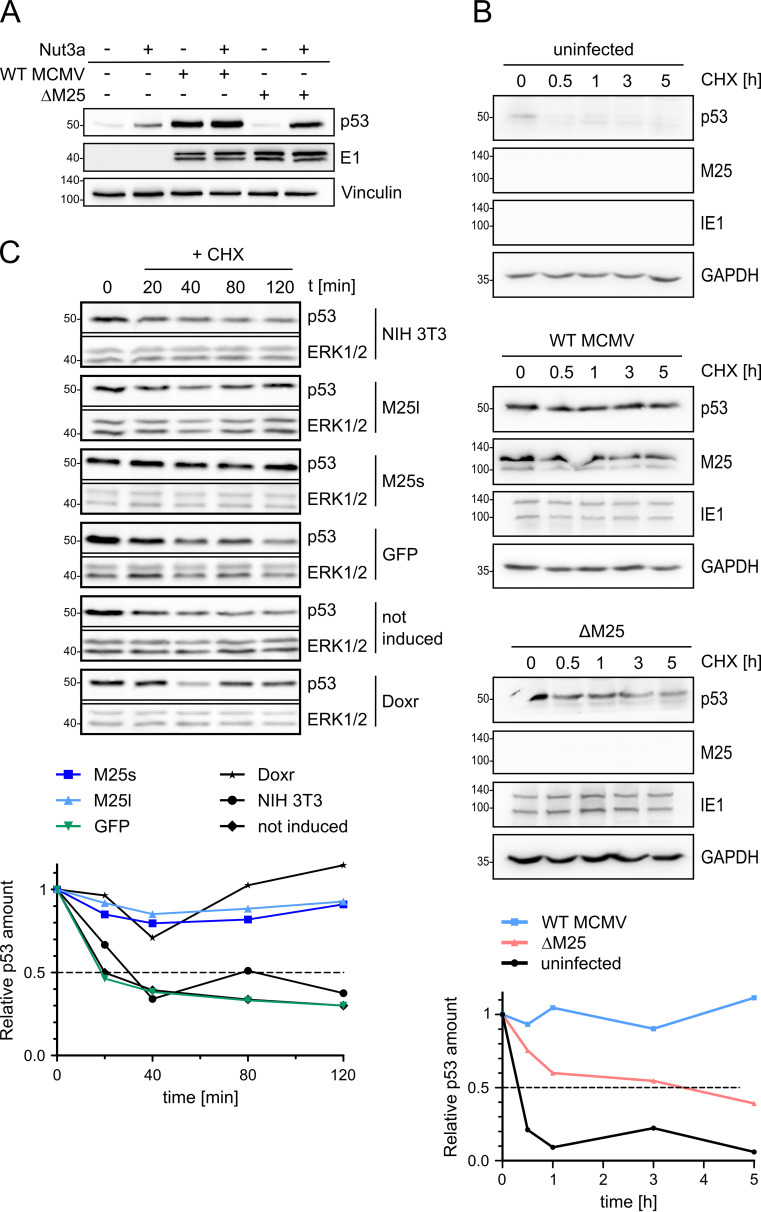
We presumed that the p53 accumulation in WT MCMV-infected cells might be due to interference with the p53 degradation pathway, in line with the mechanism described for HCMV-infected cells (38, 39). When cells were treated with Nutlin3a, an inhibitor disrupting the interaction of p53 with its negative regulator MDM2 (mouse double minute 2 protein) that normally marks p53 molecules for proteasomal degradation (40, 41), the p53 amounts in WT MCMV increased only slightly, whereas a considerable p53 increase was observed in ΔM25-infected cells, and to a lesser extent in uninfected cells (Fig. 4A). This result strongly suggested that the interaction with the M25 proteins leads to diminished turnover of p53 in infected cells. To further address this point, the half-life of p53 in uninfected cells and cells infected with WT MCMV or the M25 deletion mutant was compared (Fig. 4B). Cells at 24 h p.i., a time point when a substantial amount of p53 has accumulated in WT MCMV-infected cells, were treated with cycloheximide to stop further protein synthesis, and the decline of p53 amounts was inspected over time by Western blotting. In uninfected cells most p53 was gone after 30 min (Fig. 4B, panels at the top, first row and graph at the bottom), in line with the short half-life of 20 to 30 min reported by others (39, 42). In WT MCMV-infected cells, the p53 level at 24 h p.i. was much higher than in uninfected cells, as expected (Fig. 4B, panels in the middle, first row and graph), and the p53 amount stayed high for up to 5 h with little decline, indicating that the p53 molecules had become highly stable. Following infection with the ΔM25 mutant the decay of p53 was somewhat slower than in uninfected cells (Fig. 4B, panels at the bottom, first row and graph), which is consistent with p53 activation in ΔM25-infected cells, probably due to stress associated with infection (43). Altogether, this experiment supports the notion that interaction of p53 with the M25 proteins strongly increases its half-life.
FIG 4.
The half-life of p53 is increased in WT MCMV-infected cells and in M25-expressing cells. (A) NIH 3T3 cells were used either uninfected or were infected with WT MCMV or ΔM25 MCMV. At 18 h p.i., the cells were cultivated for another 6 h or treated with Nutlin3a (40 μM), followed by immunoblot analysis of cell lysates with the indicated antibodies. Vinculin served as a loading control, and the viral E1 early protein served as an infection marker. (B) NIH 3T3 cells either uninfected or infected as described previously (see panel A) for 24 h were then treated with cycloheximide (CHX; t = 0 h). At the specified time points thereafter, lysates of cells were prepared and analyzed by immunoblotting with the indicated antibodies. GAPDH served as a loading control, and the viral immediate early protein IE1 was used as an infection marker. (C) Parental NIH 3T3 cells and cell lines encoding the indicated proteins were treated with doxycycline for 24 h, followed by treatment with CHX for the indicated time periods (min) and analysis of p53 amounts by immunoblotting. ERK1/2 served as a loading control. The noninduced sample remained untreated (without doxycycline) and consisted of a mixture of cells transduced with M25l-, M25s-, or GFP-encoding lentiviral vectors (one third each). Doxorubicin (Doxr) treatment of NIH 3T3 cells was used for activation of p53 (bottom row). (A to C) The blots are representative of 3 independently performed experiments. (B and C) Signals were quantified by densiometric analysis using ImageJ and normalized to the respective loading control. p53 levels are depicted in relation to the amounts present at the time point when CHX was added (time point 0), and values represent the means of values from three independent experiments.
Next, we performed similar experiments with the cell lines expressing the M25l or M25s proteins. At 24 h after inducing the expression of the M25 proteins, cycloheximide was added, followed by measurement of the p53 levels. In the parental NIH 3T3 cells, the GFP-expressing cell line, and the cells not treated with doxycycline (not induced), half of the p53 amounts have disappeared after ∼30 min (Fig. 4C). In contrast, the p53 levels in the cell lines expressing pM25l or pM25s remained almost unchanged over the 2-h period analyzed (Fig. 4C and graph). Moreover, the p53 molecules displayed stability similar as in cells that experienced genotoxic stress resulting from doxorubicin treatment. Thus, both the pM25l and the pM25s protein are able to induce p53 stabilization, altering its usual rapid turnover.
M25 proteins limit transcriptional activation by p53.
Posttranslational modification such as phosphorylation and acetylation lead to the stabilization of p53 and activation of its transcriptional activity (44, 45). Thus, we examined phosphorylation of mouse p53 at serine 18 (equivalent to serine 15 of human p53) and acetylation at lysine 379 (equivalent to lysine 382 of human p53), modifications indicative for these processes (46, 47). Concomitant with the increase of total p53 in WT MCMV-infected cells, we detected serine 18-phosphorylated p53 and also lysine 379-acetylated p53 (Fig. 5A). In ΔM25-infected cells serine 18-phosphorylated p53 was present in much smaller amounts and lysine 379-acetylated p53 could be hardly detected, though this was in the presence of considerably less total p53 than in WT MCMV-infected cells (Fig. 5A, second row). These data showed that the interaction with M25 proteins did not prevent the respective posttranslational modifications of p53.
FIG 5.
Influence of M25 proteins on posttranslational modification of p53 and its transcriptional activity. (A) NIH 3T3 cells were either left uninfected or were infected with WT MCMV or the ΔM25 mutant (MOI of 3) or were treated with doxorubicin (Doxr; 0.5 μM for 24 h) or with Nutlin3a (Nut3a; 40 μM for 6 h). Cell lysates prepared at the indicated time points postinfection were analyzed by immunoblotting with antibodies for the specified proteins and the modified p53 versions. (B) p53-deficient HCT116 cells were transfected with the luciferase reporter plasmid PG13-luc, a Gaussia luciferase control plasmid and expression plasmids encoding p53, pM25s, pM25l, or the adenovirus E1B-55K protein as indicated. The luciferase activities were measured 48 h after transfection. The graph shows luciferase activities ± the standard deviations (SD) in cell lysates (ratios of firefly and Gaussia luciferase activity) of three biological replicates. A Mann-Whitney test (one-tailed) was used to analyze statistical significance. (C) Total RNA was isolated from NIH 3T3 cells infected with WT MCMV or ΔM25 (MOI of 3) for 36 h and, for comparison, from uninfected cells (either untreated or treated with Nutlin3a [5 μM]) and was subjected to qRT PCR analysis using gene-specific primers. (D) Total RNA was isolated from primary MEF infected with either WT MCMV or ΔM25 (MOI of 3) for the depicted duration and from cells that remained uninfected or were treated with doxorubicin (Doxr) for 24 h. qRT PCR was performed using Cdkn1a-specific primers. (C and D) The graphs depict the ΔΔCT values of the qRT-PCRs (log2-transformed values of expression levels in relation to the ones of uninfected cells). The means ± the SD of three to five biological replicates are shown. A t test (two-tailed) was used for statistical analysis. (E) NIH 3T3 cells were infected or treated as indicated in panel A, and cell lysates prepared at the indicated time points postinfection were analyzed by immunoblotting with antibodies specific for PUMA or BAX.
To check whether the M25 proteins influence transactivation by p53, we transfected cells with a reporter plasmid carrying a promoter with p53 response elements and performed luciferase assays. In comparison to the samples that were cotransfected with a p53 expression plasmid, the addition of plasmids coding for the pM25s or pM25l isoforms led to a reduction of the luciferase activity of about 50% (Fig. 5B) but did not completely block this p53 function, as observed for the adenoviral protein E1B-55K, a known p53 inhibitor (48, 49).
Next, we validated this finding by comparing the expression of selected p53 target genes—Cdkn1a (encoding p21), Mdm2, Bax (Bcl-2-associated X protein, an apoptosis regulator), Puma (p53 upregulated modulator of apoptosis), and Tp53 (cellular tumor antigen p53) itself—in WT MCMV- and ΔM25-infected cells at 36 h p.i. To this end, the mRNA levels of these genes were determined by quantitative reverse transcription-PCR (qRT-PCR) (Fig. 5C). The RNA of cells treated with Nutlin3a that leads to p53 accumulation and activation was included as a positive control. The mRNA amounts for the p53 target genes Cdkn1a and Mdm2 were significantly higher (∼1.5- to ∼2-fold) in ΔM25-infected cells than in WT MCMV-infected cells. No significant difference was observed for Bax, Puma, and Tp53 between cells infected with either of the two viruses. In a second experimental setup, the Cdkn1a mRNA levels were measured in primary MEF and, consistent with the first data set, ∼1.5-fold more Cdkn1a transcripts were detected in ΔM25-infected cells in comparison to WT MCMV-infected cells (Fig. 5D). We also examined the expression of Puma and Bax at the protein level over the course of infection with the two viruses (Fig. 5E). The amounts of PUMA and BAX slightly increased during the infection cycle, especially at late time points; however, there was no marked difference between cells infected with WT MCMV or the ΔM25 mutant, in line with the qPCR data for these genes. We concluded that the interaction between M25 and p53 limits the activation of a subset of the p53 target genes.
The M25 proteins affect binding of p53 to target sequences in infected cells.
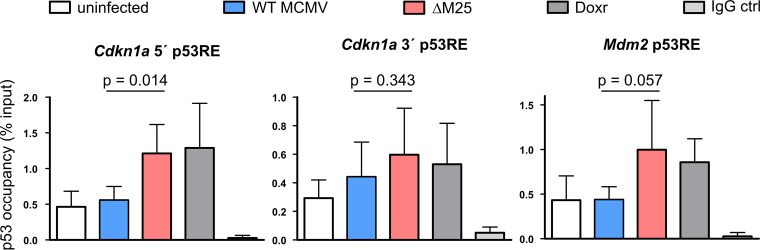
To mediate transcriptional activation, p53 binds to defined sequences within the promoters of target genes (50). Therefore, the observed effect on the Cdkn1a and Mdm2 transcript levels could result from an influence of the M25 proteins on p53 binding to such sites. To test this hypothesis, we compared the amounts of p53 associated with these promoter sequences of uninfected, WT MCMV-infected, and ΔM25-infected cells by performing chromatin immunoprecipitation (ChIP) with a p53 antibody or with control IgG. As a positive control we used cells treated by doxorubicin that results in p53 activation and binding to target sites. After infection with WT MCMV, the amount of p53 associated with the 5′-located p53 responsive element (RE) within the Cdkn1a promoter, a site known for relative strong binding of p53, was hardly different from the one in uninfected cells (Fig. 6). In ΔM25-infected cells, we observed, however, an increase in the association of p53 with this p53 RE, to a similar extent as in doxorubicin-treated cells. A trend toward higher p53 occupancy in ΔM25-infected cells was also observed for the 3′-located p53 RE of the Cdkn1a promoter, although the binding strength of this site to p53 is less pronounced, leading to overall less p53 binding. For the p53 RE in the Mdm2 promoter a 2-fold higher occupancy with p53 was observed in ΔM25-infected and doxorubicin-treated cells, whereas in WT MCMV-infected cells the p53 occupancy of this RE remained at the same level as in uninfected cells. Thus, in the presence of M25 proteins, binding of p53 to the analyzed promoter sequences was diminished, despite the overall higher p53 abundance in WT MCMV-infected cells.
FIG 6.
Effect of M25 proteins on p53 association with promoter sequences. NIH 3T3 were either used uninfected or were infected with WT MCMV or the ΔM25 mutant (MOI of 3) or were treated with doxorubicin (Doxr) for 18 h. Chromatin immunoprecipitation (ChIP) was performed using a p53 antibody or IgG for control, and the amounts of precipitated and input DNA were quantified by qPCR. The values represent the percentage of precipitated DNA (means plus the SD of four biological replicates). A t test (one-tailed) was used for statistical analysis.
The growth phenotype of the ΔM25 mutant is partially rescued in p53-negative cells.
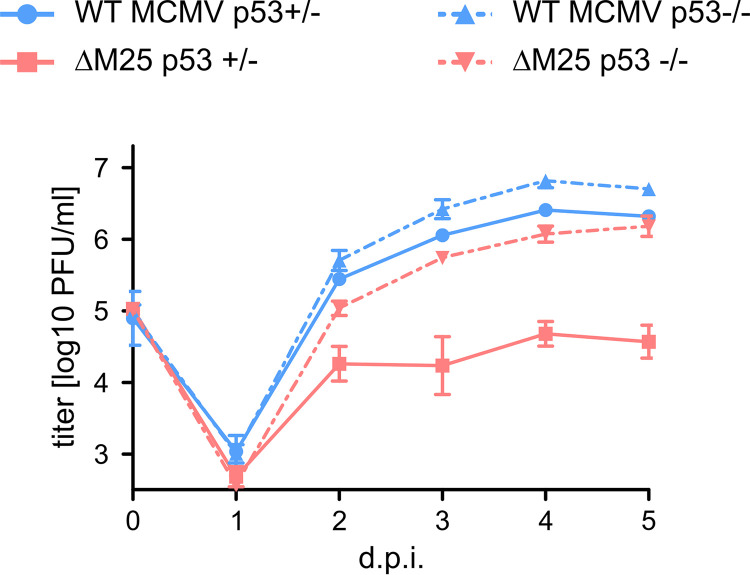
To assess the influence of p53 on the replication kinetics of WT MCMV and the ΔM25 mutant growth curve analyses were performed on p53-negative (p53–/–) and p53-positive (p53+/–) MEF (Fig. 7). WT MCMV replicated to slightly higher titers on p53-negative MEF compared to p53-positive MEF, suggesting that even the WT MCMV cannot completely counteract antiviral functions of p53. The ΔM25 mutant grew to low titers on p53-positive cells, with little increase of infectious progeny after day 2 p.i., as reported previously (32). Interestingly, the replication kinetics of the ΔM25 mutant on p53-negative MEF was similar to the one of WT MCMV on p53-positive cells, though the ΔM25 mutant did not reach the titers seen for WT MCMV on p53-negative MEF. We concluded that in the absence of M25-mediated interference with p53 the production of ΔM25 progeny is considerably impaired, while in the absence of antiviral functions of p53 this deficiency has to a large part been overcome.
FIG 7.
Growth of the ΔM25 mutant and of WT MCMV on p53-deficient MEF. p53-negative (p53–/–) and p53-positive (p53+/–) MEF were infected with the indicated viruses (MOI of 1), and titers of infectious virus in the supernatant of cultures at the indicated time points postinfection were determined by plaque assay. Average titers (means) ± the SD of triplicate cultures are depicted. The data are from one experiment representative of three independent experiments performed.
DISCUSSION
The interactome analysis detected a number of potential cellular binding partners for the MCMV M25 proteins. Since the two major protein isoforms encoded by the M25 ORF are relatively large (130 and 105 kDa [30, 32]), they are probably able to bind several proteins independently. Alternatively, the M25 proteins may be part of larger complexes, and the interaction with some of the identified proteins could be indirect. A recent comprehensive interactome study revealed a median of 9 interactions per HCMV protein, with large variation between individual viral proteins (51). Accordingly, the number of detected potential M25 interaction partners is not unusual. In this study, we focused on the tumor suppressor protein p53, while other candidate proteins are the subject of ongoing analysis.
The interaction between the M25 proteins and p53 was confirmed both during infection and following transfection of cells. The latter indicates that no other viral proteins are needed for the interaction, although it is possible that additional cellular proteins may be involved. Both the pM25l and the pM25s proteins were able to bind p53, and this finding was supported by the apparent colocalization in cell lines expressing either of the proteins. Notably, however, in infected cells no colocalization of p53 with the M25 signals at a juxtanuclear compartment in the cytoplasm was detected. Previous results suggested that mainly the pM25l protein localizes to this site to be subsequently incorporated into virions (32). Accordingly, either an additional host factor is required for the interaction with p53 in the nucleus or a putative p53 binding site is not accessible in the fraction of the pM25l molecules present at the cytoplasmic virus assembly compartment.
Further analysis revealed that p53 accumulates in MCMV-infected cells in an M25-dependent manner. In HCMV infection p53 accumulation occurs within the first 24 h p.i. (19, 52), sometimes even earlier (18), and a further increase was usually observed at later time points. In MCMV-infected cells we detected increased p53 amounts as early as 8 h p.i. when already substantial pM25s was present, and additional accumulation occurred during the late infection phase. Since MCMV and HCMV differ in their replication kinetics, with MCMV being considerably faster, it is difficult to directly compare the time courses of p53 accumulation. Also, one cannot exclude that the two viruses use different mechanisms to interfere with p53 expression and function.
The M25 proteins are the first CMV proteins discovered to mediate p53 accumulation. The cell lines expressing the M25 proteins show that each of them possesses this property. The interaction with the M25 proteins prolonged the half-life of p53 considerably, again in infected cells as well as in cells transduced with M25 expression vectors. Stabilization of p53 in HCMV-infected cells was attributed to interference with the normal p53 degradation pathway (38, 39), and it is likely that the same mechanism applies to MCMV-infected cells, since at most a minimal increase of the p53 amount was seen when we inhibited the normal p53 turnover pathway by Nutlin3a treatment. In cells infected with the ΔM25 mutant the stability of p53 was slightly increased in comparison to uninfected cells, but to a lesser extent than in WT MCMV-infected cells. It has been reported that MCMV induces a DNA damage response at an early time point of infection (43), which could explain the p53 activation and stabilization in ΔM25-infected cells. Also, other viral proteins may contribute to p53 stabilization in the absence of the M25 proteins.
In both WT MCMV- and ΔM25 mutant-infected cells we observed p53 molecules with posttranslational modification and propose therefore that the M25 proteins do not interfere with the activation of p53 but instead sequester p53 molecules. It has been postulated that p53 binding sites in herpesvirus genomes, as well as viral p53 binding proteins, may serve as a sink or “sponge” for activated p53 molecules, reducing their availability to activate cellular p53 target genes (53). The pM25-containing nuclear structures may exactly have such a role; in fact, the result of our ChIP analysis is in line with the notion that less p53 molecules are available for binding to specific p53 responsive elements. In such a scenario a small pool of p53 molecules may remain active, explaining why the transcription of p53 target genes was diminished but not completely blocked in WT MCMV-infected cells compared to infection with the ΔM25 mutant. One has to point out, however, that the reduced transcription in WT MCMV-infected cells occurred in the presence of much higher p53 levels.
Infection with many DNA viruses leads to the induction of a DNA damage response (see previously published reviews [54–56]), and this has been described for MCMV (43) and HCMV as well (57–59). Some of the pathways turned on by the DNA damage response may be beneficial for viral replication (e.g., those involved in metabolism), while others are clearly antiviral, requiring differential regulation. One of the p53-regulated genes activated early in MCMV-infected cells as a consequence of the DNA damage response is Cdkn1a (43), which could explain the increased p21 levels observed at 18 and 24 h p.i. Later during the MCMV infection course, at least some of the p53-upregulated cellular genes are counteracted (43). Multiple mechanisms have been described that explain differential regulation of p53 target genes, including p53 protein level, posttranslational modifications and interaction with cofactors (reviewed in references 44 and 60). We propose that the M25 proteins act primarily at reducing the amount of p53 molecules that can activate transcription, which could explain why we observed an effect for some p53 target genes but not for others. Besides the transcriptional level, CMVs can also counteract activation of p53-regulated genes at the protein level. Due to the faster infection cycle, this may be particularly important for MCMV. Similar to observations described for HCMV (37), Cdkn1a is probably regulated transcriptionally, as well as posttranslationally, during MCMV infection, with the latter resulting in strongly diminished p21 levels at late times. For Bax and Puma we detected no difference in the mRNA and protein levels of WT MCMV- and ΔM25-infected cells. It seems that prevention of cell death is rather mediated by a number of proteins with antiapoptotic function CMVs dedicated to this task (61). For HCMV, the proteome study by Weekes et al. (62) permits analysis of kinetic changes of proteins encoded by p53-regulated genes. Examples include PCNA, GDF15 (growth/differentiation factor 15), and BAX, which are upregulated, whereas the amounts of others (p21, CASP-1 [caspase-1], FAS [tumor necrosis factor receptor superfamily member 6], and MMP2 [matrix metalloproteinase-2]) are diminished (sometimes after transient increases). Although one must be cautious in deducing that the same regulatory mechanisms apply to MCMV infection, these data strongly suggest that CMVs regulate p53-controlled genes in a differential and probably complex manner.
Growth analysis of WT MCMV on p53-negative fibroblasts revealed replication to slightly higher titers in the absence of p53. Thus, for MCMV p53 does not seem to be beneficial, as has been described for HCMV (20, 21, 63). This does not exclude that MCMV makes use of some p53-regulated pathways; however, the advantages resulting from the absence of antiviral mechanisms seem to prevail. We will have to perform analysis of the cellular and viral transcriptome to identify the pathways induced in p53-negative and p53-positive cells after MCMV infection. In addition, it remains to be checked whether there is a p53 gene dose effect since we could not immediately obtain homozygous p53+/+ cells from littermates of the same genetic background. The ΔM25 mutant grew to substantially higher titers on p53-negative cells, which is consistent with the expectation that the phenotype of a viral mutant is rescued (or substantially ameliorated) when the cellular factor normally targeted by the respective viral protein is missing. This finding underlines the importance of M25-mediated interference with p53 for production of MCMV progeny. The mutant did, however, not reach the titers of WT MCMV on p53-negative cells, suggesting that the M25 proteins possess multiple functions. Preliminary analysis did not reveal a rescue of the cell rounding phenotype of the ΔM25 mutant, implying that this M25 function is independent of p53 sequestration. Interaction of the M25 proteins with components of the cytoskeleton is the subject of ongoing investigation in our laboratory. One of the candidates that may be involved in cytoskeletal rearrangement is the potential M25 interaction partner HAX1 identified here (see Table S1 in the supplemental material), which has been implicated with the regulation of focal adhesion and migration of cells (64).
The results of our study raise several points for follow-up investigation. For instance, it would be desirable to know additional MCMV genes interfering with p53 functions. In HCMV a number of viral proteins have been implicated in binding to p53 and modulation of its activity, including IE1, IE2, UL29/28, UL38, UL44, and UL84 (52, 65–69). Further MCMV proteins that influence the transcriptional activity of p53 could be identified by screening a library of candidate genes. Knowledge of such viral proteins would then allow to analyze the interference of MCMV with p53 activation and p53-regulated pathways in a comprehensive manner and, in particular, to study their interplay with the M25 proteins.
Another interesting question is whether the HCMV UL25 protein has a comparable function. The UL25 protein has originally been described as a structural component of the HCMV tegument, which in infected cells resides within the cytoplasm (23, 70). A more recent study suggested, however, that at least a fraction of pUL25 is present in the nucleus as well (71). Interestingly, Savaryn et al. detected pUL25 in a complex with the UL29/28 protein, a known p53 regulator (69). Other studies discovered an interaction of pUL25 with other cellular and viral proteins, including pUL26 (51, 72), implying that pUL25 fulfills multiple functions. Zimmermann et al. pointed to differences between the HCMV UL25 and MCMV M25 proteins; particularly, deletion of the UL25 gene did not affect the cytopathic effect induced by HCMV (72), as described for the MCMV M25 deletion mutant (32). The M25 ORF is almost twice as large as the HCMV UL25 ORF (73), and it is therefore not unlikely that the M25 proteins possess additional functions compared to UL25 of HCMV. The notion that members of the CMV UL25 protein family acquired different tasks during evolution is corroborated by the functions ascribed to the related UL35 proteins of human and mouse CMVs (27–29, 74, 75). On the contrary, interaction of p53 with the U14 protein of human herpesvirus 6 has been reported, supporting the idea that at least some members of the UL25 family have a similar function in modulating p53 activity (76).
In summary, we identified the first CMV gene that is responsible for accumulation of p53 in infected cells, revealing one important function of the M25 proteins. Currently, it is not completely understood how CMVs achieve differential regulation of p53 target genes and how exactly this influences the productive infection cycle. We are in the process of performing a comprehensive transcriptome analysis to get global insight into the role of the MCMV M25 proteins in modulating the expression of p53 target genes. The identification of MCMV genes involved in regulating p53 activity provides an opportunity to investigate the significance of p53 for viral pathogenesis in a relevant animal CMV model.
MATERIALS AND METHODS
Cell culture and viruses.
Mouse embryonic fibroblasts (MEF), prepared as described previously (77), the p53-deficient variant of the HCT116 cells (78), and HEK293T cells (ATCC CRL-3216) were propagated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS; PAN Biotech). NIH 3T3 fibroblasts (ECACC 93061524) were cultured in DMEM with 10% bovine serum (PAN Biotech). All culture media were supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). Cell cultures were regularly checked for the absence of mycoplasma contamination using a PCR-based mycoplasma detection kit (Minerva Biolabs). Where indicated, cells were treated with the following substances (all obtained from Sigma): cycloheximide (100 μM), doxorubicin (0.5 μM), doxycycline (4 μg/ml), and nutlin3a (concentration as indicated). For generation of p53-negative MEF homozygous p53-null (p53–/–) male mice were crossed with heterozygous (p53+/–) female mice (79). MEF were isolated from individual embryos and subsequently genotyped by PCR.
The virus referred to as WT MCMV in this study has been described previously (80). WT MCMV and the ΔM25 (32) and sΔM25 mutants express GFP, allowing us to track infected cells. The two ΔM25 mutants differ in the size of the deletion within the M25 ORF (1,632 and 2,390 bp). The genome of the vM25GFP virus was generated by en passant mutagenesis (81) using the MCMV BAC pSM3fr. vM25GFP and vM25HA (32) express M25 protein variants with a C-terminal fusion of monomeric GFP and the HA epitope, respectively. Viruses were propagated either on MEF or on conditionally immortalized MEF (82) and then purified and titrated according to established protocols (77, 83). Infection with WT MCMV and viral mutants (at the indicated MOI) and growth curve analysis using MEF obtained from individual embryos were performed as described previously (32).
Plasmids.
The expression plasmids pM25l-HA and pM25s-HA, as well as plasmids expressing myc-6×His-tagged variants of the M25 proteins and of LacZ, were as described previously (32). The HA-tagged LacZ variant was generated by PCR-mediated addition of the HA epitope coding sequences to the lacZ ORF (using plasmid placZ-myc-6×His as the template) and cloning into the pcDNA3 vector. Plasmids pG13-luc (84) and pCMV-Neo-Bam p53 wt (85) coding for human p53 are available from Addgene (catalog no. 16442 and 16434), and pcDNA3_E1B-55K (expressing the Ad5 E1B-55K protein [86]) and pGL2-p21 promoter-luc (carrying the p21 promoter; Addgene, catalog no. 33021) were kindly provided by Thomas Dobner (HPI Hamburg). Plasmid pCMV6-Entry-Trp53-Myc-DKK coding for murine p53 was purchased from OriGene (MR206086). The lentiviral vector plasmid pLIX402 was obtained from Addgene (catalog no. 41394) and was a gift from David Root (University of Colorado, Boulder, CO). To facilitate cloning, a kanamycin resistance cassette (kan) amplified from plasmid pOri6kRIT (87) with primers 5′-CCTGGAGAATTGGCTGCAGGACGCATCGTGGCCGGATCT-3′ and 5′-GTGGACCGGACGCGCTGCAGGTGACCACGTCGTGGAATGC-3′ (both providing a PstI site) was inserted into NheI/MluI-cleaved pLix-402 using the Gibson assembly master mix (NEB). The PstI sites were subsequently used to replace the kan gene with the ORFs for the M25 proteins (isolated from plasmids pM25l-HA and pM25s-HA [32]). The PCR-amplified GFP ORF (5′-GACGCTAGCCCTCACTCCTTCTCTAG-3′ and 5′-GTTACGCGTCTACTTGTACAGCTCGTCCAT-3′) was cloned into NheI/MluI-treated pLIX-402.
Antibodies.
Primary antibodies used were as follows: rabbit anti-p53 (D2H90; Cell Signaling Technology [CST]), rabbit anti-p53 (FL-393; Santa Cruz), mouse anti-p53 (1C1; CST), rabbit phospho-p53 (Ser15; catalog no. 9284; CST), rabbit acetyl-p53 (Lys379; catalog no. 2570; CST), mouse anti-p21 (SX118; BD Pharmingen), rabbit anti-HA (C29F4; CST), rabbit anti-c-Myc (sc-789; A-14; Santa Cruz), rabbit anti-GFP (D5.1; CST), rabbit anti-p44/42 MAPK (ERK1/2; 137F5; CST), rabbit anti-vinculin (HPA063777; Sigma), rabbit anti-GAPDH (14C10; CST), and rabbit IgG isotype monoclonal antibody (MAb; DA1E; CST). Mouse MAb against MCMV proteins IE1 (Chroma 101), E1 (Chroma 103), M55 (M55.01), and M25C (M25C.01) were provided by CapRi, Rijeka, Croatia. HRP HPR-conjugated goat anti-mouse (P0260) and goat anti-rabbit (P0448) secondary IgG antibodies (Dako) were used for immunoblotting, and for immunofluorescence Alexa 568-conjugated goat anti-mouse IgG (A11031), Alexa 488-conjugated donkey anti-rabbit IgG (A21206), Alexa 647-conjugated goat anti-rabbit IgG (A21245; Life Technologies), and Alexa 647-conjugated goat anti-mouse IgG (115-605-003; Jackson ImmunoResearch) served as secondary antibodies.
SILAC, affinity purification, and mass spectrometry analysis.
HEK293T cells were subjected to stable isotope labeling with amino acids in cell culture (SILAC) for at least five passages using lysine and arginine-deprived DMEM, supplemented with 2 mM l-alanyl-l-glutamine, 10% dialyzed fetal bovine serum (cutoff, 10 kDa), and heavy isotope-labeled (l-[13C6,15N2]-lysine (Lys8),l-[13C6,15N4]-arginine [Arg10]) or light isotope-labeled (natural lysine [Lys0] and arginine [Arg0]) amino acids. Labeling efficiency was checked using liquid chromatography-tandem mass spectrometry (LC-MS/MS). At 48 h posttransfection, the cells were lysed in PLC buffer (0.5% NP-40, 10% glycerol, 150 mM NaCl, 50 mM HEPES [pH 7.5], 1.5 mM MgCl2, 1 mM EGTA [supplemented with 25 μM N-ethylmaleimide]), and an aliquot of the lysate was processed as input sample (see below). For affinity purification, a μMACS c-myc isolation kit (Miltenyi Biotec) was used according to the manufacturer’s instructions with modifications as detailed previously (88). Eluted proteins and input samples were precipitated using ethanol, reduced and alkylated exactly as described previously (88). Proteins were then digested using lysyl endopeptidase (Wako Pure Chemicals) for 3 h in 6 M urea–2 M thiourea, followed by trypsin digestion overnight at 2 M urea in 50 mM ammonium bicarbonate. Enzyme activity was stopped by adding trifluoracetic acid. The peptides were desalted with C18 Stage Tips (89) prior to nano-LC-MS/MS analysis. Peptides from IP or input samples were separated on 30-min, 2-h, or 4-h gradients on an EASY-nLC system with a 250 nl/min flow rate and a 15-cm column (inner diameter, 75 μm), which was packed in-house with ReproSil-Pur C18-AQ material (Dr. Maisch HPLC GmbH, Ammerbuch, Germany). Q-Exactive Plus instruments (Thermo Fisher) were operated in data-dependent Top10 mode using higher-energy collision dissociation. The full scans were performed with a resolution of 70,000, a target value of 1 × 106 ions, and a maximum injection time of 120 ms. The MS/MS scans were performed with a 17,500 resolution, a 1 × 105 target value, and a 60-ms maximum injection time. The isolation window was set to 2 m/z, and the normalized collision energy was 26. Raw data were analyzed and processed using MaxQuant 1.5.1.2 software (90). Standard settings were kept, except that the appropriate SILAC labels were set and the requantify option was enabled. N-terminal protein acetylation and methionine oxidation were considered variable modifications, and carbamidomethylation at cysteines was considered a fixed modification. The false discovery rate was set to 1% at both peptide and protein levels and estimated by using the UniProt database containing reverted sequences. The resulting output file containing the protein groups was further processed by removing potential contaminants, reverse database hits, and proteins that were only identified based on a modified peptide. SILAC ratios were extracted as normalized by MaxQuant and log2 transformed for both input and IP runs. The intensity values of input samples were log10 transformed and averaged. Statistical significance was estimated by performing one-sample t tests (null hypothesis, μ0 = 0) of the ratios from the three IP experiments, when the proteins were quantified in at least two out of three experiments. A hyperbolic curve threshold was set (red dotted line in Fig. 1B) with empirically defined cutoff parameters (91). The plots were generated using custom in-house generated scripts using R v4.0.1, as implemented in Rstudio v1.2.5033.
Immunoblotting.
Lysates of infected cells were prepared in radioimmunoprecipitation assay (RIPA) buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0]) (Fig. 3A and B, Fig. 5), supplemented with freshly added protease inhibitors. For some experiments (Fig. 3C and D and Fig. 4), cells were lysed directly in Laemmli sample buffer (followed by sonication and boiling). Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% milk in Tris-buffered saline containing 0.1% Tween 20, followed by incubation with primary antibodies overnight at 4°C, and subsequently with appropriate horseradish peroxidase-coupled secondary antibodies for 1 h at room temperature. Signals were visualized using the Super Signal West Femto maximum sensitivity substrate (Thermo Fisher) and an LAS3000 Imager (Fujifilm) or a ChemiDoc Imaging System (Bio-Rad), and quantification of signals was done by densitometric measurement using ImageJ software. Values were normalized to the signals of the loading controls. For measuring the p53 half-life in M25-expressing cell lines, quantification of signals was done separately for each cell line or condition (Fig. 3E). For control cells (and the uninduced condition) when p53 was present at low steady-state levels (compare Fig. 3D), slightly higher protein amounts were loaded to ensure that the p53 signals did not fall below the detection limit.
Immunoprecipitation.
Infected cells were lysed in PLC buffer (containing freshly added phosphatase and protease inhibitors), and were lysates precleared by centrifugation. Samples containing equal amounts of total protein (1 mg) were incubated with GFP nanobodies (Chromotek, Munich, Germany) for 3 h at 4°C. After washing with PLC buffer, proteins were eluted in Laemmli buffer. p53-deficient HCT116 cells transfected with indicated plasmids (Fig. 2B) using Lipofectamine 3000 reagent (Thermo Fisher) were lysed in buffer containing 40 mM Tris (pH 8), 135 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM EGTA, 0.25 mM dithiothreitol, and 1% glycerol and then incubated with either anti-HA or anti-myc magnetic beads (Thermo Fisher) for 3 h at 4°C. Immunoprecipitated complexes were washed twice with 40 mM Tris (pH 8)–200 mM NaCl–0.5% NP-40, twice with 40 mM Tris (pH 8)–400 mM NaCl–0.5% NP-40, and once with Tris-buffered saline. Complexes were eluted in Laemmli buffer and further analyzed by immunoblotting.
Immunofluorescence microscopy.
Cells were grown on glass coverslips and, at the indicated time points after infection or transfection, fixed with 3% paraformaldehyde, permeabilized with 0.3% Triton X-100, and blocked with 0.2% gelatin in phosphate-buffered saline (PBS). Incubation with primary antibodies (diluted in PBS) was for 1 h at room temperature, followed by washing with PBS and then by incubation with the secondary antibodies for another hour. In case of colabeling with two primary antibodies, incubation was done successively. Cover slips were mounted using Fluoromount G (Thermo Fisher) prior to analysis with the confocal laser scanning microscope TCS SP8 (Leica Microsystems). Images were processed using ImageJ software.
Generation of stable cell lines.
Retroviral particles were produced by transfection of HEK293T cells with pLIX402-derived lentiviral vectors together with plasmids coding for gag/pol functions (p8.91 HIV gag pol) and an ecotropic envelope protein (92). NIH 3T3 cells were transduced with the resulting lentiviral vectors and subsequently subjected to puromycin selection (2 μg/ml). Cell lines were propagated in the presence of puromycin (1 μg/ml) until used for experiments.
Luciferase assays.
p53-deficient HCT116 cells were transfected with the firefly luciferase reporter plasmids (100 ng) and expression plasmids for the effector proteins (100 to 200 ng) using MIRUS reagent (Mirus Bio, Madison, WI). A Gaussia luciferase plasmid (10 ng) was included to control for variation of transfection efficiency, and DNA amounts were adjusted using the plasmid pcDNA3. At 48 h after transfection, cells were lysed in passive lysis buffer (Promega), and the relative light units of lysates were measured using a GloMax luminometer (Promega) and a 5-s measurement time.
RNA analysis and qPCR.
Total RNA was isolated from infected and uninfected cells either untreated or treated with Nutlin3a (5 μM) or doxorubicin (0.5 μM for 24 h) using an RNeasy Plus minikit (Qiagen). The isolated RNA (1 μg) was reverse transcribed using the QuantiTect reverse transcription kit (Qiagen) according to the manufacturer’s protocol. For qPCR, the cDNA was mixed with Luna Universal qPCR master mix (NEB) containing SYBR green and amplified using primers for the following genes: Trp53 (5′-TCCGACTGTGACTCCTCCAT-3′ and 5′-CTAGCATTCAGGCCCTCATC-3′), Cdkn1a (5′-ATCACCAGGATTGGACATGG-3′ and 5′-CGGTGTCAGAGTCTAGGGGA-3′), Mdm2 (5′-TGATGAGGTCTATCGGGTCA-3′ and 5′-GAAGCCAGTTCTCACGAAGG-3′), Puma (5′-CAAGAAGAGCAGCATCGACA-3′ and 5′-TAGTTGGGCTCCATTTCTGG-3′), Bax (5′-GATCAGCTCGGGCACTTTAG-3′ and 5′-TTGCTGATGGCAACTTCAAC-3′), and Gapdh (5′-TCACCACCATGGAGAAGGC-3′ and 5′-GGCATGGACTGTGGTCATGA-3′). These primers have been described previously (93–95). Real-time PCR was performed with a qTower3G device (Analytik Jena, Jena, Germany) following the conditions recommended by the qPCR master mix protocol of the manufacturer (NEB). mRNAs expression levels (CT values) were calculated in relation to the CT values measured for uninfected cells both normalized to Gapdh mRNA levels (ΔΔCT) according to the method described by Livak and Schmittgen (96).
Chromatin immunoprecipitation.
ChIP was mainly done according to the SimpleChIP chromatin immunoprecipitation protocol (CST) with some modifications. In brief, NIH 3T3 cells were either infected with WT MCMV or ΔM25 at a multiplicity of infection (MOI) of 3, mock infected, or treated with doxorubicin (0.5 μM) for 18 h. Protein-DNA complexes were cross-linked by treating cells with 1% formaldehyde for 10 min, followed by neutralization with 125 mM glycine for 5 min. Cells were lysed and nuclei isolated using SimpleChIP enzymatic cell lysis buffers A and B (catalog no. 14282; CST) following the manufacturer’s instructions. Chromatin was fragmented by treatment with micrococcal nuclease (catalog no. 10011; CST) for 20 min at room temperature. Nuclei were resuspended in SimpleChIP chromatin IP buffer (catalog no. 14231; CST) and disrupted by sonication (6 cycles high power [24 s ON, 30 s OFF] using a Bioruptor plus device [Diagenode, Liege, Belgium]). After clarification by centrifugation, chromatin fragmentation was examined by isolating the DNA from an aliquot of the sample and analysis by agarose electrophoresis. Then, 300 ng of the prepared chromatin (2% of ChIP input) was separated to serve as an input control. Chromatin (15 μg) was incubated overnight at 4°C with 30 μl of Dynabeads protein A and Dynabeads protein G (1:1; Thermo Fisher) (blocked with 7.5% bovine serum albumin and 20 μg/ml of yeast tRNA) and either anti-p53 (D2H90; CST) or the rabbit isotype control (DA1E; CST) (2.5 μg). Washing steps and elution of DNA-protein complexes were performed as suggested by the manufacturer using SimpleChIP chromatin IP buffers (catalog no. 14231, CST). Reversal of cross-linking was performed by adding NaCl (final concentration, 200 mM) and RNase A (final concentration, 100 μg/ml), followed by digestion of eluted proteins with proteinase K (250 μg/ml) for 5 h at 65°C. DNA was isolated using a QIAquick PCR purification kit (Qiagen), and qPCR was performed as described above with primers specific for the following sequences: 5′ p53 response element of the Cdkn1a promoter (5′-GGGACTAGCTTTCTGGCCTT-3′ and 5′-ATCATCAGGTCTCCACCACC-3′), 3′ p53 RE of the Cdkn1a promoter (5′-CATCACAGAAGAGGAGGCCTG-3′ and 5′-GTTCTGACATCTGCTCTCCGAT-3′), and p53 RE of the Mdm2 promoter (5′-TGGTCAAGTTGGGACACGT-3′ and 5′-GAAACGGGGCAGCGTTTAAA-3′). The amount of the immunoprecipitated DNA sequence in each sample is presented as a percentage of the amount of the same sequence in the input sample.
Statistical analysis.
Prism software version 5.02 (GraphPad Software, San Diego, CA) was used to analyze data by t test or Mann-Whitney test. The significance level was set to P ≤ 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Thomas Dobner for kindly providing cell lines and reporter plasmids and Jens Bohne for the lentiviral packaging plasmids. We are grateful to previous group members (Sarah Sengstake, Wiebke Jäger, and Kirsten Keyser) for constructing viral mutants, to Marina Marcelić for cloning one of the lentiviral vector plasmids, and to Tobias May for kindly providing conditionally immortalized fibroblasts.
This project was funded by the Deutsche Forschungsgemeinschaft (German Research Foundation) SFB900/3-158989968 (project C6).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Mocarski ES, Shenk T, Griffith PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Yue Y, Barry PA. 2008. Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv Virus Res 72:207–226. doi: 10.1016/S0065-3527(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 3.Reddehase MJ, Lemmermann NAW. 2018. Mouse model of cytomegalovirus disease and immunotherapy in the immunocompromised host: predictions for medical translation that survived the “test of time.” Viruses 10:693. doi: 10.3390/v10120693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roark HK, Jenks JA, Permar SR, Schleiss MR. 2020. Animal models of congenital cytomegalovirus transmission: implications for vaccine development. J Infect Dis 221:S60–S73. doi: 10.1093/infdis/jiz484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalejta RF. 2008. Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Curr Top Microbiol Immunol 325:101–115. doi: 10.1007/978-3-540-77349-8_6. [DOI] [PubMed] [Google Scholar]
- 6.Lane D, Levine A. 2010. p53 research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivas C, Aaronson SA, Munoz-Fontela C. 2010. Dual role of p53 in innate antiviral immunity. Viruses 2:298–313. doi: 10.3390/v2010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miciak J, Bunz F. 2016. Long story short: p53 mediates innate immunity. Biochim Biophys Acta 1865:220–227. doi: 10.1016/j.bbcan.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaux DL, Haecker G, Strasser A. 1994. An evolutionary perspective on apoptosis. Cell 76:777–779. doi: 10.1016/0092-8674(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 10.Hafner A, Bulyk ML, Jambhekar A, Lahav G. 2019. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol 20:199–210. doi: 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- 11.Fan Y, Sanyal S, Bruzzone R. 2018. Breaking bad: how viruses subvert the cell cycle. Front Cell Infect Microbiol 8:396. doi: 10.3389/fcimb.2018.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalejta RF, Shenk T. 2002. Manipulation of the cell cycle by human cytomegalovirus. Front Biosci 7:d295–d306. doi: 10.2741/A729. [DOI] [PubMed] [Google Scholar]
- 13.Spector DH. 2015. Human cytomegalovirus riding the cell cycle. Med Microbiol Immunol 204:409–419. doi: 10.1007/s00430-015-0396-z. [DOI] [PubMed] [Google Scholar]
- 14.Sato Y, Tsurumi T. 2013. Genome guardian p53 and viral infections. Rev Med Virol 23:213–220. doi: 10.1002/rmv.1738. [DOI] [PubMed] [Google Scholar]
- 15.Aloni-Grinstein R, Charni-Natan M, Solomon H, Rotter V. 2018. p53 and the viral connection: back into the future. Cancers 10:178. doi: 10.3390/cancers10060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muganda P, Mendoza O, Hernandez J, Qian Q. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J Virol 68:8028–8034. doi: 10.1128/JVI.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Speir E, Modali R, Huang ES, Leon MB, Shawl F, Finkel T, Epstein SE. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 18.Jault FM, Jault JM, Ruchti F, Fortunato EA, Clark C, Corbeil J, Richman DD, Spector DH. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol 69:6697–6704. doi: 10.1128/JVI.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortunato EA, Spector DH. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J Virol 72:2033–2039. doi: 10.1128/JVI.72.3.2033-2039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casavant NC, Luo MH, Rosenke K, Winegardner T, Zurawska A, Fortunato EA. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J Virol 80:8390–8401. doi: 10.1128/JVI.00505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuan MI, O’Dowd JM, Chughtai K, Hayman I, Brown CJ, Fortunato EA. 2016. Human cytomegalovirus nuclear egress and secondary envelopment are negatively affected in the absence of cellular p53. Virology 497:279–293. doi: 10.1016/j.virol.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldick CJ, Shenk T. 1996. Proteins associated with purified human cytomegalovirus particles. J Virol 70:6097–6105. doi: 10.1128/JVI.70.9.6097-6105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battista MC, Bergamini G, Boccuni MC, Campanini F, Ripalti A, Landini MP. 1999. Expression and characterization of a novel structural protein of human cytomegalovirus, pUL25. J Virol 73:3800–3809. doi: 10.1128/JVI.73.5.3800-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Biegalke BJ. 2002. The human cytomegalovirus UL35 gene encodes two proteins with different functions. J Virol 76:2460–2468. doi: 10.1128/jvi.76.5.2460-2468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B, Nishimura M, Tang H, Kawabata A, Mahmoud NF, Khanlari Z, Hamada D, Tsuruta H, Mori Y. 2016. Crystal structure of human herpesvirus 6B tegument protein U14. PLoS Pathog 12:e1005594. doi: 10.1371/journal.ppat.1005594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SH, Kalejta RF, Kerry J, Semmes OJ, O’Connor CM, Khan Z, Garcia BA, Shenk T, Murphy E. 2012. BclAF1 restriction factor is neutralized by proteasomal degradation and microRNA repression during human cytomegalovirus infection. Proc Natl Acad Sci U S A 109:9575–9580. doi: 10.1073/pnas.1207496109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schierling K, Stamminger T, Mertens T, Winkler M. 2004. Human cytomegalovirus tegument proteins ppUL82 (pp71) and ppUL35 interact and cooperatively activate the major immediate-early enhancer. J Virol 78:9512–9523. doi: 10.1128/JVI.78.17.9512-9523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salsman J, Jagannathan M, Paladino P, Chan PK, Dellaire G, Raught B, Frappier L. 2012. Proteomic profiling of the human cytomegalovirus UL35 gene products reveals a role for UL35 in the DNA repair response. J Virol 86:806–820. doi: 10.1128/JVI.05442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan B, Goncalves MV, Lemmermann NAW, Juranic LV, Stempel M, Bussey KA, Reimer E, Podlech J, Lienenklaus S, Reddehase MJ, Jonjic S, Brinkmann MM. 2017. The murine cytomegalovirus M35 protein antagonizes type I IFN induction downstream of pattern recognition receptors by targeting NF-κB mediated transcription. PLoS Pathog 13:e1006382. doi: 10.1371/journal.ppat.1006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu CA, Carlson ME, Henry SC, Shanley JD. 1999. The murine cytomegalovirus M25 open reading frame encodes a component of the tegument. Virology 262:265–276. doi: 10.1006/viro.1999.9942. [DOI] [PubMed] [Google Scholar]
- 31.Kattenhorn LM, Mills R, Wagner M, Lomsadze A, Makeev V, Borodovsky M, Ploegh HL, Kessler BM. 2004. Identification of proteins associated with murine cytomegalovirus virions. J Virol 78:11187–11197. doi: 10.1128/JVI.78.20.11187-11197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutle I, Sengstake S, Templin C, Glass M, Kubsch T, Keyser KA, Binz A, Bauerfeind R, Sodeik B, Čičin-Šain L, Dezeljin M, Messerle M. 2017. The M25 gene products are critical for the cytopathic effect of mouse cytomegalovirus. Sci Rep 7:15588. doi: 10.1038/s41598-017-15783-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S, Vasanji A, Pellett PE. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol 81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bühler B, Keil GM, Weiland F, Koszinowski UH. 1990. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J Virol 64:1907–1919. doi: 10.1128/JVI.64.5.1907-1919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson JG, Pereira-Smith OM. 2006. p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res 66:8356–8360. doi: 10.1158/0008-5472.CAN-06-1752. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Knutson E, Kurosky A, Albrecht T. 2001. Degradation of p21cip1 in cells productively infected with human cytomegalovirus. J Virol 75:3613–3625. doi: 10.1128/JVI.75.8.3613-3625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Utama B, Shen YH, Mitchell BM, Makagiansar IT, Gan Y, Muthuswamy R, Duraisamy S, Martin D, Wang X, Zhang MX, Wang J, Wang J, Vercellotti GM, Gu W, Wang XL. 2006. Mechanisms for human cytomegalovirus-induced cytoplasmic p53 sequestration in endothelial cells. J Cell Sci 119:2457–2467. doi: 10.1242/jcs.02974. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z, Knutson E, Wang S, Martinez LA, Albrecht T. 2007. Stabilization of p53 in human cytomegalovirus-initiated cells is associated with sequestration of HDM2 and decreased p53 ubiquitination. J Biol Chem 282:29284–29295. doi: 10.1074/jbc.M705349200. [DOI] [PubMed] [Google Scholar]
- 40.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 41.Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 42.van Leeuwen IMM, Higgins M, Campbell J, Brown CJ, McCarthy AR, Pirrie L, Westwood NJ, Laín S. 2011. Mechanism-specific signatures for small-molecule p53 activators. Cell Cycle 10:1590–1598. doi: 10.4161/cc.10.10.15519. [DOI] [PubMed] [Google Scholar]
- 43.Marcinowski L, Lidschreiber M, Windhager L, Rieder M, Bosse JB, Rädle B, Bonfert T, Györy I, de Graaf M, Prazeres da Costa O, Rosenstiel P, Friedel CC, Zimmer R, Ruzsics Z, Dölken L. 2012. Real-time transcriptional profiling of cellular and viral gene expression during lytic cytomegalovirus infection. PLoS Pathog 8:e1002908. doi: 10.1371/journal.ppat.1002908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vousden KH, Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 45.Meek DW, Anderson CW. 2009. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol 1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loughery J, Cox M, Smith LM, Meek DW. 2014. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res 42:7666–7680. doi: 10.1093/nar/gku501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed SM, Quelle DE. 2014. p53 acetylation: regulation and consequences. Cancers (Basel)) 7:30–69. doi: 10.3390/cancers7010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yew PR, Berk AJ. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 49.Pennella MA, Liu Y, Woo JL, Kim CA, Berk AJ. 2010. Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J Virol 84:12210–12225. doi: 10.1128/JVI.01442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.el Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. 1992. Definition of a consensus binding site for p53. Nat Genet 1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 51.Nobre LV, Nightingale K, Ravenhill BJ, Antrobus R, Soday L, Nichols J, Davies JA, Seirafian S, Wang EC, Davison AJ, Wilkinson GW, Stanton RJ, Huttlin EL, Weekes MP. 2019. Human cytomegalovirus interactome analysis identifies degradation hubs, domain associations and viral protein functions. Elife 8:e49894. doi: 10.7554/eLife.49894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang ES, Zhang Z, Cai H, Huang DY, Huong SM, Cha CY, Huang ES. 2009. Human cytomegalovirus IE1-72 protein interacts with p53 and inhibits p53-dependent transactivation by a mechanism different from that of IE2-86 protein. J Virol 83:12388–12398. doi: 10.1128/JVI.00304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sifford JM, Stahl JA, Salinas E, Forrest JC. 2015. Murine gammaherpesvirus 68 LANA and SOX homologs counteract ATM-driven p53 activity during lytic viral replication. J Virol 90:2571–2585. doi: 10.1128/JVI.02867-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiaofei E, Kowalik TF. 2014. The DNA damage response induced by infection with human cytomegalovirus and other viruses. Viruses 6:2155–2185. doi: 10.3390/v6052155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah GA, O’Shea CC. 2015. Viral and cellular genomes activate distinct DNA damage responses. Cell 162:987–1002. doi: 10.1016/j.cell.2015.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weitzman MD, Fradet-Turcotte A. 2018. Virus DNA replication and the host DNA damage response. Annu Rev Virol 5:141–164. doi: 10.1146/annurev-virology-092917-043534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaspar M, Shenk T. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc Natl Acad Sci U S A 103:2821–2826. doi: 10.1073/pnas.0511148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo MH, Rosenke K, Czornak K, Fortunato EA. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J Virol 81:1934–1950. doi: 10.1128/JVI.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiaofei E, Pickering MT, Debatis M, Castillo J, Lagadinos A, Wang S, Lu S, Kowalik TF. 2011. An E2F1-mediated DNA damage response contributes to the replication of human cytomegalovirus. PLoS Pathog 7:e1001342. doi: 10.1371/journal.ppat.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan KD, Galbraith MD, Andrysik Z, Espinosa JM. 2018. Mechanisms of transcriptional regulation by p53. Cell Death Differ 25:133–143. doi: 10.1038/cdd.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brune W, Andoniou CE. 2017. Die another day: inhibition of cell death pathways by cytomegalovirus. Viruses 9:249. doi: 10.3390/v9090249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang EC, Aicheler R, Murrell I, Wilkinson GW, Lehner PJ, Gygi SP. 2014. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell 157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hannemann H, Rosenke K, O’Dowd JM, Fortunato EA. 2009. The presence of p53 influences the expression of multiple human cytomegalovirus genes at early times postinfection. J Virol 83:4316–4325. doi: 10.1128/JVI.02075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H, Yue J, Huang H, Gou X, Chen SY, Zhao Y, Wu X. 2015. Regulation of focal adhesion dynamics and cell motility by the EB2 and Hax1 protein complex. J Biol Chem 290:30771–30782. doi: 10.1074/jbc.M115.671743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsai HL, Kou GH, Chen SC, Wu CW, Lin YS. 1996. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem 271:3534–3540. doi: 10.1074/jbc.271.7.3534. [DOI] [PubMed] [Google Scholar]
- 66.Bonin LR, McDougall JK. 1997. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol 71:5861–5870. doi: 10.1128/JVI.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castillo JP, Frame FM, Rogoff HA, Pickering MT, Yurochko AD, Kowalik TF. 2005. Human cytomegalovirus IE1-72 activates ataxia telangiectasia mutated kinase and a p53/p21-mediated growth arrest response. J Virol 79:11467–11475. doi: 10.1128/JVI.79.17.11467-11475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwon Y, Kim MN, Young CE, Heon KJ, Hwang ES, Cha CY. 2012. Inhibition of p53 transcriptional activity by human cytomegalovirus UL44. Microbiol Immunol 56:324–331. doi: 10.1111/j.1348-0421.2012.00446.x. [DOI] [PubMed] [Google Scholar]
- 69.Savaryn JP, Reitsma JM, Bigley TM, Halligan BD, Qian Z, Yu D, Terhune SS. 2013. Human cytomegalovirus pUL29/28 and pUL38 repression of p53-regulated p21CIP1 and caspase 1 promoters during infection. J Virol 87:2463–2474. doi: 10.1128/JVI.01926-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zini N, Santi S, Riccio M, Landini MP, Battista MC, Maraldi NM. 2000. pUL25 immunolocalization in human cytomegalovirus-infected and gene-transfected cells. Arch Virol 145:795–803. doi: 10.1007/s007050050672. [DOI] [PubMed] [Google Scholar]
- 71.Jean Beltran PM, Mathias RA, Cristea IM. 2016. A portrait of the human organelle proteome in space and time during cytomegalovirus Infection. Cell Syst 3:361–373. doi: 10.1016/j.cels.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimmermann C, Buscher N, Krauter S, Kramer N, Wolfrum U, Sehn E, Tenzer S, Plachter B. 2018. The abundant tegument protein pUL25 of human cytomegalovirus prevents proteasomal degradation of pUL26 and supports its suppression of ISGylation. J Virol 92:e01180-18. doi: 10.1128/JVI.01180-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brocchieri L, Kledal TN, Karlin S, Mocarski ES. 2005. Predicting coding potential from genome sequence: application to betaherpesviruses infecting rats and mice. J Virol 79:7570–7596. doi: 10.1128/JVI.79.12.7570-7596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maschkowitz G, Gartner S, Hofmann-Winkler H, Fickenscher H, Winkler M. 2018. Interaction of human cytomegalovirus tegument proteins ppUL35 and ppUL35A with sorting nexin 5 regulates glycoprotein B (gpUL55) localization. J Virol 92:e00013-18. doi: 10.1128/JVI.00013-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fabits M, Goncalves MV, Chan B, Girault V, Elbasani E, Rossetti E, Saeland E, Messerle M, Pichlmair A, Lisnic VJ, Brinkmann MM. 2020. The cytomegalovirus tegument protein UL35 antagonizes pattern recognition receptor-mediated type I IFN transcription. Microorganisms 8:790. doi: 10.3390/microorganisms8060790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takemoto M, Koike M, Mori Y, Yonemoto S, Sasamoto Y, Kondo K, Uchiyama Y, Yamanishi K. 2005. Human herpesvirus 6 open reading frame U14 protein and cellular p53 interact with each other and are contained in the virion. J Virol 79:13037–13046. doi: 10.1128/JVI.79.20.13037-13046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brune W, Hengel H, Koszinowski UH. 2001. A mouse model for cytomegalovirus infection, p 19.7.1–19.7.13. In Coligan JE, Bierer B, Margulies DH, Sherach EM, Strober W, Coico R (ed), Current protocols in immunology. John Wiley & Sons, New York, NY. [DOI] [PubMed] [Google Scholar]
- 78.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 79.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 80.Mathys S, Schroeder T, Ellwart J, Koszinowski UH, Messerle M, Just U. 2003. Dendritic cells under influence of mouse cytomegalovirus have a physiologic dual role: to initiate and to restrict T cell activation. J Infect Dis 187:988–999. doi: 10.1086/368094. [DOI] [PubMed] [Google Scholar]
- 81.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 82.May T, Hauser H, Wirth D. 2004. Transcriptional control of SV40 T-antigen expression allows a complete reversion of immortalization. Nucleic Acids Res 32:5529–5538. doi: 10.1093/nar/gkh887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lemmermann NAW, Podlech J, Seckert CK, Kropp KA, Grzimek NKA, Reddehase MJ, Holtappels R. 2010. CD8 T-cell immunotherapy of cytomegalovirus disease in the murine model, p 369–429. In Kabelitz D, Kaufmann SHE (ed), Methods in microbiology: immunology of infection, Academic Press, Oxford, United Kingdom. [Google Scholar]
- 84.el Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 85.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 86.Rubenwolf S, Schutt H, Nevels M, Wolf H, Dobner T. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol 71:1115–1123. doi: 10.1128/JVI.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, Heinrich F, Gasparoni G, Babic M, Tomic A, Pietra G, Nienen M, Blau IW, Hofmann J, Na IK, Prinz I, Koenecke C, Hemmati P, Babel N, Arnold R, Walter J, Thurley K, Mashreghi MF, Messerle M, Romagnani C. 2018. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol 19:453–463. doi: 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]
- 88.Bogdanow B, Schmidt M, Weisbach H, Gruska I, Vetter B, Imami K, Ostermann E, Brune W, Selbach M, Hagemeier C, Wiebusch L. 2019. Cross-regulation of viral kinases with cyclin A secures shutoff of host DNA synthesis. bioRxiv https://www.biorxiv.org/content/10.1101/856435v1. [DOI] [PMC free article] [PubMed]
- 89.Rappsilber J, Ishihama Y, Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 90.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 91.Chen JX, Cipriani PG, Mecenas D, Polanowska J, Piano F, Gunsalus KC, Selbach M. 2016. In vivo interaction proteomics in Caenorhabditis elegans embryos provides new insights into P granule dynamics. Mol Cell Proteomics 15:1642–1657. doi: 10.1074/mcp.M115.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schambach A, Galla M, Modlich U, Will E, Chandra S, Reeves L, Colbert M, Williams DA, von Kalle C, Baum C. 2006. Lentiviral vectors pseudotyped with murine ecotropic envelope: increased biosafety and convenience in preclinical research. Exp Hematol 34:588–592. doi: 10.1016/j.exphem.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 93.Thomasova D, Bruns HA, Kretschmer V, Ebrahim M, Romoli S, Liapis H, Kotb AM, Endlich N, Anders HJ. 2015. Murine Double Minute-2 prevents p53-overactivation-related cell death (podoptosis) of podocytes. J Am Soc Nephrol 26:1513–1523. doi: 10.1681/ASN.2014040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilson AM, Morquette B, Abdouh M, Unsain N, Barker PA, Feinstein E, Bernier G, Di Polo A. 2013. ASPP1/2 regulate p53-dependent death of retinal ganglion cells through PUMA and Fas/CD95 activation in vivo. J Neurosci 33:2205–2216. doi: 10.1523/JNEUROSCI.2635-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ren YA, Mullany LK, Liu Z, Herron AJ, Wong KK, Richards JS. 2016. Mutant p53 promotes epithelial ovarian cancer by regulating tumor differentiation, metastasis, and responsiveness to steroid hormones. Cancer Res 76:2206–2218. doi: 10.1158/0008-5472.CAN-15-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.