Human papillomaviruses are the major causative agents of cervical, oral, and anal cancers. The present study demonstrates that the Pyk2 tyrosine kinase phosphorylates E2 at tyrosine 131, interfering with genome replication. We provide evidence that phosphorylation of E2 prevents binding to the Brd4-CTM. Our findings add to the understanding of molecular pathways utilized by the virus during its vegetative life cycle and offers insights into the host-virus interactome.
KEYWORDS: E2, HPV, replication, tyrosine phosphorylation
ABSTRACT
The human papillomavirus (HPV) E2 protein is a key regulator of viral transcription and replication. In this study, we demonstrate that the nonreceptor tyrosine kinase Pyk2 phosphorylates tyrosine 131 in the E2 transactivation domain. Both depletion of Pyk2 and treatment with a Pyk2 kinase inhibitor increased viral DNA content in keratinocytes that maintain viral episomes. The tyrosine-to-glutamic acid (E) mutant Y131E, which may mimic phosphotyrosine, failed to stimulate transient DNA replication, and genomes with this mutation were unable to establish stable episomes in keratinocytes. Using coimmunoprecipitation assays, we demonstrate that the Y131E is defective for binding to the C-terminal motif (CTM) of Bromodomain-containing protein 4 (Brd4). These data imply that HPV replication depends on E2 Y131 interaction with the pTEFb binding domain of Brd4.
IMPORTANCE Human papillomaviruses are the major causative agents of cervical, oral, and anal cancers. The present study demonstrates that the Pyk2 tyrosine kinase phosphorylates E2 at tyrosine 131, interfering with genome replication. We provide evidence that phosphorylation of E2 prevents binding to the Brd4-CTM. Our findings add to the understanding of molecular pathways utilized by the virus during its vegetative life cycle and offers insights into the host-virus interactome.
INTRODUCTION
The activator E2 and E1 DNA helicase proteins are the earliest proteins to be expressed in human papillomavirus (HPV) infection. Small amounts of E2 mediate the initiation of viral replication, and its relative expression level and function are linked to the different stages of viral DNA replication (1, 2). The E2 protein is composed of an ∼220-amino-acid (aa) transactivation domain (TAD), a nonconserved hinge region, and a DNA-binding domain (DBD). E2 binds to a 12-nucleotide inverted palindrome strategically placed adjacent to transcriptional promoter elements and the origin of replication (ori) (3). The E2 TAD associates with cellular host proteins such as Brd4, TopBP1, TaxBP1, ChlR1, and GPS2/AMF-1 that are necessary for its genome replication, transcription of the early promoter, and tethering of viral episomes to host mitotic chromosomes during cell division (4–7).
We and others have identified several posttranslational modifications that coordinate several E2 protein functions (8–12). Our laboratory recently discovered novel regulatory interactions between E2 and tyrosine kinases belonging to fibroblast growth factor receptor family (FGFR). We have identified phosphorylation sites within E2 at tyrosine 102 (Y102) (9, 13) and Y138 (14) that control replication.
The differentiation status of the infected epithelium governs the viral replicative program (15, 16). First, infection of basal epithelial cells is where the viral genome replicates to a low copy number. In the maintenance stage in suprabasal cells, PV genomes duplicate and partition in a cell-cycle-dependent manner (17, 18). As infected cells migrate into the upper epithelial strata, in nondividing cells genomes amplify to hundreds of copies. The late capsid proteins L1 and L2 are expressed, and viral particles assemble and package episomes at this stage. We hypothesize that E2 posttranslational modifications contribute to the regulation of the switch between replication stages.
E2 host interaction network mapping using yeast two-hybrid screens and a high-throughput Gaussia princeps luciferase-based complementation assay (GPCA) detected Pyk2 as potential interacting partner of HPV-18 E2 (19). Pyk2 is a calcium-regulated nonreceptor tyrosine kinase and belongs to the focal adhesion kinase (FAK) subfamily. Pyk2 and FAK share ∼65% aa identity, similar phosphorylation sites, and domain structure (20). Pyk2 signaling regulates cellular polarity, migration, proliferation, and adhesion in response to stimuli, including hormones, growth factors, and cytokines (21). In HaCaT cells, depletion or inhibition of the proline-rich tyrosine kinase (Pyk2) was found to decrease HPV pseudovirion infection (22), and expression of L2 caused a 50% increase in Pyk2 protein levels (23).
The molecular mechanisms by which viral genome replication switches from the initial burst immediately after infection to once per cell cycle mode in the maintenance stage is unclear. Here, we provide evidence that Pyk2 controls E2 activity by phosphorylation of tyrosine 131 and limits genome replication through impairment of Brd4 C-terminal motif (CTM) association.
RESULTS
Characterization of the Pyk2 and HPV E2 interaction.
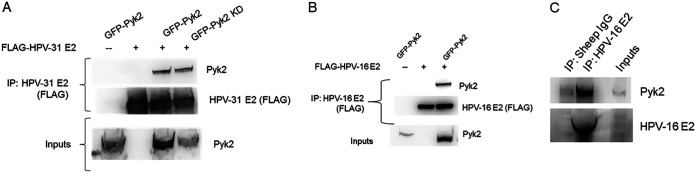
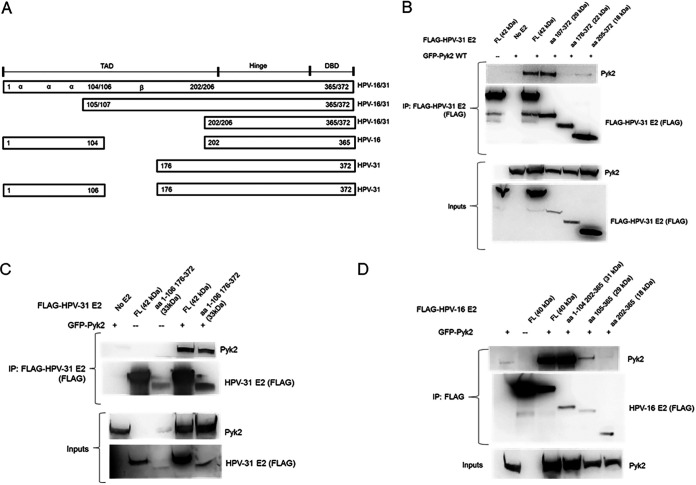
Our initial goal was to experimentally substantiate Pyk2 binding to high-risk HPV-31 and HPV-16 E2 since previous studies reported interaction with HPV-18 E2 (19). FLAG-tagged HPV-31 E2 was found to coimmunoprecipitate with both GFP-tagged wild-type (WT) and kinase-dead (KD) Pyk2 (24, 25) (Fig. 1A). FLAG-tagged HPV-16 E2 also coimmunoprecipitated with GFP-tagged Pyk2 (Fig. 1B). E2 expressed in W12 cells from the HPV-16 episomes coimmunoprecipitated with endogenous Pyk2 protein (Fig. 1C). To identify the region of binding of HPV-31 and -16 E2 to Pyk2, a series of progressive N-terminal E2 deletions were utilized (Fig. 2A). The E2 fragments express differently because of their variable protein stability. We used this assay to measure the ability of these E2 segments to bind to Pyk2 in a classical domain mapping experiment. WT Pyk2 coimmunoprecipitated with the HPV-31 E2 region of aa 107 to 372 but not to a segment that spanned aa 176 to 372 or the DBD from aa 205 to 372 (Fig. 2B), confirming that the interaction occurs within the E2 TAD. We used the 1–106/176–372 fragment to determine the importance of the excluded beta-sheet region of E2. Unexpectedly, we observed association between Pyk2 with an HPV-31 E2 truncated protein that included aa 1 to 106 linked to aa 176 to 372 (Fig. 2C). Similar binding between Pyk2 and the E2 TAD regions for HPV-16 was also observed (Fig. 2D). These results implied that two adjacent, nonoverlapping regions of the TAD can complex with Pyk2.
FIG 1.
HPV E2 interacts with Pyk2. (A) HEK293TT cells were transfected with GFP-Pyk2 WT (wild type) or GFP-Pyk2 KD (kinase dead) and FLAG-HPV-31 E2. FLAG-HPV-31 E2 was immunoprecipitated with M2 (FLAG) antibodies. Complexes were blotted with M2 (FLAG) and Pyk2 antibodies. (B) HEK293TT cells were transfected with GFP-Pyk2 WT and FLAG-HPV-16 E2. FLAG-HPV-16 E2 was immunoprecipitated with M2 (FLAG) antibodies. Complexes were blotted with M2 (FLAG) and Pyk2 antibodies. (C) HPV-16 E2 in W12 cells was immunoprecipitated using sheep anti-HPV-16 E2 serum or normal sheep IgG. Complexes were blotted with Pyk2 and HPV-16 E2 (TVG-261) antibodies.
FIG 2.
Pyk2 interacts with the TAD of E2. (A) Illustration of HPV-31/-16 E2 fragments. (B) HEK293TT cells were transfected with GFP-Pyk2 and FLAG-HPV-31 E2 or HPV-31 E2 fragments (aa 107 to 372, 176 to 372, and 205 to 372) and immunoprecipitated with M2 (FLAG) antibodies. Complexes were blotted with M2 (FLAG) and Pyk2 antibodies. (C) HEK293TT cells were transfected with GFP-Pyk2 and FLAG-HPV-31 E2 or HPV-31 E2 with the β-sheet region of the E2 TAD deleted (aa 1 to 106 and aa 176 to 372) and immunoprecipitated with M2 (FLAG) antibodies. Complexes were blotted with M2 (FLAG) and Pyk2 antibodies. (D) HEK293TT cells were transfected with GFP-Pyk2 and FLAG-HPV-16 E2 or HPV-16 E2 fragments (aa 1 to 104, 202 to 365, 105 to 365, and 202 to 365). After immunoprecipitation with M2 (FLAG) antibodies, complexes were blotted with M2 (FLAG) and Pyk2 antibodies.
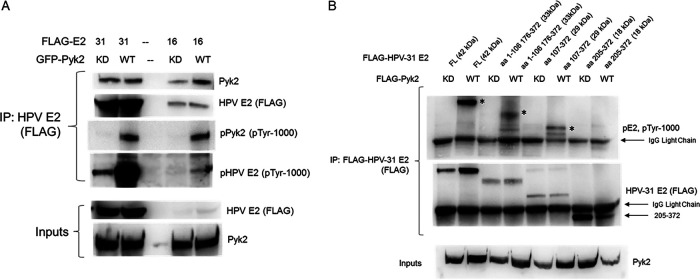
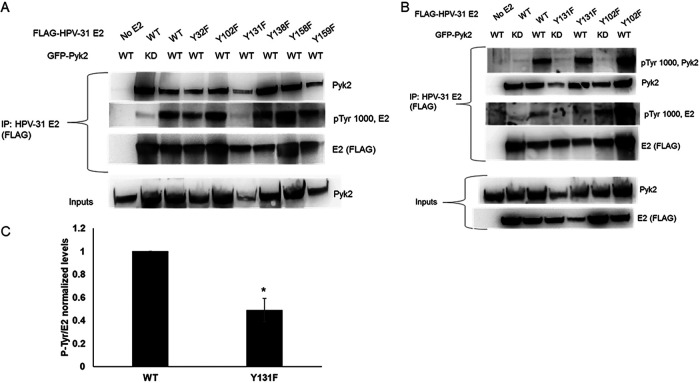
To determine whether E2 tyrosine phosphorylation increased with Pyk2 overexpression and to map the site(s) of E2 phosphorylation, HEK293TT cells were transfected with HPV-31 or HPV-16 E2 and Pyk2 constructs. E2 was immunoprecipitated and then blotted with antibodies that recognize tyrosine phosphorylation. Elevated E2 tyrosine phosphorylation levels were detected with WT Pyk2 compared to KD Pyk2 (Fig. 3A). The regions including aa 107 to 372 and including aa 1 to 106 plus 176 to 372 showed higher phosphotyrosine levels (Fig. 3B). To identify residue(s) within HPV-31 E2 phosphorylated by Pyk2, we mutated conserved tyrosines Y32, Y102, Y131, Y138, Y158, and Y159 within the E2 TAD to nonphosphorylatable phenylalanine (F). HEK293TT cells were cotransfected with GFP-Pyk2 and these FLAG-tagged E2 mutant expression plasmids. Using FLAG antibodies to immunoprecipitate the HPV-31 E2 proteins, which all coimmunoprecipitated Pyk2, we found reduced phosphotyrosine signal with Y131F (49%) but not with Y102F (Fig. 4). This led us to postulate that Y131 is a target for Pyk2 phosphorylation.
FIG 3.
Pyk2 phosphorylates the E2. (A) HEK293TT cells were transfected with GFP-Pyk2 WT or GFP-Pyk2 KD and FLAG-HPV-31 or FLAG-HPV-16 E2. FLAG-HPV-E2 was immunoprecipitated with M2 (FLAG) antibodies, and complexes were blotted with pTyr-1000, M2 (FLAG), and Pyk2 antibodies. (B) HEK293TT cells were transfected with FLAG-Pyk2 WT or FLAG-Pyk2 KD and FLAG-HPV-31 E2 fragments (aa 1 to 106, 176 to 372, 107 to 372, and 205 to 372). FLAG-HPV-31 E2 and FLAG-Pyk2 were immunoprecipitated with M2 (FLAG) antibodies, and complexes were blotted with pTyr-1000 and M2 (FLAG) antibodies. Phospho-E2 fragments are identified by an asterisk (*).
FIG 4.
Pyk2 induced tyrosine phosphorylation of HPV-31 E2 Y to F mutants. HEK293TT cells were transfected with GFP-Pyk2 and FLAG-HPV-31 E2 phenylalanine F mutants (A) or with GFP-Pyk2 WT or GFP-Pyk2 KD and FLAG-HPV-31 F mutants (B). Lysates were immunoprecipitated with M2 (FLAG), and bound proteins were blotted with Pyk2, pTyr-1000, and M2 (FLAG) antibodies. (C) Quantification of phosphotyrosine levels of Y131F. Values are means ± the SEM (n = 3). *, P < 0.05 (two-way t test).
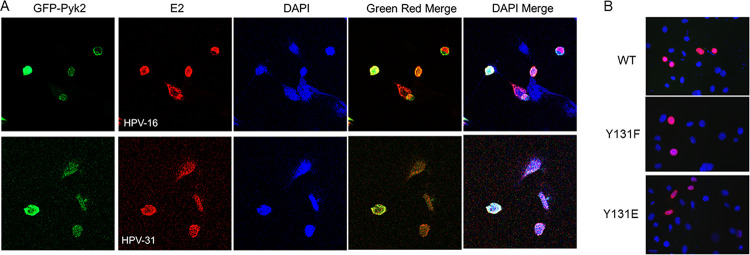
Previous studies have shown that Pyk2 localizes in the nuclei of human epidermal keratinocytes (26). The localization of Pyk2 and E2 was analyzed using confocal microscopy, and we observed Pyk2 and both HPV-16 and HPV-31 E2 in the nucleus and nuclear membrane (Fig. 5A). We then tested the nuclear localization of the FLAG-HPV-31 E2 WT, Y131F, and Y131E proteins in HeLa cells and confirmed that these mutants localized to the nucleus (Fig. 5B).
FIG 5.
Nuclear localization of HPV E2 and Pyk2. (A) CV-1 cells were transfected with GFP-Pyk2 WT and FLAG-HPV-31 or -16 E2 constructs. Immunofluorescence staining for HPV-E2 proteins with M2 (FLAG) antibodies and DAPI (blue) was performed. Cells were visualized under ×60 magnification using confocal microscopy. Pyk2 (Green), E2 (red), and DAPI (blue) are indicated. (B) HeLa cells were transfected with FLAG-HPV-31 E2 WT or Y131 mutants. Immunofluorescence staining was done with M2 (FLAG, red) antibodies or DAPI (blue). Magnification, ×60.
Reduction of Pyk2 increases HPV episome copy number.
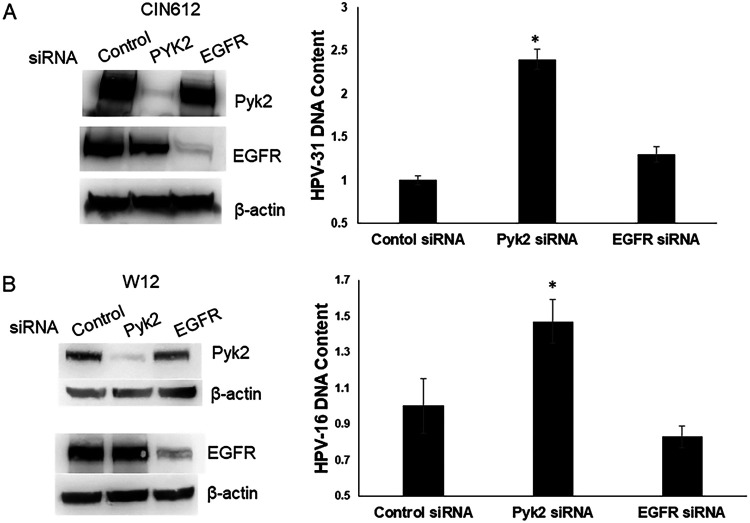
To determine the effects of Pyk2 reduction on viral genome replication, we used small interfering RNA (siRNA) to deplete Pyk2 levels in CIN612-9E cells, which maintain HPV-31 episomes, and in W12 cells with HPV-16 episomes, followed by quantitative PCR (qPCR) to measure the HPV DNA content. As a control for these experiments, siRNA directed to the tyrosine kinase epidermal growth factor receptor (EGFR) was utilized since we previously showed that its silencing did not alter HPV-31 DNA content in CIN612-9E cells (27). HPV DNA was significantly increased for HPV-31 (∼2.5-fold, Fig. 6A) and HPV-16 genomes (∼1.5-fold, Fig. 6B) after depletion of Pyk2 but not of EGFR.
FIG 6.
Pyk2 siRNA increases HPV DNA content. (A) CIN612-9E cells were transfected with a scrambled control, Pyk2, or EGFR siRNA. After 72 h, lysates were immunoblotted with Pyk2, EGFR, or β-actin antibodies. CIN612-9E cells were transfected with a control, Pyk2, or EGFR siRNA. Real-time PCR was performed for the HPV-31 long control region (LCR) and normalized to the level of β-actin. Values are expressed as means ± the SEM (n = 6). *, P < 0.05. (B) W12 cells were transfected with a control, Pyk2, or EGFR siRNA. After 72 h, the lysates were immunoblotted with Pyk2, EGFR, and β-actin antibodies. W12 cells were transfected with a control, Pyk2, or EGFR siRNA. DNA was isolated for real-time PCR for the HPV16 E6 DNA region and normalized to the levels of β-actin. Values are expressed as means ± the SEM (n = 12). *, P < 0.05.
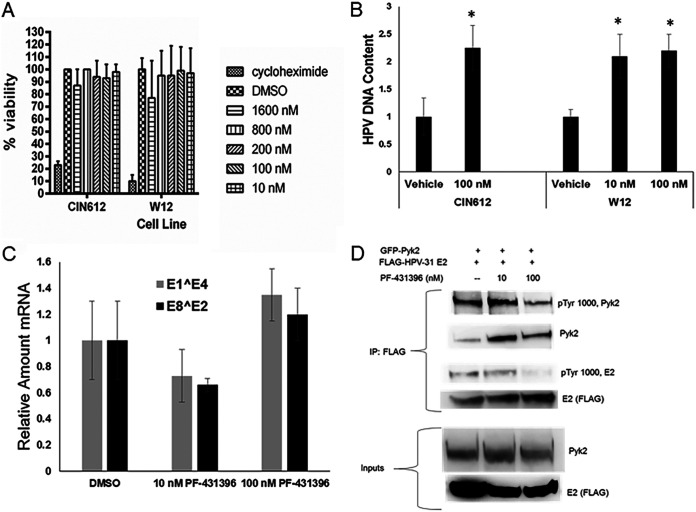
The viability of CIN612 and W12 cells treated with up to 1.2 μM concentrations of the Pyk2 kinase inhibitor PF-431396 (28) was not altered (Fig. 7A). HPV DNA context increased 2-fold after inhibitor treatment with 10 or 100 nM PF-431396 for 72 h in both cell lines (Fig. 7B), without a significant change in HPV transcripts in CIN612 cells (Fig. 7C). To assess whether Pyk2 inhibition altered E2 phosphorylation, HEK293TT cells were transfected with WT Pyk2 and HPV-31 E2 constructs and exposed to PF-431396 for 24 h. Immunoblotting showed that 100 nM PF-431396 decreased the phospho-E2 and phospho-Pyk2 levels (Fig. 7D).
FIG 7.
Pyk2 inhibition increases HPV DNA content by decreasing E2 tyrosine phosphorylation. (A) CIN612-9E and W12 cells were treated with the Pyk2 inhibitor PF-431396 for 24 h, and cell viability was quantified by an MTS cell proliferation assay. Values are expressed as means ± the SEM (n = 8). *, P < 0.05. (B) CIN612-9E cells were incubated with 100 nM PF-431396, and W12 cells were incubated with 10 or 100 nM PF-431396 for 72 h. DNA was isolated, and real-time-PCR was performed for HPV-31 or HPV-16 DNA region near the LCR and normalized to the levels of β-actin. Values are expressed as means ± the SEM (n = 6). *, P < 0.05. (C) CIN612-9E cells were treated with 10 or 100 nM PF-431396 for 72 h. RNA was isolated and converted to cDNA using reverse transcription-PCR. Real-time PCR was carried out with primers to HPV-31 E1̂E4 and HPV-31 E2̂E8 mRNA and normalized to 18S transcripts. Values are means ± the SEM (n = 3). *, P < 0.05. (D) HEK293TT cells were transfected with GFP-Pyk2 and FLAG-HPV-31 E2. Cells were treated with 10 or 100 nM PF-431396 for 24 h. E2 was immunoprecipitated with M2 (FLAG) antibodies. Complexes were blotted with pTyr-1000, Pyk2, and M2 (FLAG) antibodies.
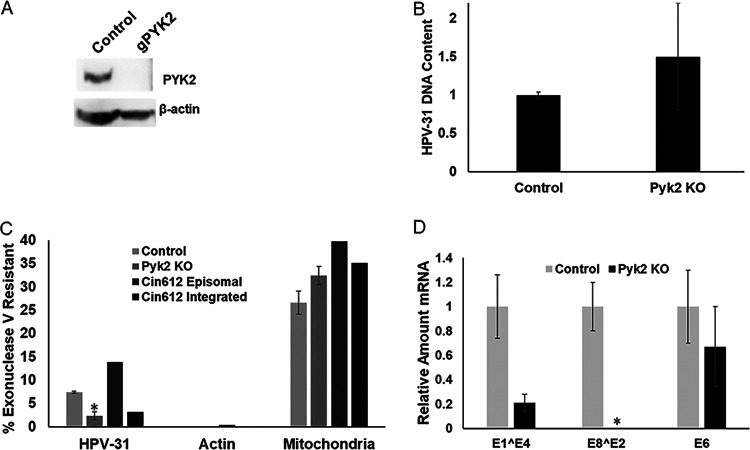
Pyk2 knockout (KO) CIN612-9E cells were created using lentivirus-expressed CRISPR-Cas9 and confirmed by immunoblotting with Pyk2 antibodies (Fig. 8A). We did not observe any changes in the phenotype of the Pyk2 KO cells. The HPV-31 DNA content was increased in these cells; however, the effect was not significant (Fig. 8B). To test whether these Pyk2-null cell lines maintained episomes or promoted HPV DNA integration, we used an exonuclease V assay (29, 30). Exonuclease V does not digest double-stranded circular DNA such as mitochondrial DNA or HPV episomes. This assay was used to quantify episomal maintenance. DNA isolated from control and Pyk2 KO cells was subjected to exonuclease V digestion and analyzed by qPCR. Actin DNA was completely digested after treatment with exonuclease V (Fig. 8C), while circular mitochondrial DNA was resistant in all samples (Fig. 8C). Pyk2 KO cells had significantly lower episomal HPV DNA (Fig. 8C), as well as spliced (E1̂E4 and E8̂E2) but not E6 transcripts (Fig. 8D) compared to the control. These results were observed in early passages of Pyk2 KO cell lines, because after passage 4 the CRISPR-Cas9 control cell lines had increased integrated HPV-31 genomes.
FIG 8.
Pyk2 knockout increases HPV genome integration. (A) CIN612-9E cells infected with pLentiCRISPR v2 (control) or pLentiCRISPR v2-Pyk2 guide constructs. Lysates were immunoblotted with Pyk2 and β-actin antibodies. (B) DNA from control and Pyk2 knockout (KO) CIN612-9E cell lines were analyzed by qPCR for the HPV-31 LCR and normalized to β-actin. Values are expressed as means ± the SEM (n = 9). (C) DNA from control and Pyk2 KO CIN612-9E cell lines were subjected to exonuclease V digestion. Resistant HPV-31 DNA was quantified by qPCR. Actin DNA was used as a positive control for digestion, and mitochondrial DNA was used as a resistant control. Values are expressed as means ± the SEM (n = 9) *, P < 0.05. (D). Real-time PCR was performed using primers for HPV-31 E1̂E4, E8̂E2, and E6 transcripts and normalized to actin transcripts. Values are means ± the SEM (n = 3). *, P < 0.05.
Characterization of E2 Y131.
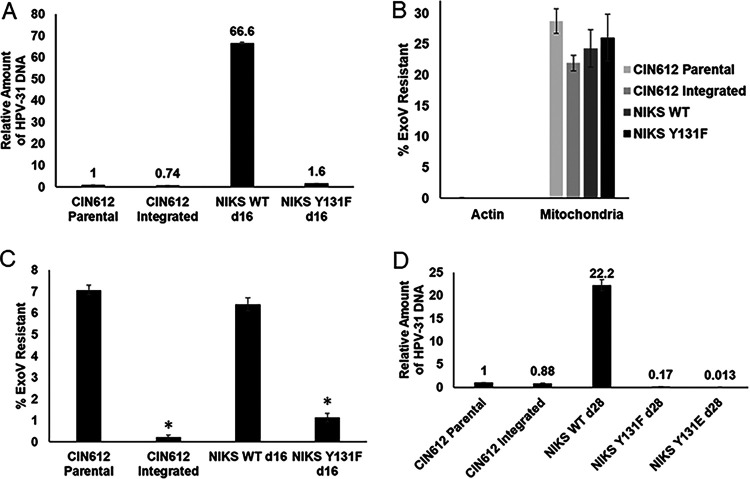
To test replication of genomes with E2 Y131 mutations, we generated quasiviruses packaged with G418-selectable HPV-31 genomes for infection of NIKS keratinocytes (31). At 2 weeks postinfection and G418 selection, colonies formed with WT and Y131F genomes. while only one small, slowly proliferating colony was observed with Y131E genomes. HPV Y131F genome copy (1.6-fold) and WT (66.6-fold) were increased compared to CIN612 cells with episomal HPV (Fig. 9A). WT genomes were maintained as episomes but Y131F displayed integration, as indicated by exonuclease V assay. To distinguish between circular and linear HPV DNA, CIN612 cells with HPV-31 episomal and integrated genomes were utilized as controls (29, 30) (Fig. 9B and C). At 28 days postinfection, genomic DNA was isolated from NIKS cells with WT, Y131F, and Y131E genomes. Both Y131F genome copy (0.17-fold compared to CIN612 cells with episomal HPV) and WT (22.2-fold) were lower than at 14 days (Fig. 9D). The HPV-31 Y131E genome copy was detected at very low levels (0.013-fold compared to CIN612, Fig. 9D).
FIG 9.
E2 Y131 is important for episomal maintenance. (A) NIKS cells were infected with HPV-31 WT and mutant quasiviruses and selected with G418 for 2 weeks. Quantification of viral HPV-31 DNA was carried out using qPCR in NIKS cells infected with HPV-31 WT and Y131F quasiviruses. Values are expressed as means ± the SEM (n = 3). (B) CIN612 episomal and integrated cell lines served as controls in the exonuclease V digestion assay. HPV-31 DNA from NIKS WT and Y131F quasiviruses was quantified using real-time PCR after exonuclease V digestion for actin and mitochondrial DNA. Values are means of the percent exonuclease V resistance ± the SEM (n = 3). (C) Percent exonuclease V resistance for HPV-31 DNA in CIN612 and NIKS WT and Y131F cell lines. Values are means ± the SEM (n = 3) *, P < 0.05 (compared to CIN612 cells by one-way t test). (D) NIKS cells were infected with HPV-31 WT and mutant quasiviruses and selected with G418 for 28 days. Quantification of HPV-31 DNA using qPCR in NIKS cells infected with HPV-31 WT and Y131 mutant quasiviruses. Values expressed as means ± the SEM (n = 3).
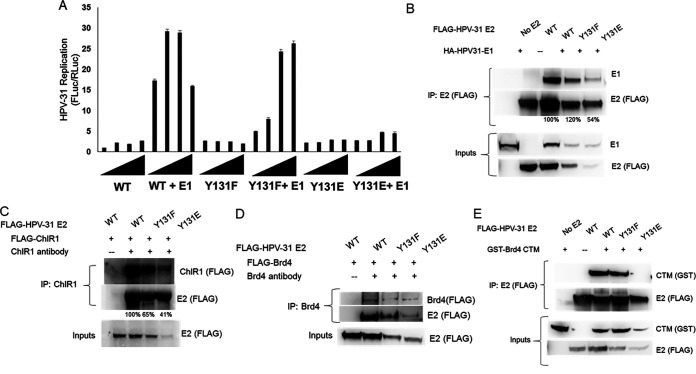
Because the E2 mutations affected genome copy number, it was necessary to independently test these in a transient-replication assay using CMV-based expression vectors for HPV-31 E1 and E2 mutants, along with an HPV ori-luciferase plasmid replicon (32). E2 Y131F but not Y131E activated E1-dependent replication equal to WT (Fig. 10A). Coimmunoprecipitation experiments demonstrated that Y131F maintained ability to bind to the E1 protein, as did Y131E, albeit at a lower efficiency (54%; Fig. 10B). An HPV-16 E2 mutation of this tyrosine to an alanine (Y131A) displayed reduced binding to the helicase ChlR1, which is necessary for mitotic segregation of HPV-16 episomes (33, 34). HEK293TT cells were transfected with FLAG-ChlR1 and FLAG-E2 Y131 mutants. While ChlR1 associated with the E2 mutants, Y131F and Y131E were reduced to 65 and 41% of the WT, respectively (Fig. 10C). We recently reported that another tyrosine in close proximity within the beta-strand region in the TAD, Y138, was important for binding E2 to the Brd4 CTM (aa 1224 to 1362) (14). Full-length Brd4 protein coimmunoprecipitated with both HPV-31 E2 Y131F and Y131E (Fig. 10D), which is known to occur independent of the CTM (35). Although Y131F complexed with the CTM, Y131E binding was not detectable (Fig. 10E).
FIG 10.
Y131E is unable to stimulate transient replication and is defective for Brd4 CTM binding. (A) C33A cells were transfected with HPV-31 E1 and HPV-31 Y131 mutants, along with pFLORI31 and pRL constructs. After 72 h later, the firefly and Renilla luciferase levels were measured using Dual-Glo luciferase reagent. The firefly luciferase levels were normalized to the Renilla luciferase levels. Values are expressed as means ± the SEM (n = 8). (B) HEK293TT cells transfected with HA-HPV-31 E1 and FLAG-HPV-31 E2 Y131 mutants. HPV-31 E2 was immunoprecipitated with M2 (FLAG) antibodies. Complexes were blotted with E1 and M2 (FLAG) antibodies. (C) HEK293TT cells were transfected with FLAG-ChlR1 and FLAG-HPV-31 E2 Y131 mutants. Immunoprecipitations were performed with goat ChlR1 antibodies and immunoblotted with M2 (FLAG) antibodies. (D) HEK293TT cells transfected with FLAG-Brd4 full-length and FLAG-HPV-31 E2 Y131 mutants. Immunoprecipitations were performed with Brd4 antibodies and immunoblotted with M2 (FLAG) antibodies. (E) HEK293TT cells transfected with GST-CTM and FLAG-HPV-31 E2 Y131 mutants. HPV-31 E2 was immunoprecipitated with M2 (FLAG) antibodies. Complexes were blotted with GST and M2 antibodies.
DISCUSSION
Several tyrosine kinases have been proposed to be E2 binding partners, including FGFR3 (27), FGFR2 (36), BAZ1B (37), and Pyk2 (19). A yeast two-hybrid screen revealed Pyk2 as an interacting partner of HPV-39 E2 and GPCA-based assay found that high-risk HPV-18 E2 interacted with Pyk2 (19). Pyk2 is distributed in the cytoplasm and the nucleus. It was previously reported that Pyk2 depletion by siRNA resulted in the retention of HPV-16 pseudoviruses in the trans-Golgi network (22). We describe the significance of E2 association with Pyk2 on HPV-16 and HPV-31 genome replication and identify the probable amino acid modified by this kinase as tyrosine 131.
Coimmunoprecipitations of HPV-16 and -31 E2 revealed that the interaction with Pyk2 is independent of its kinase activity. Experiments with E2 deletions suggest that Pyk2 binds to two distinct sites within aa 1 to 175 of the TAD and phosphorylates tyrosine(s) in this region. Tyrosine 131 is among one of several Y phosphorylations identified within HPV-31 E2 using mass spectrometry (MS) (13). However, MS studies with Pyk2 overexpression are not possible since this technique is not quantitative. Instead, we used a series of tyrosine-to-phenylalanine mutants in the TAD and were able to narrow down a potential target residue for Pyk2 phosphorylation at Y131. Interestingly, HPV-16 E2 Y131 mediates binding to the cellular DNA helicase ChlR1 (33). An alanine substitution at this E2 residue significantly reduced but did not completely abrogate this protein-protein interaction, which is essential for viral episomal association to mitotic chromosomes (6). HFKs harboring HPV-16 Y131A mutant genomes replicated initially but were undetectable at later passages (33). Y131E, similar to the nearby glutamate mutant at Y138, failed to replicate, and while it bound E1 with less efficiency, still recruited E1 to the viral origin (14), which therefore is unlikely to explain the inability of Y131E to initiate replication.
We analyzed the effects of Y131E in context of the whole genome. In this experiment, we used a physiologically relevant genome infection model (38) coupled with G418 gene to improve colony selection (39). We observed that infection efficiency was equivalent between the Y131 genome mutants compared to the WT in HEK293 and HeLa cells after G418 treatment. This suggests normal uptake of the virus, cellular processing, and transport of genomes to the nucleus. HPV DNA content was significantly lower and showed integration in cell lines infected with Y131 mutant genomes compared to cells infected with WT genomes. These data suggest that the dephosphorylated status at Y131 is essential for viral episomal establishment.
We previously reported that siRNA depletion of FGFR3 in CIN612 cells resulted in a modest 1.5-fold increase in viral copy number. Similarly, HPV genome content significantly increased after transient Pyk2 depletion in cell lines that maintain HPV episomes. Culture of CIN612 cells with 100 nM PF-431396, a FAK/Pyk2 inhibitor (28), reduced E2 phosphorylation and produced an increase in viral DNA content comparable to the levels achieved using Pyk2 siRNA knockdown. These data suggest a model in which Pyk2 activation is inhibitory for E2 induced HPV replication.
We expected that CRISPR-Cas9-mediated KO of Pyk2 in HPV genome containing cells would increase the viral genome content several fold. We observed a modest increase in HPV DNA content in these cells, but it was not significant. Exonuclease V assay revealed that Pyk2 knockout leads to viral genome integration. We postulate that Pyk2 knockout increased replication initially, but as the cells were passaged, the genomes became integrated or were lost due to failed mitotic segregation. Transient depletion or inhibition of Pyk2 results in increase in copy number, whereas permanent loss of Pyk2 leads to viral genome integration.
Pyk2 immunohistochemistry from normal skin tissues in the Protein Atlas Database (v18) displays high Pyk2 expression in the epidermal basal layer (40). We posit that Pyk2 kinase activity in basal cells restricts viral replication by tyrosine phosphorylation of E2 Y131 to maintain a low copy number. During differentiation-dependent genome amplification in upper epithelial strata, Pyk2 levels are low, and this would allow robust E2 activity. The mechanism(s) by which phosphorylation modulates E2 function appears to be the inability to bind to the Brd4-CTM, a phenotype observed with both Y131E and Y138E. Future studies are needed to understand how the association between E2 and the Brd4-CTM enables initiation of HPV DNA replication.
MATERIALS AND METHODS
Plasmids and antibodies.
The following plasmids were used. Codon-optimized pCDNA3-FLAG HPV-31 E2 and pCDNA3-FLAG-HPV-16 E2 (27, 41), FLAG-Pyk2 WT and KD (42), GFP-Pyk2 WT and KD (24), and FLAG-HPV-31 E2 fragments were described previously (14). FLAG-HPV-16 E2 fragments (aa 105 to 365 and aa 202 to 365) were created by PCR with oligonucleotides to the codon-optimized HPV-16 E2 (from A. McBride) and cloned into the HindIII and BamHI sites of pCDNA3. The FLAG-HPV-16 E2 1–104/202–365 fragment was created by PCR as described above. Amino acids 1 to 104 were cloned into the HindIII and BamHI sites of pCDNA3 and aa 202 to 365 into this vector using the EcoRI and ApaI sites. Y131 was mutated in pCDNA3-HPV-31 E2 using the Q5 site-directed mutagenesis kit (New England BioLabs) and confirmed by sequencing. pLentiCRISPR v2 was from Addgene (catalog no. 52961) (43), and pLentiCRISPR v2 human Pyk2 guide 1 was from GenScript. Reporter constructs pFLORI31 (ori FLuc) and pRL (RLuc) were previously described (32).
The antibodies included mouse anti-FLAG M2, anti-β-actin (sigma), rabbit anti-Pyk2 (Sigma), rabbit anti-Brd4 (Cell Signaling), goat-ChlR1 (6), rat anti-16E1 (44), rabbit-GST (14), and rabbit anti-pTyr-1000 (Cell Signaling). Mouse anti-HPV-16-E2 TVG261 and HPV-16 E2 sheep antiserum (45) were used to identify HPV-16 E2 proteins.
Cell culture.
All cell lines were maintained at 37°C and 5% CO2. HEK293TT (from J. Schiller and C. Buck), HeLa, and CV-1 cells were cultured in Dulbecco modified Eagle medium (DMEM; Life Technologies) with 10% fetal bovine serum (FBS; Perk Serum) and penicillin/streptomycin (100 U/ml; Life Technologies). CIN612-9E cells (from L. Laimins) were grown in E medium with J23T3 fibroblast feeders (from H. Green). W12 (from M. Stanley and P. Lambert) and NIKS cells were grown in F medium with J23T3 fibroblast feeders.
Immunofluorescence.
CV-1 cells were transfected with Lipofectamine 2000 and seeded onto coverslips. After 24 h, coverslips were fixed in 4% paraformaldehyde for 15 min, followed by phosphate-buffered saline (PBS) washes. Coverslips were placed in 10% goat serum in PBS/Triton X-100 (0.2%) for 30 min, followed by primary antibody in PBS-T overnight, washed with PBS, incubated in a 1:5,000 dilution of goat anti-mouse 594 (Invitrogen) for 1 h, washed with PBS, and mounted onto slides with Vectashield mounting media with DAPI (4′,6′-diamidino-2-phenylindole). Cells were analyzed with an Olympus FV1000 MPE confocal microscope. HeLa cells were transfected with polyethyleneimine (PEI; 2 mg/ml). After 48 h, the coverslips were fixed in 4% paraformaldehyde for 15 min, followed by PBS washes. The coverslips were treated as described above and analyzed with a Nikon microscope.
Coimmunoprecipitations and immunoblotting.
Cells were transfected with PEI (2 mg/ml) and lysed, and immunoprecipitation and immunoblotting were performed as previously described (14). The HPV-16 E2 coimmunoprecipitation methods were as described previously (36).
HPV DNA content PCR.
CIN612-9E and W12 cells without feeders were transfected using Lipofectamine 2000 (Invitrogen) and either control siRNA (Santa Cruz, sc-37007), PYK2 siRNA (Santa Cruz, sc-36332), or EGFR siRNA (Santa Cruz, sc-29301) at a final concentration 15 nM in E or F medium. After 72 h, the cells were lysed in Tris-EDTA with 0.1% SDS with 10 ng/μl RNase. DNA was isolated, and qPCR was performed using Sso Fast Evagreen Mastermix (Bio-Rad). The HPV-31 DNA content was measured using primers to HPV-31 LCR (5′-GTTCTGCGGTTTTTGGTTTC and 5′-TGTTGGCAAGGTGTGTTAGG-3′), and HPV-16 DNA was analyzed using the primers 5′-AAACTGCACATGGGTGTGTG-3′ and 5′-TTCGGTTACGCCCTTAGTTTT-3′. HPV DNA was normalized to β-actin DNA using the primers 5′-GAGGCACTCTTCCAGCCTTC-3′ and 5′-CGGATGTCCACGTCACACTT-3′ (44).
Pyk2 inhibitor PF-431396.
PF-431396 (Aobious, Inc.) stock solution (10 mM) was prepared in dimethyl sulfoxide. To test the inhibition of Pyk2 phosphorylation of E2 in HEK293TT, the cells were transfected and, 24 h later, the medium was replaced with DMEM plus 2.5% FBS with 10 or 100 nM PF-431396 for 24 h.
CIN612-9E and W12 cells, without feeders, were grown to 40% confluence and then treated with 10 or 100 nM PF-431396 in E or F media. After 72 h, the cells were lysed in Tris-EDTA plus 0.1% SDS with 10 ng/μl RNase. DNA was isolated with phenol-chloroform-isoamyl alcohol (25:24:1; Fisher Scientific), and qPCR was performed using Sso Fast Evagreen Mastermix (Bio-Rad). The HPV-31 and HPV-16 DNA contents were measured using the primers described above and normalized to β-actin DNA. RNA was isolated using TRIzol (Invitrogen). RNA was converted to cDNA using SuperScript III reverse transcriptase (Life Technologies). Real-time PCR was performed using Sso Fast Evagreen Mastermix (Bio-Rad). HPV-31 E1̂E4 transcripts were measured using previously described primers (46). HPV-31 E8̂E2 transcripts were detected using 5′-GTGGAAACGCAGCAGATGGTA-3′ and 5′-TTCGATGTGGTGGTGTTGTTG-3′, and HPV-31 E6 was measured using 5′-AAGACCGTTGTGTCCAGAAGA-3′ and 5′-CTGTCCACCTTCCTCCTATGTT-3′. Gene expression was normalized to 18S (29) or the actin transcripts with primers listed above. Cell viability against PF-431396 (10 nM to 1.6 μM) was tested in CIN612 and W12 cells using a CellTiter96 AQueous One solution cell proliferation assay (Promega).
Generation of Pyk2-null cell lines.
HEK293TT cells were transfected with pLentiCRISPRV2 or pLentiCRISPRV2 Pyk2 guide and Pax2 and VSVG constructs using PEI (2 mg/ml). The purified virus and Polybrene was added to CIN612-9E cells with puromycin-resistant J23T3 feeders. At 48 h after infection, the cells were treated with 1 μg/ml puromycin to generate polyclonal cell lines.
Exonuclease V digestion.
Cell pellets were resuspended in PBS and isolated by using the DNeasy blood and tissue kit (Qiagen). Then, 500 ng of DNA was either treated with exonuclease V (New England Biolabs) or left untreated for 2 to 4 h at 37°C. The enzyme was inactivated at 70°C for 20 min. Next, 50 ng of the digested or undigested DNA was quantified by real-time PCR. HPV-31 LCR and actin primers and the human mitochondrial DNA primers and exonuclease V protocol were described (27, 29, 44). Percent exonuclease V-resistant DNA was calculated as 2–(Digested CT – Undigested CT) × 100.
Transient DNA replication assays.
C33A cells were seeded into a 96-well plate, and each well was transfected with 0.5 ng of pRL (RLuc), 2.5 ng of pFLORI31, 10 ng of codon-optimized triple FLAG-HPV-31-E1 (32), and 10 ng of FLAG HPV-31 E2 WT, Y131F, or Y131E using Lipofectamine 2000. After 72 h, the cells were lysed, and the luciferase activity was measured using Dual Glo (Promega). The firefly luciferase levels were normalized to the Renilla luciferase levels.
Production of quasiviruses.
A QuikChange mutagenesis kit (Agilent) was used to create the Y131F and Y131E mutations of E2 in the pBR322-HPV31neoHC plasmid (13) using EcoRI and PvuI. Mutations were confirmed by sequencing. Wild-type and mutant genomes were digested overnight with HindIII, and the 8-kb HPV-31 genome was extracted from the vector using a PureLink quick gel extraction kit (Invitrogen). Genomes were ligated overnight with T4 DNA ligase (New England Biolabs).
Quasiviruses were produced as previously described (47) with the following modifications. HEK293TTF cells (from R. Roden), which express furin, were transfected with 10 μg of ligated genomes and 10 μg of HPV-16 capsid codon modified L1 and L2 genes (pShell L1L2, from J. Schiller) using 2 μg/ml PEI. The cells were washed in PBS with 10 mM MgCl2 and lysed in PBS containing 10 mM MgCl2, 0.5% Triton X-100, 25 mM ammonium sulfate (pH 9), and 5 mM CaCl2. Lysates were incubated at 37°C 48 h for maturation and spun at 7,000 rpm for 5 min (48).
To make extracellular matrix (ECM), HaCaT cells were seeded grown to confluence and washed in PBS and ECM buffer containing 0.5% Triton X-100 and 20 mM ammonium hydroxide in PBS (31). Virus stock was added to the well and placed at 37°C. After 6 h, the medium containing the virus particles was removed and replaced with NIKS cells. Then, after 48 h, 250 μg/ml of G418 was added to the NIKS cells.
Statistical analysis.
A two-way or one-way t test was used for analysis. Means are expressed ± the standard errors of the mean (SEM). The appearance of an asterisk (*) in the figures indicates a P value of ≤0.05.
ACKNOWLEDGMENTS
We thank the following individuals for their provision of plasmids. FLAG-PYK2 constructs were obtained from Joseph C. Loftus, Mayo Clinic, Phoenix, AZ. GFP-PYK2 constructs were obtained from Kristiina Vuori, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA. Brd4 constructs were provided by Cheng-Ming Chiang (University of Texas Southwestern Medical Center, Dallas, TX).
This study was supported by National Cancer Institute (NIH)/National Institutes of Health grants R01CA58376 (E.J.A.) and T32AI060519 (M.D.). M.D. is also supported by an award from the Ralph W. and Grace M. Showalter Research Trust at the Indiana University School of Medicine.
The content is solely the responsibility of the authors and does not represent the official views of NIH or the Indiana University School of Medicine. The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Hubert WG, Kanaya T, Laimins LA. 1999. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J Virol 73:1835–1845. doi: 10.1128/JVI.73.3.1835-1845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stubenrauch F, Lim HB, Laimins LA. 1998. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J Virol 72:1071–1077. doi: 10.1128/JVI.72.2.1071-1077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBride AA. 2013. The papillomavirus E2 proteins. Virology 445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Naidu SR, Sverdrup F, Androphy EJ. 2009. Tax1BP1 interacts with papillomavirus E2 and regulates E2-dependent transcription and stability. J Virol 83:2274–2284. doi: 10.1128/JVI.01791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parish JL, Bean AM, Park RB, Androphy EJ. 2006. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol Cell 24:867–876. doi: 10.1016/j.molcel.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Peng Y-C, Breiding DE, Sverdrup F, Richard J, Androphy EJ. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J Virol 74:5872–5879. doi: 10.1128/jvi.74.13.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinlan EJ, Culleton SP, Wu SY, Chiang CM, Androphy EJ. 2013. Acetylation of conserved lysines in bovine papillomavirus E2 by p300. J Virol 87:1497–1507. doi: 10.1128/JVI.02771-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culleton SP, Kanginakudru S, DeSmet M, Gilson T, Xie F, Wu SY, Chiang CM, Qi G, Wang M, Androphy EJ. 2017. Phosphorylation of the bovine papillomavirus E2 protein on tyrosine regulates its transcription and replication functions. J Virol 91:e01854-16. doi: 10.1128/JVI.01854-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang SW, Liu WC, Liao KY, Tsao YP, Hsu PH, Chen SL. 2014. Phosphorylation of HPV-16 E2 at serine 243 enables binding to Brd4 and mitotic chromosomes. PLoS One 9:e110882. doi: 10.1371/journal.pone.0110882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas Y, Androphy EJ. 2017. Human papillomavirus replication regulation by acetylation of a conserved lysine in the E2 protein. J Virol 92:e01912-17. doi: 10.1128/JVI.01912-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuck S, Ruse C, Stenlund A. 2013. CK2 phosphorylation inactivates DNA binding by the papillomavirus E1 and E2 proteins. J Virol 87:7668–7679. doi: 10.1128/JVI.00345-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilson T, Culleton S, Xie F, DeSmet M, Androphy EJ. 2020. HPV-31 tyrosine 102 regulates interaction with E2 binding partners and episomal maintenance. J Virol 94:e00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeSmet M, Jose L, Isaq N, Androphy EJ. 2019. Phosphorylation of a conserved tyrosine in the papillomavirus E2 protein regulates Brd4 binding and viral replication. J Virol 93doi:10.1128/jvi.01801–18. doi: 10.1128/JVI.01801-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinney C, Hussmann K, McBride A. 2015. The role of the DNA damage response throughout the papillomavirus life cycle. Viruses 7:2450–2469. doi: 10.3390/v7052450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong S, Laimins LA. 2013. Regulation of the life cycle of HPVs by differentiation and the DNA damage response. Future Microbiol 8:1547–1557. doi: 10.2217/fmb.13.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann R, Hirt B, Bechtold V, Beard P, Raj K. 2006. Different modes of human papillomavirus DNA replication during maintenance. J Virol 80:4431–4439. doi: 10.1128/JVI.80.9.4431-4439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinson T, Henno L, Toots M, Ustav M Jr, Ustav M. 2015. The cell cycle timing of human papillomavirus DNA replication. PLoS One 10:e0131675. doi: 10.1371/journal.pone.0131675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller M, Jacob Y, Jones L, Weiss A, Brino L, Chantier T, Lotteau V, Favre M, Demeret C. 2012. Large-scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions. PLoS Pathog 8:e1002761. doi: 10.1371/journal.ppat.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genna A, Lapetina S, Lukic N, Twafra S, Meirson T, Sharma VP, Condeelis JS, Gil-Henn H. 2018. Pyk2 and FAK differentially regulate invadopodia formation and function in breast cancer cells. J Cell Biol 217:375–395. doi: 10.1083/jcb.201702184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu SS, Jácamo RO, Vong SK, Rozengurt E. 2006. Differential regulation of Pyk2 phosphorylation at Tyr-402 and Tyr-580 in intestinal epithelial cells: roles of calcium, Src, Rho kinase, and the cytoskeleton. Cell Signal 18:1932–1940. doi: 10.1016/j.cellsig.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Gottschalk EY, Meneses PI. 2015. A dual role for the nonreceptor tyrosine kinase Pyk2 during the intracellular trafficking of human papillomavirus 16. J Virol 89:9103–9114. doi: 10.1128/JVI.01183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An X, Hao Y, Meneses PI. 2017. Host cell transcriptome modification upon exogenous HPV16 L2 protein expression. Oncotarget 8:90730–90747. doi: 10.18632/oncotarget.21817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao M, Finlay D, Zharkikh I, Vuori K. 2016. Novel role of Src in priming Pyk2 phosphorylation. PLoS One 11:e0149231. doi: 10.1371/journal.pone.0149231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong W, Parsons JT. 1997. Induction of apoptosis after expression of PYK2, a tyrosine kinase structurally related to focal adhesion kinase. J Cell Biol 139:529–539. doi: 10.1083/jcb.139.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindler EM, Baumgartner M, Gribben EM, Li L, Efimova T. 2007. The role of proline-rich protein tyrosine kinase 2 in differentiation-dependent signaling in human epidermal keratinocytes. J Invest Dermatol 127:1094–1106. doi: 10.1038/sj.jid.5700662. [DOI] [PubMed] [Google Scholar]
- 27.Xie F, DeSmet M, Kanginakudru S, Jose L, Culleton SP, Gilson T, Li C, Androphy EJ. 2017. Kinase activity of fibroblast growth factor receptor 3 regulates activity of the papillomavirus E2 protein. J Virol 91:e01066-17. doi: 10.1128/JVI.01066-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S, Mistry A, Chang JS, Cunningham D, Griffor M, Bonnette PC, Wang H, Chrunyk BA, Aspnes GE, Walker DP, Brosius AD, Buckbinder L. 2009. Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design. J Biol Chem 284:13193–13201. doi: 10.1074/jbc.M809038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bienkowska-Haba M, Luszczek W, Myers JE, Keiffer TR, DiGiuseppe S, Polk P, Bodily JM, Scott RS, Sapp M. 2018. A new cell culture model to genetically dissect the complete human papillomavirus life cycle. PLoS Pathog 14:e1006846. doi: 10.1371/journal.ppat.1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers JE, Guidry JT, Scott ML, Zwolinska K, Raikhy G, Prasai K, Bienkowska-Haba M, Bodily JM, Sapp MJ, Scott RS. 2019. Detecting episomal or integrated human papillomavirus 16 DNA using an exonuclease V-qPCR-based assay. Virology 537:149–156. doi: 10.1016/j.virol.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jose L, Androphy EJ, DeSmet M. 2020. Phosphorylation of the human papillomavirus E2 protein at tyrosine 138 regulates episomal replication. J Virol 94:e00488-20. doi: 10.1128/JVI.00488-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fradet-Turcotte A, Morin G, Lehoux M, Bullock PA, Archambault J. 2010. Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication. Virology 399:65–76. doi: 10.1016/j.virol.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris L, McFarlane-Majeed L, Campos-León K, Roberts S, Parish JL. 2017. The cellular DNA helicase ChlR1 regulates chromatin and nuclear matrix attachment of the human papillomavirus 16 E2 protein and high-copy-number viral genome establishment. J Virol 91:e01853-16. doi: 10.1128/JVI.01853-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ. 2006. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J Cell Sci 119:4857–4865. doi: 10.1242/jcs.03262. [DOI] [PubMed] [Google Scholar]
- 35.Wu S-Y, Nin DS, Lee AY, Simanski S, Kodadek T, Chiang C-M. 2016. BRD4 phosphorylation regulates HPV E2-mediated viral transcription, origin replication, and cellular MMP-9 expression. Cell Rep 16:1733–1748. doi: 10.1016/j.celrep.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeSmet M, Kanginakudru S, Jose L, Xie F, Gilson T, Androphy EJ. 2018. Papillomavirus E2 protein is regulated by specific fibroblast growth factor receptors. Virology 521:62–68. doi: 10.1016/j.virol.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang MK, Anderson DE, van Doorslaer K, McBride AA. 2015. A proteomic approach to discover and compare interacting partners of papillomavirus E2 proteins from diverse phylogenetic groups. Proteomics 15:2038–2050. doi: 10.1002/pmic.201400613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2004. Efficient intracellular assembly of papillomaviral vectors. J Virol 78:751–757. doi: 10.1128/jvi.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Doorslaer K, Porter S, McKinney C, Stepp WH, McBride AA. 2016. Novel recombinant papillomavirus genomes expressing selectable genes. Sci Rep 6:37782. doi: 10.1038/srep37782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. 2015. Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 41.Sakakibara N, Mitra R, McBride AA. 2011. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J Virol 85:8981–8995. doi: 10.1128/JVI.00541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riggs D, Yang Z, Kloss J, Loftus JC. 2011. The Pyk2 FERM regulates Pyk2 complex formation and phosphorylation. Cell Signal 23:288–296. doi: 10.1016/j.cellsig.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeSmet M, Kanginakudru S, Rietz A, Wu WH, Roden R, Androphy EJ. 2016. The replicative consequences of papillomavirus E2 protein binding to the origin replication factor ORC2. PLoS Pathog 12:e1005934. doi: 10.1371/journal.ppat.1005934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddiqa A, Leon KC, James CD, Bhatti MF, Roberts S, Parish JL. 2015. The human papillomavirus type 16 L1 protein directly interacts with E2 and enhances E2-dependent replication and transcription activation. J Gen Virol 96:2274–2285. doi: 10.1099/vir.0.000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terhune SS, Hubert WG, Thomas JT, Laimins LA. 2001. Early polyadenylation signals of human papillomavirus type 31 negatively regulate capsid gene expression. J Virol 75:8147–8157. doi: 10.1128/jvi.75.17.8147-8157.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 119:445–462. doi: 10.1385/1-59259-982-6:445. [DOI] [PubMed] [Google Scholar]
- 48.Wang JW, Matsui K, Pan Y, Kwak K, Peng S, Kemp T, Pinto L, Roden RBS. 2015. Production of furin-cleaved papillomavirus pseudovirions and their use for in vitro neutralization assays of L1- or L2-specific antibodies. Curr Protoc Microbiol 38:14B.5.1–5.26. doi: 10.1002/9780471729259.mc14b05s38. [DOI] [PMC free article] [PubMed] [Google Scholar]