CD8 TCR repertoires responding to chronic viral infections (HIV, hepatitis C virus [HCV], Epstein-Barr virus [EBV], and cytomegalovirus [CMV]) have limited breadth and diversity. How these repertoires change and are maintained throughout the chronic infection are unknown. We thus characterized the LCMV-specific CD8 TCR repertoires of stem-like and terminally exhausted subsets generated during chronic LCMV infections. During chronic LCMV infections, the repertoires started as diverse but became more clonal at the late time point. Further, the exhausted subset was composed of dominant clonotypes that were shared with the stem-like subset. Together, we demonstrate that the TCR repertoire contracts over time and is almost exclusively derived from the stem-like subset late during the persistent viral infection. Our data suggest that dominant clonotypes in the exhausted subset are derived from a diverse pool of stem-like clonotypes, which may be contributing to the clonality observed during chronic viral infections.
KEYWORDS: chronic viral infection, T cell exhaustion, T cell immunity, T cell receptor, lymphocytic choriomeningitis virus
ABSTRACT
Recent studies on chronic viral infections have defined a novel programmed cell death 1-positive (PD-1+) T cell factor 1-positive (TCF1+) stem-like CD8 T cell subset that gives rise to the terminally differentiated exhausted CD8 T cells. In this study, we performed T cell receptor beta (TCRβ) sequencing of virus-specific CD8 T cells during chronic lymphocytic choriomeningitis virus (LCMV) infection to examine the TCR diversity and lineage relationship of these two functionally distinct subsets. We found that >95% of the TCR repertoire of the exhausted CD8 T cell subset was shared with the stem-like CD8 T cells. The TCR repertoires of both CD8 T cell subsets were composed mostly of a few dominant clonotypes, but there was slightly more breadth and diversity in the stem-like CD8 T cells than their exhausted counterpart (∼40 versus ∼15 GP33+ clonotypes; ∼20 versus ∼7 GP276+ clonotypes). Interestingly, the breadth of the TCR repertoire was broader during the early stages (day 8) of the chronic infection than the later stages (days 45 to 60), showing that there was a narrowing of the TCR repertoire during chronic infection (∼2-fold GP33+ and GP276+ stem-like subset; ∼10-fold GP33+ and ∼5-fold GP276+ exhausted subset). In contrast, during acute LCMV infection, the TCR repertoire was much broader in both GP33-specific effector (∼160 clonotypes) and memory CD8 T cells (∼160 clonotypes). Overall, our data demonstrate that the virus-specific CD8 T cell TCR repertoire is broad and remains stable after acute LCMV infection, but it contracts and is narrower during chronic infection. Our study also shows that the repertoire of the exhausted CD8 T cell subset is almost completely derived from the stem-like CD8 T cell subset during established chronic LCMV infection.
IMPORTANCE CD8 TCR repertoires responding to chronic viral infections (HIV, hepatitis C virus [HCV], Epstein-Barr virus [EBV], and cytomegalovirus [CMV]) have limited breadth and diversity. How these repertoires change and are maintained throughout the chronic infection are unknown. We thus characterized the LCMV-specific CD8 TCR repertoires of stem-like and terminally exhausted subsets generated during chronic LCMV infections. During chronic LCMV infections, the repertoires started as diverse but became more clonal at the late time point. Further, the exhausted subset was composed of dominant clonotypes that were shared with the stem-like subset. Together, we demonstrate that the TCR repertoire contracts over time and is almost exclusively derived from the stem-like subset late during the persistent viral infection. Our data suggest that dominant clonotypes in the exhausted subset are derived from a diverse pool of stem-like clonotypes, which may be contributing to the clonality observed during chronic viral infections.
INTRODUCTION
CD8 T cells play a vital role in the antiviral response by recognizing and killing virally infected cells through their T cell receptors (TCRs). If the antigenic stimulus is cleared, as in an acute viral infection, a subset of the heterogenous pool of effector CD8 T cells will survive to become long-lived memory cells that are longitudinally maintained independently of TCR stimulation (1–5). In contrast, T cells that endure persistent antigenic stimulation induced by chronic viral infection or cancer eventually become dysfunctional or “exhausted.” T cell exhaustion is associated with the upregulation of various inhibitory receptors, most notably programmed cell death 1 (PD-1) (6–8), and can be defined as the subsequent inability of the immune system to completely clear the antigen due to functional impairments (9–12). It is now appreciated that “exhausted” CD8 T cells are a heterogenous population with regard to their gene expression profiles and dysfunctional states (13–16). This heterogeneity was definitively outlined by the recent discovery of a novel subset of antigen-specific PD-1-positive (PD-1+) T cell factor 1-positive (TCF1+) CD8 T cells that act as resource cells to sustain virus-specific CD8 T cell responses during a chronic viral infection (17–20). These self-renewing resource cells, henceforth called stem-like CD8 T cells, do not have cytolytic activities but have proliferative potential. Upon TCR and costimulatory signals (21), stem-like cells differentiate into terminally differentiated CD8 T cells, henceforth called exhausted, which harbor limited proliferative and cytotoxic potential. In addition to their transcriptional and epigenetic differences (17–19, 22), these two distinct subsets differ greatly in their tissue distribution. During chronic lymphocytic choriomeningitis virus (LCMV) infection, stem-like CD8 T cells preferentially reside in specialized niches within lymphoid organs, while exhausted cells are present in both lymphoid and nonlymphoid organs at sites of active infection (17, 23). Stem-like CD8 T cells have been recently also detected in both murine and human cancers and have been shown to play a vital role in antitumoral immunity (24–26). Importantly, stem-like CD8 T cells provide the proliferative burst observed upon initiation of PD-1-targeted therapies. This emphasizes the importance of these cells not only in maintaining CD8 T cell responses in the presence of chronic antigen but also contributing greatly in the clinical context of immunotherapy. However, the TCR relationships, as well as other characteristics of the TCR repertoire, such as breadth and diversity, of the two functionally distinct subsets are still unexplored. In this study, we used TCR beta (TCRβ) sequencing of LCMV-specific, tetramer-sorted CD8 T cells to study the diversity and lineage relationship between stem-like and exhausted subsets during chronic LCMV infection.
RESULTS
LCMV-specific TCR repertoires show substantial overlap between stem-like and exhausted CD8 T cells within mice.
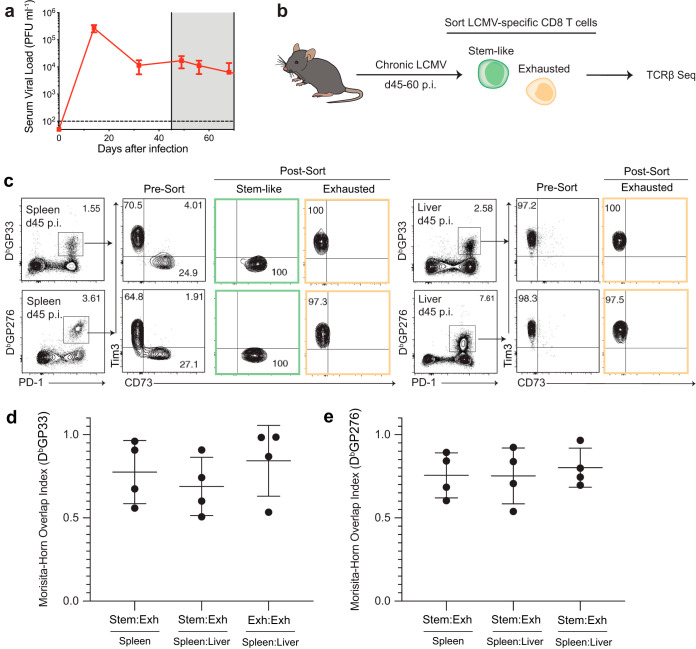
In this study, we investigated the TCR relationship between stem-like and exhausted LCMV-specific CD8 T cells during chronic LCMV infection by sequencing their TCRβ locus. We employed a stringent chronic LCMV infection model that results in lifelong viremia through transient depletion of CD4 T cells at the time of infection with the LCMV clone 13 strain (9). At the chronic time point (45 to 60 days postinfection [p.i.]) (Fig. 1a), PD-1-expressing LCMV GP33- and GP276-specific CD73+ Tim3-negative (Tim3−) stem-like CD8 T cells and CD73− Tim3+ exhausted CD8 T cells from the spleen and liver were sorted (Fig. 1b and c) (17) for TCRβ sequencing. We first assessed the overall overlap of the TCR repertoire between the stem-like CD8 T cells and the exhausted CD8 T cells using the Morisita-Horn overlap index. The Morisita-Horn overlap index measures the presence or absence of identical TCR sequences and also incorporates the frequency of shared clones. An overlap index of 0 corresponds to completely different repertoires, and 1 corresponds to identical repertoires. We observed that within individual mice, a high degree of overlap was observed between stem-like and exhausted T cells in the spleen (Fig. 1d and e). Furthermore, a comparable overlap was observed comparing the TCR repertoire of exhausted T cells from the spleen and liver. These data support our previously established lineage relationship model that stem-like CD8 T cells maintain the antiviral CD8 T cell response during a chronic infection by self-renewal and differentiation into the exhausted T cell subset (17). The high overlap between the two subsets, as well as exhausted cells from lymphoid and nonlymphoid organs at this time point, suggests that the LCMV-specific TCR repertoire is derived from the stem-like subset. Furthermore, since the stem-like CD8 T cells are not present in nonlymphoid organs such as the liver at this time point (17), our data suggest that the differentiation of stem-like CD8 T cells seeds the repertoire responding to systemic sites of infection.
FIG 1.
High clonal overlaps between the stem-like and exhausted subsets are observed within an individual mouse. (a) Serum viral load during chronic LCMV infection. (b) Mice were sacrificed at days 45 to 60 p.i. to isolate LCMV-specific stem-like and/or exhausted CD8 T cells from the spleen and liver for TCRβ sequencing. (c) Antigen-specific stem-like and exhausted CD8 T cells were sorted using pMHC tetramers, Tim3, and CD73. (d and e) The Morisita-Horn index was used to quantify the clonal overlap between the GP33-specific and GP276-specific stem-like (Stem) and exhausted (Exh) subsets within an individual mouse. Shown are the mean and standard deviation (SD) from experiments with 4 mice.
LCMV-specific TCR repertoires are private among inbred mice.
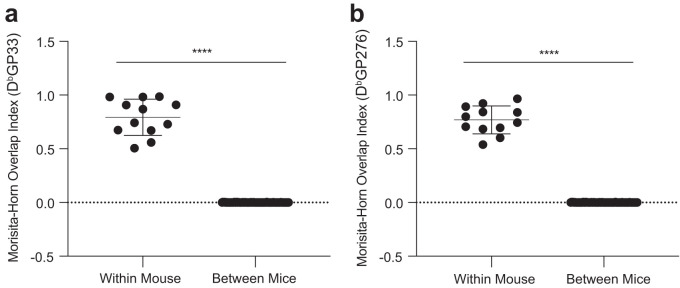
We then compared the TCR repertoire overlap of virus-specific CD8 T cells between individual mice using the Morisita-Horn overlap index. In both GP33+ and GP276+ CD8 T cell responses, we did not observe any significant overlap when comparing the TCR repertoires of virus-specific CD8 T cells between mice (Fig. 2a and b), while significant TCR overlap was present within an individual mouse (Fig. 1d and e). These data demonstrate that, despite analyzing inbred mice, individual GP33+ and GP276+ CD8 TCR repertoires are primarily composed of private, nonshared clonotypes. The private nature of the analyzed TCR repertoires (nonoverlapping TCRβ amino acid sequences) thus requires the separate analysis of individual TCR repertoires and does not allow pooling of LCMV-specific CD8 T cells for TCR repertoire analysis.
FIG 2.
LCMV-specific TCR overlaps are not observed between inbred mice. (a and b) The Morisita-Horn index was used to quantify the clonal overlap of GP33-specific (a) and GP276-specific (b) CD8 T cells within and between individual mice. Stem-like and exhausted subsets were compared within each individual mouse, and all possible pairwise comparisons between the stem-like and exhausted subsets were performed between mice. Shown are the mean and SD from experiments with 4 mice. ****, P ≤ 0.0001 (all determined using Student's t test).
Dominant clonotypes within the exhausted subset are shared with the stem-like population.
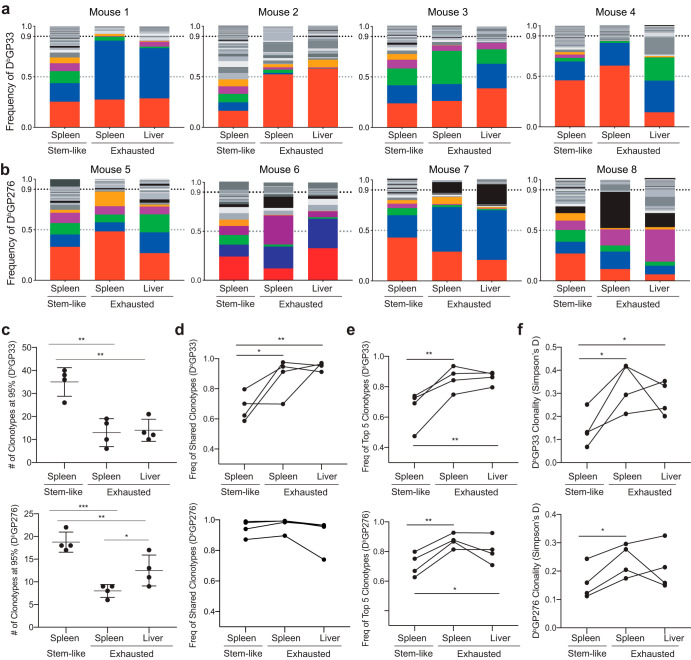
To address the question of how each clonotype contributed to the repertoire in each of the subsets, we next analyzed the frequency distribution of every unique clonotype in each subset within an individual mouse for GP33+ (Fig. 3a) and GP276+ (Fig. 3b) CD8 T cells. The exhausted subset was more oligoclonal than the stem-like subset, where the clones were more evenly distributed. Even though we observed a skewing of a few dominant clones in the stem-like subset in mouse 4, these dominant clones accounted for ∼60% of the stem-like repertoire versus ∼85% in the exhausted subset (Fig. 3a). Conversely, 1 to 3 major clonotypes dominated the exhausted repertoire in every mouse analyzed. These 1 to 3 dominant clonotypes within the exhausted subsets were also present in the stem-like subset in significant proportions (Fig. 3a). These data suggest that there is a bias toward certain dominant clonotypes capable of responding to the viral infection.
FIG 3.
The TCR repertoire of the stem-like subset is more broad and diverse compared to the exhausted. (a and b) GP33+ (n = 4) (a) and GP276+ (n = 4) (b) frequency distribution of each clonotype within individual mice. Each color indicates the same TCR clonotype only within an individual mouse. (c) Number of clonotypes that make up 95% of the repertoire in each of the subsets. (d) Cumulative frequency of clonotypes that are shared among all subsets in an individual mouse. (e) Cumulative frequency of the 5 most dominant clonotypes in each subset. (f) Quantification of diversity using Simpson’s D where D = 1 is monoclonal and D = 0 is completely polyclonal. Shown are the mean and SD from experiments with 4 mice. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 (all determined using paired analysis of variance [ANOVA] with multiple comparisons).
We next determined the number of unique TCR clonotypes, or the richness, present in each subset and accounting for 95% of detected sequences. Before assessing the richness, it is important to note that the frequency of LCMV-specific stem-like CD8 T cells in the spleen is ∼4-fold lower (10 to 30%) than the exhausted CD8 T cells (70 to 90%) (Fig. 1c) (17). Despite the lower cell numbers, both GP33+ and GP276+ stem-like CD8 T cells displayed greater richness than the exhausted subsets in the spleen and liver (Fig. 3c). The two exhausted subsets obtained from different organs showed similar richness (Fig. 3c). Additionally, the GP276-specific repertoire was slightly (∼1.5-fold) less broad than the GP33-specific repertoire. Due to the increased richness in the stem-like repertoire (Fig. 3c), we next asked what proportion of clonotypes in each subset was shared among all 3 analyzed subsets (stem-like cells from the spleen as well as exhausted cells from spleen and liver). In the GP276-specific repertoire, almost all clonotypes detected were shared by all subsets, and in the GP33-specific repertoire, ∼60 to 80% of the stem-like repertoire was composed of shared clonotypes (Fig. 3d). These data suggest that the majority of the repertoire in the exhausted subsets are derived from the stem-like subset and that there are unique stem-like clonotypes, particularly in the GP33-specific repertoire, that are not detectable in the exhausted subsets. We next wanted to quantify the evenness, or the diversity, of each repertoire by quantifying the cumulative frequencies of the top 5 clonotypes and the Simpson’s D index, respectively. The top 5 clonotypes accounted for <80% of the stem-like subset but >80% of the exhausted subset (Fig. 3e). Using Simpson’s D index, a diversity index that incorporates the richness, evenness, and the frequency of each clone, we observed that the greatest TCR diversity was evident in the stem-like subset (Fig. 3f). Together, these data demonstrate that there is a high overlap of the TCR repertoires and a clear clonal relationship between both subsets. Furthermore, it appears that the stem-like repertoire is broader and more diverse than the exhausted subset during chronic viral infection.
Virus-specific TCR repertoire diversity contracts during chronic viral infection.
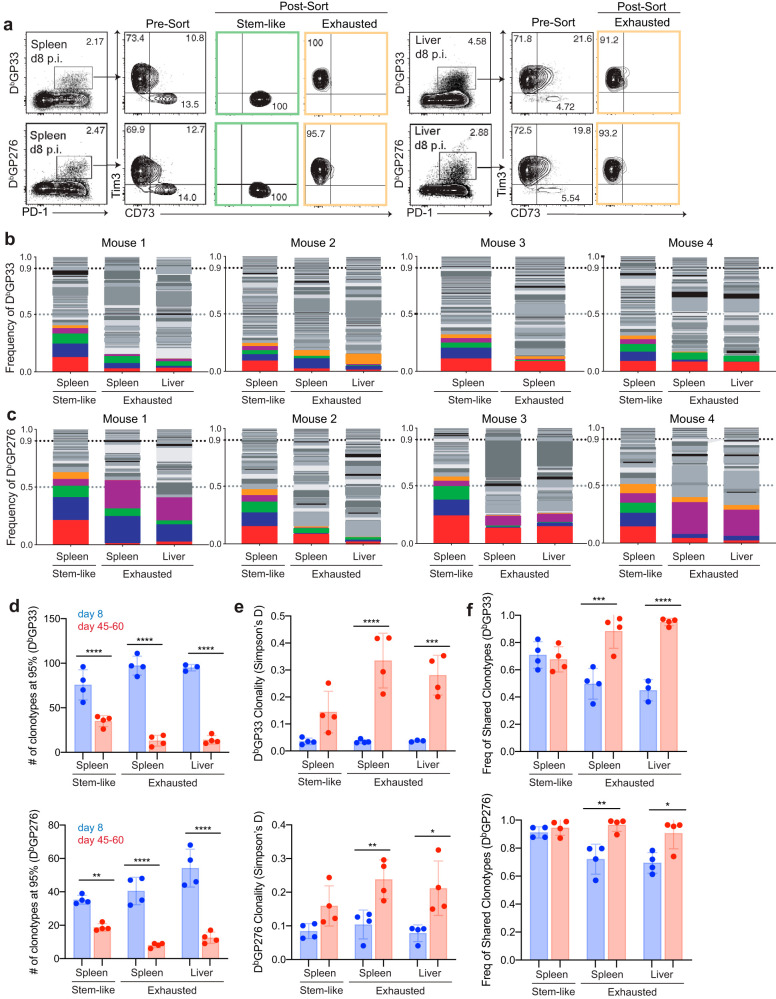
Because the two subsets showed a high TCR overlap at a late time point when the chronic infection was firmly established, we wanted to examine the repertoires early (day 8 p.i.) during chronic viral infection when the peak of the antiviral CD8 T cell response occurs (10, 27). We sorted tetramer-positive stem-like (CD73+ Tim3−) and exhausted (CD73− Tim3+) cells from spleen and liver 8 days after infection (Fig. 4a). We first examined the distribution of individual clonotypes. The analysis of the clonotype frequency distribution showed that in all analyzed mice, both subsets of GP33+, as well as GP276+, CD8 T cells had a broad and evenly distributed TCR repertoire at this early time point (Fig. 4b and c). No obvious dominant clonotypes (>30%) were present at this early stage of chronic viral infection.
FIG 4.
TCR repertoire diversity contracts during chronic viral infection. (a) Day 8 p.i. after chronic viral infection, LCMV-specific stem-like and/or exhausted CD8 T cells from the spleen and liver were sorted using pMHC tetramers, Tim3, and CD73 for TCRβ sequencing. (b and c) GP33+ (n = 4) (b) and GP276+ (n = 4) (c) specific frequency distribution of each clonotype within individual mouse. Each color indicates the same TCR clonotype only within an individual mouse. (d) Comparison of clonotype numbers between the two time points. (e) Comparison of the Simpson’s D diversity index between the two time points. (f) Comparison of the cumulative frequency of clonotypes shared among all subsets in an individual mouse. Shown are the mean and SD from experiments with 4 mice. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 (all determined using Student's t test).
We next compared the TCR repertoires observed at early and late time points postinfection in terms of the number of detected clonotypes and diversity and frequency of shared clonotypes between subsets. The TCR repertoire was broader at the early time point than the late time point (Fig. 4d). Importantly, there was a large contraction of richness in all subsets of both specificities, especially the exhausted subset (∼4- to 10-fold) from early to the late time point (Fig. 4d). Although a striking increase in the clonality (decrease in the diversity) of the exhausted subset was observed between the two time points, no significant changes in diversity were detected among stem-like subsets (Fig. 4e). Our data suggest that the diversity of the stem-like subset seems to change minimally over time, whereas the diversity of the exhausted subset diminishes over time.
We then wanted to address whether differences in the clonal overlap between the two subsets were present between two time points. To answer this question, we analyzed the cumulative frequency of the shared clonotypes present in all subsets (the stem-like subset in the spleen and the exhausted subsets in the spleen and liver). The cumulative frequencies of shared clonotypes were not different in the stem-like subsets between the two time points (Fig. 4f). However, in the exhausted subset, the shared clonotypes accounted for about 50% (GP33 specific) to 70% (GP276 specific) of the repertoire 1 week after the infection, whereas at the later time point, the majority, if not all, of the exhausted repertoire was composed of clonotypes derived from the stem-like subset (Fig. 4f). These data suggest that there are unique clonotypes in the stem-like subset that are not present in the exhausted subset both early and late during a chronic infection. Furthermore, these data suggest that a wide breadth of naive precursors are recruited during the initial antigen encounter but contract during chronic viral infections. Importantly, at the late time point, the vast majority of the TCR repertoire is derived from the stem-like population acting as resource cells that maintain the CD8 T cell response against the virus.
Comparison of TCR repertoire diversity between acute and chronic viral infection models.
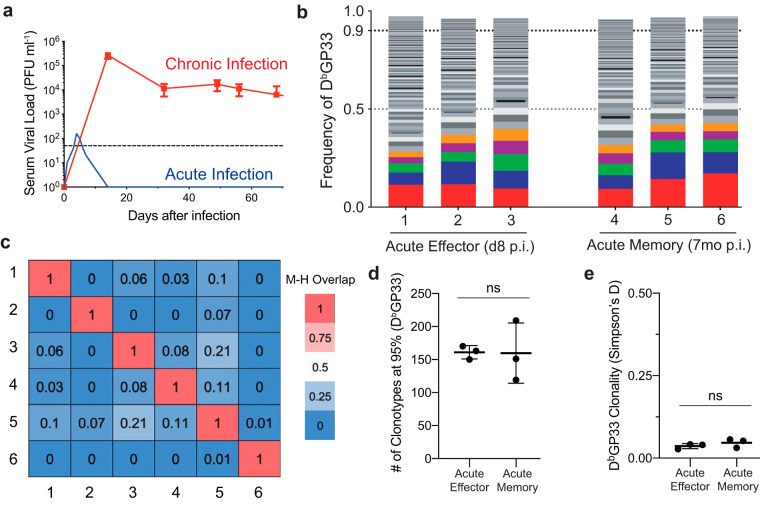
We next wanted to compare the TCR repertoire diversity between acute and chronic LCMV infections. We thus infected mice with the LCMV Armstrong strain, which is cleared within a week (Fig. 5a) and generates polyfunctional immune memory (2, 10, 27, 28). This is in contrast to the chronic viral infection model where viral burden is lifelong and canonical memory is not generated (27–29). The acute and chronic strains of LCMV differ only by two amino acids in the glycoprotein and viral polymerase but do not differ in their epitope specificities recognized by CD8 T cells (10, 30). Because CD8 T cell epitopes are identical between the two contrasting strains, the LCMV model provides an ideal system to compare and contrast the LCMV-specific TCR repertoire in an acute and chronic infection setting. We wanted to establish the repertoire in acutely infected mice by sorting GP33-specific splenic CD8 T cells from a set of mice at the peak of the effector phase (day 8 p.i.) and a set of different mice at a late memory time point (7 months p.i.) after viral clearance (Fig. 5b). We first assessed the clonal overlap between different mice and observed that the GP33-specific CD8 T cell repertoires were private even during the acute infection (Fig. 5c). GP33-specific CD8 T cells obtained at a late-memory time point retained a very broad TCR repertoire indistinguishable from the TCR repertoire observed at the peak of the effector phase (Fig. 5d). When comparing the clonality index, no significant changes in the diversity were observed between the two time points (Fig. 5e). These data suggest that, although the CD8 T cell response undergoes a major contraction (∼90 to 95%) (31, 32) in terms of total cell numbers during the formation of long-term memory, this contraction does not significantly affect the breadth of the TCR repertoire. Furthemore, the number of unique GP33-specific clonotypes (100 to 200) that are recruited and maintained are in line with the previously reported GP33-specific naive precursor frequency (51–53).
FIG 5.
A very broad and diverse TCR repertoire is maintained during and after an acute viral infection. (a) Longitudinal assessment of serum viral titer in acute LCMV Armstrong (cartoon illustration based on historical data [30, 50, 58]) and chronic LCMV Cl-13 (n = 15) infection models. (b) Six individual mice were sacrificed to analyze TCR frequency distribution of GP33-specific clonotypes 8 days (effector phase) and 7 months (memory phase) after an acute infection. Each color indicates the same TCR clonotype only within an individual mouse. (c) The Morisita-Horn (M-H) index was used to quantify the clonal overlap between mice. (d) Cross-sectional comparison of clonotype numbers between different time points. (e) Cross-sectional comparison of clonality measurements between different time points. Shown are the mean and SD from experiments with 3 mice. ns, nonsignificant (determined using Student's t test).
Because the repertoire between the effector and memory phases were congruous, we wondered how the TCR repertoires of LCMV-specific CD8 T cells responding to an acute infection compared to those of a chronic infection. We first compared the richness of GP33-specific CD8 T cells during acute and chronic LCMV infection. Even early in a chronic infection, the stem-like and exhausted populations exhibited a reduced TCR richness compared to both effector and memory populations following an acute infection (Fig. 6a). At the late time point, both the chronic stem-like and the exhausted subsets showed reduced richness (5- to 10-fold) compared to the acute response as well as the early chronic response (Fig. 6a). It is possible that both antigen chronicity and chronic inflammation may play a role in reducing the breadth of the TCR repertoire. In terms of the clonality measurement, no statistical differences were observed between the acute effector, acute memory, and the chronic stem-like subsets, but a dramatic increase in the clonality was observed in the exhausted subset at late stages of chronic infection (Fig. 6b). Overall, these data demonstrate that even during the early phase of a chronic viral infection, a narrowing of the repertoire of virus-specific CD8 T cells can be observed and that chronic antigen stimulation drastically reduces the breadth and increases the clonality of the exhausted TCR repertoire, which is supported by the stem-like cells that maintain a diverse TCR repertoire over time.
FIG 6.
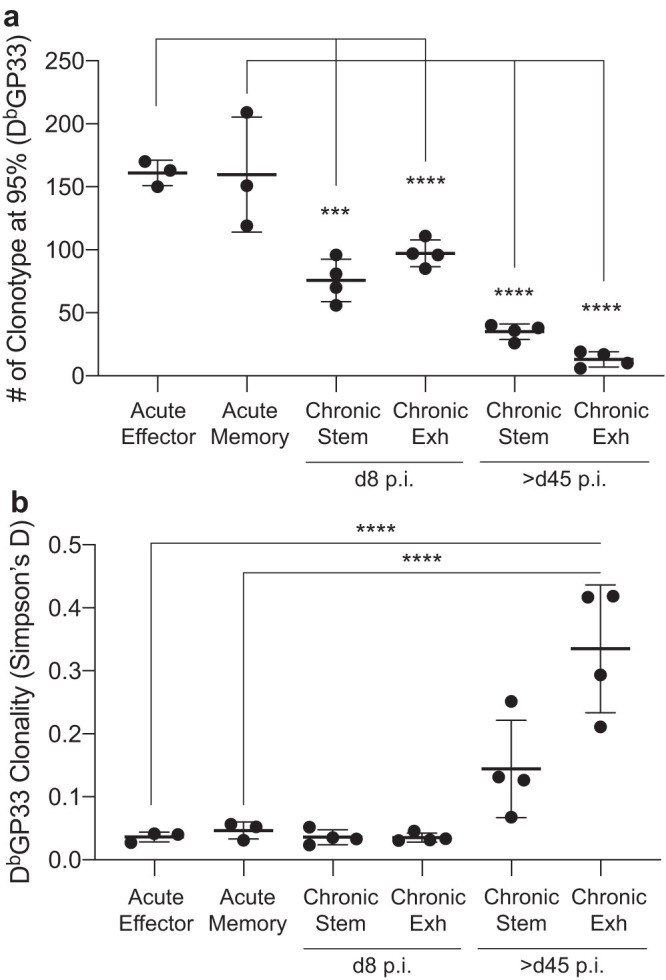
Comparison of GP33-specific TCR repertoire during acute and chronic LCMV infections. (a) Comparison of clonotype numbers between different subsets during acute and chronic infections. (b) Comparison of clonality between different subsets during acute and chronic infections. Shown are the mean and SD from experiments with 3 to 4 mice. ***, P ≤ 0.001; ****, P ≤ 0.0001 (all determined using ANOVA with multiple comparisons).
DISCUSSION
T cell immunity is equipped with vast TCR specificities that allow responses to a wide number of foreign and altered-self antigens. Because the TCR plays a crucial role in T cell immunity, it is important to establish the clonal relationship between various CD8 T cell subsets that respond to antigenic stimulation. Using the murine model of chronic LCMV infection, we demonstrate that PD-1+ TCF1+ stem-like CD8 T cells are clonally related to the PD-1+ TCF1− exhausted CD8 T cells. Despite analyzing the same epitope specificities in inbred mice, minimal TCR overlap was observed in LCMV-specific CD8 T cells between different mice in both chronic and acute models. This suggests that a highly private CD8 T cell repertoire is elicited against LCMV similar to those directed against an influenza nucleic acid polymerase-derived epitope (33). It is thus important to analyze individual TCR repertoire without pooling to prevent overestimation of richness and misinterpretation of clonal overlap. Interestingly, at the late time point, the stem-like subset exhibited a broader and more diverse TCR repertoire, with nearly all of the clonotypes present in the exhausted subsets in both the spleen and the liver being shared with the stem-like subset. At the earlier time point, the overlap between the stem-like and the exhausted cells is lower. It is possible that there are unique clonotypes in each subset. However, it is important to note that at this early time point, the CD8 T cell response is derived from a very diverse pool of naive CD8 T cells, and thus, lower overlap between the two subsets may be observed due to insufficient sampling of the TCR repertoire. Overall, the confirmation of this clonal relationship using TCR sequencing is important, as stem-like cells provide the proliferative burst upon PD-1 blockade, and changes in the TCR repertoire of unknown specificities have been used in the clinic to predict clinical outcome upon cancer immunotherapy (34–37). Furthermore, it would be pertinent to study the TCR relationship between the additional transitory CD8 T cell subset (38, 39) that emerges after PD-1 blockade and the two subsets described herein.
Finally, our results demonstrate that chronic antigenic stimulation narrows the TCR repertoire of antigen-specific CD8 T cells. This contraction is also observed in antigen-specific CD8 responses against HIV, Epstein-Barr virus (EBV), cytomegalovirus (CMV), and hepatitis C virus (HCV) in humans (40–47). The narrowing of the TCR repertoire was observed most obviously in the exhausted subset where both the richness and the diversity were dramatically reduced from early to late time points postinfection. The stem-like subset did have a small decrease in the richness, but no significant decrease in the diversity was observed over time. However, in an acute infection setting where, after antigen clearance, activated CD8 T cells differentiate into long-lived memory cells (48, 49), the TCR repertoire was very broad and diverse with no apparent differences in the richness or the evenness over time, suggesting that the breadth and diversity of the TCR repertoire is efficiently maintained during the contraction phase of an acute viral infection.
Interestingly, even during the early phases of chronic viral infection, a reduction in breadth was observed, which was followed by a loss of both richness and evenness, particularly in the exhausted subset, as antigen persisted. This contraction observed even at the early stages of LCMV Cl-13 compared to the Armstrong infection can be explained by the differences in the initial antigen exposure. The Armstrong strain is minimally disseminated and mostly cleared within the first week of infection. However, the Cl-13 strain is highly disseminated with high viral loads that persist in multiple organs (50). It would be of interest to study the TCR relationship between the stem-like and exhausted CD8 T cell subsets in the CD4-helped model of LCMV Cl-13 specifically after 4 to 6 weeks, when the mice start clearing the virus from many tissues.
Further investigation is needed to explain why dichotomous repertoire differences exist early on between the acute and chronic models and why diversity differences between the two clonally related subsets at the chronic phase of the infection are observed. The contraction of diversity observed during chronic viral infection may be due to the generation of clonotypes with differing capacity for persistence. The differing capacity of clonotypes to persist in the presence of chronic antigen stimulation may be linked to their intrinsic TCR affinity (46), induction of positive costimulatory signals, and/or cytokine milieu in which they differentiate. Another possibility is that stochastic recruitment, differentiation, and persistence of certain clonotypes contributes to the contraction of diversity across time in the setting of chronic antigen stimulations (54). Deciphering the mechanisms contributing to the persistence of specific clonotypes might provide novel insights and opportunities to improve the efficacy of immunotherapeutic interventions. Overall, using a clonal relationship approach, we confirmed in this study that stem-like CD8 T cells sustain the immune response during a chronic antigen setting and that antigen chronicity results in a contraction of the TCR repertoire, which is primarily observed in the exhausted subset.
MATERIALS AND METHODS
Mice and viruses.
All animal experiments were performed in accordance with Emory University Institutional Animal Care and Use Committee. C57BL/6J female mice were purchased from Jackson Laboratory. Mice were infected with LCMV Armstrong strain (2 × 105 PFU, intraperitoneally [i.p.]) for acute infection or with LCMV clone 13 strain (2 × 106 PFU, intravenously) after the transient depletion of CD4+ T cells by treatment with 300 μg anti-CD4 antibody (GK1.5 clone) i.p. twice, establishing a lifelong chronic viral infection (9).
Flow cytometry, cell sorting, gDNA isolation, and TCRβ sequencing.
Surface staining was performed using fluorochrome-conjugated antibodies against CD8, CD4, CD19, PD-1, Tim-3, CD73, and CD44 in phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS) on ice for 30 min. For detecting LCMV-specific CD8 T cell responses, peptides bound to major histocompatibility complex (pMHC) tetramers were prepared as described previously (32, 55). Cell viability was determined with the Live/Dead fixable aqua dead cell stain kit (Invitrogen). Flow cytometric sorting was performed on a FACSAria II (BD Biosciences). Lymphocytes from the spleen and liver were isolated as described previously (10). Fluorescence-activated cell sorter (FACS) data were analyzed with FlowJo software (TreeStar). The sorted cells were lysed, and genomic DNA (gDNA) was isolated using the QIAmp DNA micro kit (Qiagen) according to the manufacturer’s instructions. The isolated gDNA was sent to Adaptive Biotechnologies (Seattle, WA, USA) for TCRβ sequencing.
TCR repertoire analysis.
Complementarity determining region 3 (CDR3) sequences were called and quantified using immunoSEQ analyzer (Adaptive Biotechnologies). Clonotype number, or richness, was calculated as the number of clonotypes that made up 95% of the detected sequences. TCR overlap was defined as two clonotypes that share identical CDR3 amino acid sequences. The Morisita-Horn index (56) was used to quantify the clonal overlap as previously described. Simpson’s D (57) was used to quantify the diversity of the repertoire as previously described, where D = 1 is monoclonal and D = 0 is completely polyclonal.
Data analysis.
All experiments were analyzed using Prism 8 (GraphPad Software). Statistical differences were assessed using a two-tailed unpaired or paired Student's t test with 95% confidence interval. P values of <0.05 indicated the significant difference between relevant groups.
Data availability.
All TCRβ sequencing data are publicly available through the immune ACCESS portal via Adaptive Biotechnologies at https://doi.org/10.21417/YMC2020JV.
ACKNOWLEDGMENTS
This work was supported by grants RO1 AI030048 and P01 AI056299 to R.A. and a Adaptive Biotechnologies Young Investigator Award to Y.M.C.
We thank Robert Karaffa and Kametha Fife of the Emory Flow Cytometry Core for sorting assistance.
We declare no conflict of interest.
REFERENCES
- 1.Kaech SM, Wherry EJ. 2007. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity 27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 3.Kaech SM, Wherry EJ, Ahmed R. 2002. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol 2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 4.Murali-Krishna K, Lau LL, Sambhara S, Lemonnier F, Altman J, Ahmed R. 1999. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science 286:1377–1381. doi: 10.1126/science.286.5443.1377. [DOI] [PubMed] [Google Scholar]
- 5.Lau LL, Jamieson BD, Somasundaram T, Ahmed R. 1994. Cytotoxic T-cell memory without antigen. Nature 369:648–652. doi: 10.1038/369648a0. [DOI] [PubMed] [Google Scholar]
- 6.Wherry EJ. 2011. T cell exhaustion. Nat Immunol 12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, Araki K, Ahmed R. 2018. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med 69:301–318. doi: 10.1146/annurev-med-012017-043208. [DOI] [PubMed] [Google Scholar]
- 8.Sharma P, Allison JP. 2015. The future of immune checkpoint therapy. Science 348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 9.Matloubian M, Concepcion RJ, Ahmed R. 1994. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol 68:8056–8063. doi: 10.1128/JVI.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77:4911–4927. doi: 10.1128/jvi.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahan SM, Wherry EJ, Zajac AJ. 2015. T cell exhaustion during persistent viral infections. Virology 479-480:180–193. doi: 10.1016/j.virol.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao C, Sun HW, Lacey NE, Ji Y, Moseman EA, Shih HY, Heuston EF, Kirby M, Anderson S, Cheng J, Khan O, Handon R, Reilley J, Fioravanti J, Hu J, Gossa S, Wherry EJ, Gattinoni L, McGavern DB, O'Shea JJ, Schwartzberg PL, Wu T. 2019. Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nat Immunol 20:890–901. doi: 10.1038/s41590-019-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paley MA, Kroy DC, Odorizzi PM, Johnnidis JB, Dolfi DV, Barnett BE, Bikoff EK, Robertson EJ, Lauer GM, Reiner SL, Wherry EJ. 2012. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. 2012. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol 86:8161–8170. doi: 10.1128/JVI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, Wherry EJ. 2009. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, Sharpe AH, Freeman GJ, Germain RN, Nakaya HI, Xue HH, Ahmed R. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537:417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, Ye L. 2016. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 537:412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 19.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, Pradervand S, Thimme R, Zehn D, Held W. 2016. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity 45:415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 20.Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, Wu W, Lang PA, Gattinoni L, McGavern DB, Schwartzberg PL. 2016. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1:eaai8593. doi: 10.1126/sciimmunol.aai8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, Sica GL, Sharpe AH, Freeman GJ, Blazar BR, Turka LA, Owonikoko TK, Pillai RN, Ramalingam SS, Araki K, Ahmed R. 2017. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355:1423–1427. doi: 10.1126/science.aaf0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jadhav RR, Im SJ, Hu B, Hashimoto M, Li P, Lin JX, Leonard WJ, Greenleaf WJ, Ahmed R, Goronzy JJ. 2019. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci U S A 116:14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Im SJ, Konieczny BT, Hudson WH, Masopust D, Ahmed R. 2020. PD-1+ stemlike CD8 T cells are resident in lymphoid tissues during persistent LCMV infection. Proc Natl Acad Sci U S A 117:4292–4299. doi: 10.1073/pnas.1917298117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, Freeman SS, Reuben A, Hoover PJ, Villani AC, Ivanova E, Portell A, Lizotte PH, Aref AR, Eliane JP, Hammond MR, Vitzthum H, Blackmon SM, Li B, Gopalakrishnan V, Reddy SM, Cooper ZA, Paweletz CP, Barbie DA, Stemmer-Rachamimov A, Flaherty KT, Wargo JA, Boland GM, Sullivan RJ, Getz G, Hacohen N. 2018. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175:998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, Luther SA, Speiser DE, Held W. 2019. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50:195–211.e10. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 26.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, Manos M, Gjini E, Kuchroo JR, Ishizuka JJ, Collier JL, Griffin GK, Maleri S, Comstock DE, Weiss SA, Brown FD, Panda A, Zimmer MD, Manguso RT, Hodi FS, Rodig SJ, Sharpe AH, Haining WN. 2019. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20:326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wherry EJ, Barber DL, Kaech SM, Blattman JN, Ahmed R. 2004. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc Natl Acad Sci U S A 101:16004–16009. doi: 10.1073/pnas.0407192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 29.Utzschneider DT, Legat A, Fuertes Marraco SA, Carrie L, Luescher I, Speiser DE, Zehn D. 2013. T cells maintain an exhausted phenotype after antigen withdrawal and population reexpansion. Nat Immunol 14:603–610. doi: 10.1038/ni.2606. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. 1984. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med 160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busch DH, Pilip IM, Vijh S, Pamer EG. 1998. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity 8:353–362. doi: 10.1016/s1074-7613(00)80540-3. [DOI] [PubMed] [Google Scholar]
- 32.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 33.Kedzierska K, Day EB, Pi J, Heard SB, Doherty PC, Turner SJ, Perlman S. 2006. Quantification of repertoire diversity of influenza-specific epitopes with predominant public or private TCR usage. J Immunol 177:6705–6712. doi: 10.4049/jimmunol.177.10.6705. [DOI] [PubMed] [Google Scholar]
- 34.Han J, Duan J, Bai H, Wang Y, Wan R, Wang X, Chen S, Tian Y, Wang D, Fei K, Yao Z, Wang S, Lu Z, Wang Z, Wang J. 2020. TCR repertoire diversity of peripheral PD-1(+)CD8(+) T cells predicts clinical outcomes after immunotherapy in patients with non-small cell lung cancer. Cancer Immunol Res 8:146–154. doi: 10.1158/2326-6066.CIR-19-0398. [DOI] [PubMed] [Google Scholar]
- 35.Hogan SA, Courtier A, Cheng PF, Jaberg-Bentele NF, Goldinger SM, Manuel M, Perez S, Plantier N, Mouret JF, Nguyen-Kim TDL, Raaijmakers MIG, Kvistborg P, Pasqual N, Haanen J, Dummer R, Levesque MP. 2019. Peripheral blood TCR repertoire profiling may facilitate patient stratification for immunotherapy against melanoma. Cancer Immunol Res 7:77–85. doi: 10.1158/2326-6066.CIR-18-0136. [DOI] [PubMed] [Google Scholar]
- 36.Hopkins AC, Yarchoan M, Durham JN, Yusko EC, Rytlewski JA, Robins HS, Laheru DA, Le DT, Lutz ER, Jaffee EM. 2018. T cell receptor repertoire features associated with survival in immunotherapy-treated pancreatic ductal adenocarcinoma. JCI Insight 3:e122092. doi: 10.1172/jci.insight.122092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, Gupta RK, Curtis C, Bucktrout SL, Davis MM, Chang ALS, Chang HY. 2019. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 25:1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin JX, Konieczny BT, Im SJ, Freeman GJ, Leonard WJ, Kissick HT, Ahmed R. 2019. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1(+) stem-like CD8(+) T cells during chronic infection. Immunity 51:1043–1058.e4. doi: 10.1016/j.immuni.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, Cui W. 2019. CD4(+) T cell help is required for the formation of a cytolytic CD8(+) T cell subset that protects against chronic infection and cancer. Immunity 51:1028–1042.e4. doi: 10.1016/j.immuni.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Annels NE, Callan MF, Tan L, Rickinson AB. 2000. Changing patterns of dominant TCR usage with maturation of an EBV-specific cytotoxic T cell response. J Immunol 165:4831–4841. doi: 10.4049/jimmunol.165.9.4831. [DOI] [PubMed] [Google Scholar]
- 41.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, Nayak L, Moss PA. 2002. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol 169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 42.Kolowos W, Schmitt M, Herrman M, Harrer E, Low P, Kalden JR, Harrer T. 1999. Biased TCR repertoire in HIV-1-infected patients due to clonal expansion of HIV-1-reverse transcriptase-specific CTL clones. J Immunol 162:7525–7533. [PubMed] [Google Scholar]
- 43.Levitsky V, de Campos-Lima PO, Frisan T, Masucci MG. 1998. The clonal composition of a peptide-specific oligoclonal CTL repertoire selected in response to persistent EBV infection is stable over time. J Immunol 161:594–601. [PubMed] [Google Scholar]
- 44.Lim A, Trautmann L, Peyrat MA, Couedel C, Davodeau F, Romagne F, Kourilsky P, Bonneville M. 2000. Frequent contribution of T cell clonotypes with public TCR features to the chronic response against a dominant EBV-derived epitope: application to direct detection of their molecular imprint on the human peripheral T cell repertoire. J Immunol 165:2001–2011. doi: 10.4049/jimmunol.165.4.2001. [DOI] [PubMed] [Google Scholar]
- 45.Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. 2004. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med 200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schober K, Voit F, Grassmann S, Müller TR, Eggert J, Jarosch S, Weißbrich B, Hoffmann P, Borkner L, Nio E, Fanchi L, Clouser CR, Radhakrishnan A, Mihatsch L, Lückemeier P, Leube J, Dössinger G, Klein L, Neuenhahn M, Oduro JD, Cicin-Sain L, Buchholz VR, Busch DH. 2020. Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection. Nat Immunol 21:434–441. doi: 10.1038/s41590-020-0628-2. [DOI] [PubMed] [Google Scholar]
- 47.Wilson JD, Ogg GS, Allen RL, Goulder PJ, Kelleher A, Sewell AK, O'Callaghan CA, Rowland-Jones SL, Callan MF, McMichael AJ. 1998. Oligoclonal expansions of CD8(+) T cells in chronic HIV infection are antigen specific. J Exp Med 188:785–790. doi: 10.1084/jem.188.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, Alexe G, Nagar S, McCausland MM, Gupta S, Tata P, Haining WN, McElrath MJ, Zhang D, Hu B, Greenleaf WJ, Goronzy JJ, Mulligan MJ, Hellerstein M, Ahmed R. 2017. Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, Dogra P, Davis CW, Konieczny BT, Antia R, Cheng X, Ahmed R. 2017. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 552:404–409. doi: 10.1038/nature25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matloubian M, Kolhekar SR, Somasundaram T, Ahmed R. 1993. Molecular determinants of macrophage tropism and viral persistence: importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J Virol 67:7340–7349. doi: 10.1128/JVI.67.12.7340-7349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. 2008. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol 181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obar JJ, Khanna KM, Lefrancois L. 2008. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity 28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blattman JN, Antia R, Sourdive DJ, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R. 2002. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med 195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Graf P, Verschoor A, Schiemann M, Hofer T, Busch DH. 2013. Disparate individual fates compose robust CD8+ T cell immunity. Science 340:630–635. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 55.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 56.Horn HS. 1966. Measurement of “overlap” in comparative ecological studies. The Am Nat 100:419–424. doi: 10.1086/282436. [DOI] [Google Scholar]
- 57.Simpson EH. 1949. Measurement of diversity. Nature 163:688–688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 58.Ahmed R, Byrne JA, Oldstone MB. 1984. Virus specificity of cytotoxic T lymphocytes generated during acute lymphocytic choriomeningitis virus infection: role of the H-2 region in determining cross-reactivity for different lymphocytic choriomeningitis virus strains. J Virol 51:34–41. doi: 10.1128/JVI.51.1.34-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All TCRβ sequencing data are publicly available through the immune ACCESS portal via Adaptive Biotechnologies at https://doi.org/10.21417/YMC2020JV.