In the present study, the role of CLDN4 in PRRSV infection was studied. The results showed that CLDN4 blocked absorption into cells and restricted extracellular viral particles via the interaction between the CLDN4 small extracellular loop, ECL2, and the viral surface protein GP3. GP3 was found to downregulate CLDN4 through ubiquitination of the transcription factor SP1 to facilitate viral entry. The mechanism by which highly pathogenic PRRSV infects the epithelium is suggested. A novel function of GP3 in regulating gene transcription was discovered. Moreover, ECL2 could block PRRSV absorption and infection, as well as neutralizing the virus in the supernatant, which may lead to the development of novel therapeutic antiviral strategies.
KEYWORDS: GP3, blocking, claudin 4, porcine reproductive and respiratory syndrome virus, transcriptional regulation
ABSTRACT
Claudins (CLDN) are a family of proteins that represent the most important components of tight junctions, where they establish the paracellular barrier that controls the flow of molecules in the intercellular space between epithelial cells. Several types of viruses make full use of CLDN to facilitate entry into cells. Porcine reproductive and respiratory syndrome virus (PRRSV) is one of the most important pathogens in the swine industry. In this study, we found that CLDN4 functions as an anti-PRRSV factor by blocking its absorption during the early stages of infection. The small extracellular loop (ECL2) of CLDN4 restricted the viral particles outside cells by binding to GP3. A novel function of GP3-mediated regulation of CLDN4 transcription was suggested. CLDN4 can be decreased through downregulating the level of CLDN4 transcription by ubiquitinating the transcription factor, SP1. The mechanism by which highly pathogenic PRRSV infects the epithelium was proposed. Importantly, ECL2 was found to block PRRSV absorption and infection and neutralize the virus. A more in-depth understanding of PRRSV infection is described, and novel therapeutic antiviral strategies are discussed.
IMPORTANCE In the present study, the role of CLDN4 in PRRSV infection was studied. The results showed that CLDN4 blocked absorption into cells and restricted extracellular viral particles via the interaction between the CLDN4 small extracellular loop, ECL2, and the viral surface protein GP3. GP3 was found to downregulate CLDN4 through ubiquitination of the transcription factor SP1 to facilitate viral entry. The mechanism by which highly pathogenic PRRSV infects the epithelium is suggested. A novel function of GP3 in regulating gene transcription was discovered. Moreover, ECL2 could block PRRSV absorption and infection, as well as neutralizing the virus in the supernatant, which may lead to the development of novel therapeutic antiviral strategies.
INTRODUCTION
Tight junctions (TJs) are highly specialized membrane domains that regulate paracellular permeability by forming intercellular barriers between epithelial cells. TJs are also highly involved in a variety of cellular signaling transduction pathways (e.g., cell proliferation, differentiation, cell-cell adhesion, gene expression, and apoptosis) (1–5). TJs are part of a large family composed of more than 40 different proteins, of which occludins (OCLNs), claudins (CLDNs), and junctional adhesion molecules (JAMs) represent three integral proteins (6, 7). Of these, CLDNs are thought to be the most crucial for mediating TJ formation and permeability (8). As the tight physical barriers between cells, CLDNs help to defend against infection and can also be hijacked by viruses to facilitate their infection, including several medically important human pathogens (e.g., hepatitis C virus, West Nile virus, human immunodeficiency virus, respiratory syncytial virus, and other viruses) (9–14).
Porcine reproductive and respiratory syndrome (PRRS) is one of the most economically significant infectious diseases affecting pigs worldwide. PRRS virus (PRRSV) is the causative agent of PRRS and can result in late‐term reproductive failure in pregnant sows, as well as respiratory distress in pigs of all ages. In the United States, PRRSV infection causes an estimated annual loss of $600 million (15). PRRSV has a single-stranded positive-sense RNA genome, which undergoes rapid genetic evolution. Since the identification of the first strain in Europe in 1991, several highly pathogenic PRRSV strains have evolved to cause numerous acute disease outbreaks in different regions (16–19). In particular, PRRSV infection in China is highly complex, and rapid genetic evolution frequently occurs due to mutations, insertions, and deletions in the viral genome or recombination between different strains. Since the emergence of low-pathogenic PRRSV (LP-PRRSV) in 1996, a highly pathogenic PRRSV (HP-PRRSV) emerged in 2006 (19–21), and NADC30-like strains have also been detected since 2013 (22). To date, HP-PRRSV represents the dominant strain in the field, and recombination events between HP-PRRSV and NADC30-like strains are readily detected, which contributes to the inability to completely contain this disease (23–26). Unfortunately, there are currently no effective vaccines or drugs against PRRS.
PRRSV belongs to the family Arteriviridae and has a genome that is approximately 15 kb in length and encodes eight structural proteins (GP2a, E, GP3, GP4, GP5, ORF5a, M, and N) and 14 nonstructural proteins (27–29). In vitro, only the African green monkey kidney cell line MA-104 or its derivatives (e.g., MARC-145) can fully support PRRSV replication (30). Classical PRRSV enters cells via endocytosis mediated by cellular receptors. CD163 has been determined to be the major receptor that plays a key role in viral internalization and disassembly (31, 32). The virus spreads by cell-to-cell communication (33). Classical PRRSV has a very restricted tropism for cells of the monocytic lineage, and the fully differentiated porcine alveolar macrophage serves as a primary cell target for PRRSV infection (34). However, an increasing number of studies have reported an expanding tissue tropism of HP-PRRSV. It has been confirmed that with the exception of porcine alveolar macrophages (PAMs), HP-PRRSV infection can also be detected in vascular endothelial cells, epithelium cells, and other cell types (35–39). However, how the virus infects these cells remains unclear.
Due to the disease severity, high pathogenicity, and genetic variability of HP-PRRSV, digital gene expression profiling (DGE) was performed to identify the mRNA changes that occur following a PRRSV infection in MARC-145 cells. The CLDN4 gene was identified as a differentially expressed gene. Therefore, this study aimed to elucidate the role of CLDN4 in PRRSV infection.
RESULTS
CLDN4 plays a role in PRRSV infection.
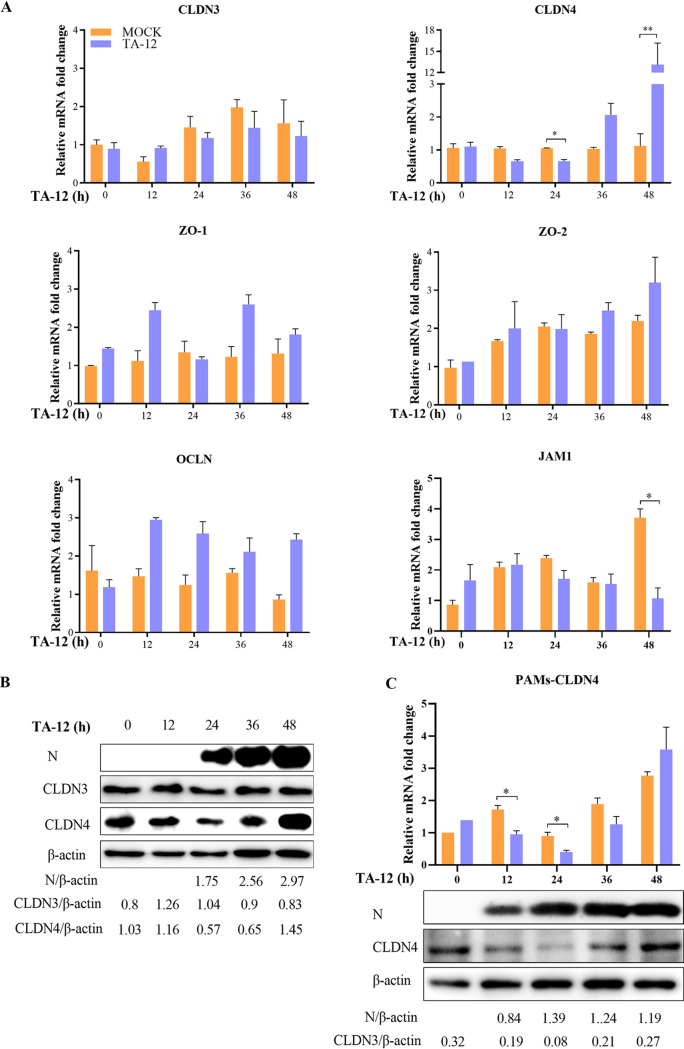
TJs can be divided into transmembrane proteins and cytoplasmic actin-binding proteins according to their distribution and function. CLDNs, OCLNs, and JAMs are the main members of the transmembrane group, and zonula occludens (ZO) proteins are cytoplasmic actin-binding proteins (40). In our previous DGE analysis, the level of CLDN4 mRNA expression decreased by approximately 1-fold at 24 h postinfection (hpi) by HP-PRRSV strain TA-12, whereas there was a >3-fold increase at 48 hpi (data not shown). Thus, we hypothesized that CLDN4 may play a role in PRRSV infection. To study the specific role of CLDN4 in PRRSV infection, the levels of OCLN, CLDN4, JAM-1, ZO-1, and ZO-2 mRNA expression were detected in PRRSV-infected cells. CLDN3, which showed the highest nucleotide similarity to CLDN4, was also detected as a control. After PRRSV infection, the level of CLDN4 expression decreased at 24 hpi and increased at 48 hpi (Fig. 1A). The expression of JAM-1 was decreased only at 48 hpi. No changes in the levels of CLDN3, ZO-1, ZO-2, and OCLN mRNA expression were observed. The level of CLDN4 protein expression also decreased at 24 hpi, followed by an increase at 48 hpi by Western blotting (Fig. 1B). Changes in CLDN4 in the primary target cells, PAMs, were also detected by quantitative real-time PCR (qPCR) and Western blotting. There was a similar decreasing trend at 12 and 24 hpi, after which an increasing trend was observed (Fig. 1C). These results indicate that CLDN4 plays a role during PRRSV infection.
FIG 1.
CLDN4 plays a role in PRRSV infection. (A) Changes in TJ mRNA expression during TA-12 infection of MARC-145 cells. MARC-145 cells were infected with PRRSV TA-12, and the mRNAs were collected at 0, 12, 24, 36, and 48 hpi. The levels of CLDN3, CLDN4, OCLN, ZO-1, ZO-2, and JAM-1 mRNA were measured by relative qPCR. (B) Western blot analysis of the effects of PRRSV infection on CLDN4. MARC-145 cells were infected with TA-12, and the cells were collected at 12, 24, 36, and 48 hpi. The cellular proteins were analyzed by Western blotting using anti-CLDN4, anti-CLDN3, and anti-N antibodies. (C) Changes in CLDN4 expression in PAMs during TA-12 infection. PAMs were infected with PRRSV TA-12, and the cells were collected at 0, 12, 24, 36, and 48 hpi. The levels of mRNA and protein expression of CLDN4 were measured by relative qPCR and Western blotting, respectively. β-Actin was used as an internal control. The asterisks indicate significant differences relative to the control values: *, P < 0.05; **, P < 0.01 (Student's t test).
CLDN4 functions as an anti-PRRSV factor.
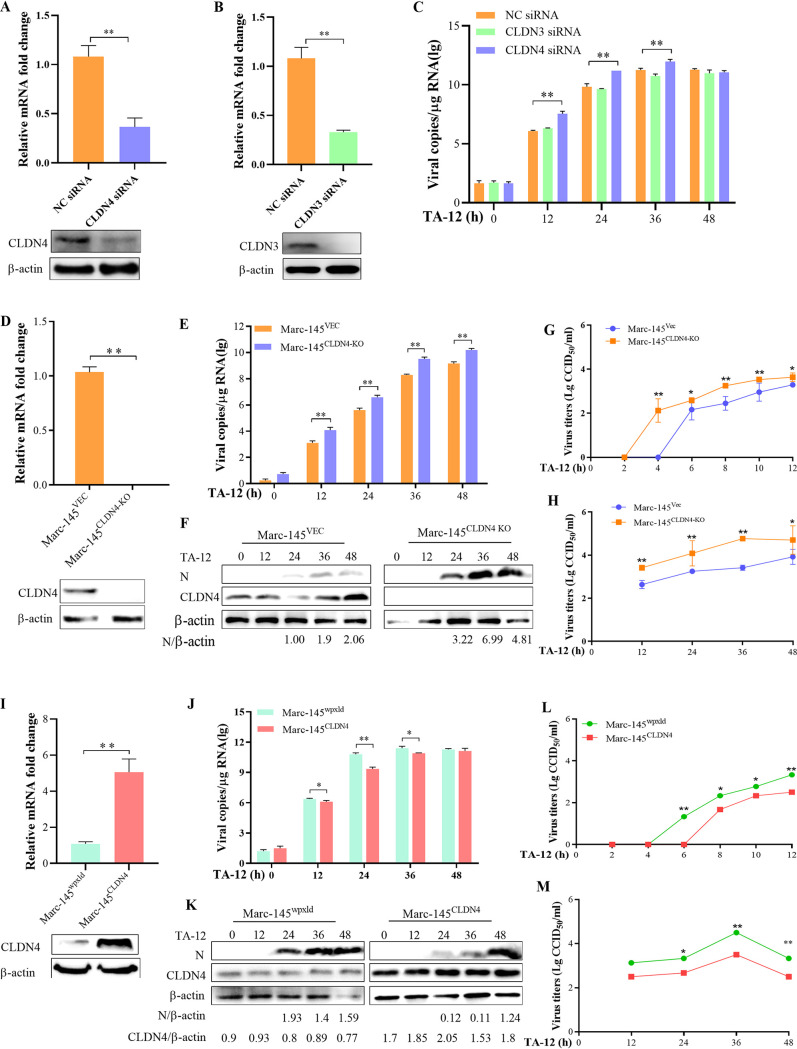
To identify the role of CLND4 in a PRRSV infection, the CLDN4 and CLDN3 genes were knocked down (Fig. 2A and B) in the PRRSV-permissive MARC-145 cell line. The qPCR results showed that a knockdown of CLDN4 could increase TA-12 infection, whereas the CLDN3 knockdown had no effect on TA-12 infection (Fig. 2C). To further confirm this effect, the CLDN4 knockout cell line MARC-145CLDN4-KO was developed (Fig. 2D). The numbers of viral genome copies and viral proteins were significantly increased in MARC-145CLDN4-KO cells (Fig. 2E and F). The kinetic growth of TA-12 in the cell culture supernatants was also detected via one-step or multistep titration methods. TA-12 was first detected at 2 hpi in MARC-145CLDN4-KO cells, which were detected at 4 hpi in the control MARC-145VEC cells (Fig. 2G) and maintained a higher level until 48 hpi (Fig. 2H). The CLDN4-overexpressing cell line MARC-145CLDN4 was also developed (Fig. 2I). The numbers of TA-12 genome copies and viral proteins both significantly decreased compared with the corresponding control cells (Fig. 2J and K). The appearance of TA-12 in the cell culture supernatant was detected at 6 hpi in MARC-145CLDN4 cells, which was delayed by 2 h compared with that of the control cell line MARC-145wpxld (Fig. 2L), and TA-12 was maintained at a lower level until 48 hpi (Fig. 2M). The knockdown of CLDN4 or CLDN3, as well as the knockout (MARC-145CLDN4-KO) or overexpression (MARC-145CLDN4) of CLDN4, had no effect on cell growth (data not shown). All the results showed that knocking down or knocking out CLDN4 could significantly increase PRRSV infection, whereas overexpressing CLDN4 significantly decreased PRRSV infection.
FIG 2.
CLDN4 functions as an anti-PRRSV factor. To confirm the role of CLDN4 in an PRRSV infection, CLDN4 was knocked down, knocked out, or overexpressed and PRRSV infection was evaluated by qPCR, Western blotting, or viral titration. (A) The CLDN4 knockdown was verified by qPCR and Western blotting. (B) The knockdown of CLDN3 was identified by qPCR and Western blotting. (C) MARC-145 cells were transfected with siRNA targeted to CLDN4, CLDN3, or control siRNA NC and then infected with TA-12. Cell samples were collected at 0, 12, 24, 36, and 48 hpi. The PRRSV genome was detected by qPCR. (D) Identification of the CLDN4 knockout cell line, MARC-145CLDN4-KO, by qPCR and Western blotting. TA-12 was inoculated onto MARC-145CLDN4-KO cells, and the cell samples were collected at 0, 12, 24, 36, and 48 hpi. (E and F) Infection with TA-12 was evaluated based on the N gene and N protein by qPCR (E) and Western blotting (F). MARC-145CLDN4-KO cells were infected with TA-12, and the cellular supernatants were collected at 0, 2, 4, 6, 8, 10, 12, 24, 36, and 48 hpi. The viruses in the supernatant were titrated as a one-step (G) and multistep (H) kinetic growth curve. (I) The CLDN4-overexpressing MARC-145 cell line, MARC-145CLDN4, was confirmed by qPCR and Western blotting. MARC-145CLDN4 cells were infected with TA-12, and the cell samples were collected at 0, 12, 24, 36, and 48 hpi. PRRSV infection were detected by qPCR (J) and Western blotting (K). The MARC-145CLDN4 cells were infected with TA-12, and the cellular supernatant was collected at the same time point as that of the MARC-145CLDN4-KO cells for virus titration as a one-step (L) and multistep (M) kinetic growth curve. GAPDH was used as an internal control. Error bars indicate the SDs of three experimental replicates. The asterisks indicate significant differences from the control value: *, P < 0.05; **, P < 0.01 (Student's t test).
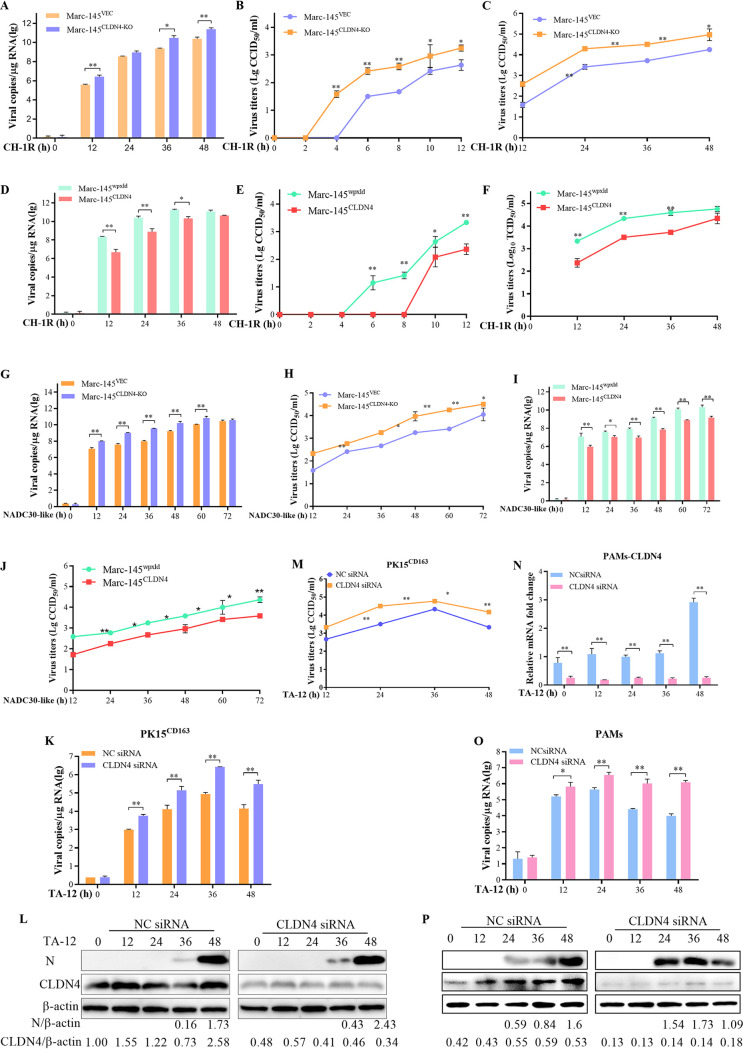
To study the specific role of CLDN4 in PRRSV infection, MARC-145CLDN4-KO and MARC-145CLDN4 cells were infected with a low-pathogenic PRRSV strain (CH-1R) and a NADC30-like strain (TA01). The viral genome copies were significantly increased in MARC-145CLDN4-KO cells compared to that in MARC-145VEC cells, except at 24 hpi (Fig. 3A). CH-1R in the cell culture supernatant was first detected at 2 hpi in MARC-145CLDN4-KO cells, which were detected at 4 hpi in MARC-145VEC cells (Fig. 3B). The titer remained higher until 48 hpi (Fig. 3C). The number of CH-1R viral genome copies was significantly decreased in MARC-145CLDN4 cells (Fig. 3D), and virus genome appearance in the cell culture was delayed by 4 h compared with that of the MARC-145wpxld control cell lines (Fig. 3E and F). Similar results of increasing NADC30-like infection in MARC-145CLDN4-KO cells and decreasing NADC30-like infection in MARC-145CLDN4 cells were also detected (Fig. 3G to J). To further confirm the antiviral role of CLDN4, cells of another PRRSV replication-supporting line, PK-15CD163 (PK15 cells stably overexpressing CD163), were transfected with CLDN4-specific small interfering RNA (siRNA) and control NC siRNA. Compared with the NC siRNA, the viral genome, protein expression, and titer of TA-12 were significantly increased in the CLDN4 knockdown cells (Fig. 3K to M). The siRNA targeted to CLND4 works effectively in PAMs (Fig. 3N). The knockdown of CLDN4 in PAMs could also increase the viral genome and proteins in PAMs (Fig. 3O and P). Together, these results suggest that CLDN4 functions as an anti-PRRSV infection factor.
FIG 3.
Confirming the role of CLDN4 PRRSV infection. CLDN4 knockout or CLDN4-overexpressing cells were infected with either a lower-pathogenic CH-1R strain or a NADC30-like strain. qPCR and viral titration were employed to detect PRRSV infection. The qPCR results showed that knocking down CLDN4 could increase the number of viral genome copies (A) and viral titer (B) of CH-1R and the viral genome (C) and viral titer (D) of the NADC30-like PRRSV strain. CLDN4 overexpression in MARC-145 cells could decrease the number of CH-1R genomic copies (E) and titer (F) and those of the NADC30-like strain (G and H). The level of CLDN4 in the PK15CD163 cells was knocked down by siRNA, the TA-12 genome was detected by qPCR (I), the viral protein was measured by Western blotting (J), and viral titer was determined (K). The level of CLDN4 in the PAMs was knocked down by siRNA, after which TA-12 was inoculated onto the cells. The cell samples were collected at 0, 12, 24, 36, and 48 hpi. The level of CLDN4 mRNA was tested by qPCR (N). The TA-12 genome was detected by qPCR (O), and the viral protein was measured by Western blotting (P).
CLDN4 blocked PRRSV absorption.
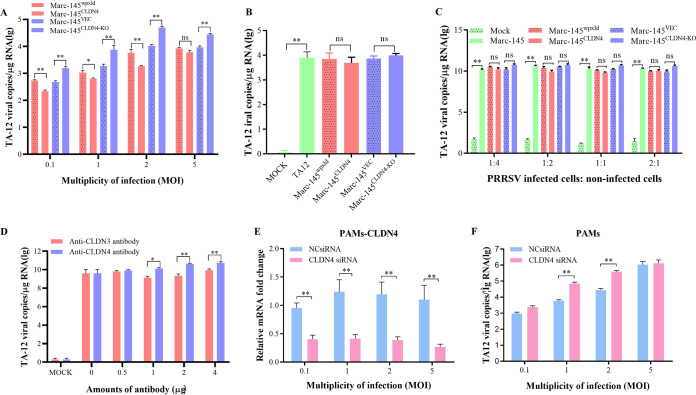
CLDN4 is a transmembrane protein that is distributed along the cell surface and may function as a PRRSV receptor that may play a role in viral absorption or internalization. To determine whether CLDN4 plays a role in PRRSV absorption, TA-12 was mixed with the MARC-145CLDN4 and the MARC-145CLDN4-KO cell lines at 4°C for 2 h to permit only the binding of viral particles with cellular proteins. The cells were incubated for a further 4 h at 37°C to achieve the full internalization of the binding viral particles. The qPCR results showed that the number of PRRSV genomic copies in the absorption stage increased significantly in the MARC-145CLDN4-KO cells and decreased significantly in the MARC-145CLDN4 cells (Fig. 4A), but there was no effect on PRRSV internalization (Fig. 4B). It has also been reported that PRRSV can utilize cell-to-cell communication for efficient spread and that CLDN can inhibit the cell-to-cell spread of viruses (33, 41). Therefore, the possibility of transmission between cells using CLDN4 was also confirmed. PRRSV-infected normal MARC-145 cells were cocultured with MARC-145CLDN4 or MARC-145CLDN4-KO cells. The qPCR results showed that there were no differences in the cocultured cells (Fig. 4C). These results suggest that CLDN4 did not play a role in virus transmission. To further confirm this finding, CLDN4 was blocked with an anti-CLDN4 antibody, and then cells were infected with PRRSV. The anti-CLDN3 antibody was also applied as a control. The results showed that anti-CLDN4 could promote virus infection, whereas anti-CLDN3 had no effect (Fig. 4D). To uncover the role of the CLDN4 in PAMs, CLDN4 was knocked down in PAMs (Fig. 4E). The TA-12 strain was added to the cells at 4°C for 2 h at different multiplicities of infection (MOI). The qPCR results showed that a CLDN4 knockdown could promote TA-12 absorption at MOI of 1 and 2, whereas no significant difference was observed in the absorption at MOI of 0.1 and 5 (Fig. 4F). Collectively, these results indicate that CLDN4 plays a role in virus absorption.
FIG 4.
CLDN4 could block PRRSV absorption. (A) Confirmation of the role of CLDN4 in absorption in MARC-145 cells. MARC-145CLDN4-KO and MARC-145CLDN4 cells and their corresponding control cell lines were infected with TA-12 at MOI of 0.1, 1, 2, and 5 and absorbed at 4°C for 1 h. After a washing with cold PBS, the cells were collected and the viral RNA was extracted. The attached PRRSV particles were evaluated by qPCR. (B) Confirmation of the role of CLDN4 in viral internalization. MARC-145CLDN4-KO and MARC-145CLDN4 cells and their corresponding control cell lines were infected with TA-12 at an MOI of 1, absorbed at 4°C for 1 h, and then shifted to 37°C for 4 h to allow internalization of the attached viruses. After a washing with low-pH PBS buffer, the cells were collected and the viral RNAs were extracted. The internalized PRRSV particles were evaluated by qPCR. (C) Confirmation of the role of CLDN4 in cell-to-cell transmission. Normal MARC-145 cells were infected with TA-12 at an MOI of 1 and collected at 24 h postinfection. After extensive washing in a low-pH buffer, they were mixed with MARC-145CLDN4-KO and MARC-145CLDN4 cells and their corresponding control cell lines at ratios of 1:4, 1:2, 1:1, and 2:1 before incubation for another 24 h. The cells were then collected, and the viral RNA was extracted. The PRRSV genome was evaluated by qPCR. (D) The blocking effects of an anti-CLDN4 antibody on PRRSV absorption. Anti-CLDN4 antibodies were incubated onto a monolayer of MARC-145 cells at 37°C for 1 h. After being washed, the cells were inoculated with TA-12 at an MOI of 0.1 and incubated at 37°C for 24 h. The cells were collected, and the viral RNAs were extracted. The genomic PRRSV was evaluated by qPCR. The anti-CLDN3 antibody was used as a negative control. The role of CLDN4 in absorption was confirmed in PAMs. PAMs were transfected with CLDN4 siRNAs for 24 h and incubated with TA-12 at MOI of 0.1, 1, 2, and 5 for 2 h at 4°C. After a washing with cold PBS, the cells were collected, and the total RNA was extracted. The CLDN4 knockdown was verified by qPCR (E); The PRRSV genome was evaluated by qPCR (F). All data are means and standard deviations (error bars) for three samples per group (*, P < 0.05; **, P < 0.01; ns, no significance). GAPDH was used as an internal control. All experiments were repeated at least three times with consistent results.
The small extracellular loop, ECL2, of CLDN4 blocks PRRSV infection by binding to the C-terminal domain of GP3.
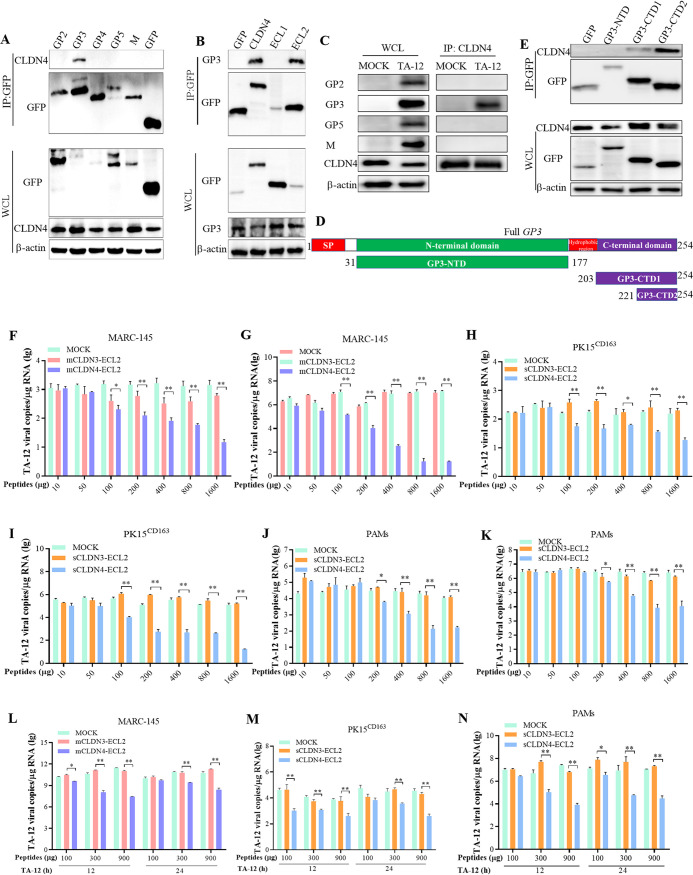
The above results indicate that CLDN4 plays a role in PRRSV absorption. Therefore, it was deduced that CLDN4 may interact with the structural viral proteins lining the surface of the PRRSV particle. The viral surface proteins GP2, GP3, GP4, GP5, and M were expressed in HEK293T cells and the corresponding interactors were pulled down via a green fluorescent protein (GFP) trap. The Western blotting results revealed that only GP3 could interact with CLDN4 (Fig. 5A). Moreover, CLDNs are four-transmembrane-domain proteins composed of intracellular N-terminal and C-terminal tails, four transmembrane helices, two extracellular loops (large ECL1 and small ECL2), and one cytoplasmic loop (42). Thus, it is possible that the interaction between GP3 and CLDN4 may be by binding with one of the two extracellular loops, ECL1 or ECL2, or both. To study the binding domain for CLDN4 and GP3, cells were cotransfected with constructs expressing ECL1, ECL2, and GP3, respectively. Coimmunoprecipitation (co-IP) was performed, and the Western blotting results showed that GP3 bound to ECL2 (Fig. 5B). To identify which viral protein interacted with CLDN4, the viral proteins were immunoprecipitated from TA-12-infected MARC-145 cells using anti-CLDN4 antibodies. Only GP3 was detected in the immunoprecipitated products (Fig. 5C). GP3 contains N- and C-terminal domains, which are connected by a short hydrophobic region (Fig. 5D). To uncover the specific binding domain associated with CLDN4, the N- and C-terminal domains of GP3 were cloned and expressed in 293T cells. A GFP pulldown was carried out, and the results showed that the C-terminal domain containing 33 amino acids interacted with CLDN4 (Fig. 5E).
FIG 5.
The small extracellular loop ECL2 of CLDN4 blocks PRRSV infection by binding with C-terminal domain of GP3. (A) Western blot analysis of GFP trap pulldown assays using GFP-tagged PRRSV major surface structural proteins of GP2, GP3, GP4, GP5, and M as the bait in HEK293T cells. Bound CLDN4 was immunoblotted with an anti-CLDN4 monoclonal antibody (MAb). GFP was used as a negative control. WCL, whole-cell lysate. (B) Western blot analysis of GFP trap pulldown assays using GFP-tagged CLDN4 and its two extracellular loops, ECL1 and ECL2. Bound GP3 was immunoblotted with an anti-GP3 polyclonal antibody. GFP was used as a negative control. (C) MARC-145 cells were inoculated with TA-12, and the cell lysates were prepared at 24 hpi. The potential viral proteins that interacted with CLDN4 were coimmunoprecipitated with an anti-CLDN4 monoclonal antibody. The co-IP products were analyzed by Western blotting using mouse anti-GP2, -GP3, -GP5, and -M polyclonal antibodies. (D) Schematic diagram of the GP3 protein. GP3 contains an SP, N-terminal domain, hydrophobic region, and C-terminal domain. The names and sizes of truncated GP3 are indicated. SP, signal peptide. (E) Western blot analysis of GFP trap pulldowns using GFP-tagged GP3-NTD, GP3-CTD1, and GP3-CTD2 and an empty vector as the bait in HEK293T cells. Bound CLDN4 was immunoblotted with an anti-CLDN4 antibody. TA-12 was incubated with monkey-originated ECL2 (mCLDN4-ECL2) or swine-originated ECL2 (sCLDN4-ECL2 peptide) for 1 h and inoculated on normal MARC-145 or PK-15CD163 cells at 4°C for another 1 h. After being washed in cold PBS, the cells were collected and the viral RNA was extracted. The attached PRRSV particles were evaluated by qPCR in MARC-145 (F), PK-15CD163 cells (H) or PAMs (J). TA-12 was incubated with mCLDN4-ECL2 or sCLDN4-ECL2 for 1 h and inoculated onto MARC-145 cells, PK-15CD163 cells, or PAMs at 4°C for another 1 h. After washing with cold PBS, the cells were incubated at 37°C for 24 h. The cells were collected, and the viral RNA was extracted. Genomic PRRSV was evaluated by qPCR in MARC-145 cells (G), PK-15CD163 cells (I), or PAMs (K). MARC-145 or PK-15CD163 cells were infected with TA-12. At 12 h and 24 h postinfection, mCLDN4-ECL2 and sCLDN4-ECL2 were added to the corresponding cells. After another incubation at 37°C for 12 h (a total of 36 hpi) All cells were collected, and the genomic PRRSV was evaluated by qPCR in MARC-145 cells (L), PK-15CD163 cells (M), and PAMs (N).
To further confirm this interaction, peptides targeting the ECL2 domains of monkey-derived CLDN3 (mCLDN3-ECL2) and CLDN4 (mCLDN4-ECL2) were synthesized, incubated with TA-12, and then inoculated into MARC-145 cells. The results showed that mCLDN4-ECL2 could block PRRSV absorption (Fig. 5F) and corresponding replication (Fig. 5G), whereas mCLDN3-ECL2 had no effect on TA-12 infection (Fig. 5F and G). Moreover, the blocking effect was dose dependent. The corresponding ECL2 domains of swine CLDN4 and CLDN3 were also synthesized. The ECL2 domains of swine CLDN3 (sCLDN3-ECL2) and CLDN4 (sCLDN4-ECL2) were also incubated with TA-12 and subsequently used to infected PK15CD163 cells and PAMs. The results showed that only sCLDN4-ECL2 could block the absorption (Fig. 5H and J) and infection (Fig. 5I and K) of TA-12 on both PK15CD163 cells and PAMs.
The above results demonstrate that mCLDN4-ECL2 or sCLDN4-ECL2 can block PRRSV infection by binding with GP3, which lines the surface of the PRRSV particle. This indicates that ECL2 may inhibit PRRSV infection by binding with the PRRSV particle in the supernatant. To further confirm this hypothesis, mCLDN4-ECL2 or sCLDN4-ECL2 was added to the cell culture supernatant of PRRSV-infected cells after 12 or 24 hpi. All of the samples were collected at 36 hpi. The results showed that mCLDN4-ECL2 or sCLDN4-ECL2 could significantly decrease PRRSV infection in MARC-145 cells, PK15CD163 cells, and PAMs (Fig. 5L to N). Together, these results confirm that ECL2 can effectively block PRRSV infection.
GP3 downregulates the level of CLDN4 transcription.
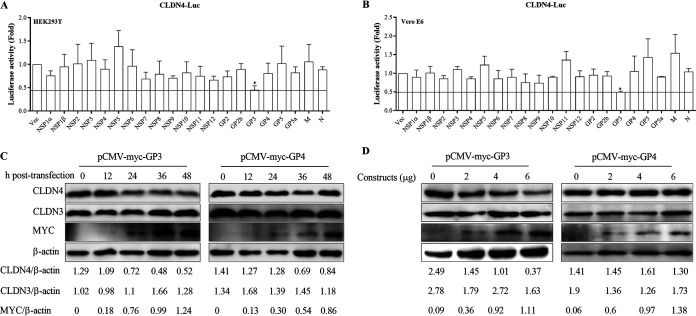
As shown in Fig. 1, the expression of CLDN4 decreased during the early stages (before 24 h) of PRRSV infection. One possible explanation for this is that after virus particles enter a cell, the viral protein(s) decreases the level of CLDN4 expression to help other viral particles attach to the cells and gain entry. To confirm this effect and determine which viral protein downregulates CLDN4 expression, the promoter region of CLDN4 was cloned and cotransfected with all the viral proteins. The results showed that only GP3 could downregulate the level of transcription by more than 50% of CLDN4 in both the HEK293T and Vero E6 cell lines (Fig. 6A and B). To further confirm this effect, the recombinant pCMV-myc-GP3 and pCMV-myc-GP4 constructs were transfected into MARC-145 cells. The Western blotting results showed that with an increasing amount of GP3, a greater amount of CLDN4 was decreased in a time- and dose-dependent manner, whereas GP4 had no influence on CLDN4 expression (Fig. 6C and D). These results suggest that GP3 downregulated the level of CLDN4 transcription.
FIG 6.
GP3 downregulates the level of CLDN4 transcription. Role of viral proteins on CLDN4 transcription. The constructs containing viral proteins and pGL3-CLDN4-luc containing the CLDN4 promoter were cotransfected into HEK293T and Vero E6 cells. At 24 hpt, the transfected HEK293T cells (A) and Vero E6 cells (B) were collected and analyzed for firefly and Renilla luciferase activity using a dual-luciferase reporter assay. (C) MARC-145 cells were transfected with pCMV-myc-GP3, and the cells were collected at 12, 24, 36, and 48 hpi and analyzed by Western blotting with anti-Myc and anti-CLDN4 antibodies. (D) MARC-145 cells were transfected with 2, 4, and 6 μg of pCMV-myc-GP3, and the cells were collected at 24 hpi and analyzed by Western blotting. pCMV-myc-GP4 and blank plasmids were also transfected as negative controls. Actin was used as an internal control.
GP3 decreases the transcription factor SP1 by ubiquitination.
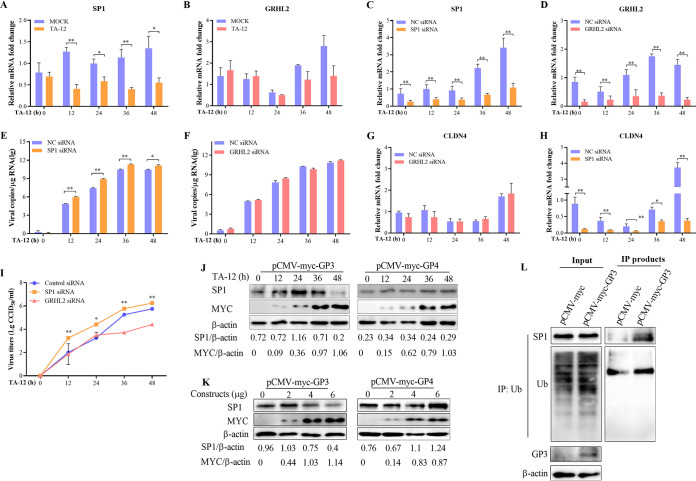
The transcriptional factors Grainyhead-like 2 (GRHL2) and specificity protein 1 (SP1) have been reported to regulate the transcription of CLND4 (43, 44). First, the level of GRHL2 or SP1 mRNA expression was detected following infection with TA-12. The qPCR results revealed that a TA-12 infection could downregulate the level of SP1 mRNA, whereas there was no effect on that of GRHL2 (Fig. 7A and B). To further confirm these results, SP1 and GRHL2 were knocked down in MARC-145 cells using specific siRNA. A knockdown of SP1 significantly increased the TA-12 genomic copies and titer (Fig. 7C to E), and the expression of CLDN4 was also significantly decreased (Fig. 7F). However, the TA-12 infection and level of CLDN4 expression were not affected in GRHL2-knockdown MARC-145 cells (Fig. 7E and G to I). These results show that the downregulation of the level of CLDN4 transcription is under the control of SP1. To confirm whether CLND4 downregulation may be the result of SP1 inhibition by GP3, different amounts (2, 4, and 6 μg) of pCMV-myc-GP3 were used to transfect MARC-145 cells, and the cells were collected at 12, 24, and 36 h posttransfection (hpt). Western blotting results showed that GP3 could decrease SP1 in a time- and dose-dependent manner, whereas GP4 could not (Fig. 7J). The function of SP1 is highly regulated by posttranslational modifications (45, 46). One direct method of decreasing SP1 is via ubiquitin-proteasome cleavage. To confirm this, the ubiquitination of SP1 was detected. The results indicated that GP3 can ubiquitinate SP1 (Fig. 7K).
FIG 7.
GP3 decreases the level of the transcription factor SP1 by ubiquitination. MARC-145 cells were infected with PRRSV TA-12, and the mRNA was collected at 0, 12, 24, 36, and 48 hpi. The levels of SP1 (A) and GRHL2 (B) mRNA were measured by relative qPCR. MARC-145 cells were transfected with siRNA targeted to SP1 or control siRNA (NC) and then infected with TA-12. The cell samples were collected at 0, 12, 24, 36, and 48 hpi. The levels of SP1 mRNA (C), PRRSV genome (D), and CLDN4 mRNA (E) were detected by qPCR. MARC-145 cells were transfected with siRNA targeted to GRHL2 or control siRNA NC and then infected with TA-12. The cell samples were collected at 0, 12, 24, 36, and 48 h postinfection. The level of GRHL2 mRNA (E), PRRSV genome (F), and level of CLDN4 mRNA (G) were detected by qPCR. (I) Viral titers in the supernatant of SP1 and GRHL2 knockdown cells were measured in normal MARC-145 cells. GAPDH was used as an internal control. The error bars indicate the SDs for three experimental replicates. Asterisks indicate a significant difference from the control value: *, P < 0.05; **, P < 0.01 (Student's t test). (J and K) Western blot analysis of the effects of GP3 on SP1 with anti-Myc and anti-SP1 antibodies. The analyzed samples were prepared as described for Fig. 6C and D. (L) HEK293T cells were transfected with pCMV-myc-GP3 or a blank control. The cells were collected at 24 hpt, and the whole-cell lysates were prepared. The cell lysates were incubated with an antiubiquitin (Ub) antibody and then precipitated using protein A agarose beads. The immunoprecipitated products were analyzed by Western blotting using anti-SP1, anti-Ub, and anti-GP3 antibodies. Actin was used as an internal control.
DISCUSSION
In vivo, PAMs are the primary target cells of classical PRRSV (47). HP-PRRSV has been reported to infect the epithelium (35–39). Moreover, CLDNs are abundant in epithelial cells. Therefore, this study focused on the role of CLDN4 during infection with the HP-PRRSV strain (TA-12).
TJs are thought to function as a physical barrier between the apical and basolateral domains of polarized epithelial cells, which restrict pathogens to a cellular compartment. As a key component of TJs, CLDNs are hijacked by viruses as cellular receptors (e.g., hepatitis C virus and dengue virus) (9, 48, 49) or by degrading TJs to facilitate viral entry and dissemination (e.g., West Nile virus, Japanese encephalitis virus, norovirus, and human rhinovirus) (11, 50–52). In this study, we found that during the early stages of infection, PRRSV could degrade CLDN4 in both the primary PAMs and cell lines (Fig. 1). Moreover, CLDN4 overexpression could decrease TA-12 replication, and knocking down or knocking out CLDN4 could increase TA-12 replication (Fig. 2). The low-pathogenic strain CH-1R and the NADC30-like strain TA01 were inoculated into MARC-145CLDN4-KO, PK15CD163, and MARC-145CLDN4 cells. All of the results showed that CLDN4 overexpression could increase the infectivity of both strains, whereas knocking down or knocking out CLDN4 decreased the infectivity of both strains. These results indicate that CLDN4 was specifically degraded during PRRSV infection.
In addition to the receptor or physical barrier role of TJs, the property of inhibiting the cell-to-cell spread of viruses has also been suggested (41, 53). Thus, the role of CLDN4 in the TA-12 infection process of absorption, internalization, or cell-to cell transmission was determined. It was interesting to find that CLDN4 played a role only in absorption (Fig. 4), since the results indicated the binding of the small extracellular loop, ECL2, of CLDN4 with the last 33 amino acids in the C-terminal domain of the PRRSV structural glycoprotein, GP3 (Fig. 5A and B). Based on these results, it was concluded that CLDN4 restricted the PRRSV particle outside the cells by binding with GP3, which prompted us to study whether ECL2 can block PRRSV infection. Two blocking methods were used: (i) TA-12 was mixed with ECL2 for 1 h and then inoculated into MARC-145 cells, PK15CD163 cells, or PAMs or (ii) ECL2 was placed onto MARC-145 cells or PAMs for 1 h and then TA-12 was added to elucidate the blocking effects of ECL2 on TA-12 infection. No changes in TA-12 infection were detected with the latter method (data not shown). With the former method, ECL2 could block TA-12 absorption and infection in all MARC-145 cells, PK15CD163 cells, or PAMs (Fig. 5F to K). Even after PRRSV infection for 12 h or 24 h, in which viral particles were abundant in the supernatant, ECL2 could also significantly block TA-12 infection (Fig. 5L to N). This indicated that ECL2 may act as an inhibitor to neutralize PRRSV viremia.
Figure 1 suggests that PRRSV degraded CLDN4 at an early stage, and further experiments showed that GP3 could downregulate the level of CLDN4 transcription, as well as decreasing the expression of CLDN4 (Fig. 6) by decreasing the transcriptional factor SP1. SP1 is a transcription factor that can activate or suppress transcription in response to physiological and pathological stimuli and has been reported to regulate CLDN transcription (44). The function of SP1 is highly regulated by posttranslational modifications (e.g., phosphorylation, O‐linked glycosylation, acetylation, SUMOylation, and ubiquitylation) (45, 54, 55). In this study, GP3 expression could decrease SP1 by ubiquitination and then targeting proteasome‐mediated degradation pathways (Fig. 7K). However, the detailed mechanism requires further deciphering.
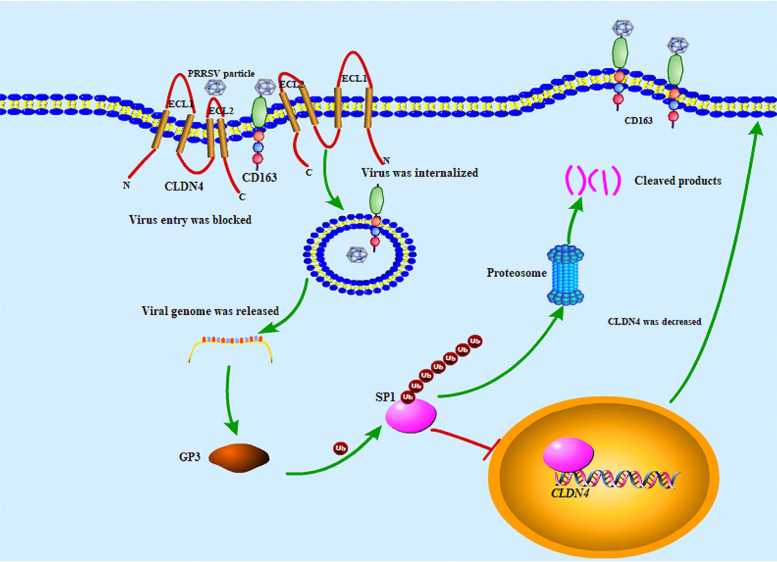
CLDNs represent a large family comprising at least 27 different members in mammals and are roughly classified as either barrier or pore claudins (42). CLDN4 is classified as a barrier claudin (56). Based on the present results, a proposed model for the PRRSV and CLDN4 interaction was established (Fig. 8). After the inoculation of susceptible cells with PRRSV, most viral particles were kept out of the cells by (i) the abundance of the barrier claudin CLDN4 lining the cell surface and blocking the contact of viral particles with their receptors and (ii) the interaction between the GP3 and the ECL2 domain of CLDN4. Only a portion of viral particles bind to the cellular receptor CD163 and enter the cells. This can explain why not all cells were infected during the early stage, even after inoculation with an MOI of 10 (data not shown).
FIG 8.
Proposed model for the interaction of PRRSV and CLDN4 based on the results of this study. After the inoculation of PRRSV onto susceptible cells, most viral particles were kept out of the cells by an interaction of GP3 and the ECL2 domain of CLDN4. Only parts of viral particles bind to the cellular receptor CD163 and enter the cells. GP3 is produced early following viral entry into the cells and ubiquitinates the transcription factor SP1 via an unknown mechanism. The SP1 was then degraded in a proteasome‐mediated manner, the level of CLDN4 transcription was downregulated, and the level of CLDN4 was decreased. A large number of viral particles could enter the cells.
GP3 is produced following viral entry into host cells and ubiquitinates the transcription factor SP1 using an unknown mechanism. SP1 was degraded in a proteasome‐mediated manner, followed by the downregulation of CLDN4 transcription level and decreased levels of CLDN4. A substantial number of viral particles were able to enter the cells. However, the present study did not determine why there was an increase in CLDN4 at 48 hpi, when severe cytopathic effects develop. The reason for this may be that PRRSV infection greatly disturbed the normal cellular transcription and translation process, and CLDN4 was substantially increased. GP3 is a minor structural glycoprotein and forms a disulfide-linked heterotrimeric complex with viral GP2 and GP4, which is essential for cell entry (57–59). GP3 contains neutralizing epitopes and is the primary candidate for vaccine development (60). In this study, evidence that GP3 regulates gene transcription was confirmed.
Several mechanisms of PRRSV entry into susceptible PAMs or MARC-145 cells have been discovered. The data presented here explain a mechanism of PRRSV entry into the epithelium. In the epithelium-derived MARC-145 and PK15CD163 cells, CLDN4 was able to restrict extracellular viral particles by binding to GP3. In contrast, GP3 downregulated the expression of CLDN4 to facilitate additional viral entry. PRRSV infection could be blocked by the ELC2 domain of CLDN4. These findings provide a greater understanding of the GP3 function in PRRSV infection and identify potential novel therapeutic antiviral strategies. However, there still remains a lack of details regarding cellular receptor expression on epithelial cells, which could be used by PRRSV. Thus, further research is needed in this area.
MATERIALS AND METHODS
Cell lines and virus.
A PRRSV-permissive cell line derived from African monkey kidney (MARC-145), Vero E6 cells, and human embryonic kidney 293T (HEK293T) cells were obtained from the China Center for Type Culture Collection (Wuhan, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Langley, OK, USA) supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Beit HaEmek, Israel) at 37°C in 5% CO2 in a humidified incubator. PAMs were isolated from five healthy 5-week-old crossbred weaned pigs (Landrace × Yorkshire) as previously described (61). The low-pathogenic PRRSV strain CH-1R and PK15CD163 (PK15 cell line stably expressing CD163) cells were kindly provided by Enmin Zhou (Northwest A&F University, China). The HP-PRRSV strain TA-12 (GenBank no. HQ416720) and NADC30-like strain TA01 were previously isolated and stored in our lab.
Constructs.
The PRRSV structural protein genes of GP2, GP3, GP4, GP5, and M (GenBank accession number HQ416720.1), CLDN4 (GenBank accession number NM_001194564), and the ECL1 and ELC2 loops of CLDN4 were amplified by reverse transcription PCR (RT-PCR) or annealed and cloned into the pEGFP-C1 vector (Clontech Laboratories, Inc., Mountain View, CA, USA) together with the PRRSV GP2, GP3, GP4, GP5, and M genes. GP3 contains a signal peptide (SP), N-terminal domain, hydrophobic region, and C-terminal domain (62). The names and sizes of truncated GP3 are shown in Fig. 5D. The PRRSV NSP1α, NSP1β, NSP3 to -12, GP2, E, GP3, GP4, GP5, M, and N genes were cloned into pCMV-Myc (Clontech Laboratories, Inc., Mountain View, CA, USA) and identified by sequencing and Western blotting. The sequences containing the CLDN4 promoter (between −1500 bp and −1 bp upstream of the transcription start sites) was cloned into a pGL3 Enhancer vector (Promega, USA) to create expression constructs. All of the primers used in this research are listed in Table 1.
TABLE 1.
Primers used for constructs and qPCR
| Vector or gene | Primer | Sequence (5′-3′) |
|---|---|---|
| pEGFP-C1 | PRRSV-GP2-F | CCGCTCGAGCTATCAAATGGGGTCTATGC |
| PRRSV-GP2-R | CGCGGATCCTTATCACCATGAGTTCAAAAG | |
| PRRSV-GP3-F | GAAGATCTATGGCTAATAGCTGTACATTCCTCC | |
| PRRSV-GP3-R | TTGGTACCTTACTAATCGCCGTGCGGCACTG | |
| PRRSV-GP4-F | GAAGATCTATGGCTGCGCCCTTTC | |
| PRRSV-GP4-R | GGGGTACCTTATCAAATTGCCAGTAGGATG | |
| PRRSV-GP5-F | CCGCTCGAGCTATGTTGGGGAAGTGCTTG | |
| PRRSV-GP5-R | CGCGGATCCTAACTAGAGACGACCCCATTG | |
| PRRSV-M-F | GAAGATCTATGGGGTCGTCTCTAGAC | |
| PRRSV-M-R | GGGGTACCTTATTATTTGGCATATTTAAC | |
| GP3-NTD-F | AAGATCTATGGGTGGCGGTGGCTCCACGTTCTGTTTTTGG | |
| GP3-NTD-R | GGGGTACCTTAATTGCCGCCGTCGACCTGG | |
| GP3-CTD1-F | GAAGATCTATGGGTGGCGGTGGCTCCTCGCCTGCAAGC | |
| GP3-CTD1-R | GGGGTACCTTACTATCGCCGTGCGGCACTG | |
| GP3-CTD2-F | GAAGATCTATGGGTGGCGGTGGCTCCTCCAGGACATCAG | |
| GP3-CTD2-R | GGGGTACCTTACTATCGCCGTGCGGCACTG | |
| Monkey-CLDN4-F | GAAGATCTATCGGCAGCAGCCATCATC | |
| Monkey-CLDN4-R | GGGGTACCTTATTACACGTAGTTGCTGGCG | |
| Swine-CLDN4-F | CAAGATCTATCGCTTCCATGGGGCTGC | |
| Swine-CLDN4-R | GGGGTACCTTATTACACGTAGTTGCTGGCG | |
| Monkey-CLDN4-ECL1-F | GAAGATCTCCCATGTGTCGCGTGAC | |
| Monkey-CLDN4-ECL1-R | CGCGGATCCTTACTGCAGGTCCTGTG | |
| Monkey-CLDN4-ECL2-F | GGAAGATCTACGGCCCACAACATACTCCAAGACTTCTACAACCCGCTGGTGGCCTCCGGGCAGAAGCGGGAGATGTAAGGATCCGCG | |
| Monkey-CLDN4-ECL2-R | CGCGGATCCTTACATCTCCCGCTTCTGCCCGGAGGCCACCAGCGGGTTGTAGAAGTCTTGGATGATGTTGTGGGCCGTAGATCTGAA | |
| pCMV-Myc | PRRSV-GP2-F | ACGCGTCGACCATGAAATGGGGTCTATGC |
| PRRSV-GP2-R | GGGGTACCTTATCAGCATGAGTTCAAAAG | |
| PRRSV-GP3-F | GAAGATCTCTATGGCTAATAGCTGTACAT | |
| PRRSV-GP3-R | TTGGTACCTTACTATCGCTGTGCGGCACTG | |
| PRRSV-GP4-F | GAAGATCTCTATGGCTGCGCCCTTTC | |
| PRRSV-GP4-R | GGGGTACCTTATCAGATTGCCAGTAGGATG | |
| PRRSV-GP5-F | ACGCGTCGACCATGTTGGGGAAGTGCTTG | |
| PRRSV-GP5-R | TTGGTACCTTACTAGAGACGAGCCCATTG | |
| PRRSV-M-F | GAAGATCTCTATGGGGTCGTCTCTAGAC | |
| PRRSV-M-R | GGGGTACCTTATTATTTGGCATATTTAAC | |
| Monkey-CLDN4-F | TTGTCGACCATGGGCAGCAGCCATCATC | |
| Monkey-CLDN4-R | GGGGTACCTTATTACACGTAGTTGCTGGCG | |
| Swine-CLDN4-F | CAAGATCTCTATGGCTTCCATGGGGCTGC | |
| Swine-CLDN4-R | GGGGTACCTTATTAGACGTAGTTGCTGGCG | |
| pGL3-Enhancer | CLDN4-F | TTGGTACCGCTTCGGCTCCGCATCAGAG |
| CLDN4-R | GAAGATCTGAATACAGCATGGGCAAGTGGG | |
| PRRSV-N | Forward | AGATCATCGCCCAACTAAAC |
| Reverse | GACACAATTGCCGCTCACTA | |
| Monkey-CLDN4 | Forward | CTCGTCATCATCAGCATCA |
| Reverse | GGCAGAGTAAGCCTTGTC | |
| Swine-CLDN4 | Forward | GTGCTAGGTGTGCTGCTGTC |
| Reverse | ATTGTGGGCGGTCCAGG | |
| Monkey-CLDN3 | Forward | CACCATCGTGTGCTGCGC |
| Reverse | TTTGGCCGTGTCGTCCTGC | |
| Monkey-ZO-1 | Forward | GGGACAACAGCATCCTTCCA |
| Reverse | ATCACAGTGTGGTAAGCGCA | |
| Monkey-ZO-2 | Forward | GGATGATGACCCTGAAGAC |
| Reverse | CTCCTCGGCTGGCTCATC | |
| Monkey-OCLN | Forward | CAGGCCTCTTGAATGTCCACC |
| Reverse | AGGCTGGCTGAGAGAGCATT | |
| Monkey-JAM1 | Forward | GGCATTGGGCAGCGTTACAG |
| Reverse | GTCTCCTTGGTCAAACTTC | |
| Monkey-GADPH | Forward | ACCCACTCTTCCACCTTCGACGCT |
| Reverse | TGTTGTTGTAGCCAAATTCG | |
| Swine-GAPDH | Forward | CCTTCCGTGTCCCTACTGCCAAC |
| Reverse | GACGCCTGCTTCACCACCTTCT |
Western blotting.
The total cellular proteins or samples from the pulldown and IP assay were separated by 10% to 15% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore Corporation, Bedford, MA, USA) using a Bio-Rad semidry transfer apparatus (Bio-Rad Laboratories, Hercules, CA, USA) in accordance with standard procedures. The primary antibodies used to detect viral and host proteins included the anti-CLDN3 (AF0129; Affinity Biosciences), anti-CLDN4 (ab53156; Abcam), anti-β-actin (AP0060; Bioworld Technology), anti-Myc (AT0023; CMCTAG), anti-GFP (AT0028; CMCTAG), anti-SP1 (21962-1-AP; Proteintech), and antiubiquitin (P4D1) (no. 3936; Cell Signaling Technology) antibodies. Mouse polyclonal antibodies against the PRRSV structural GP2, GP3, GP5, and M proteins (prepared in our lab) and monoclonal antibodies against the PRRSV nucleocapsid 6D10 protein were also used (36). Horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin antibodies were purchased from Jackson (West Grove, PA, USA) for use as secondary antibodies. Protein bands were visualized using the Clarity Western ECL substrate (Bio-Rad).
Quantitative real-time PCR (qPCR).
Total RNA was isolated from the cells using a GeneJET RNA purification kit (Thermo Scientific, MA, USA) and was then reverse transcribed using a ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan) in accordance with the manufacturer’s instructions. Cellular genes were quantified by relative-quantification PCR (qPCR), with the abundance of GAPDH mRNA being used as an internal reference. Primers targeting the PRRSV ORF7 mRNA were designed to detect viral genes by absolute qPCR. All qPCR assays were performed using the ABI qPCR System (Applied Biosystems, Foster City, CA, USA) with SYBR green qPCR master mix (Toyobo). The primers used for qPCR assays in this study are listed in Table 1.
Transfection.
For the pulldown and IP assays, constructs were transfected into a monolayer of HEK293T cells in 10-cm dishes using X-tremeGENE HP DNA transfection reagent (Roche, Basel, Switzerland) in accordance with the manufacturer’s instructions. The cells were collected at 24 h posttransfection (hpt) for further analysis. For siRNA, monolayers of MARC-145 cells, PK15CD163 cells, and PAMs in 24-well plates were transfected with siRNA (Table 2) using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. The cells were collected at 24 hpt. The knockdown efficiency was determined by Western blotting and qPCR. To elucidate the role of the knocked-down target on virus infection, TA-12 was inoculated onto the siRNA-transfected cells at a multiplicity of infection (MOI) of 0.1 at 24 hpt. The cells and supernatants were harvested at 0, 12, 24, 36, and 48 hpi. The transfected cells were mock infected with DMEM as a control.
TABLE 2.
siRNAs
| Gene | Nucleotide position | Sequence (5′-3′) | Antisense sequence (5′-3′) |
|---|---|---|---|
| Negative control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT | |
| CLDN4 | 243 | CCCUCGUCAUCAUCAGCAUTT | AUGCUGAUGAUGACGAGGGTT |
| 560 | CGCACAGACAAGCCUUACUTT | AGUAAGGCUUGUCUGUGCGTT | |
| CLDN3 | 422 | CCAUUAUCCGGGAAUUCUATT | UAGAAUUCCCGGAUAAUGGTT |
| 566 | AGAAGUACAUGCCCACCAATT | UUGGUGGGCAUGUACUUCUTT | |
| SP1 | 333 | GCAAGUUCUGACAGGACUATT | UAGUCCUGUCAGAACUUGCTT |
| 1355 | GCACCACUCUUACACCCAUTT | AUGGGUGUAAGAGUGGUGCTT | |
| GRHL2 | 393 | GGAGAAUUCCAAGCGGGAATT | UUCCCGCUUGGAAUUCUCCTT |
| 1004 | CCGAUUACAAGGAGAGCUUTT | AAGCUCUCCUUGUAAUCGGTT |
Cell viability.
To analyze the effect of siRNA knockdown, knockout, and overexpression on cells, the viability of the transfected cells was measured using a cell counting kit 8 (CCK-8; Beyotime, Nanjing, China) assay in accordance with the manufacturer’s instructions. Cells were grown in each well of 96-well plates to form monolayers and further cultured for 48 h, after which the CCK-8 reagent was added to each well. Following incubation for 2 h at 37°C, cell viability was evaluated by measuring the absorbance at 450 nm. The optical density of the wells containing untreated control cells was defined as indicating 100% viability.
Establishment of stable cell lines with lentivirus infection.
CLDN4 genes were amplified from the cDNA derived from MARC-145 cells and subcloned into a modified pWPXLd vector (Addgene, Cambridge, MA, USA) containing a puromycin resistance gene. The following primers were used for cloning the CLDN4 genes: Lenti-CLDN4F (5′-CGACGCGTATGGGTAGCAGCCATCATCATC-3′) and Lenti-CLDN4R (5′-CTTCCCGGGTTACACGTAGTTGCTGGCGGC-3′). These constructs (or a pWPXLd empty vector) were cotransfected with the lentiviral packaging plasmids psPAX2 and pMD2.G (at a 3:2:1 ratio) into HEK293T cells in six-well plates (40% to 50% confluence) using X-tremeGENE HP DNA transfection reagent in accordance with the manufacturer’s instructions. At 48 hpt, lentiviruses were harvested, filtered through a 0.45-μm filter, mixed with an equivalent volume of complete medium, and used to infect monolayers of MARC-145 cells. After 12 h of incubation, the spent medium was replaced with fresh medium. At 48 hpi, CLDN4-overexpressing cells and pWPXLd empty-vector-infected cells were screened using 10 μg/ml puromycin. MARC-145 cells exhibiting stable expression of CLDN4 were obtained by subcloning in 96-well plates and termed MARC-145CLDN4 cells. Cells containing the pWPXLd empty vector were termed MARC-145wpxld cells.
Generation of CLDN4 knockout cells.
A clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 vector system carrying two guide RNA-expressing cassettes was used to construct recombinant plasmids: (i) target 1 (CAS9CLDN4F1 [CACCGTGCCTCGCTCTACGTCGGCTGGG] and CAS9CLDN4R1 [AAACCCCAGCCGACGTAGAGCGAGGCAC]), which was inserted into PX459M, and (ii) target 2 (CAS9CLDN4F2 [CACCGCCCGCCCCGTACAGACAAGCCTT] and CAS9CLDN4R2 [AAACCAAGGCTTGTCTGTACGGGGCGGG]), which was inserted into EZ-GuideXH. Target 1 and target 2 were ligated to form the CRISPR-Cas9 knockout (KO) plasmid. The identified recombinant CRISPR-Cas9 KO plasmid was transfected into MARC-145 cells and subsequently sorted by replacing selective medium with 10 μg/ml of puromycin (TaKaRa, Beijing, China) approximately every 2 to 3 days for a minimum of 3 days. The knockout was then confirmed by Western blotting and qPCR, and the cells were termed MARC-145CLDN4-KO cells. The blank vectors also were also treated with the same procedure, and the corresponding cell line was termed MARC-145VEC.
Virus absorption assay.
Monolayers of MARC-145wpxld, MARC-145CLDN4, MARC-145VEC, and MARC-145CLDN4-KO cells were washed in ice-cold phosphate-buffered saline (PBS) and subsequently incubated with TA-12 at MOI of 0.1, 1, 2, and 5 for 2 h at 4°C, respectively. For PAMs, the cells were transfected with CLDN4 siRNAs (Table 1) using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions for 24 h. The cells were incubated with MOI of 0.1, 1, 2, and 5 of TA-12 for 2 h at 4°C. The exposed cells were then washed five times with ice-cold PBS to remove excess virions. The cells were washed extensively with chilled PBS and then lysed. Viral RNA was extracted using a GeneJET RNA purification kit (Thermo Scientific, MA, USA) and quantified by qPCR.
PRRSV internalization.
The kinetics of TA-12 attachment was studied by adding the virus at an MOI of 0.1 to chilled normal MARC-145, MARC-145wpxld, MARC-145CLDN4, MARC-145VEC, and MARC-145CLDN4-KO cells. After incubation for 2 h at 4°C, the cells were washed extensively with chilled PBS and shifted to 37°C to allow internalization of the attached viruses. At 4 h postinternalization, the cells were washed extensively with acidic PBS (pH 2.5) and PBS to remove any viruses attached to the cell surface. The viral RNA of each lysate sample was extracted and quantified by qPCR.
PRRSV cell-to-cell transmission.
Normal MARC-145 cells were inoculated with TA-12 at an MOI of 2 for 24 h. The infected cells were digested with trypsin and washed with acidic PBS (pH 2.5) and PBS to remove any viruses attached to the cell surface. The cells were mixed with MARC-145CLDN4, MARC-145wpxld, MARC-145CLDN4-KO, and MARC-145VEC cells at ratios of 1:4, 1:2, 1:1, and 2:1. The cells and supernatants were collected at 36 h postmixing. The viral RNA of the cells was extracted and quantified by qPCR. The viral titer in the supernatant was determined on normal MARC-145 cells.
PRRSV virus blocking assay.
The cells were treated with different concentrations of CLDN4-specific antibodies or CEL2 peptides for 1 h at 37°C, and the effects of the antibodies and peptides on virus binding and viral transmission between cells were verified by infecting cells with TA-12 at an MOI of 0.1. The MARC-145 cells were treated with different concentrations of anti-CLDN4 antibodies (0.5, 1, 2, or 4 μg) for 1 h at 37°C. After a washing with PBS, TA-12 was inoculated onto the cells at an MOI of 0.1 for 24 h. The cells were collected, and the viral RNA was extracted and quantified by qPCR. Anti-CLDN3 antibodies were applied as a control. mCLDN4-ECL2 (NIIQDFYNPLVASGQKREMGAS) and sCLDN4-ECL2 (AHNVIRDFYNPLVASGQKRE) peptides (10, 50, 100, 200, 400, 800, and 1,600 μg) were incubated with TA-12 for 1 h at 37°C and then inoculated onto MARC-145 or PK15CD163 cells for 2 h at 4°C, respectively. The cells were collected immediately or incubated further for 24 h at 37°C to detect the role of ECL2 in viral absorption and infection. The viral RNA of all the collected cells was extracted and quantified by qPCR. The roles of mCLDN3-ECL2 (NTIIREFYNPVVPEAQKREMGTS) and sCLDN3-ECL2 (VGAQCTNCVQDDTAKAKILY) in a TA-12 infection were also detected as a control. All of the peptides were synthesized by GenScript (Nanjing, China).
GFP pulldown assay and coimmunoprecipitation.
To determine the interactions between the PRRSV structural proteins with CLDN4, a GFP pulldown assay was performed. Briefly, monolayers of HEK293T cells in 10-cm dishes were transfected with recombinant plasmids GFP-GP2, GFP-GP3, GFP-GP4, GFP-GP5, GFP-M, GP3-NTD, GP3-CTD1, GP3-CTD2, and pEGFP-C1 (four dishes/plasmid). At 24 hpt, the cells were harvested and lysed. Clarified cell lysates were incubated with GFP-Trap beads (Chromotek, Munich, Germany) for 2 h at 4°C, and the bound proteins were eluted with 100 μl of 2× SDS sample buffer. For co-IP, the cell lysates were prepared and incubated with 2 μg anti-ubiquitin antibodies with the cell lysates at 4°C for 12 h under constant rotation. Then, 50 μl of protein G resin was added, and incubation was continued for 2 h. After five washes with lysis buffer, the bound proteins were eluted with 100 μl of 2× SDS sample buffer. To detect the interaction between viral proteins and endogenous CLDN4, normal MARC-145 cells were inoculated with TA-12 at an MOI of 0.1 for 24 h. The cells were lysed in 250 μl cell lysis buffer. The cell lysates were incubated with 3 μg of an anti-CLDN4 monoclonal antibody (ab53156; Abcam) at 4°C for 2 h with rotation. To assess the potential interaction between cellular proteins, proteins were pulled down by 100 μl prewashed protein A resin (L00210; GenScript) and eluted with 2× SDS loading buffer. The samples were loaded onto an SDS-PAGE gel for immunoblotting analysis.
Luciferase reporter assays.
HEK293T cells and Vero E6 cells were seeded into 24-well plates at a cell density of 4 × 104 cells/well. At 14 h to 16 h after plating, the cells were transfected with either a control plasmid or plasmids expressing viral nonstructural and structural proteins along with pGL3-CLDN4-luc and pRL-TK using X-tremeGENE HP DNA transfection reagent. A PRL-TK plasmid was used to expresses Renilla luciferase and normalize the transfection efficiency. At 24 h posttransfection, the cellular extracts were prepared and analyzed for firefly and Renilla luciferase activity using a dual-luciferase reporter assay kit (Beyotime) according to the manufacturer's instructions.
Viral titration.
MARC-145 cells were seeded into 96-well plates and incubated for 24 h at 37°C in 5% CO2. Virus supernatants were 10-fold serially diluted and added to each well (100 μl per well) in six replicates. After adsorption for 1 h at 37°C in 5% CO2, the medium was replaced with fresh medium containing difopein at specific concentrations. At 2 days postinfection, the 50% cell culture-infective dose (CCID50) was calculated using the Reed-Muench method.
Statistical analysis.
Statistical analyses were performed using a one-way analysis of variance when more than two groups were being compared and Student's t test when only two groups were being compared. The analyses were performed using the SPSS 20.0 software package (version 20.0; SPSS Inc., Chicago, IL, USA). The data were expressed as means and standard deviations (SD) from at least three independent experiments. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31772708) and the Funds of Shandong “Double Tops” Program. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
We are grateful to Enmin Zhou (Northwest A&F University, China), who kindly provided PK15CD163 cells and the PRRSV strain CH-1R.
We declare that there is no conflict of interest.
REFERENCES
- 1.Anderson JM, Van Itallie CM. 2009. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 1:a002584. doi: 10.1101/cshperspect.a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.González-Mariscal L, Miranda J, Raya-Sandino A, Domínguez-Calderón A, Cuellar-Perez F. 2017. ZO-2, a tight junction protein involved in gene expression, proliferation, apoptosis, and cell size regulation. Ann N Y Acad Sci 1397:35–53. doi: 10.1111/nyas.13334. [DOI] [PubMed] [Google Scholar]
- 3.Ikari A, Sato T, Takiguchi A, Atomi K, Yamazaki Y, Sugatani J. 2011. Claudin-2 knockdown decreases matrix metalloproteinase-9 activity and cell migration via suppression of nuclear Sp1 in A549 cells. Life Sci 88:628–633. doi: 10.1016/j.lfs.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Ren Y, Wu Q, Liu Y, Xu X, Quan C. 2013. Gene silencing of claudin6 enhances cell proliferation and migration accompanied with increased MMP2 activity via p38 MAPK signaling pathway in human breast epithelium cell line HBL100. Mol Med Rep 8:1505–1510. doi: 10.3892/mmr.2013.1675. [DOI] [PubMed] [Google Scholar]
- 5.Ding L, Lu Z, Foreman O, Tatum R, Lu Q, Renegar R, Cao J, Chen YH. 2012. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology 142:305–315. doi: 10.1053/j.gastro.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A, Ruco L, Villa A, Simmons D, Dejana E. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J Cell Biol 142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Mariscal L, Betanzos A, Nava P, Jaramillo BE. 2003. Tight junction proteins. Prog Biophys Mol Biol 81:1–44. doi: 10.1016/s0079-6107(02)00037-8. [DOI] [PubMed] [Google Scholar]
- 8.Lal-Nag M, Morin PJ. 2009. The claudins. Genome Biol 10:235. doi: 10.1186/gb-2009-10-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Yin P, Zhang L. 2019. COPII cargo claudin-12 promotes hepatitis C virus entry. J Viral Hepat 26:308–312. doi: 10.1111/jvh.13026. [DOI] [PubMed] [Google Scholar]
- 11.Medigeshi GR, Hirsch AJ, Brien JD, Uhrlaub JL, Mason PW, Wiley C, Nikolich-Zugich J, Nelson JA. 2009. West nile virus capsid degradation of claudin proteins disrupts epithelial barrier function. J Virol 83:6125–6134. doi: 10.1128/JVI.02617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo X, Guo L, Zhang J, Xu Y, Gu W, Feng L, Wang Y. 2017. Tight junction protein occludin is a porcine epidemic diarrhea virus entry factor. J Virol 91:e00202-17. doi: 10.1128/JVI.00202-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kast JI, McFarlane AJ, Głobińska A, Sokolowska M, Wawrzyniak P, Sanak M, Schwarze J, Akdis CA, Wanke K. 2017. Respiratory syncytial virus infection influences tight junction integrity. Clin Exp Immunol 190:351–359. doi: 10.1111/cei.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J, Xie Y, Campbell R, Song J, Massachi S, Razi M, Chiu R, Berenson J, Yang OO, Chen IS, Pang S. 2005. Involvement of claudin-7 in HIV infection of CD4(-) cells. Retrovirology 2:79. doi: 10.1186/1742-4690-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 16.Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F. 1991. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 17.Mengeling WL, Lager KM, Vorwald AC. 1998. Clinical consequences of exposing pregnant gilts to strains of porcine reproductive and respiratory syndrome (PRRS) virus isolated from field cases of “atypical” PRRS. Am J Vet Res 59:1540–1544. [PubMed] [Google Scholar]
- 18.Karniychuk UU, Geldhof M, Vanhee M, Van Doorsselaere J, Saveleva TA, Nauwynck HJ. 2010. Pathogenesis and antigenic characterization of a new East European subtype 3 porcine reproductive and respiratory syndrome virus isolate. BMC Vet Res 6:30. doi: 10.1186/1746-6148-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P. 2007. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J 174:577–584. doi: 10.1016/j.tvjl.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 21.An TQ, Tian ZJ, Leng CL, Peng JM, Tong GZ. 2011. Highly pathogenic porcine reproductive and respiratory syndrome virus, Asia. Emerg Infect Dis 17:1782–1784. doi: 10.3201/eid1709.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou L, Wang Z, Ding Y, Ge X, Guo X, Yang H. 2015. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis 21:2256–2257. doi: 10.3201/eid2112.150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang HM, Liu YG, Tang YD, Liu TX, Zheng LL, Wang TY, Liu SG, Wang G, Cai XH. 2018. A natural recombinant PRRSV between HP-PRRSV JXA1-like and NADC30-like strains. Transbound Emerg Dis 65:1078–1086. doi: 10.1111/tbed.12852. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L, Kang R, Xie B, Tian Y, Yang X, Yu J, Wang H. 2018. Complete genome sequence of a recombinant NADC30-like strain, SCnj16, of porcine reproductive and respiratory syndrome virus in southwestern China. Genome Announc 6:e00004-18. doi: 10.1128/genomeA.00004-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Ji G, Wang J, Tan F, Zhuang J, Li X, Tian K. 2016. Complete genome sequence of an NADC30-like porcine reproductive and respiratory syndrome virus characterized by recombination with other strains. Genome Announc 4:e00330-16. doi: 10.1128/genomeA.00330-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bian T, Sun Y, Hao M, Zhou L, Ge X, Guo X, Han J, Yang H. 2017. A recombinant type 2 porcine reproductive and respiratory syndrome virus between NADC30-like and a MLV-like: genetic characterization and pathogenicity for piglets. Infect Genet Evol 54:279–286. doi: 10.1016/j.meegid.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 27.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res 154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 29.Fang Y, Treffers EE, Li Y, Tas A, Sun Z, van der Meer Y, de Ru AH, van Veelen PA, Atkins JF, Snijder EJ, Firth AE. 2012. Efficient -2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci U S A 109:E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HS, Kwang J, Yoon IJ, Joo HS, Frey ML. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch Virol 133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- 31.Kreutz LC, Ackermann MR. 1996. Porcine reproductive and respiratory syndrome virus enters cells through a low pH-dependent endocytic pathway. Virus Res 42:137–147. doi: 10.1016/0168-1702(96)01313-5. [DOI] [PubMed] [Google Scholar]
- 32.Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch SK. 2007. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J Virol 81:7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo R, Katz BB, Tomich JM, Gallagher T, Fang Y. 2016. Porcine reproductive and respiratory syndrome virus utilizes nanotubes for intercellular spread. J Virol 90:5163–5175. doi: 10.1128/JVI.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan X, Nauwynck HJ, Pensaert MB. 1997. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch Virol 142:2483–2497. doi: 10.1007/s007050050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu SP, Zhang Z, Liu YG, Tian ZJ, Wu DL, Cai XH, He XJ. 2013. Pathogenicity and distribution of highly pathogenic porcine reproductive and respiratory syndrome virus in pigs. Transbound Emerg Dis 60:351–359. doi: 10.1111/j.1865-1682.2012.01354.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Qiu H, Zhang M, Cai X, Qu Y, Hu D, Zhao X, Zhou E, Liu S, Xiao Y. 2015. Distribution of highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) in different stages of gestation sows: HP-PRRSV distribution in gestation sows. Vet Immunol Immunopathol 166:88–94. doi: 10.1016/j.vetimm.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Zhao Q, Ge X, Teng K, Kuang Y, Chen Y, Guo X, Yang H. 2012. Chinese highly pathogenic porcine reproductive and respiratory syndrome virus exhibits more extensive tissue tropism for pigs. Virol J 9:203. doi: 10.1186/1743-422X-9-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Do TD, Park C, Choi K, Jeong J, Vo MK, Nguyen TT, Chae C. 2015. Comparison of pathogenicity of highly pathogenic porcine reproductive and respiratory syndrome virus between wild and domestic pigs. Vet Res Commun 39:79–85. doi: 10.1007/s11259-015-9628-3. [DOI] [PubMed] [Google Scholar]
- 39.Wang T, Wang X, Li XA, Nie L, Zhang M, Liu S, Zhao X, Shang Y, Zhou EM, Hiscox JA, Xiao Y. 2015. Intranasal inoculation of sows with highly pathogenic porcine reproductive and respiratory syndrome virus at mid-gestation causes transplacental infection of fetuses. Vet Res 46:142. doi: 10.1186/s13567-015-0283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell HK, Maiers JL, DeMali KA. 2017. Interplay between tight junctions & adherens junctions. Exp Cell Res 358:39–44. doi: 10.1016/j.yexcr.2017.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yumine N, Matsumoto Y, Ohta K, Fukasawa M, Nishio M. 2019. Claudin-1 inhibits human parainfluenza virus type 2 dissemination. Virology 531:93–99. doi: 10.1016/j.virol.2019.01.031. [DOI] [PubMed] [Google Scholar]
- 42.Gunzel D, Fromm M. 2012. Claudins and other tight junction proteins. Compr Physiol 2:1819–1852. doi: 10.1002/cphy.c110045. [DOI] [PubMed] [Google Scholar]
- 43.Senga K, Mostov KE, Mitaka T, Miyajima A, Tanimizu N. 2012. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol Biol Cell 23:2845–2855. doi: 10.1091/mbc.E12-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikari A, Atomi K, Takiguchi A, Yamazaki Y, Miwa M, Sugatani J. 2009. Epidermal growth factor increases claudin-4 expression mediated by Sp1 elevation in MDCK cells. Biochem Biophys Res Commun 384:306–310. doi: 10.1016/j.bbrc.2009.04.120. [DOI] [PubMed] [Google Scholar]
- 45.Beishline K, Azizkhan-Clifford J. 2015. Sp1 and the 'hallmarks of cancer'. FEBS J 282:224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 46.Black AR, Black JD, Azizkhan-Clifford J. 2001. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol 188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 47.Teifke JP, Dauber M, Fichtner D, Lenk M, Polster U, Weiland E, Beyer J. 2001. Detection of European porcine reproductive and respiratory syndrome virus in porcine alveolar macrophages by two-colour immunofluorescence and in-situ hybridization-immunohistochemistry double labelling. J Comp Pathol 124:238–245. doi: 10.1053/jcpa.2000.0458. [DOI] [PubMed] [Google Scholar]
- 48.Che P, Tang H, Li Q. 2013. The interaction between claudin-1 and dengue viral prM/M protein for its entry. Virology 446:303–313. doi: 10.1016/j.virol.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Meertens L, Bertaux C, Cukierman L, Cormier E, Lavillette D, Cosset FL, Dragic T. 2008. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J Virol 82:3555–3560. doi: 10.1128/JVI.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agrawal T, Sharvani V, Nair D, Medigeshi GR. 2013. Japanese encephalitis virus disrupts cell-cell junctions and affects the epithelial permeability barrier functions. PLoS One 8:e69465. doi: 10.1371/journal.pone.0069465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeo NK, Jang YJ. 2010. Rhinovirus infection-induced alteration of tight junction and adherens junction components in human nasal epithelial cells. Laryngoscope 120:346–352. doi: 10.1002/lary.20764. [DOI] [PubMed] [Google Scholar]
- 52.Troeger H, Loddenkemper C, Schneider T, Schreier E, Epple HJ, Zeitz M, Fromm M, Schulzke JD. 2009. Structural and functional changes of the duodenum in human norovirus infection. Gut 58:1070–1077. doi: 10.1136/gut.2008.160150. [DOI] [PubMed] [Google Scholar]
- 53.Sufiawati I, Tugizov SM. 2014. HIV-associated disruption of tight and adherens junctions of oral epithelial cells facilitates HSV-1 infection and spread. PLoS One 9:e88803. doi: 10.1371/journal.pone.0088803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spengler ML, Guo LW, Brattain MG. 2008. Phosphorylation mediates Sp1 coupled activities of proteolytic processing, desumoylation and degradation. Cell Cycle 7:623–630. doi: 10.4161/cc.7.5.5402. [DOI] [PubMed] [Google Scholar]
- 55.Wang YT, Yang WB, Chang WC, Hung JJ. 2011. Interplay of posttranslational modifications in Sp1 mediates Sp1 stability during cell cycle progression. J Mol Biol 414:1–14. doi: 10.1016/j.jmb.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 56.Van Itallie C, Rahner C, Anderson JM. 2001. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J Clin Invest 107:1319–1327. doi: 10.1172/JCI12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Vries AA, Chirnside ED, Horzinek MC, Rottier PJ. 1992. Structural proteins of equine arteritis virus. J Virol 66:6294–6303. doi: 10.1128/JVI.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Q, Yoo D. 2015. PRRS virus receptors and their role for pathogenesis. Vet Microbiol 177:229–241. doi: 10.1016/j.vetmic.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, Duan X, Nauwynck HJ. 2010. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J Gen Virol 91:1659–1667. doi: 10.1099/vir.0.020503-0. [DOI] [PubMed] [Google Scholar]
- 60.Vu HL, Kwon B, Yoon KJ, Laegreid WW, Pattnaik AK, Osorio FA. 2011. Immune evasion of porcine reproductive and respiratory syndrome virus through glycan shielding involves both glycoprotein 5 as well as glycoprotein 3. J Virol 85:5555–5564. doi: 10.1128/JVI.00189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao S, Cong F, Tan M, Ding G, Liu J, Li L, Zhao Y, Liu S, Xiao Y. 2019. 14–3-3epsilon acts as a proviral factor in highly pathogenic porcine reproductive and respiratory syndrome virus infection. Vet Res 50:16. doi: 10.1186/s13567-019-0636-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M, Krabben L, Wang F, Veit M. 2018. Glycoprotein 3 of porcine reproductive and respiratory syndrome virus exhibits an unusual hairpin-like membrane topology. J Virol 92:e00660-18. doi: 10.1128/JVI.00660-18. [DOI] [PMC free article] [PubMed] [Google Scholar]