Our study revealed that the virome was very stable across all developmental stages of both lab-derived and field-collected Aedes albopictus. The data representing the core virome in lab A. albopictus proved the vertical transmission route of these viruses, forming a “vertically transmitted core virome.” Field mosquitoes also contained this stable vertically transmitted core virome as well as additional viruses, which probably represented “environment-derived core virome” and which therefore were less stable over time and geography. By further screening publicly available SRA viral metagenomic data sets from mosquitoes belonging to the genus Aedes, some of the identified core ISVs were shown to be present in the majority of SRAs, such as Phasi Charoen-like phasivirus and Guadeloupe mosquito virus. How these core ISVs influence the biology of the mosquito host and arbovirus infection and evolution deserves to be further explored.
KEYWORDS: Aedes mosquito, Guadeloupe mosquito virus, Phasi Charoen-like phasivirus, core virome, environment-derived core virome, vertically transmitted core virome
ABSTRACT
Aedes mosquitoes can efficiently transmit many pathogenic arboviruses, placing a great burden on public health worldwide. In addition, they also carry a number of insect-specific viruses (ISVs), and it was recently suggested that some of these ISVs might form a stable species-specific “core virome” in mosquito populations. However, little is known about such a core virome in laboratory colonies and if it is present across different developmental stages. In this study, we compared the viromes in eggs, larvae, pupae, and adults of Aedes albopictus mosquitoes collected from a lab colony and compared each to the virome of different developmental stages collected in the field. The virome in lab-derived A. albopictus was very stable across all stages, consistent with a vertical transmission route of these viruses, and formed a possible “vertically transmitted core virome.” The different stages of field-collected A. albopictus mosquitoes also contained this stable vertically transmitted core virome, as well as another set of viruses (e.g., viruses distantly related to Guadeloupe mosquito virus, Hubei virga-like virus 2, and Sarawak virus) shared by mosquitoes across different stages, which might represent an “environment-derived core virome.” To further study this core set of ISVs, we screened 48 publicly available SRA viral metagenomic data sets of mosquitoes belonging to the genus Aedes, showing that some of the identified ISVs were identified in the majority of SRAs and providing further evidence supporting the core-virome concept.
IMPORTANCE Our study revealed that the virome was very stable across all developmental stages of both lab-derived and field-collected Aedes albopictus. The data representing the core virome in lab A. albopictus proved the vertical transmission route of these viruses, forming a “vertically transmitted core virome.” Field mosquitoes also contained this stable vertically transmitted core virome as well as additional viruses, which probably represented “environment-derived core virome” and which therefore were less stable over time and geography. By further screening publicly available SRA viral metagenomic data sets from mosquitoes belonging to the genus Aedes, some of the identified core ISVs were shown to be present in the majority of SRAs, such as Phasi Charoen-like phasivirus and Guadeloupe mosquito virus. How these core ISVs influence the biology of the mosquito host and arbovirus infection and evolution deserves to be further explored.
INTRODUCTION
Mosquitoes are highly diverse and widely disseminated. They occupy numerous biotopes and are potential vectors for several pathogenic arboviruses. Specifically, Aedes spp. pose a great threat to global public health. This is due to their traits of ecological plasticity that include egg diapause (1), versatility in using natural and/or urban breeding spots (2), and opportunistic feeding patterns (3, 4), which might have promoted their dispersion and adaptation to new, uncolonized territories that range from tropical to temperate regions (5). Moreover, a large number of pathogenic arboviruses, such as dengue virus (DENV), yellow fever virus (YFV), Zika virus (ZIKV), and chikungunya virus (CHIKV), are efficiently transmitted by Aedes mosquitoes, mainly A. aegypti and A. albopictus (6). In particular, DENV epidemics are a major public health concern in Guangdong Province of southern China, especially in its capital city, Guangzhou (GZ). The dengue fever cases in Guangzhou represented 50% of the national incidence between 1978 and 2011 (7), and there was an explosive outbreak in 2014 with 45,224 reported dengue fever cases (8). A. albopictus is among the most invasive mosquitoes and is widely distributed in China. It is the main vector for DENV in China and the sole vector in Guangzhou (9, 10).
In light of the holobiont concept developed in recent years, mosquito-associated microbial communities play an important role in host biology, which may provide new strategies for mosquito and arbovirus control (11). Mosquitoes are holometabolous insects whose life cycle goes through four separate stages, including egg, larva, and pupa stages in an aquatic habitat and a subaerial adult stage. A continuum of bacteria from the aquatic environment to immature stages and adult mosquitoes has been suggested (12), but bacterial clearance occurs during mosquito metamorphosis from pupae to adults (13). In addition, the nutrients produced by symbiotic bacteria are very important for larval development (14) and larval bacteria can influence immune responses and vector competence in adult mosquitoes (15).
However, little is known about the virome community dynamics in mosquitoes during holometabolous development. Some studies have performed viral metagenomics on field mosquitoes collected from different countries (including the United States, Puerto Rico, China, Kenya, Australia, Sweden, etc.), mainly focusing on novel virus discovery (16–24). Some newly discovered mosquito-specific viruses (MSVs) have been proposed as future biological control agents against arboviruses (25–27) or as novel vaccine platforms (27) or used in diagnostic assays in chimeric virus formation with structural protein genes from arboviruses (27). In a recent study, we showed the presence of a relatively stable “core eukaryotic virome” in adult field A. aegypti and Culex quinquefasciatus mosquitoes collected in Guadeloupe (28). However, the presence of insect-specific viruses (ISVs) in mosquito lab colonies is poorly studied, which limits our understanding of their role in the development and physiology of mosquitoes. On the other hand, knowledge of the normal healthy background virome as a reference will allow us to distinguish between inherent vertically transmitted components and components acquired from the environment of field mosquitoes, which will enable the identification of potential viral pathogens and improve the reliability of risk assessment using lab mosquitoes.
The first aim of this study was to analyze the virome structure of A. albopictus mosquitoes during distinct developmental stages in a laboratory-derived colony originally obtained in Jiangsu Province, China. Second, we compared these lab colony-derived viromes with the viromes of field-collected A. albopictus mosquitoes from Guangzhou (Guangdong Province, China), representing locations that are approximately 1,300 kilometers apart. Third, we conducted a comparative meta-analysis using comparisons between the viruses identified in this study and 48 publicly available virome SRA data sets of Aedes mosquitoes. Finally, phylogenetic analyses were performed on the following three selected viruses: Aedes phasmavirus (APV), which was highly present in both field and lab A. albopictus from China; Guadeloupe mosquito virus (GMV)-related viruses; and Phasi Charoen-like phasivirus (PCLPV), which was found in the majority of investigated virome data sets.
RESULTS
Comparison of the eukaryotic viromes in lab- and field-collected A. albopictus.
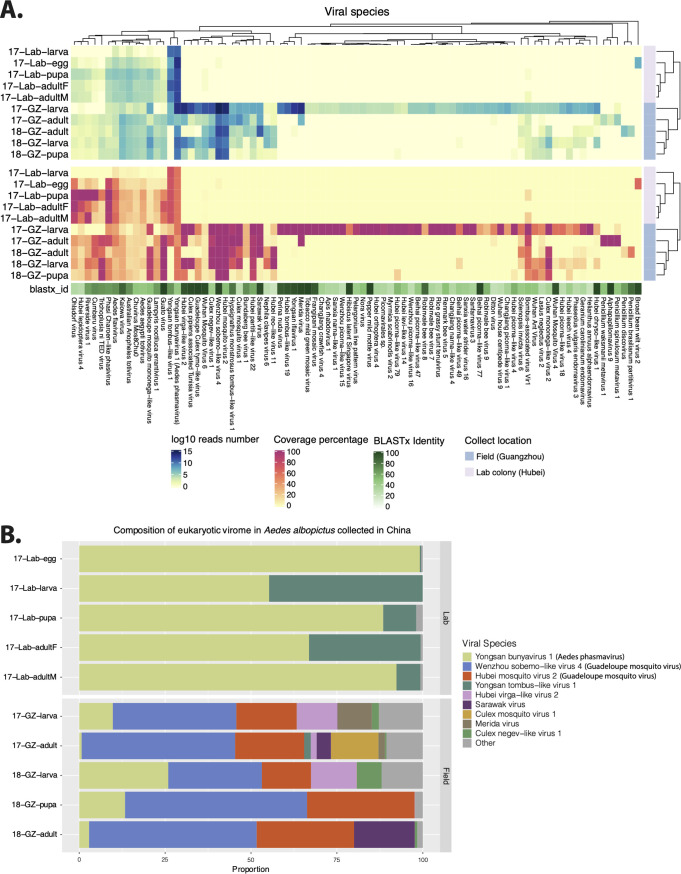
Pools of eggs, larvae, pupae, and (male/female) adults from a lab colony in Wuhan, as well as pools of larvae, pupae, and adults from the field in Guangzhou underwent metagenomic sequencing. Averages of 26.2 million trimmed reads were obtained per pool (Table 1) and were de novo assembled into 1,657,229 contigs in total. The clustering of 71,303 contigs longer than 500 bp from all samples at 95% nucleotide (nt) identity over 80% of the read length resulted in 56,419 representative contigs. According to BLASTx annotation results obtained using DIAMOND, the majority (93%) of the representative contigs belonged to Eukaryota (mainly derived from the mosquito host genome) and 179 contigs were assigned as eukaryotic viruses. Eukaryotic viral reads in each pool occupied 0.2% to 1.8% of the trimmed reads, except for the 17-GZ-larva pool, which contained 15.8% viral reads. Most of the eukaryotic viral contigs belonged to 80 different viruses (including several viruses with segmented genomes), although some of these had very low similarities to known viruses in the database (Fig. 1A). No known pathogenic arboviruses were identified, and the closest relatives of the identified viruses were generally found in insects or plants. Only 20 of the 80 viral species belonged to an established viral genus or family (e.g., Flavivirus, Iflavirus, Phasivirus, Quaranjavirus, Rhabdoviridae, Virgaviridae, Totiviridae, Nodaviridae), whereas the remaining viral genomes were most closely related to viruses discovered in recent years that current lack a formal taxonomical classification.
TABLE 1.
Aedes albopictus from China used for viral metagenomic sequencing
| Sample name |
Mosquito species |
Yr of collection |
Habitat | Location | No. of mosquitoes used for sequencing |
No. of trimmed reads obtained |
|---|---|---|---|---|---|---|
| 17-Lab-egg | A. albopictus | 2017 | Lab | Wuhan, Hubei, China | 200 | 23,801,510 |
| 17-Lab-larva | A. albopictus | 2017 | Lab | Wuhan, Hubei, China | 250 | 22,990,930 |
| 17-Lab-pupa | A. albopictus | 2017 | Lab | Wuhan, Hubei, China | 250 | 18,066,298 |
| 17-Lab-adultF | A. albopictus | 2017 | Lab | Wuhan, Hubei, China | 250 | 20,109,482 |
| 17-Lab-adultM | A. albopictus | 2017 | Lab | Wuhan, Hubei, China | 250 | 19,227,638 |
| 17-GZ-larva | A. albopictus | 2017 | Field | Liwan district, Guangzhou, China | 2,000 | 29,564,646 |
| 17-GZ-adult | A. albopictus | 2017 | Field | Liwan district, Guangzhou, China | 1,000 | 37,766,072 |
| 18-GZ-larva | A. albopictus | 2018 | Field | Liwan district, Guangzhou, China | 2,000 | 35,580,984 |
| 18-GZ-pupa | A. albopictus | 2018 | Field | Liwan district, Guangzhou, China | 500 | 25,744,126 |
| 18-GZ-adult | A. albopictus | 2018 | Field | Liwan district, Guangzhou, China | 1,100 | 29,644,914 |
FIG 1.
Eukaryotic viral genomes in wild and lab-derived Aedes albopictus pools. (A) The heat maps show the total number of mapped reads on a log10 scale (upper panel) and length coverage (lower panel) of assembled contigs of each virus. The hierarchical clustering is based on the Bray-Curtis distance matrix calculated from the number of log10 reads. The viral species names shown in the heat map are from the taxonomic annotation by DIAMOND and KronaTools. For each of the contigs assigned to a particular species, the open reading frame (ORF) with the highest BLASTx identity to a reference sequence was taken, and the median identity of these different ORFs is shown in the shaded green bar. (B) Relative abundances of the viral species in each pool based on the number of reads.
The heat map shown in Fig. 1A displays the number of reads mapped against each of the representative (partial) viral genomes as well as the percentage of coverage of each virus per sample (see Table S3 and Fig. S1 in the supplemental material). The virome in field-collected A. albopictus from both 2017 and 2018 showed significantly higher richness and diversity than that in lab-derived mosquito pools. The adjusted P values of the Wilcoxon test determined for the Chao1 and Shannon indices were 0.012 and 0.008, respectively (Fig. S1). The virome profiles of the different pools of lab colonies were relatively stable, except that several viral species were absent from or were present in low abundance in the larva pool (Fig. 1A). The field-collected larva, pupa, and adult pools collected in GZ in 2018 and the adult pool collected in 2017 also displayed very similar viral communities. However, the larval pool collected in GZ in 2017 contained more than 30 additional unique viral species with almost 100% length coverage that were almost completely absent in the adults collected at the same time and in the same place. Furthermore, the lab and field A. albopictus pools had 20 viral species in common. Among them, some contigs distantly related (47% BLASTx identity) to Yongsan bunyavirus 1 (YBV1) were present in all lab- and field-derived mosquito pools with high abundance and length coverage (Fig. 1). Some viruses such as Yongsan tombus-like virus 1 and Phasi Charoen-like phasivirus appeared to be present at higher levels in the lab-derived A. albopictus samples. In contrast, reads of several viruses appeared to be present in higher numbers in the field samples, as was the case for Guato virus and Guadeloupe mosquito mononega-like virus (Fig. 1A). Furthermore, field mosquito pools contained many more viruses which were absent in lab-derived samples (Fig. 1A), as exemplified by Wenzhou sobemo-like virus 4 and Hubei mosquito virus 2 (both with close relationship to Guadeloupe mosquito virus) (Fig. 1B).
Alpha diversity of viromes in the lab and field Aedes albopictus samples. Download FIG S1, PDF file, 0.2 MB (160.4KB, pdf) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of reads of each eukaryotic viral species in each sequencing sample. Download Table S3, XLSX file, 0.01 MB (15.7KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Prevalence of viruses in public SRA virome data sets derived from Aedes mosquitoes.
All 48 available sets of viral metagenomic data representing Aedes sp. from the public database (SRA) (see Table S1 in the supplemental material) were further analyzed to determine the conserved prevalence of ISVs in various Aedes mosquitoes. These samples were collected from locations on different continents, including the United States, Puerto Rico, Australia, Thailand, Guadeloupe, China, and Kenya. The mosquito species in the majority of the samples was A. aegypti, except for one sample from China (A. albopictus) and five from southwestern Australia (A. alboannulatus or A. camptorhynchus).
SRA datasets used in this study. Download Table S1, XLSX file, 0.01 MB (13KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
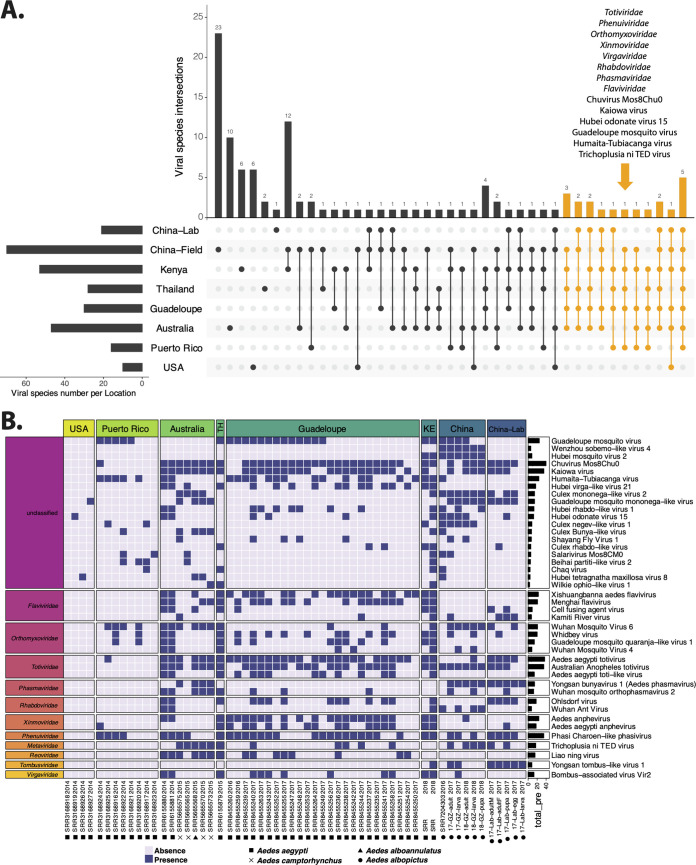
In order to investigate the potential presence of a “core virome,” we analyzed the presence/absence of each virus across the geographic origins reflected in the available SRA entries. The number of viral species shared among different locations is displayed in Fig. 2A, and virus names corresponding to each overlapping category can be found in Table S5. Twenty viruses belonging to the families Totiviridae, Phenuiviridae, Orthomyxoviridae, Xinmoviridae, Virgaviridae, Rhabdoviridae, Phasmaviridae, and Flaviviridae and six unclassified viruses (Chuvirus Mos8Chu0, Kaiowa virus, Hubei odonate virus 15, Guadeloupe mosquito virus, Humaita-Tubiacanga virus, Trichoplusia ni TED virus) were identified from at least five locations. Notably, the total number of viral species present in samples from the United States and Puerto Rico was much lower than that in samples from other locations, which could have been due to the presence of different viruses in these samples, less optimal VLP enrichment procedures used, and/or the lower sequencing depths.
FIG 2.
Conservation of viral species in Aedes sp. virome data sets. (A) The number of shared viral species among Aedes mosquitoes from different locations. (B) The viral species shown in the heat map present in samples from at least three locations and containing at least one contig that is longer than 1,500 bp. The specific viral species was considered to be present in the sample as long as the sample had one contig (>500 bp) assigned to the species.
Coverage of each eukaryotic viral species in each sequencing sample. Download Table S4, XLSX file, 0.01 MB (15.5KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Presence and absence of viral species in different locations. Download Table S5, CSV file, 0.00 MB (4.8KB, csv) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The presence or absence of each virus in each of the analyzed virome data sets is shown in the heat map in Fig. 2B. Only the 42 viral species present in a minimum of three locations and containing at least one contig longer than 1.5 kb are displayed in the figure, and they were grouped by collection locations and viral families. Totiviridae was the most prevalent viral family, containing two highly abundant species (Aedes aegypti totivirus and Australian Anopheles totivirus) with positivity rates across samples of 63.8% and 60.3%, respectively. Phasi Charoen-like phasivirus belonging to the Phenuiviridae was present in all four Aedes species and at all locations (except SRAs from the United States, in which only three of the investigated viruses were identified). Among the unclassified viral species, Chuvirus Mos8Chu0 virus and Kaiowa virus were present in 40 and 35 of 58 samples, respectively, and were found in all four Aedes species. Guadeloupe mosquito virus was detected in field A. aegypti and A. albopictus mosquitoes from Puerto Rico, Thailand, Guadeloupe, Kenya, and China. Humaita-Tubiacanga virus (absent in lab-derived and wild A. albopictus from this study) was found in field A. albopictus from Yunnan (China) and field A. aegypti from 5 locations.
Phylogeny of three selected viruses.
Considering the abundance, variance, universality, and number of complete coding genomes of viruses obtained, phylogenetic analysis was conducted on the following three selected viruses: (i) Aedes phasmavirus (APV; see below), a novel virus distantly related to YBV1 with high abundance in both field and lab A. albopictus samples from China; (ii) Phasi Charoen-like phasivirus (PCLPV), present in all four Aedes species and all locations (except the United States) of screened virome data sets; and (iii) Guadeloupe mosquito virus (GMV), occupying the majority of reads in the field-collected A. albopictus samples from Guangzhou, widely distributed in SRA data sets and showing a high level of variance. Complete coding viral genomes of APV, PCLPV, and GMV recovered from analyzed virome data sets of Aedes mosquitoes as well as corresponding reference genomes from GenBank were used for phylogeny. YBV1 is a newly identified virus reported in 2019 that showed high prevalence in A. vexans nipponii from Korea (29) and is classified in the family Phasmaviridae. We identified a novel virus, APV, in this family in both lab and field A. albopictus samples from China whose closest reference was YBV1. PCLPV, a widely distributed mosquito virus, has been identified from lab colonies, mosquito cell lines, and wild Aedes mosquitoes in eight counties or regions (China, Puerto Rico, Thailand, Kenya, the United States, Australia, Guadeloupe, and Brazil) (Table 2). It has been suggested that it is maintained in nature through vertical transmission (30). A recent study has reported that GMV is closely related to Wenzhou sobemo-like virus 4 and Hubei mosquito virus 2 (28). GMV-related viruses were detected only in mosquito samples collected from the field as listed in Table 2 (i.e., were not detected in the mosquito cell lines or lab colonies), indicating that it was likely acquired horizontally from the environment.
TABLE 2.
Distribution of APV-, PCLPV-, and GMV-related viruses
| Source | Region | Origin | APV (related to YBV1) |
PCLPV | GMV-related virus |
|---|---|---|---|---|---|
| Lab | China | 2017-Wuhan-lab | + | + | |
| Belgium | 2018-Belgium-lab cell line | + | |||
| United Kingdom | 2017-Bristol-lab cell line | + | |||
| Field | China | 2016-Zhejiang | + | + | |
| 2017-Guangzhou | + | + | + | ||
| 2018-Guangzhou | + | + | + | ||
| Puerto Rico | 2014-Puerto Rico | + | + | ||
| Thailand | 2008-Thailand | + | |||
| 2015-Thailand | + | + | |||
| Kenya | 2018-Kenya1 | + | + | ||
| 2018-Kenya2 | + | + | |||
| United States | 2014-United States | ||||
| 2015-United States | + | ||||
| Australia | 2014-Australia | + | |||
| 2015-Australia | + | + | |||
| 2016-Australia | + | ||||
| Korea | 2016-Korea | + | |||
| Guadeloupe | 2016-Guadeloupe | + | + | ||
| 2017-Guadeloupe | + | + | |||
| Brazil | 2012-Brazil | + | |||
Phylogeny of Aedes phasmavirus.
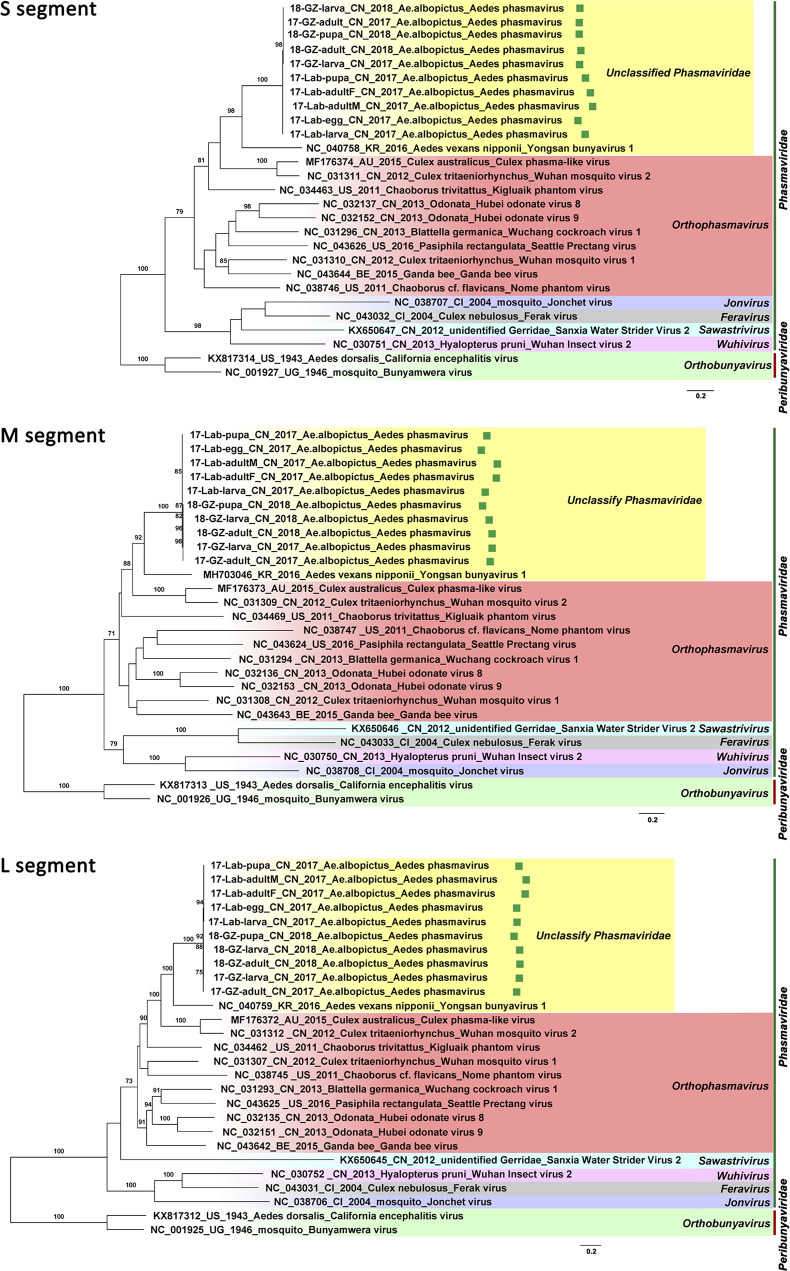
In both lab- and field-derived A. albopictus sequencing data from samples collected in China in 2017 and 2018, three contigs of APV in each sample were found to share the highest amino acid identities of 50.8%, 48.1%, and 43.6% with the S, M, and L segments of YBV1 (GenBank accession no. MH703047.1, MH703046.1, and MH703045.1), respectively. In total, 10 viral genomes of APV with three segments were recovered from all sequenced A. albopictus samples (17-lab-egg, 17-lab-larva, 17-lab-pupa, 17-lab-adultM, 17-lab-adultF, 17-GZ-larva, 17-GZ-adult, 18-GZ-larva, 18-GZ-pupa, and 18-GZ-adult). The longest lengths were of 1,185, 2,022, and 6,468 nt, which corresponded to the nucleocapsid, glycoprotein, and RNA-dependent RNA polymerase (RdRp) genes, respectively. For all three segments, the nucleotide identity among the 10 assembled viral genomes ranged between 96% and 100%, which indicated that they represent closely related variants. In a separate phylogenetic analysis of the three segments, the 10 viral genomes of APV clustered together within the genus Orthophasmavirus in the family Phasmaviridae (Fig. 3). Their closest relative was YBV1, which was identified in A. vexans mosquitoes collected in South Korea in 2016, but with low levels of nucleotide similarity (ranging from 52% to 57%) in the coding regions of the three segments. Thus, the APVs present in the lab and field A. albopictus samples collected in China appeared to represent a novel species in the genus Orthophasmavirus.
FIG 3.
Maximum-likelihood phylogenetic tree based on the nucleotide level of complete coding region in the S, M, and L segments of the newly found Aedes phasmaviruses (APVs) in Aedes mosquitoes from China (highlighted with green squares) and other representative members in the family Phasmaviriade. Representatives in the genus Orthobunyavirus belonging to the family Peribunyaviridae were used as the outgroup.
Phylogeny of Phasi Charoen-like phasivirus.
Since the abundances of PCLPV reads in the A. albopictus samples from Guangzhou and lab-derived mosquitoes were relatively low, no complete genome was recovered from these pools. All three segments of PCLPV with a complete coding region were successfully recovered in one A. aegypti sample from Puerto Rico collected in 2014 and two A. aegypti samples from Kenya collected in 2018. The phylogenetic analysis was performed on these newly identified PCLPVs, on all other PCLPV genomes in GenBank (including those identified in A. aegypti mosquitoes collected in Thailand in 2008, in the United States in 2015, in Australia in 2016, in Zhejiang Province of China in 2016, in Guadeloupe in 2017, and in Aag2 cells from the United Kingdom), and on two more genomes obtained from lab Aag2 cell lines in Belgium (Fig. 4). It was interesting that all of the PCLPVs collected in distant geographic locations, in different years, and from field mosquitoes versus lab cell lines clustered very closely in the three maximum-likelihood (ML) trees of each segment. The nucleotide similarities among the genomes of PCLPVs were very high, ranging from 94% to 98% for the S segment, from 95% to 99% for M segment, and from 94% to 99% for L segment.
FIG 4.
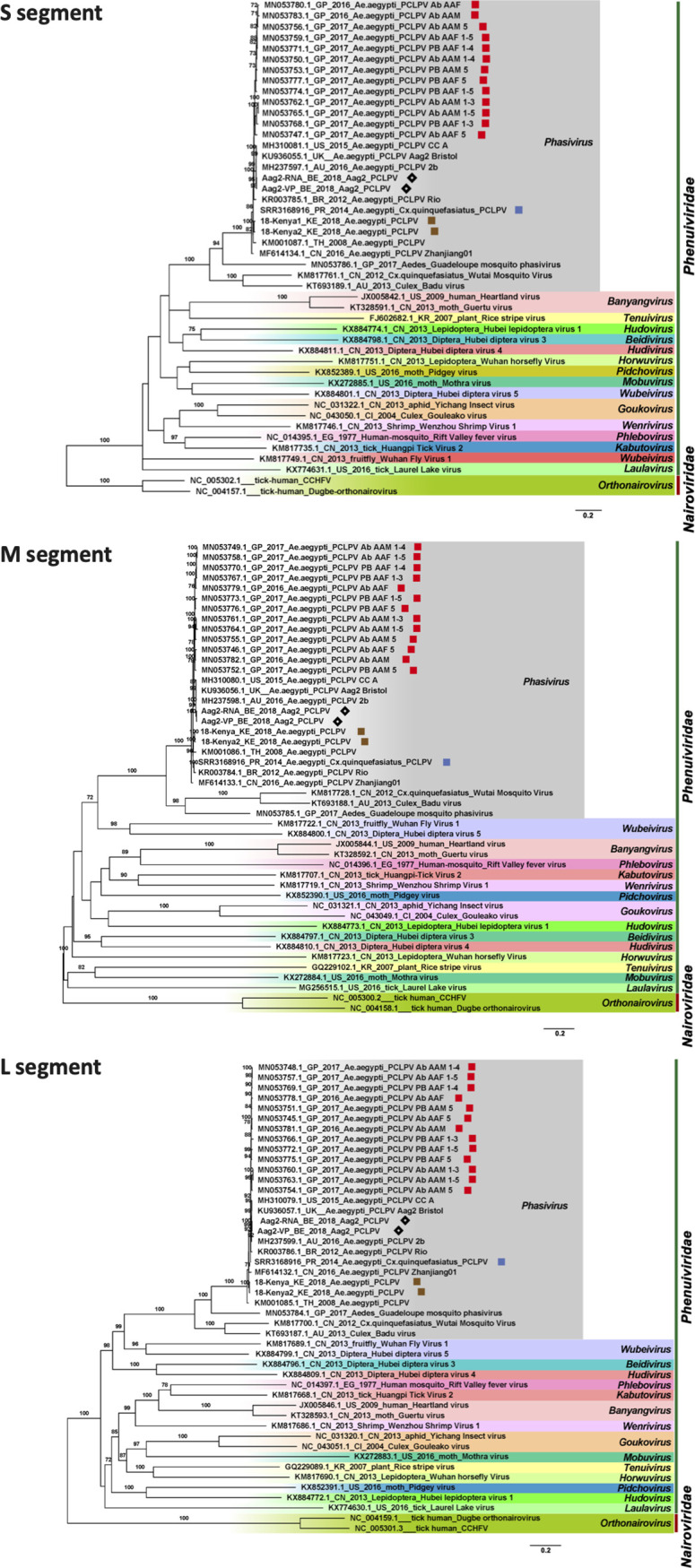
Maximum-likelihood phylogenetic tree based on the nucleotide sequence of complete coding regions of the S, M, and L segments of Phasi Charoen-like phasivirus (PCLPV) identified from Aedes mosquitoes and cells, together with other representative members of the family Phenuiviriade. The strains from Guadeloupe are highlighted with red squares, strains derived from Belgian Aag2 cells with black diamonds, strains from Kenya with brown squares, and strains from Puerto Rico with blue squares. Representative Orthonairovirus strains belonging to the family Nairoviridae were used as the outgroup.
Phylogeny of Guadeloupe mosquito virus-related viruses.
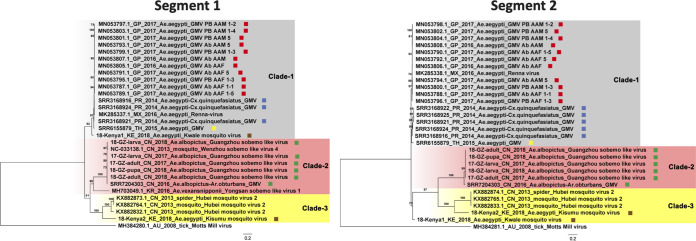
Twelve viral genomes (with complete coding regions) similar to GMV, Wenzhou sobemo-like virus 4, and Hubei mosquito virus 2 were obtained from the analyzed virome data sets, which included six pools of field A. albopictus collected in China (17-GZ-larvae, 17-GZ-adult, 18-GZ-larva, 18-GZ-pupa, 18-GZ-adult, and SRR7204303-CN-2016), three pools of an A. aegypti and Cx. quinquefasciatus mixture collected in Puerto Rico (SRR3168916-PR-2014, SRR3168921-PR-2014, and SRR3168924-PR-2014), and two A. aegypti pools collected in Kenya (SRR12102797-KE-2018 and SRR12102798-KE-2018) and one collected in Thailand (SRR6155879-TH-2015). The viral genomes contained two segments encoding an RdRp and one hypothetical protein on segment 1 (1,500 to 1,600 nt) and a capsid and one hypothetical protein encoded by segment 2 (3,000 to 3,200 nt). GMV, Wenzhou sobemo-like virus 4, and Hubei mosquito virus 2 are all newly described and currently unclassified viruses with a distant relationship to the families Luteoviridae and Sobemoviridae (28).
The phylogenetic analysis performed on the basis of segments 1 and 2 indicated that these sequences fell into three clades (Fig. 5). The genomes identified in samples collected in Thailand in 2015, Puerto Rico in 2014, and Kenya in 2018 clustered together with the Guadeloupe mosquito viruses found in Guadeloupe in 2016 and 2017, forming clade 1, with the nucleotide identity in this clade ranging from 90% to 99% for both segments. Five genomes recovered from A. albopictus collected in Guangzhou in 2017 and 2018, one recovered from the same host collected in Yunnan in 2016, and Wenzhou sobemo-like virus 4 clustered together in clade 2, with nucleotide identities among them ranging from 94% to 98%. The third clade consisted of Hubei mosquito virus 2 and one sequence from a sample collected in Kenya in 2018, with nucleotide identities ranging from 70% to 100%. The nucleotide similarities for the two segments among these three clades ranged from 54% to 69%.
FIG 5.
Maximum-likelihood phylogenetic tree based on the nucleotide sequence of the complete coding regions in segments 1 and 2 of Guadeloupe mosquito virus (GMV)-related viruses. The red, blue, yellow, brown, and green squares after the sequence names highlight the strains from Guadeloupe, Puerto Rico, Thailand, Kenya, and China, respectively. Motts Mill virus identified from ticks was used as the outgroup.
DISCUSSION
In this study, we first characterized the eukaryotic virome across different developmental stages of lab-derived and field-collected A. albopictus from China. Five pools from lab mosquitoes (containing 1,250 individuals in total) and five pools from field-collected mosquitoes (6,600 individuals in total) were analyzed (Table 1). Considering that the rearing conditions were stable in the lab and that the colony has been reared in the lab for many years, we collected lab-derived samples only in 2017 as no scientifically significant changes over time were anticipated. To take potential yearly virome fluctuations for field mosquitoes into account, they were collected in two consecutive years (2017 and 2018). The proportion of the eukaryotic viral reads in each sample (ranging from 0.2% to 15.8%) was comparable to that reported from previous mosquito virome studies (24, 28), suggesting that the sequencing depth was sufficient for virome analysis.
As expected, the virome diversity in lab A. albopictus was lower than that in field mosquitoes (Fig. 1), probably due to the less complex environment and the availability of clean water and food resources. Unlike the bacterial communities seen in field-collected mosquitoes (13, 31–35), it is interesting that the virome in lab-derived A. albopictus was very stable across all developmental stages, consistent with vertical transmission of these viruses. For the larvae only, relatively few viral reads were identified. This finding suggested that the virus might remain dormant at a very low concentration without or with very low rates of replication. The fact that these viruses were also present in field mosquitoes suggested that A. albopictus seems to contain a “vertically transmitted core virome” as was described before for A. aegypti and Culex quinquefasciatus from Guadeloupe (28), for which stable distinct core eukaryotic viromes were identified which possessed nearly identical viruses across time and space (28). In addition, another set of viruses was found to be shared by the field-collected A. albopictus mosquitoes across different stages, suggesting that they have a “core virome” with higher richness and diversity (Fig. S1). Whether these additional viruses were acquired from the environment or the lab-derived mosquitoes lost these viruses in captivity remains to be determined. All these vertically transmitted core viromes in A. albopictus deserve more attention with respect to their effects on vector competence for important medically relevant arboviruses. Due to the mosquito samples having originated from only one lab colony or one location in the field, further surveillance over a larger geographic range and longer periods of time will be needed to confirm the stability of the virome profile across different developmental stages. In addition, the larval pool collected in the wild in 2017 was an outlier, as it contained many unique viruses, which probably originated from the particular water environment they lived in, which could have been contaminated by other coinhabiting insects or larvae. Alternatively, it could be the case that a number of mosquitoes other than A. albopictus were included in the pool by accident. These results further suggest a significant influence of the breeding site ecology on the mosquito viral community.
Since some viruses (e.g., PCLPV, GMV, and Aedes aegypti totivirus) identified in A. albopictus from China were also identified as part of the core virome in A. aegypti from Guadeloupe (28), we were interested in determining how these core viruses in Aedes mosquitoes were further distributed around the world. Therefore, we explored 48 Aedes sp. viral metagenomic data sets from a public database (SRA). The samples were from different countries on different continents (China, the United States, Australia, Kenya, Thailand, Puerto Rico, and Guadeloupe) and from different mosquito species (A. aegypti, A. albopictus, A. alboannulatus, and A. camptorhynchus) and were treated with different wet-lab and sequencing procedures using different amounts of pooled mosquitoes and sequencing depths, but a highly prevalent set of widely distributed viruses, such as Totiviridae, Orthomyxoviridae, Xinmoviridae, Flaviviridae, PCLPV, and GMV, were able to be identified on the family or species level (Fig. 2). How these conserved viruses in Aedes mosquitoes interact with or influence an arbovirus infection is an interesting point for further studies. A previous study explored the effect of coinfections of PCLPV and cell-fusing agent virus (CFAV) on the replication of arboviruses in cell line Aa23 derived from A. albopictus (36). CFAV-PCLPV-positive Aa23 cells strongly inhibited the growth of two flaviviruses (ZIKV and DENV) and completely blocked infection by La Crosse virus (bunyavirus). Although the exact blocking mechanism was not known, on the one hand, the results suggested that the data generated from laboratory cell lines persistently infected by mosquito-specific viruses (MSVs) should be interpreted with caution. On the other hand, the intra-MSV interactions need to be considered when the influence of MSVs on vector competence is explored.
Although many viruses were very prevalent in Aedes mosquitoes, such as Chuvirus Mos8Chu0, Kaiowa virus, Wuhan mosquito virus 6, Whidbey virus, Aedes aegypti totivirus, and Australian Anopheles totivirus, phylogenetic analysis was further performed on GMV and PCLPV, which had the greatest number of complete coding genomes, and on APV, which was highly abundant in both field and lab A. albopictus from China. APV was found to be distantly related to the YBV1 identified from A. vexans from South Korea forming a separate clade. All the Chinese strains from both lab- and field-collected mosquitoes were very similar (Fig. 3). Also, for PCLPV, the genomes found in samples from different years, locations, and habitats were very similar for all three segments (Fig. 4). Interactions between bunyaviruses and arthropods occurring over 20 million years had been previously demonstrated (37–41). The findings indicating that APV and PCLPV seem to be very closely related were puzzling and might suggest a rather relatively recent spread of this virus or a very low evolutionary rate, which would be unexpected for RNA viruses. However, the effects of these viruses on their host are very poorly understood. A recent study revealed that preexisting infection of Aag2 cells with PCLPV did not affect the infection and growth of the mosquito-borne viruses in genus Flavivirus, Alphavirus, and Rhabdovirus in cell culture (42). GMV is a recently described two-segmented, currently unclassified virus. Three separate clades were observed for GMVs and GMV-related viruses, largely clustering according to location. This group of viruses should be proposed as a new family, and their role in arbovirus infection needs to be further studied.
In summary, our results reveal that the virome profile was very stable across different stages in both lab- and field-derived Aedes albopictus and that a number of possible core viruses exist in Aedes mosquitoes of at least four species in multiple locations in seven countries. Since the number of available Aedes virome data sets is still rather limited and since wet-lab procedures, sequencing depths, and pool sizes differed greatly among the analyzed data sets, the core viruses need to be further confirmed by next-generation sequencing (NGS) or PCR. To fully characterize and understand the genetic and phenotypic diversity of mosquito-specific viruses, samples from individual Aedes mosquitoes of additional species collected from additional locations and at additional time points, processed and sequenced with the same method, should be analyzed.
MATERIALS AND METHODS
Sample collection.
An A. albopictus colony was established in our laboratory (Wuhan, China) in 2017 which originated from a stable colony from the National Institute for Communicable Disease Control and Prevention (China CDC; Beijing, China). The adult mosquitoes were maintained at 27 to 30°C with 60% to 85% relative humidity using a photoperiod of a 12-h:12-h light-dark cycle. The larvae were fed with a mixture of yeast powder and wheat mill. Adult mosquitoes were placed into cages (30 cm by 30 cm by 30 cm), and the males were fed with 10% sucrose solution, while the females were fed with blood from mice. Egg, larva, pupa, and male and female adult samples of lab colony mosquitoes were collected in August 2017 (Table 1).
Field larva, pupa, and adult samples of A. albopictus were trapped in Guangzhou (Liwan district), Guangdong Province, China, in July to November of 2017 and 2018. In Kenya, adult mosquitoes of A. aegypti were trapped during July and August of 2018 in Ukunda and Kisumu. All samples were collected from residential quarters in urban areas, transported to the laboratory using an appropriate cold chain, and stored at −80°C. Mosquito species were determined by morphological identification, and samples were assigned into pools according to the date of collection (Table 1).
Sample preparation for NGS.
The collected mosquitoes were pooled according to developmental stage for sequencing. The pooled samples were triturated by the use of a Tgrinder OSE-Y30 electric tissue grinder (Tiangen, China) on ice using sterile pestles with 200 μl of RPMI medium. Mosquito homogenates were clarified by centrifugation at 20,000 × g (4°C for 30 min) and filtered through a 0.45-μm-pore-size membrane filter (Millipore, Billerica, USA). Supernatants were collected and stored at −80°C. RNA was extracted from the supernatants with an RNeasy minikit (Qiagen, Germany) according to the manufacturer’s instructions. Then, strand-specific libraries were prepared using a TruSeq stranded total RNA sample preparation kit (Illumina, USA). TruSeq PE (paired-end) cluster kit v3 (Illumina, USA) and TruSeq SBS kit v3-HS (Illumina, USA) (300 cycles) were used for sequencing with 150 bp per read (PE 2 × 150 bp), which was performed on an Illumina HiSeq 2500 platform (Illumina, USA) by Shanghai Biotechnology Corporation.
Bioinformatic analysis of viral metagenomic data.
The obtained raw paired-end reads were trimmed for quality and adapters using Trimmomatic (43), and the remaining reads were de novo assembled into contigs using metaSPAdes (44). Contigs from all pools longer than 500 bp were filtered for redundancy at 95% nucleotide identity over 80% of the length using ClusterGenomes (45). The collection of representative contigs was classified using DIAMOND (46) against the nr database (updated 29 September 2019) using the sensitive mode for taxonomic annotation. KronaTools (47) was used to parse the output file of DIAMOND, which was used to find the least common ancestor of the best 25 DIAMOND hits (based on BLASTx score) for each contig. All contigs annotated as eukaryotic viruses were extracted using an in-house Python script. The abundance and length coverage of eukaryotic virus contigs in individual pools were obtained by aligning trimmed and decontaminated reads of each sample to the collection of representative contigs using BBMap (48). Abundance tables were extracted from eukaryotic viruses and further used for making heat maps in R with the ComplexHeatmap (49), ggplot2 (50), and phyloseq (51) packages. The length coverage of each viral species per sample was calculated by the following formula: contig length covered by at least one read/the total length of the corresponding contig.
Eukaryotic virus screening of Aedes mosquito virome data in SRA.
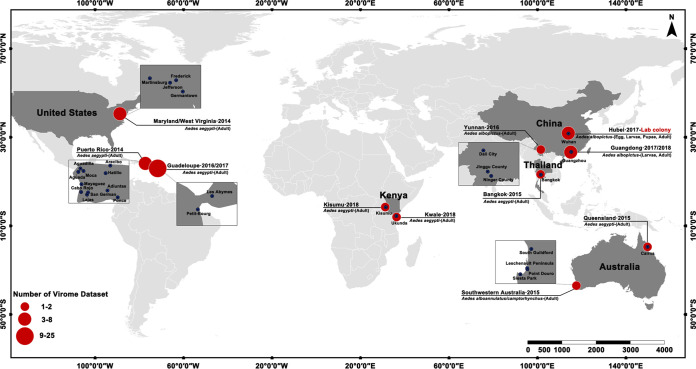
To investigate the level of conservation of the eukaryotic core viruses identified in Chinese samples from this study with those of other Aedes mosquitoes worldwide, we retrieved 48 published SRA data sets (including 25 data sets of Aedes sp. from Guadeloupe 28], 8 from Puerto Rico [16], 4 from the United States [16], 7 from Australia [52, 53], 2 from Kenya, 1 from Thailand [53], and 1 from China [54]) (Fig. 6; see also Table S1 in the supplemental material). The world map displaying the geographic origin of all Aedes virome data sets used in this study was drawn with ArcGI (ArcMap 10.5). The raw reads of the SRA data sets were de novo assembled using SKESA (55) with default settings, which was less computationally intensive than metaSPAdes. These obtained contigs were taxonomically annotated using DIAMOND against the nr database (updated 29 September 2019) in sensitive mode. KronaTools (47) and an in-house Python script were used to parse the output file of DIAMOND and extract eukaryotic virus contigs.
FIG 6.
Global distribution of Aedes mosquitoes virome data sets used in this study.
The presence of eukaryotic viruses in all Aedes virome data sets.
Taxonomically identified eukaryotic virus contigs longer than 500 bp were extracted from all 58 Aedes virome data sets and placed together in a single file. Viral species that contained at least one contig longer than 1,500 bp were used for visualization purposes. A viral species was considered present in the sample as long as the sample had one contig (>500 bp) assigned to the species. The viral species shared among different countries were visualized with an UpSet plot (Fig. 2A) using the UpSetR package (56). The viral species are shown in the heat map (Fig. 2B) only if they were present in samples from at least three countries.
Phylogenetic analysis.
To obtain the complete genomes related to Yongsan bunyavirus 1 (YBV1), Phasi Charoen-like phasivirus (PCLPV), and Guadeloupe mosquito virus (GMV) from all Aedes virome data sets, the trimmed reads of each sample were individually mapped with BWA (57) against the following selected reference genomes: (i) 17-Lab-egg-L (MT361040), 17-Lab-egg-M (MT361040), and 17-Lab-pupa-S (MT361044) as the reference for YBV1; (ii) PCLPV Rio strain (KR003786.1, KR003784.1, and KR003785.1) for PCLPV; and (iii) 18-GZ-larva-seg1 (MT361057) and 18-GZ-pupa-seg2 (MT361060) for GMV. The consensus sequences of these viruses were generated from the bam files using SAMtools and bcftools (58).
The nucleotide sequences of the complete genomes or complete coding regions in the genomes (segments) of these viruses were used to determine their evolutionary history together with appropriate reference strains from GenBank. Alignments of the sequences were performed with MAFFT v7.222 (59) using the most accurate algorithm, L-INS-I, with 1,000 cycles of iterative refinement. Ambiguously aligned regions were removed by trimAl v1.2 (60) using an automated trimming heuristic, which was optimized for maximum-likelihood (ML) phylogenetic tree reconstruction. The phylogenetic trees for each segment were reconstructed from 1,000 ultrafast bootstrap ML tree replicates using IQ-TREE v1.6.11 (61) with best-fit model selection by ModelFinder (62). FigTree v1.4.3 (63) was used for phylogenetic tree visualization.
Data availability.
Accession numbers of the viruses obtained from our data set are listed in Table S2, and viral genome sequences recovered from the SRA data sets can be found at https://github.com/Matthijnssenslab/AedesVirome. All viral sequences used to construct phylogenetic trees can be found at https://github.com/Matthijnssenslab/AedesVirome.
Accession numbers of YBV1-, PCLPV-, and GMV-related viruses identified in this study. Download Table S2, XLSX file, 0.01 MB (11.4KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Science and Technology of the People’s Republic of China (2018ZX10101004); the National Health Commission of the People’s Republic of China (2018ZX10711001-006); the Chinese Academy of Sciences (153211KYSB20160001); the Wuhan Institute of Virology, China (WIV-135-PY2); and the Health Commission of Hubei Province (WJ2019Q060).
C.S. performed the bioinformatics analysis of NGS data with support from J.M. L.Z., H.X., and A.E. designed the experiments, conducted the wet lab experiments, and performed the phylogenetic analysis. W.Z. collected the field mosquito samples. C.S. and H.X. drafted the manuscript. L.Z., A.E., X.H., J.M., and Z.Y. reviewed the manuscript.
We declare that we have no competing interests.
REFERENCES
- 1.Focks DA, Linda SB, Craig GB, Jr, Hawley WA, Pumpuni CB. 1994. Aedes albopictus (Diptera: Culicidae): a statistical model of the role of temperature, photoperiod, and geography in the induction of egg diapause. J Med Entomol 31:278–286. doi: 10.1093/jmedent/31.2.278. [DOI] [PubMed] [Google Scholar]
- 2.Paupy C, Delatte H, Bagny L, Corbel V, Fontenille D. 2009. Aedes albopictus, an arbovirus vector: from the darkness to the light. Microbes Infect 11:1177–1185. doi: 10.1016/j.micinf.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Delatte H, Desvars A, Bouetard A, Bord S, Gimonneau G, Vourc'h G, Fontenille D. 2010. Blood-feeding behavior of Aedes albopictus, a vector of chikungunya on La Reunion. Vector Borne Zoonotic Dis 10:249–258. doi: 10.1089/vbz.2009.0026. [DOI] [PubMed] [Google Scholar]
- 4.Kamgang B, Nchoutpouen E, Simard F, Paupy C. 2012. Notes on the blood-feeding behavior of Aedes albopictus (Diptera: Culicidae) in Cameroon. Parasit Vectors 5:57. doi: 10.1186/1756-3305-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonizzoni M, Gasperi G, Chen XG, James AA. 2013. The invasive mosquito species Aedes albopictus: current knowledge and future perspectives. Trends Parasitol 29:460–468. doi: 10.1016/j.pt.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. 2020. Vector-borne diseases. https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases.
- 7.Lai S, Huang Z, Zhou H, Anders KL, Perkins TA, Yin W, Li Y, Mu D, Chen Q, Zhang Z, Qiu Y, Wang L, Zhang H, Zeng L, Ren X, Geng M, Li Z, Tatem AJ, Hay SI, Yu H. 2015. The changing epidemiology of dengue in China, 1990–2014: a descriptive analysis of 25 years of nationwide surveillance data. BMC Med 13:100. doi: 10.1186/s12916-015-0336-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao JP, He JF, Deng AP, Lin HL, Song T, Peng ZQ, Wu XC, Liu T, Li ZH, Rutherford S, Zeng WL, Li X, Ma WJ, Zhang YH. 2016. Characterizing a large outbreak of dengue fever in Guangdong Province. Infect Dis Poverty 5:44. doi: 10.1186/s40249-016-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng HJ, Lai HB, Zhang QL, Xu BY, Zhang H, Liu WH, Zhao W, Zhou YP, Zhong XG, Jiang S, Duan JH, Yan GY, He JF, Chen XG. 2012. A local outbreak of dengue caused by an imported case in Dongguan China. BMC Public Health 12:83. doi: 10.1186/1471-2458-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Kamara F, Zhou G, Puthiyakunnon S, Li C, Liu Y, Zhou Y, Yao L, Yan G, Chen XG. 2014. Urbanization increases Aedes albopictus larval habitats and accelerates mosquito development and survivorship. PLoS Negl Trop Dis 8:e3301. doi: 10.1371/journal.pntd.0003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guegan M, Zouache K, Demichel C, Minard G, Tran Van V, Potier P, Mavingui P, Valiente Moro C. 2018. The mosquito holobiont: fresh insight into mosquito-microbiota interactions. Microbiome 6:49. doi: 10.1186/s40168-018-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimonneau G, Tchioffo MT, Abate L, Boissiere A, Awono-Ambene PH, Nsango SE, Christen R, Morlais I. 2014. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect Genet Evol 28:715–724. doi: 10.1016/j.meegid.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Moll RM, Romoser WS, Modrzakowski MC, Moncayo AC, Lerdthusnee K. 2001. Meconial peritrophic membranes and the fate of midgut bacteria during mosquito (Diptera: Culicidae) metamorphosis. J Med Entomol 38:29–32. doi: 10.1603/0022-2585-38.1.29. [DOI] [PubMed] [Google Scholar]
- 14.Correa MA, Matusovsky B, Brackney DE, Steven B. 2018. Generation of axenic Aedes aegypti demonstrate live bacteria are not required for mosquito development. Nat Commun 9:4464. doi: 10.1038/s41467-018-07014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Garcia M, Vargas V, Ramirez-Bello I, Hernandez-Martinez G, Lanz-Mendoza H. 2015. Bacterial exposure at the larval stage induced sexual immune dimorphism and priming in adult Aedes aegypti mosquitoes. PLoS One 10:e0133240. doi: 10.1371/journal.pone.0133240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey KG, Biser T, Hamilton T, Santos CJ, Pimentel G, Mokashi VP, Bishop-Lilly KA. 2016. Bioinformatic characterization of mosquito viromes within the eastern United States and Puerto Rico: discovery of novel viruses. Evol Bioinform Online 12:1–12. doi: 10.4137/EBO.S38518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia H, Wang Y, Shi C, Atoni E, Zhao L, Yuan Z. 2018. Comparative metagenomic profiling of viromes associated with four common mosquito species in China. Virol Sin 33:59–66. doi: 10.1007/s12250-018-0015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi C, Liu Y, Hu X, Xiong J, Zhang B, Yuan Z. 2015. A metagenomic survey of viral abundance and diversity in mosquitoes from Hubei province. PLoS One 10:e0129845. doi: 10.1371/journal.pone.0129845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atoni E, Wang Y, Karungu S, Waruhiu C, Zohaib A, Obanda V, Agwanda B, Mutua M, Xia H, Yuan Z. 2018. Metagenomic virome analysis of Culex mosquitoes from Kenya and China. Viruses 10:30. doi: 10.3390/v10010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadeghi M, Altan E, Deng X, Barker CM, Fang Y, Coffey LL, Delwart E. 2018. Virome of >12 thousand Culex mosquitoes from throughout California. Virology 523:74–88. doi: 10.1016/j.virol.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 21.Fauver JR, Grubaugh ND, Krajacich BJ, Weger-Lucarelli J, Lakin SM, Fakoli LS, Bolay FK, Diclaro JW, Dabiré KR, Foy BD, Brackney DE, Ebel GD, Stenglein MD. 2016. West African Anopheles gambiae mosquitoes harbor a taxonomically diverse virome including new insect-specific flaviviruses, mononegaviruses, and totiviruses. Virology 498:288–299. doi: 10.1016/j.virol.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 22.Hameed M, Liu K, Anwar MN, Wahaab A, Li C, Di D, Wang X, Khan S, Xu J, Li B, Nawaz M, Shao D, Qiu Y, Wei J, Ma Z. 2020. A viral metagenomic analysis reveals rich viral abundance and diversity in mosquitoes from pig farms. Transbound Emerg Dis 67:328–343. doi: 10.1111/tbed.13355. [DOI] [PubMed] [Google Scholar]
- 23.Ohlund P, Hayer J, Lunden H, Hesson JC, Blomstrom AL. 2019. Viromics reveal a number of novel RNA viruses in Swedish mosquitoes. Viruses 11:1027. doi: 10.3390/v11111027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersson JH, Shi M, Eden JS, Holmes EC, Hesson JC. 2019. Meta-transcriptomic comparison of the RNA viromes of the mosquito vectors Culex pipiens and Culex torrentium in northern Europe. Viruses 11:1033. doi: 10.3390/v11111033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelman ZN, Blair CD, Carlson JO, Beaty BJ, Olson KE. 2001. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol Biol 10:265–273. doi: 10.1046/j.1365-2583.2001.00267.x. [DOI] [PubMed] [Google Scholar]
- 26.Bolling BG, Weaver SC, Tesh RB, Vasilakis N. 2015. Insect-specific virus discovery: significance for the Arbovirus community. Viruses-Basel 7:4911–4928. doi: 10.3390/v7092851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolling BG, Vasilakis N, Guzman H, Widen SG, Wood TG, Popov VL, Thangamani S, Tesh RB. 2015. Insect-specific viruses detected in laboratory mosquito colonies and their potential implications for experiments evaluating Arbovirus vector competence. Am J Trop Med Hyg 92:422–428. doi: 10.4269/ajtmh.14-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi C, Beller L, Deboutte W, Yinda KC, Delang L, Vega-Rua A, Failloux AB, Matthijnssens J. 2019. Stable distinct core eukaryotic viromes in different mosquito species from Guadeloupe, using single mosquito viral metagenomics. Microbiome 7:121. doi: 10.1186/s40168-019-0734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanborn MA, Klein TA, Kim HC, Fung CK, Figueroa KL, Yang Y, Asafo-Adjei EA, Jarman RG, Hang J. 2019. Metagenomic analysis reveals three novel and prevalent mosquito viruses from a single pool of Aedes vexans nipponii collected in the Republic of Korea. Viruses 11:222. doi: 10.3390/v11030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Huang S, Jin T, Lin P, Huang Y, Wu C, Peng B, Wei L, Chu H, Wang M, Jia Z, Zhang S, Xie J, Cheng J, Wan C, Zhang R. 2018. Discovery and high prevalence of Phasi Charoen-like virus in field-captured Aedes aegypti in South China. Virology 523:35–40. doi: 10.1016/j.virol.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Chavshin AR, Oshaghi MA, Vatandoost H, Yakhchali B, Zarenejad F, Terenius O. 2015. Malpighian tubules are important determinants of Pseudomonas transstadial transmission and longtime persistence in Anopheles stephensi. Parasit Vectors 8:36. doi: 10.1186/s13071-015-0635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coon KL, Brown MR, Strand MR. 2016. Gut bacteria differentially affect egg production in the anautogenous mosquito Aedes aegypti and facultatively autogenous mosquito Aedes atropalpus (Diptera: Culicidae). Parasit Vectors 9:375. doi: 10.1186/s13071-016-1660-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Favia G, Ricci I, Damiani C, Raddadi N, Crotti E, Marzorati M, Rizzi A, Urso R, Brusetti L, Borin S, Mora D, Scuppa P, Pasqualini L, Clementi E, Genchi M, Corona S, Negri I, Grandi G, Alma A, Kramer L, Esposito F, Bandi C, Sacchi L, Daffonchio D. 2007. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci U S A 104:9047–9051. doi: 10.1073/pnas.0610451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Bagdasarian M, Walker ED. 2015. Elizabethkingia anophelis: molecular manipulation and interactions with mosquito hosts. Appl Environ Microbiol 81:2233–2243. doi: 10.1128/AEM.03733-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coon KL, Vogel KJ, Brown MR, Strand MR. 2014. Mosquitoes rely on their gut microbiota for development. Mol Ecol 23:2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz MJ, Frydman HM, Connor JH. 2018. Dual insect specific virus infection limits Arbovirus replication in Aedes mosquito cells. Virology 518:406–413. doi: 10.1016/j.virol.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ballinger MJ, Bruenn JA, Hay J, Czechowski D, Taylor DJ. 2014. Discovery and evolution of bunyavirids in Arctic phantom midges and ancient Bunyavirid-like sequences in insect genomes. J Virol 88:8783–8794. doi: 10.1128/JVI.00531-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, Qin XC, Xu J, Holmes EC, Zhang YZ. 2015. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin XC, Shi M, Tian JH, Lin XD, Gao DY, He JR, Wang JB, Li CX, Kang YJ, Yu B, Zhou DJ, Xu JG, Plyusnin A, Holmes EC, Zhang YZ. 2014. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc Natl Acad Sci U S A 111:6744–6749. doi: 10.1073/pnas.1324194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokarz R, Sameroff S, Leon MS, Jain K, Lipkin WI. 2014. Genome characterization of Long Island tick rhabdovirus, a new virus identified in Amblyomma americanum ticks. Virol J 11:26. doi: 10.1186/1743-422X-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tokarz R, Williams SH, Sameroff S, Leon MS, Jain K, Lipkin WI. 2014. Virome analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. J Virol 88:11480–11492. doi: 10.1128/JVI.01858-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fredericks AC, Russell TA, Wallace LE, Davidson AD, Fernandez-Sesma A, Maringer K. 2019. Aedes aegypti (Aag2)-derived clonal mosquito cell lines reveal the effects of pre-existing persistent infection with the insect-specific bunyavirus Phasi Charoen-like virus on arbovirus replication. PLoS Negl Trop Dis 13:e0007346. doi: 10.1371/journal.pntd.0007346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anonymous. 2016. docker-clustergenomes. https://bitbucket.org/MAVERICLab/docker-clustergenomes.
- 46.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 47.Ondov BD, Bergman NH, Phillippy AM. 2011. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anonymous. 2020. BioInfoTools/BBMap. https://github.com/BioInfoTools/BBMap.
- 49.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 50.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 51.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi M, Neville P, Nicholson J, Eden JS, Imrie A, Holmes EC. 2017. High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J Virol 91:e00680-17. doi: 10.1128/JVI.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zakrzewski M, Rasic G, Darbro J, Krause L, Poo YS, Filipovic I, Parry R, Asgari S, Devine G, Suhrbier A. 2018. Mapping the virome in wild-caught Aedes aegypti from Cairns and Bangkok. Sci Rep 8:4690. doi: 10.1038/s41598-018-22945-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao P, Han J, Zhang Y, Li C, Guo X, Wen S, Tian M, Li Y, Wang M, Liu H, Ren J, Zhou H, Lu H, Jin N. 2018. Metagenomic analysis of Flaviviridae in mosquito viromes isolated from Yunnan Province in China reveals genes from dengue and Zika viruses. Front Cell Infect Microbiol 8:359. doi: 10.3389/fcimb.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Souvorov A, Agarwala R, Lipman DJ. 2018. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol 19:153. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conway JR, Lex A, Gehlenborg N. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anonymous. 2016. https://github.com/rambaut/figtree/releases.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alpha diversity of viromes in the lab and field Aedes albopictus samples. Download FIG S1, PDF file, 0.2 MB (160.4KB, pdf) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Number of reads of each eukaryotic viral species in each sequencing sample. Download Table S3, XLSX file, 0.01 MB (15.7KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SRA datasets used in this study. Download Table S1, XLSX file, 0.01 MB (13KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Coverage of each eukaryotic viral species in each sequencing sample. Download Table S4, XLSX file, 0.01 MB (15.5KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Presence and absence of viral species in different locations. Download Table S5, CSV file, 0.00 MB (4.8KB, csv) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession numbers of YBV1-, PCLPV-, and GMV-related viruses identified in this study. Download Table S2, XLSX file, 0.01 MB (11.4KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Accession numbers of the viruses obtained from our data set are listed in Table S2, and viral genome sequences recovered from the SRA data sets can be found at https://github.com/Matthijnssenslab/AedesVirome. All viral sequences used to construct phylogenetic trees can be found at https://github.com/Matthijnssenslab/AedesVirome.
Accession numbers of YBV1-, PCLPV-, and GMV-related viruses identified in this study. Download Table S2, XLSX file, 0.01 MB (11.4KB, xlsx) .
Copyright © 2020 Shi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.