Bcl-2 proteins are molecular gatekeepers of the mitochondrial pathway of apoptosis that emerged early during metazoan evolution.
Abstract
In metazoans, Bcl-2 family proteins are major regulators of mitochondrially mediated apoptosis; however, their evolution remains poorly understood. Here, we describe the molecular characterization of the four members of the Bcl-2 family in the most primitive metazoan, Trichoplax adhaerens. All four trBcl-2 homologs are multimotif Bcl-2 group, with trBcl-2L1 and trBcl-2L2 being highly divergent antiapoptotic Bcl-2 members, whereas trBcl-2L3 and trBcl-2L4 are homologs of proapoptotic Bax and Bak, respectively. trBax expression permeabilizes the mitochondrial outer membrane, while trBak operates as a BH3-only sensitizer repressing antiapoptotic activities of trBcl-2L1 and trBcl-2L2. The crystal structure of a trBcl-2L2:trBak BH3 complex reveals that trBcl-2L2 uses the canonical Bcl-2 ligand binding groove to sequester trBak BH3, indicating that the structural basis for apoptosis control is conserved from T. adhaerens to mammals. Finally, we demonstrate that both trBax and trBak BH3 peptides bind selectively to human Bcl-2 homologs to sensitize cancer cells to chemotherapy treatment.
INTRODUCTION
Apoptosis represents one of the hallmarks of multicellularity in the animal kingdom (1). This process shapes the body during embryonic development, maintains tissue homeostasis, and eliminates undesirable or potentially harmful cells from the organism (2). Proteins belonging to the Bcl-2 family are critical arbiters of apoptosis. Found originally in chromosomal translocations in follicular lymphomas, Bcl-2 homologs have now been found in almost all metazoans (3). Genetic screening of cell death mutants performed initially in nematodes and subsequent studies in cnidarians, arthropods, platyhelminthes, and vertebrates uncovered a number of Bcl-2–related proteins (4–8). These proteins are characterized by the presence of one to four conserved Bcl-2 (BH) motifs, as well as a hydrophobic C-terminal transmembrane (TM) motif, allowing them to be localized to intracellular membranes (9). Bcl-2 proteins are divided into members that either promote or oppose apoptosis. Proapoptotic Bcl-2 proteins are further subdivided into multimotif Bcl-2 homologs and the phylogenetically separate BH3-only proteins, which have only the BH3 motif (10).
It is generally accepted that Bcl-2 family proteins were evolutionarily selected to control cell survival in metazoans (11). However, the control of apoptosis differs substantially between roundworms and vertebrates. Seminal experiments conducted in Caenorhabditis elegans demonstrated that the Bcl-2 protein Ced-9 binds to the caspase-activating Apaf-1 homolog Ced-4. Apoptosis ensues when the BH3-only protein Egl-1 is up-regulated to competitively bind to Ced-9, thus releasing Ced-4 and leading to the activation of the caspase Ced-3. In vertebrates, however, Bcl-2 proteins are found to control a key step in the induction of apoptosis called mitochondrial outer membrane permeabilization (MOMP) (12). Antiapoptotic Bcl-2 homologs including Bcl-2, Bcl-w, Bcl-xL, Mcl-1, and A1/Bfl-1 repress proapoptotic Bax and Bak, which the effect release of cytochrome c and subsequent caspase activation through their oligomerization and perforation of the MOM (13). Proapoptotic BH3-only proteins tip the balance toward cell death. At the functional level, BH3-only proteins can be divided into activators and sensitizers based on whether they bind and directly activate Bax and Bak (“activators”) or antagonize antiapoptotic Bcl-2 proteins by binding to their conserved ligand binding groove (“sensitizers”), respectively (14).
Although highly debated, the origin and the evolution of the Bcl-2 protein family remain unresolved (11). Structural studies of distantly related Bcl-2 homologs identified a conserved interaction between the ligand binding groove of antiapoptotic Bcl-2 and the BH3 motif of proapoptotic multimotif proteins in sponges, albeit with subtle differences in the detailed mechanism of binding that rendered the sponge antiapoptotic Bcl-2 protein exquisitely specific for its sponge-encoded proapoptotic counterpart (15). In several animals from phylogenetically more basal branches, the repertoire of Bcl-2 family members is exclusively composed of multimotif Bcl-2 proteins, with BH3-only proteins being absent (6, 16, 17). Furthermore, Bcl-2–regulated apoptosis proceeds in the human HCT-116 cell line when all BH3-only proteins have been removed genetically (18, 19). Combined, these findings raise the possibility that the regulation of MOMP in the early stages of the metazoan evolution might be dependent solely on homo- and heteroassociation between anti and proapoptotic multimotif Bcl-2 proteins.
Multicellular animals appeared during the late Precambrian period around 780 million years ago (20). Placozoans represent one of the most basal branches of metazoans, with Trichoplax adhaerens being one of three living representatives of this phylum (21). It has the simplest animal morphology, is composed of only six cell types, rarely undergoes embryonic (sexual) development, and instead divides vegetatively by fission (22, 23). The genome of T. adhaerens was sequenced (24), divulging the presence of putative modulators of apoptosis including caspases and Bcl-2 family members (25). Here, we report the identification and the molecular characterization of four T. adhaerens Bcl-2–like (trBcl-2L1 to trBcl-2L4) proteins. Our results reveal the existence of ancient and conserved functional interplay of T. adhaerens–encoded members of the Bcl-2 family of proteins during MOMP regulation and suggest that BH3-only proteins may have originated from Bcl-2 multimotif homologs.
RESULTS
In silico identification and characterization of Bcl-2–like proteins
We performed extensive tBlastN screening of the genome of the T. adhaerens Grell-BS-1999 strain publicly available on the National Center for Biotechnology Information (NCBI). Using the full-length human Bcl-2 protein and individual BH and TM motifs as query sequences, we identified four protein-encoding sequences potentially corresponding to Bcl-2 homologs. We named this group of proteins T. adhaerens Bcl-2–like proteins or trBcl-2L (Fig. 1A). We further confirmed the expression and the sequence of the four trbcl-2–like genes in T. adhaerens by performing reverse transcriptase polymerase chain reaction (PCR) using specific primers targeting the first exon of each gene (fig. S1A). Sequences were deposited into the NCBI databank under the following accession numbers KU500586.1, KU500587.1, KU500588.1, and KU500589.1 corresponding to trBcl-2L1, trBcl-2L2, trBcl-2L3, and trBcl-2L4, respectively. Multiple alignments coupled with secondary structure prediction showed that all four sequences presented the typical multimotif BH organization with a TM motif located at the C terminus, as well as an all α-helical secondary structure (Fig. 1A and fig. S1B). Analysis of the primary structure of trBcl-2L3 also highlighted a unique S/T-rich region between α helices 1 and 2. This region was predicted to be a mucin-like domain, which is often found in secreted proteins. Notably, over the course of our survey, we were not able to detect BH3-only protein-encoding genes in the T. adhaerens genome.
Fig. 1. Sequence and expression analysis of Bcl-2 homologs found in T. adhaerens.
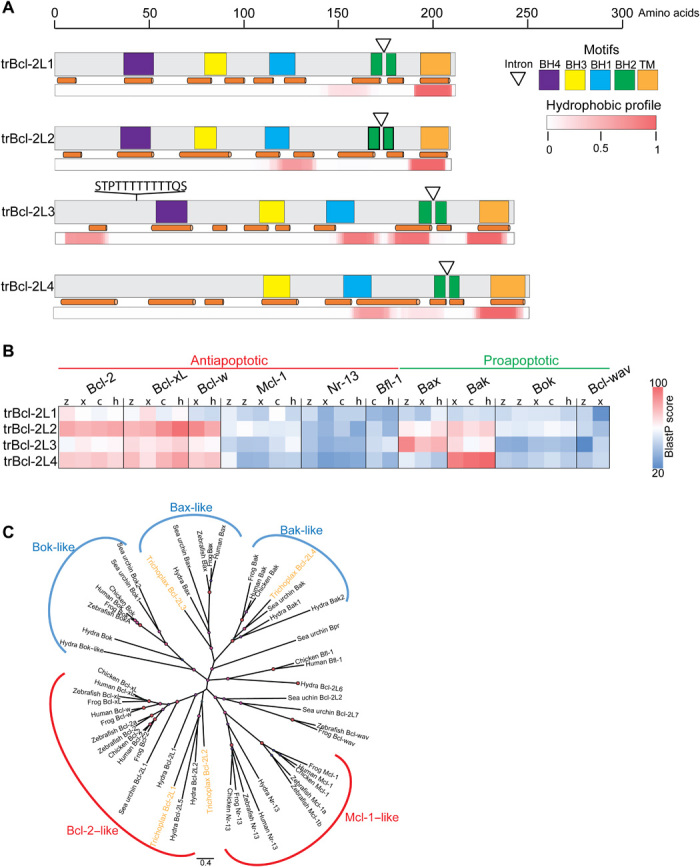
(A) Schematic representation of the domain structures of four multimotif T. adhaerens Bcl-2–like (trBcl-2L1 to trBcl-2L4) proteins. The positions of conserved BH1-BH4 motifs and the C-terminal TM motif and the locations of the predicted α helices are indicated with colored boxes and orange cylinders, respectively. The white triangles represent the positions of the conserved BH2 splitting intron. The hydrophobic profile for each protein is presented below. An amino acid scale bar is presented above. The mucin-like site is indicated on trBcl-2L3. (B) In silico hybridization matrix representing the alignment of four trBcl-2–like proteins versus Bcl-2–related multimotif proteins found in Danio rerio [zebrafish (z)], Gallus gallus (chicken, c), Xenopus tropicalis (xenopus, x), and Homo sapiens (human, h). Colored boxes refer to the total alignment score [blue, poor alignment; red, good alignment (>60)]. The trBcl-2L1 and trBcl-2L2 cluster with antiapoptotic Bcl-2 members, whereas trBcl-2L3 and trBcl-2L4 cluster with the proapoptotic Bax and Bak, respectively. (C) Phylogenetic tree using the neighbor-joining clustering method. trBcl-2L1 and trBcl-2L2 cluster with the Bcl-2–like subgroup whereas trBcl-2L3 and trBcl-2L4 cluster with Bax and Bak orthologs, respectively.
The four trBcl-2 homologs cluster in two groups at both the genomic and protein level. For instance, all four trBcl-2–like genes have a unique BH2 splitting intron. However, this intron was relatively long for trbcl-2l1 and trbcl-2l2 with 1991 and 1725 nucleotide (nt), respectively, whereas it was substantially shorter in trbcl-2l3 and trbcl-2l4 with 463 and 280 nt, respectively. Furthermore, trBcl-2L1 and trBcl-2L2 proteins displayed similar molecular weights and calculated isoelectric points (pI) of 24,766 and 24,282 Da and pI of 8.65 and 8.85, respectively, whereas trBcl-2L3 and trBcl-2L4 were slightly larger, 27,382 and 27,944 Da with lower pI (6.96 and 6.45, respectively). Finally, using local alignment matrices and neighbor-joining phylogenetic tree generation, we showed that trBcl-2L1 and trBcl-2L2 were highly divergent and clustered with the antiapoptotic subgroup of the Bcl-2 family, whereas trBcl-2L3 and trBcl-2L4 clustered with Bax and Bak homologs, respectively (Fig. 1, B and C). We, hereafter, named these proteins trBax and trBak, respectively.
Subcellular localization of trBcl-2 proteins
In vertebrates, members of the Bcl-2 family can be localized both to the mitochondria and the endoplasmic reticulum (ER) (26). To analyze the subcellular localization of Trichoplax Bcl-2 homologs, we performed a series of immunofluorescence experiments on mammalian cells transiently expressing N-terminally Flag-tagged trBcl-2–like proteins. Under these conditions, we were able to detect trBcl-2L1 and trBcl-2L2 in mitochondrial and ER membranes (Fig. 2A). Notably, cells expressing trBcl-2L2 proteins presented clustered perinuclear mitochondria. In addition, intracellular trBcl-2L2 localization was more dispersed than trBcl-2L1, suggesting that this protein could also be localized in the cytosol. We confirmed these observations by performing subcellular fractionation on baby mouse kidney (BMK) cells lacking endogenous bax and bak [double knockout (DKO)] followed by immunoblotting. We found that trBcl-2L1 was exclusively membrane-bound, whereas a fraction of trBcl-2L2 was also found in the cytosol (Fig. 2B). When we expressed trBax and trBak, we detected these proteins primarily at the level of the mitochondria, with minor localization in the microsomal fraction (Fig. 2, A and B).
Fig. 2. Subcellular localization of trBcl-2–like proteins.
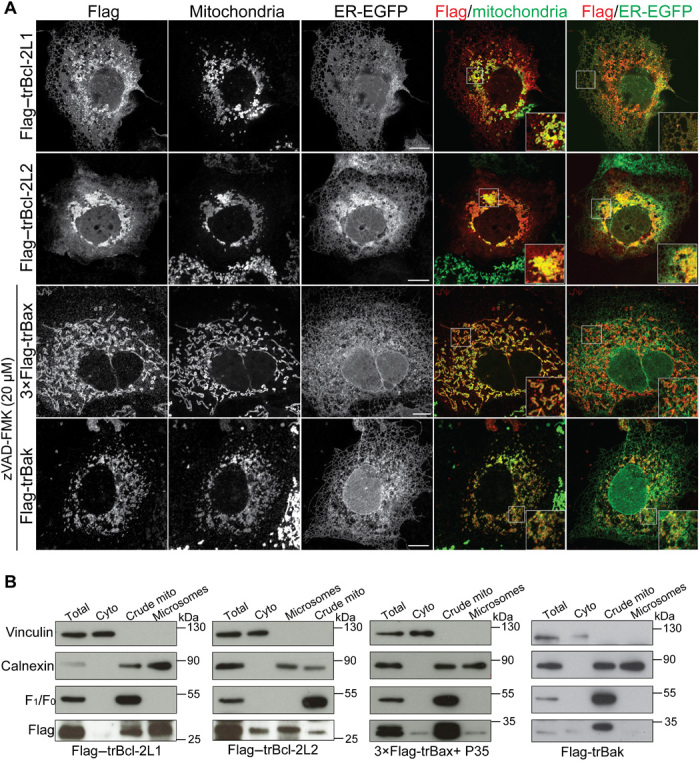
(A) Representative confocal images of Cos-7 cells expressing trBcl-2–like proteins. Subcellular localization of each protein was analyzed using ER-targeted enhanced green fluorescent protein (EGFP) for staining ER membranes, anti-Tom20 for labeling mitochondria, and the anti-Flag antibody for detection of Flag-tagged Bcl-2–like proteins. Merged channels between Flag and ER-GFP and between Flag and mitochondria are shown on the right. White boxes represent the enlarged area of each merged image. Scale bars, 20 μm. (B) Subcellular fractionation of BMK DKO cells expressing Flag-tagged trBcl-2–like proteins. trBcl-2L1 and trBcl-2L2 proteins were found in crude mitochondrial and microsomal fractions. Full-length trBcl-2L2 was also detected in the cytosolic fraction. trBax and trBak were found mostly in the crude mitochondrial fraction. Anti-F1F0 ATPase (adenosine triphosphatase), calnexin, and vinculin antibodies were used as mitochondrial, ER, and cytosolic markers, respectively. P35 cotransfection with trBax was used to delay cell detachment.
We next performed experiments in the budding yeast Saccharomyces cerevisiae. Treatment of purified yeast mitochondria with Na2CO3 revealed that trBcl-2L1 is tightly inserted in the mitochondrial membrane, whereas trBcl-2L2 and trBak are more loosely associated (fig. S2, A and B). Coexpression of the human Bax mutant P168A, which is primarily localized to mitochondria, leads to trBcl-2L2 and trBak stabilization at the MOM. Notably, expression of trBax was highly cytotoxic to the yeast, hampering the detection of the protein.
Bax localization to the MOM represents a dynamic process involving translocation and retrotranslocation that depends on internal cues not only the hydrophobic α9 helix containing the TM motif but also the α1 helix and a proline residue located after the BH2 motif (27–30). Expression of enhanced green fluorescent protein (EGFP)–tagged wild-type (WT) trBax in BMK cells resulted in rapid translocation to internal membranes, suggesting that trBax does not have the same translocation-retrotranslocation dynamics as human Bax (fig. S2C). To understand how trBax localizes to the MOM, we generated mutants lacking either helix α1 (∆α1), α9 (∆α9), or both helices α1and α9 (∆α1∆α9), and two site-directed mutants, P212A and N(8)T, in which we replaced the eight threonine residues located in an S/T-rich region between α helix 1 and α helix 2 that were predicted to act as a mucin-like motif (fig. S2D). Expression of trBax mutants lacking the α9 helix but not α1 resulted in clear cytosolic localization. However, both the P212A and the N(8)T mutant did not reveal any change in the subcellular distribution compared to WT trBax, suggesting that the α9 helix encompassing the TM motif is critical for trBax localization to the MOM (fig. S2E).
Next, we investigated the importance of the TM motif for the subcellular localization of the other three trBcl-2 proteins. To this end, we generated mutants lacking the C-terminal hydrophobic α helix (∆TM), as predicted in silico. When expressed in Cos-7 cells, trBcl-2L1∆TM, trBcl-L2∆TM, and trBak∆TM displayed a mainly cytosolic distribution, further emphasizing the role of the TM motif in the subcellular localization of trBcl-2 proteins (fig. S2F).
Role of trBcl-2 proteins in the control of MOMP
To gain further mechanistic insight into the molecular activity of trBcl-2–like proteins, we performed apoptosis assays in HeLa cells by analyzing the activation of Caspase 3 using a specific antibody. Transient expression of trBcl-2L1 (2.2 ± 1.3%) and trBcl-2L2 (2.4 ± 1.2%) for 24 hours did not lead to a significant increase in Caspase 3 activation compared to control cells expressing ER-targeted EGFP (6.3 ± 5.2%). Conversely, expression of trBax (89.5 ± 4.5%) and trBak (88.7 ± 4.5%) led to substantial Caspase 3 activation (Fig. 3A and fig. S3A), similar to the expression of the zebrafish Bax ortholog (zBax) (87.0 ± 4.7%), suggesting that both trBak and trBax are bona fide proapoptotic Bcl-2 proteins (Fig. 3A and fig. S3A). We further analyzed the antiapoptotic activity of trBcl-2L1 and trBcl-2L2 proteins using HeLa cells treated with three apoptosis-inducing agents: thapsigargin, etoposide, and staurosporine. Expression of trBcl-2L1 and trBcl-2L2 led to a decreased percentage of Caspase 3–positive cells compared to control cells transfected with empty vector, demonstrating that these two proteins are indeed antiapoptotic Bcl-2 members (Fig. 3B). We next used zebrafish embryos as a model system for studying the activity of trBcl-2 homologs. During zebrafish development, embryos are resistant to apoptotic stimuli before the gastrulation stage, which starts approximately 6 hours post fertilization (hpf). The forced expression of proapoptotic Bcl-2 proteins has been shown to induce apoptosis in these embryos before this stage (7, 31). We thus injected at the one cell stage in vitro–transcribed mRNA encoding for each T. adhaerens Bcl-2 homolog. Expression of the red fluorescent protein mCherry, trBcl-2L1, and trBcl-2L2 modestly increased embryo mortality. In contrast, expression of trBax and trBak was highly lethal, with embryo death reaching almost 100% at 6 hpf (Fig. 3C). Coexpression of trBcl-2L1 and trBcl-2L2 in combination with either trBax or trBak proteins led to the complete inhibition of trBax or trBak proapoptotic activity. These results suggest that trBax and trBak are proapoptotic Bcl-2 members whose activity may be antagonized by either trBcl-2L1 or trBcl-2L2.
Fig. 3. Control of MOMP by trBcl-2–like proteins.
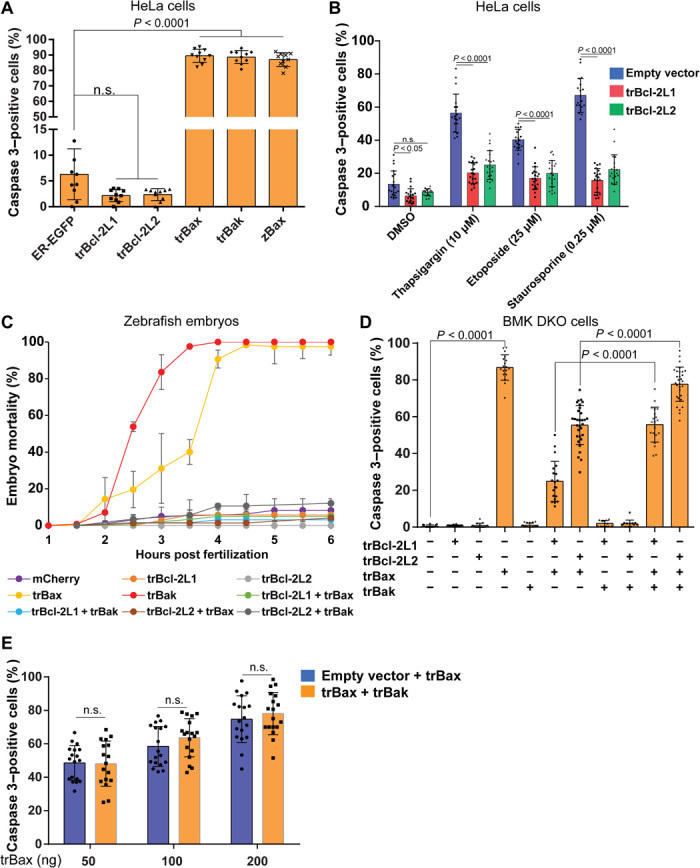
(A) Quantification of Caspase 3 activation in HeLa cells transiently expressing trBcl-2–like proteins (means ± SD; three independent experiments; one-way ANOVA statistical analysis; n.s., not significant). (B) Histogram showing the antiapoptotic effect of trBcl-2L1 and trBcl-L2 proteins on HeLa cells treated with thapsigargin (10 μM), etoposide (25 μM), or staurosporine (0.25 μM) for 24 hours. Activated Caspase 3 was used as a marker for monitoring apoptosis (means ± SD; three independent experiments, two-way ANOVA statistical analysis). DMSO, dimethyl sulfoxide. (C) Graph presenting the effect of trBcl-2–like protein expression on the mortality of zebrafish embryos. In contrast to trBcl-2L1 and trBcl-2L2, which had no effect, mRNA injection (100 ng/μl) of trBax and trBak into one cell–stage embryos induced early mortality during gastrulation (means ± SD; three independent experiments). (D) Histogram showing the effect of trBcl-2–like protein expression on Caspase 3 activation in BMK DKO cells. In comparison to HeLa cells, trBak protein is unable to induce Caspase 3 activation in BMK DKO cells (means ± SD; three independent experiments; one-way ANOVA statistical analysis). (E) Histogram showing the effect of the expression of trBak protein on the proapoptotic activity of trBax in BMK DKO cells. Cells were transfected with increasing amounts of trBax expressing vector (50, 100, and 200 ng) in the presence of a constant amount of pCS2+ empty vector or pCS2 + trBak. trBak coexpression did not lead to an increase of the percentage of Caspase 3–positive cells compared to transfection with a trBax-expressing vector and empty vector (means ± SD; three independent experiments, two-way ANOVA statistical analysis. The spread of the data is indicated by black dots and squares.
We then evaluated the capacity of trBax to induce apoptosis independently of the presence of endogenous Bax and Bak by performing a series of apoptosis assays using BMK Bax/Bak DKO cells (fig. S3B). trBax expressed in these cells rapidly translocated to the mitochondria, where it oligomerized and induced mitochondrial fragmentation and cytochrome c release (fig. S3, C to E). Furthermore, most of the trBax-expressing cells were Caspase 3–positive (86.8 ± 3.1%) in comparison to the control cells (0.5 ± 0.45%), demonstrating that trBax is a bona fide Bax-like protein (Fig. 3D). Notably, the mitochondrial localization of trBax was tightly correlated with its capacity to induce Caspase 3 activation, since mutants lacking the TM motif displayed a reduced capacity to induce cell death in BMK DKO cells (fig. S3F). Furthermore, coexpression of trBcl-2L1 (24.8 ± 3.8%) and, to a lesser extent, trBcl-2L2 (54.9 ± 3.1%) counteracted trBax proapoptotic activity (Fig. 3D).
Expression of trBak on its own in BMK DKO cells did not result in cytochrome c release or significant Caspase 3 activation (Fig. 3D, and fig. S3, C to E). Instead, trBak expression inhibited the antiapoptotic activity of trBcl-2L1 and trBcl-2L2 against trBax, without increasing its proapoptotic potential, suggesting that trBak may act as a BH3-only sensitizer (Fig. 3, D and E).
Structural insights into trBcl-2 interactions
The above results suggested the existence of a regulatory network controlled by interactions between T. adhaerens–encoded Bcl-2 proteins that regulate MOMP. To test this hypothesis, we performed fluorescence resonance energy transfer (FRET)–based interaction analyses using an EGFP donor and an Alexa Fluor 568 acceptor in BMK DKO cells. We expressed EGFP-tagged trBax and trBak in combination with Flag-tagged trBcl-2L1 and trBcl-2L2 and performed immunofluorescence imaging using anti-Flag and secondary antibodies coupled with the Alexa Fluor 568 fluorophore. Interactions between trBcl-2–like proteins were detected by measuring the dequenching of EGFP emissions induced by Alexa Fluor 568 photobleaching. As shown in fig. S4, FRET efficacy between EGFP-trBax/Flag–trBcl-2L1 (5.84%) and EGFP-trBak/Flag–trBcl-2L2 (5.45%) was similar to control cells expressing EGFP fused to human Bax and to Flag-tagged human Bcl-2 (4.92%). FRET efficacy between EGFP-trBak/Flag–trBcl-2L1 (11.18%) was close to the positive-control NeonGreen-Flag-Nrh (11.20%), whereas EGFP-trBax and Flag–trBcl-2L2 interacted only weakly (2.81%) (fig. S4, A and B). These observations were further confirmed by performing coimmunoprecipitation experiments between Flag-tagged trBcl-2L1 and trBcl-2L2, and EGFP-tagged trBax and trBak proteins. Under these conditions, trBcl-2L1 and trBcl-L2 proteins interacted with trBax and trBak (fig. S4C). Notably, interactions between antiapoptotic trBcl-2 proteins and trBax were disrupted in the presence of a trBak protein, supporting the notion that trBak is acting as a sensitizer BH3-only protein (fig. S4D).
We next examined the ability of recombinant trBcl-2L1 and trBcl-2L2 to engage peptides spanning the BH3 motif of trBak and trBax using isothermal titration calorimetry (ITC) (Fig. 4A). Our analysis revealed that trBcl-2L1 only engaged trBax with nanomolar affinity (KD of 426 nM), while trBak was bound with micromolar affinity (KD of 1.43 μM). In contrast, trBcl-2L2 bound both trBak and trBax with nanomolar affinities (KD of 563 and 728 nM, respectively). We then established the structural basis of apoptosis inhibition by determining the crystal structure of a trBcl-2L2:trBak BH3 complex. The structure was refined to a resolution of 1.6 Å, with clear and continuous electron density obtained for trBcl-2L2 residues 34 to 182 and trBak BH3 103 to 127 (table S1). trBcl-2L2 adopts the classical Bcl-2 globular helical bundle fold comprising eight α-helices, which is also found in human and sponge Bcl-2 homologs (Fig. 4, B to D). trBcl-2L2 uses the canonical Bcl-2 ligand binding groove formed by helices α2 to α5 to engage BH3 motif ligands (Fig. 4B). In the trBcl-2L2:trBak BH3 complex (Fig. 4E), trBak residues I109, L113, L116, and Y120 protrude into four hydrophobic pockets of trBcl-2L2. In addition, the hallmark ionic interaction observed across the Bcl-2 family is formed between trBcl-2L2 R97 and trBak D118. Two additional ionic interactions are found between trBcl-2L2 R69 and trBak E108 and trBcl-2L2 E54 and trBak R123. These three ionic interactions are supplemented by an additional two hydrogen bonds between the trBcl-2L1 E54 carboxyl group and the trBak Y120 hydroxyl group and trBcl-2L2 N94 aspartate group and trBak D118 carboxyl group.
Fig. 4. Molecular interplay of T. adhaerens Bcl-2 proteins.
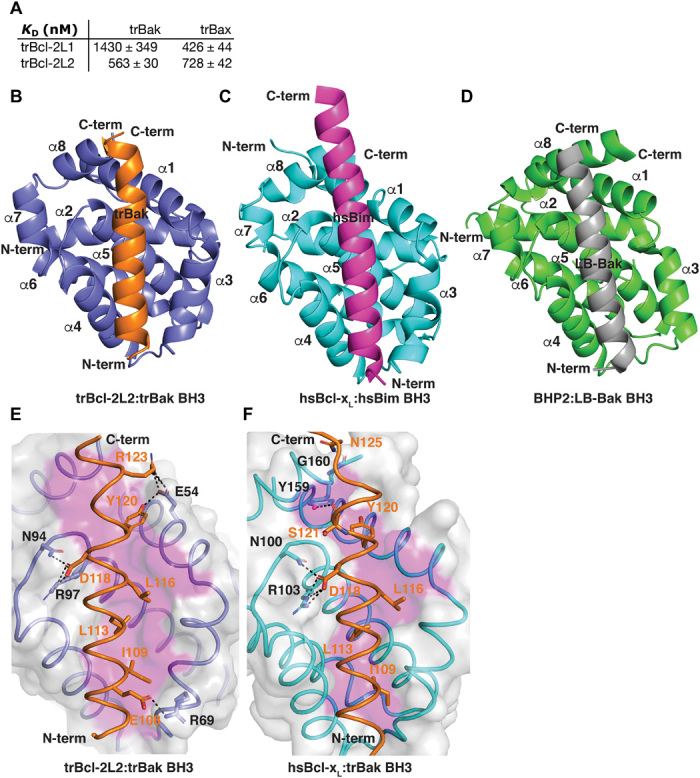
(A) trBcl-2L1 and trBcl-2L2 bind peptides spanning the BH3 motif of trBak and trBax. Affinities were measured using ITC, and KD values (in nM) are the means of three experiments ± SD. (B) trBcl-2L2 (slate) in complex with the trBak BH3 motif peptide (orange). trBcl-2L2 helices are labeled α1 to α8. The view is into the hydrophobic binding groove formed by helices α2 to 5. (C) Human Bcl-xL (cyan) bound to Bim BH3 (magenta) (PDB ID 1PQ1) (49). The view is as in (B). (D) Geodia cydonium BHP2 (green) bound to LB-Bak BH3 (gray) (PDB ID 5TWA) (15). The view is as in (B). (E) Detailed view of the trBcl-2L2:trBak BH3 complex. The trBcl-2L2 surface, backbone, and floor of the binding groove are shown in gray, slate, and magenta, respectively, while trBak BH3 is shown as an orange ribbon with sticks. The four key hydrophobic residues of trBak BH3 (I109, L113, L116, and Y120) are protruding into the binding groove, and key ionic interactions and hydrogen bonds are labeled and shown as black dotted lines. (F) Detailed view of the human Bcl-xL:trBak BH3 complex. The Bcl-xL surface, backbone, and floor of the binding groove are shown in gray, cyan, and magenta, respectively, while trBak BH3 is shown as an orange ribbon with sticks. The four key hydrophobic residues of trBak BH3 (I109, L113, L116, and Y120) are protruding into the binding groove, and key ionic interactions and hydrogen bonds are labeled and shown as black dotted lines.
A DALI protein similarity analysis indicated that rat Bcl-xL [Protein Data Bank (PDB) ID 4QNQ] is the closest structural homolog to trBcl-2L2 with a root mean square deviation value of 1.3 Å over 142 Cα atoms. We thus determined the crystal structure of human (hs) Bcl-xL bound to the BH3 motif of trBak. Although the trBak BH3 motif binds in the conserved ligand binding groove of hsBcl-xL formed by helices α2 to α5, a detailed comparison of the interactions between trBcl-2L2 and hsBcl-xL with trBak revealed that only the canonical interactions are conserved, including the ionic interaction of the trBak Asp118 residue with the conserved Arg in trBcl-2–L2 and hsBcl-xL and engagement of the four hydrophobic trBak residues I109, l113, L116, and Y120 by the four pockets in the grooves of trBcl-2L2 and hsBcl-xL (Fig. 4, E and F).
Interplay between trBcl-2 proteins and human Bcl-2 homologs
The existence of a conserved ligand binding groove that binds BH3 motif ligands in a trBcl-2 homolog suggested the possibility of conservation of interactions between T. adhaerens–encoded Bcl-2 proteins and their mammalian counterparts. To examine the interaction network between T. adhaerens and human Bcl-2 proteins, we performed a series of ITC measurements. Both trBcl-2L1 and trBcl-L2 bound to hsBax BH3 peptide (7 and 15 nM, respectively) and to the BH3-only proteins hsPuma (5 and 81 nM), hsBim (83 and 41 nM), and hsBid (124 and 170 nM) with high affinity. Conversely, using recombinant human Bcl-2, Bcl-w, Bcl-xL, Mcl-1, and A1/Bfl-1 with BH3-motif peptides from trBak, we showed that all five human antiapoptotic proteins displayed nanomolar affinity for trBak BH3 peptide, with hsA1/Bfl-1, hsBcl-2, and hsBcl-xL, the tightest binders with KD values of 13, 45, and 197 nM, respectively (Fig. 5A).
Fig. 5. Interplay between trBcl-2 proteins and human Bcl-2 homologs.
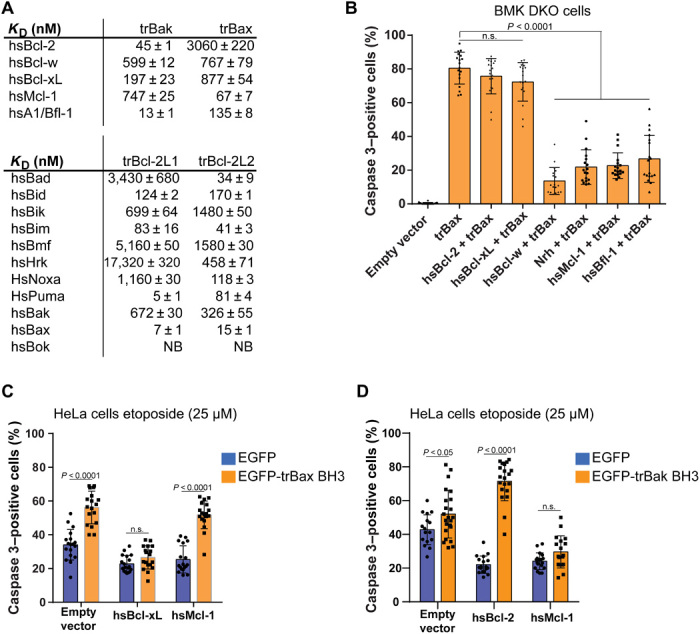
(A) Human and T. adhaerens antiapoptotic Bcl-2 proteins bind peptides spanning the BH3 motif of proapoptotic Bcl-2 proteins. Affinities were measured using ITC, and KD values (in nM) are the means of three experiments ± SD. NB, no binding detected. (B) Histogram showing the effect of the expression of human antiapoptotic Bcl-2 homologs on the proapoptotic activity of trBax in BMK DKO cells. Proteins belonging to the Mcl-1 subgroup and Bcl-w antagonize trBax proapoptotic activity, whereas Bcl-2 and Bcl-xL have no effect (means ± SD; three independent experiments; one-way ANOVA statistical analysis). (C) Histogram showing the effect of the expression of EGFP or EGFP-tagged trBax BH3 peptide alone (empty vector) or in combination with hsMcl-1 or hsBcl-xL in HeLa cells treated with etoposide (25 μM) for 24 hours. trBax BH3 peptide increases apoptosis in HeLa cells, overexpressing hsMcl-1 but not hsBcl-xL (means ± SD; three independent experiments, two-way ANOVA statistical analysis). (D) Histogram showing the effect of the expression of EGFP or EGFP-tagged trBak BH3 peptide alone (empty vector) or in combination with hsBcl-2 or hsMcl-1 in HeLa cells treated with etoposide (25 μM) for 24 hours. trBak BH3 peptide increases apoptosis in HeLa cells, overexpressing hsBcl-2 but not hsMcl-1 (means ± SD; three independent experiments, two-way ANOVA statistical analysis). The spread of the data is indicated by black squares.
We next performed ITC experiments using the trBax BH3 peptide and human Bcl-2 homologs. We observed that only hsMcl-1 and hsA1/Bfl-1 displayed high affinity (KD of 67 and 135 nM, respectively), whereas hsBcl-w, hsBcl-xL, and hsBcl-2 bound to trBax were more weakly (KD of 767, 877, and 3060 nM, respectively) (Fig. 5A). Coimmunoprecipitation assays confirmed that trBax binds more efficiently to hsMcl-1 compared to hsBcl-2 or hsBcl-xL (fig. S5A). Considering the putative sensitizer activity of trBak, we next examined whether trBak BH3 was able to displace trBax BH3 from human antiapoptotic Bcl-2 proteins using a competition ITC approach. Consistent with our measured affinities (Fig. 5A), we observed that trBak BH3 is able to displace trBax from existing complexes with Bcl-2, Bcl-xL, Bcl-w, and A1/Bfl-1 but not with Mcl-1 (fig. S6).
To test whether the observed interactions correlated with a functional interplay between these distantly related homologs, we performed cell death assays in BMK DKO cells expressing human Bcl-2 antiapoptotic proteins in combination with trBax. We detected a significant decrease in the number of Caspase 3–positive cells when coexpressing trBax with Mcl-1, A1/Bfl-1, Nrh, and Bcl-w, whereas coexpression of Bcl-2 and Bcl-xL had no effect on trBax proapoptotic activity (Fig. 5B and fig. S5B). This prompted us to examine whether peptides corresponding to the BH3 motif of trBax or trBak may discriminate between human Bcl-2 homologs in a cell-based assay. To this end, we expressed EGFP-tagged trBax or trBak BH3 peptides in HeLa cells alone or in combination with hsMcl, hsBcl-xL, or hsBcl-2 and treated them with the topoisomerase II inhibitor etoposide. Overexpression of EGFP-trBax BH3 alone or in combination with hsMcl-1 but not with hsBcl-xL sensitizes cells toward etoposide treatment, suggesting that this peptide is indeed specific to hsMcl-1 overexpression (Fig. 5C). In contrast, trBak BH3 peptide was able to sensitize cells overexpressing hsBcl-2 but not hsMcl-1 (Fig. 5D).
DISCUSSION
We report the identification and characterization of four T. adhaerens–encoded Bcl-2 homologs. All four genes harbor a single conserved BH2 splitting intron, which is located specifically after the TGG codon encoding a tryptophan residue. This specific intron is not restricted to placozoans but has also been found in almost all bcl-2 homologous genes in multicellular animals (32). This observation suggests that Bcl-2 multimotif members arose from a common bcl-2 ancestral gene, which already had this intron. However, it remains unclear whether this ancestral gene harbored pro- and/or antiapoptotic function or functions unrelated to the regulation of MOMP or, indeed, apoptosis regulation. All four trBcl-2 proteins contain a C-terminal TM motif that allows trBcl-2L1 and trBcl-L2 to localize not only to the MOM but also to the ER.
Our results show that trBax is a bona fide proapoptotic Bax-like protein that is primarily localized to the MOM, where it induces MOMP and cytochrome c release in cells lacking endogenous Bax and Bak. Unlike its human counterpart that dynamically shuttles between the cytosol and the MOM via translocation and retrotranslocation (28), trBax appears to directly target the mitochondrion. This mitochondrial localization is dependent on the C-terminal TM motif since mutants lacking the C-terminal hydrophobic helix mainly displayed a cytosolic distribution and decreased proapoptotic activity.
In mining the Trichoplax genome, we did not detect a gene encoding a BH3-only protein. One possibility is that Trichoplax encodes for a highly divergent BH3-only protein, which remains elusive and not detectable using sequence-based searches. Alternatively, it is possible that trBak, although structurally related to multimotif Bcl-2 proteins, adopts the role of a functional BH3-only protein. When expressed in BMK DKO cells, trBak failed to induce cytochrome c release and Caspase 3 activation in comparison to trBax, which promoted MOMP independent of the presence of endogenous Bax and Bak. Furthermore, trBak binds to both antiapoptotic trBcl-2L1 and trBcl-2L2 and represses heterodimer formation between antiapoptotic trBcl-2 proteins and trBax, thus acting as a BH3-only sensitizer that neutralizes antiapoptotic Bcl-2 activity. BH3-only proteins represent a diverse subgroup of the Bcl-2 protein family, with both convergent and divergent evolutionary origins. The BH3-only protein Bid represents a good example for divergent evolution since it clusters with multimotif Bcl-2 family members such as Bcl-rambo, Bcl-G, and Bfk and adopts the canonical Bcl-2 fold (33–35). Our observations argue that at least some BH3-only proteins may have arisen from an ancestral Bak-like protein, which lost its MOMP activity during the early steps of metazoan evolution.
Notwithstanding the question of a role for trBak as a BH3-only-like sensitizer or more executioner-like proapoptotic protein, our results demonstrate the existence of a conserved interplay between Trichoplax and mammalian Bcl-2 homologs. Trichoplax-encoded antiapoptotic proteins are able to directly bind and inhibit both Trichoplax and human proapoptotic Bax and Bak. Furthermore, it is not only the ability to directly bind Bak and Bax that is conserved, with conservation of both mode of engagement and specific atomic interactions. This suggests that a Bcl-2 family–mediated network of protein-protein interactions that controls MOMP arose early in metazoan evolution and that key features including interaction patterns, mode of engagement, and detailed atomic interactions carried through to the modern Bcl-2–mediated signaling pathways seen in mammals. While the mode of engagement is also conserved for another ancient metazoan organism, the sponge, subtle but significant differences were observed at the level of atomic interactions, which allowed for exquisite specificity of sponge-encoded Bcl-2 proteins for each other over their modern human counterparts (15). In the cnidarian Hydra vulgaris, a number of Bcl-2 family proteins have been identified, including several putative BH3-only proteins. While most of these proteins did not display a clear apoptosis regulatory function, antiapoptotic hyBcl-2-4 was identified, which together with proapoptotic hyBH3-only-2 and hyBak may constitute a core Bcl-2 machinery to control MOMP (6). However, affinity measurements and structural studies remain to be performed to establish how closely such a hydra encoded Bcl-2 pathway mimics the mechanics of more modern mammalian Bcl-2–regulated apoptosis machineries.
In our hands, BH3 peptides generated from trBax and trBak were able to bind with high affinity and specificity to mammalian Bcl-2 antiapoptotic proteins. The analysis of the crystal structure of an hsBcl-xL:trBak BH3 complex revealed that except for five core binding sites, no other interactions are conserved. This suggests that except for these fixed anchor points, the remainder of the interaction surface can be used flexibly to tune the required affinity and specificity. These binding sites have been conserved down to the level of intermolecular interactions from the basal metazoans Placozoa and Porifera (sponges), to Cnidaria and Bilateria (including the higher or “modern” animals). The importance of these Bcl-2:BH3 interactions in cancer has recently been demonstrated by the discovery of venetoclax, a member of a new class of drug, the BH3 mimetics, which directly targets the binding groove of antiapoptotic Bcl-2 and is now being used as an anticancer treatment (36). In the case of trBax, its BH3 motif was able to discriminate between hsBcl-xL and hsMcl-1. HeLa cells overexpressing hsMcl-1 but not hsBcl-xL were sensitized by this peptide for treatment with the chemotherapeutic drug etoposide. Mcl-1 is frequently overexpressed in a number of human malignancies and represents a potential therapeutic target (37–40). Current efforts are directed toward the development of BH3 mimetic drugs that can specifically target Mcl-1, sparing platelet cytotoxicity because of concomitant Bcl-xL inhibition (41).
Together, our results support the hypothesis that Bcl-2 proteins acquired the ability to control MOMP early in metazoan evolution and point to the existence of functional interplay between distantly related Bcl-2 proteins. Consequently, the mining and molecular characterization of ancient Bcl-2 homologs may represent a valuable avenue for the generation of new molecules with antitumor activity.
MATERIALS AND METHODS
trBcl-2 ORF cloning and expression analysis
Approximately 100 randomly picked animals of T. adhaerens “Grell” were used for total RNA isolation [for animal culture conditions, see (42)]. The animals were initially transferred into a clean glass petri dish, washed three times with 3.5% artificial seawater (ASW), and subsequently starved overnight to prevent algae contamination. The animals were transferred into a 1.5-ml Eppendorf tube, and the ASW was removed after a short centrifugation. Animals were lysed in 500 μl of prewarmed homogenization buffer [HOM-I: 100 mM tris-HCl, 10 mM EDTA, 100 mM NaCl, 2.5 mM dithiothreitol (DTT), 0.5% SDS, and 0.1% diethyl pyrocarbonate (DEPC) in ultrapure water at pH 8.0 and 65°C]. Proteins were digested with 25 μl of DEPC-treated proteinase K (10 mg/ml) for 20 min at 65°C. The lysate was further homogenized by 2× suction through a hypodermic needle connected to a 2.5-ml syringe. Nucleic acids were isolated from the lysate via a standard phenol/chloroform/isoamyl alcohol (25:24:1) extraction and a subsequent precipitation with isopropanol. Subsequent DNA digestion with deoxyribonuclease I (Thermo Fisher Scientific) was conducted according to the manufacturer’s recommendations. The quality and purity of total RNA were examined using agarose gel electrophoresis. First-strand complementary DNA (cDNA) synthesis with oligo(dT) primers was conducted using the SuperScript II Reverse Transcriptase (Thermo Fisher Scientific) following the manufacturer’s protocol.
The open reading frames of the four Bcl-2 homologs in Trichoplax were cloned using cDNA from the T. adhaerens Grell clone. PCR amplicons were cloned into the pJET1.2/blunt cloning vector (Thermo Fisher Scientific) and verified by sequencing. Verified sequences were deposited in the NCBI databank using the following accession numbers: KU500586.1 (trBcl-2L1), KU500587.1 (trBcl-2L2), KU500588.1 (trBcl-2L3), and KU500589.1 (trBcl-2L4). For expression analysis, internal primers targeting exon 1 of each bcl-2 homolog were used for PCR amplification.
Flag-tagged sequences were manufactured using gBlocks synthesis (IDT) and cloned into the pCS2+ expression vector using Bam HI/Xho I restriction sites. The trBax (trBcl-2L3) sequence was codon optimized (trBax-opt) for expression in eukaryote cell system, and synthetic cDNA was used. Hemagglutinin (HA)–tagged trBcl-2L1, trBcl-2L2, trBak ∆TM mutants, and 3×Flag-tagged trBax sequences were cloned into the pCS2+ expression vector using Bam HI/Xho I and Eco RI/Xho I restriction sites, respectively. trBax and associated mutants and BH3 trBax and BH3 trBak sequences were cloned into the pEGFP-C1 vector using Bgl II/Eco RI restriction sites. trBaxN8T mutant cDNA was obtained using gBlock synthesis and cloned into pEGFP-C1 using Bgl II/Eco RI restriction sites. Primers used for cloning were summarized into table S2.
Cell lines and transfection
BMK WT or bax/bak DKO (provided by the Eileen White lab, Rutgers Cancer Institute of New Jersey), HeLa, and Cos-7 cell lines were grown under standard cell culture conditions (37°C, 5% CO2) in Dulbecco’s modified Eagle’s medium high-glucose medium (Gibco) supplemented with 10% fetal bovine serum (Sigma-Aldrich), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Gibco). Cell transfection was performed using X-tremeGENE HP DNA Transfection Reagent (Roche) according to the manufacturer’s protocol. Six hours after transfection, cell medium was removed, and cells were incubated for 24 hours with a new medium. When required, cell detachment was prevented using Z-VAD-FMK (10 to 20 μM; Sigma-Aldrich) and Q-VD-Oph (10 μM; Sigma-Aldrich), which was added before the transfection or 6 hours after transfection. Alternatively and when stated, trBax cells were cotransfected with baculovirus caspase inhibitor p35.
Subcellular fractionation
All steps were carried out at 4°C. Transfected BMK DKO cells were placed into two 15-cm-diameter plates, washed twice in phosphate-buffered saline (PBS), and topped up to 1.5 ml using ice-cold MB buffer [210 mM mannitol, 70 mM sucrose, 1 mM EDTA, and 10 mM Hepes (pH 7.5) containing protease inhibitors; Roche]. Cells were disrupted by shearing with a dounce homogenizer 20 to 30 times. The disrupted cells were then centrifuged twice at 600g for 5 min to remove nuclei and centrifuged at 7000g to harvest crude mitochondria. The resultant pellet was washed twice with fresh MB buffer and centrifuged at 10,000g. Mitochondria were resuspended in an appropriate volume of MB buffer for further analyses. The supernatant was centrifuged at 100,000g for 1 hour, and the pellet containing the microsomal fraction was suspended in radioimmunoprecipitation assay buffer [150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM tris-HCl (pH 7.4) containing protease inhibitors]. Fifteen micrograms of each subcellular fraction and 50 μg of the total fraction were analyzed by Western blot.
Western blot analysis and antibodies
Western blot analysis was performed, as previously described (43). The following antibodies were used: F1F0 ATPase (adenosine triphosphatase) (1:1000; BD, 612518), vinculin (1:2000; Santa Cruz Biotechnology, sc-55465), calnexin (1:1000; Cell Signaling Technology, 2679), Flag M2 (1/1000; Sigma-Aldrich, F3165), and HA (1:1000; Ozyme, BLE901501), cytochrome c (1:1000; BD Bioscience, 556433). Horseradish peroxidase–conjugated goat anti-mouse, rabbit anti-mouse, and goat anti-rabbit secondary antibodies (DAKO) were used as secondary antibodies. Western blot analysis was performed according to standard procedures.
Immunofluorescence staining, FRET measurements, cytochrome c release, and apoptosis detection
Cells were placed on a glass coverslip in 12-well plates at 50% confluence and left to attach overnight. Cells were then transfected with trBcl-2–like encoding vectors in combination or with pCS2 + EGFP-CytB5 (ER-localized EGFP) for ER staining. For trBax and trBak subcellular localization, 20 μM zVAD-FMK (Sigma-Aldrich) was added 6 hours after transfection to delay cell detachment. Cells were then fixed with a 4% paraformaldehyde solution for 20 min, followed by three washes with PBS. Following cell permeabilization using 0.1% PBS Triton X-100 supplemented with 3% bovine serum albumin, cells were incubated with the primary antibodies Flag M2 (1:2500; Sigma-Aldrich, F3165), cytochrome c (1:1000; BD Bioscience, 556432), or Tom20 (1:500; Santa Cruz Biotechnology, sc-11415). Goat anti-mouse or goat anti-rabbit immunoglobulin G Alexa Fluor 488 (green fluorescent) or 568 (red fluorescent) were used as secondary antibodies (Molecular Probes) at 1:1000 dilution for 1 hour. Nuclei were visualized using Hoechst 33342 dye (Invitrogen H3570) at 10 μg/ml.
For FRET-based interaction analyses, BMK DKO cells were transfected with Flag-tagged trBcl-2L1 and trBcl-L2 in combination with EGFP-tagged trBax and trBak. Twenty-four hours later, cells were fixed and immunostained using mouse anti-Flag M2 antibody coupled to an anti-mouse Alexa Fluor 568 secondary antibody. FRET was measured, as previously described (44). Briefly, EGFP fluorescent intensities were measured before and after photobleaching with the Alexa Fluor 568 fluorophore. FRET efficacy was calculated using the following formula:
For apoptosis detection, HeLa and BMK DKO cells at 80 to 90% confluence were transiently transfected for 24 hours with the Flag-tagged or EGFP-tagged trBcl-2, hsBcl-2, or BH3 proteins. For treatment with apoptosis-inducing agents, cells were incubated for an additional 24 hours with etoposide (25 μM; Sigma-Aldrich), thapsigargin (10 μM; Sigma-Aldrich), or staurosporine (0.25 μM; Sigma-Aldrich). Cells were fixed and permeabilized, as described above, and incubated with anti-activated Caspase 3 antibody (1:500; BD Bioscience). Nuclei were labeled using Hoechst 33342 dye. Cells were observed under a fluorescent microscope, and the percentage of Caspase 3– and Hoechst-positive cells was determined.
Protein expression and purification
Synthetic cDNA encoding for codon-optimized WT trBcl-2L1 (Uniprot accession number A0A369S2N9) lacking the 33 N-terminal and 29 C-terminal residues, as well as trBcl-2L2 (Uniprot A0A369SIM9) lacking the 7 N-terminal and 34 C-terminal residues, was cloned into the bacterial expression vector pGEX-6P-1 (GenScript). Recombinant trBcl-2L1 and trBcl-2L2 were expressed in C41(DE3) cells in 2YT medium supplemented with ampicillin (1 mg/ml) at 37°C in a shaking incubator until an OD600 (optical density at 600 nm) of 0.6 was reached. The protein expression was induced using the autoinduction method (45) for 18 hours at 22°C. Bacterial cells were harvested by centrifugation at 5000 rpm (JLA 9.1000 rotor; Beckman Coulter Avanti J-E) for 20 min and resuspended in lysis buffer A [50 mM tris (pH 8.0), 300 mM NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol]. The cells were homogenized using an Avestin EmulsiFlex homogenizer and lysed using sonication (Fisher Scientific Model 705 Sonic Dismembrator). Bacterial lysates were transferred into SS34 tubes and centrifuged at 18,000 rpm (JA-25.50 rotor; Beckman Coulter Avanti J-E) for 30 min. The supernatant was filtered using a 0.45 μM syringe filter, loaded onto 5 ml of glutathione Sepharose 4B (GE Healthcare) equilibrated with buffer A, and the column was washed with 150 ml of buffer A. Target proteins were liberated from the fusion tag on-column by adding HRV 3C protease overnight at 4°C. Cleaved proteins were eluted using buffer A and concentrated using a centrifugal concentrator with 3-kDa molecular weight cutoff (Amicon Ultra 15). Concentrated target proteins were subjected to size-exclusion chromatography using a Superdex S200 increase 10/300 column mounted on an ÄKTA pure system (GE Healthcare) and equilibrated in 25 mM Hepes (pH 7.0), 150 mM NaCl, and 5 mM DTT for trBcl-2L2 and 25 mM sodium acetate (pH 5.0) and 200 mM NaCl for trBcl-2L1. Fractions were analyzed using SDS–polyacrylamide gel electrophoresis, with the final sample purity estimated to be higher than 95%. Appropriate fractions were pooled and concentrated using a centrifugal concentrator with 3-kDa molecular weight cutoff (Amicon Ultra 15). Synthetic cDNA encoding for human Bcl-2 (residues 10 to 203; Uniprot P10415), Bcl-xL (residues 1 to 209 Δloop45 to 84; Uniprot Q07817), Bcl-w (residues 9 to 152; Uniprot Q92843), Mcl-1 (residues 171 to 320; Uniprot Q07820), and A1/Bfl-1 (residues 1 to 151; Uniprot Q16548) were cloned into the expression vector PGEX-6P1. Bcl-w and Mcl-1 were expressed using BL21 Star cells under conditions as described above, whereas Bcl-2, Bcl-xL and A1 were expressed as described above for trBcl-2L1 and trBcl-L2. All proteins were purified as described above and concentrated in 25 mM Hepes (pH 7.0) and 150 mM NaCl to a concentration of 5 mg/ml for measurement of dissociation constants.
Measurement of dissociation constants
Binding affinities were measured by ITC using a MicroCal iTC200 system (GE Healthcare) at 25°C using WT trBcl-2L1, trBcl-2L2, Bcl-2, Bcl-xL, Bcl-w, Mcl-1, and A1/Bfl-1 in 25 mM Hepes (pH 7.5), 150 mM NaCl, and 5 mM TCEP [tris(2-carboxyethyl)phosphine] at a final concentration of 30 μM. BH3 motif peptide ligands were used at a concentration of 300 μM and titrated using 19 injections of 2.0 μl of ligand. All affinity measurements were performed in triplicate, with affinities calculated using a one-site binding model using the Origin software package. Protein concentrations were measured using a NanoDrop UV-Vis spectrophotometer (Thermo Scientific) at a wavelength of 280 nm. Peptide concentrations were calculated on the basis of the dry peptide weight after synthesis. Human BH3-motif peptides used were commercially synthesized and purified to a final purity of 95% (GenScript) and previously described (46). T. adhaerens BH3 peptides, trBak BH3 peptide (Uniprot A0A369S334; residues 102 to 128, PSSPTSEIGRHLAQLGDSYSVRFQNE) and trBax BH3 peptide (Uniprot A0A369RYN3; residues 70 to 98, IEPTSPTIQVARTIRQIGDELDSDPHLN), were commercially synthesized and purified to a final purity of 95% (GenScript).
Competition ITC measurements were performed by titrating trBax BH3 peptide into Bcl-2, Bcl-xL, Bcl-w, Mcl-1, and A1/Bfl-1 in 25 mM Hepes (pH 7.5), 150 mM NaCl, and 5 mM TCEP, followed by a second titration of trBak BH3 into the same reaction chamber. The apparent affinity of trBak BH3 (Kapp) was calculated using the competitive binding model using the Origin software package.
Crystallization and structure determination
Crystals for the trBcl-2L2:trBak BH3 and hsBcl-xL:trBak BH3 complexes were obtained by mixing trBcl-2L2 or hsBcl-xL with trBak BH3 26-nucleotide oligomer peptide using a 1:1.25 molar ratio, as described previously (15), and concentrated using a centrifugal concentrator with 3 kDa-molecular weight cutoff (Amicon Ultra 0.5). Initial high-throughput -sparse matrix screening was performed using 96-well sitting-drop trays (Swissic, Neuheim, Switzerland).
trBcl-2L2:trBak BH3 complex crystals were grown using the sitting-drop vapor diffusion method at 20°C in 0.2 M ammonium sulfate, 0.1 M sodium citrate (pH 5.8), and 29% (w/v) of PEG-4000 (polyethylene glycol, molecular weight 4000) using a protein concentration of 15 mg/ml. The crystals were flash cooled at −173°C in mother liquor supplemented with 20% ethylene glycol. The trBcl-2L2:trBak BH3 complex formed single rectangular-shaped crystals belonging to space group P1 with a = 34.30 Å, b = 40.86 Å, c = 56.72 Å, α = 97.30 Å, β = 90.09 Å, and γ = 101.09 Å in the triclinic crystal system.
Diffraction data were collected at the Australian Synchrotron MX2 beamline using an Eiger detector with an oscillation range 0.1° per frame with a wavelength of 0.9537 Å, integrated using x-ray detector software (XDS) (47), and scaled using AIMLESS. Molecular replacement was carried out using PHASER (48) with the previously solved structure of Bcl-xL [PDB ID, 1PQ1 (49)] as a search model. trBcl-2L2:trBak BH3 crystals contained two molecules of trBcl-2L2 and two trBak BH3 peptides in the asymmetric unit, with 33.56% solvent content and final TFZ and LLG values of 23.9 and 598, respectively. The trBcl-2L2:trBak BH3 model was built manually over several cycles using Coot (50) and refined using PHENIX (51).
HsBcl-xL:trBak BH3 complex crystals were grown using the sitting-drop vapor diffusion method at 20°C in 0.2 M ammonium sulfate and 30% (w/v) of PEG 8000 using a protein concentration of 13.4 mg/ml. The crystals were flash cooled at −173°C in mother liquor supplemented with 30% MPD (2-methyl-2,4-pentanediol), and diffraction data were collected and processed, as described above. Molecular replacement was carried out using PHASER (48) with the previously solved structure of Bcl-xL [PDB ID, 1PQ1 (49)] as a search model. HsBcl-xL:trBak BH3 complex crystals contained two molecules of hsBcl-xL and two trBak BH3 peptides in the asymmetric unit, with 35.55% solvent content and final TFZ and LLG values of 20.9 and 448, respectively. The hsBcl-xL:trBak BH3 model was built manually over several cycles using Coot (50) and refined using PHENIX (51). All images were generated using the PyMOL Molecular Graphics System, version 1.8, Schrödinger LLC. All softwares were accessed using the SBGrid suite (52).
Statistical analysis
Error bars displayed on graphs represent the means ± SD of three independent experiments. Statistical significance was analyzed using the one-way analysis of variance (ANOVA) and two-way ANOVA tests. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We would like to thank G. Ichim, V. Daubin, and N. Peyrieras for all the fruitful discussions. We would like to thank L. Coudert, T. Hoffman, K. Radkova, and C. Céré for technical assistance. We thank the staff at the MX beamlines at the Australian Synchrotron for help with the x-ray data collection. This research was undertaken, in part, using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector. We thank the Comprehensive Proteomics Platform at La Trobe University for core instrument support. Funding: This research was supported by Ligue nationale contre le cancer (Comité du Rhône) (to N.P.), Fondation ARC (grant no. PGA1 RF20180206799 to G.G.), France BioImaging infrastructure ANR-10-INBS-04, Morphoscope ANR11-EQPX-0029, and InterDIM 2011 Région Paris Ile-de-France and the Australian Research Council (DP190103591 to M.K.). Author contributions: N.P., L.J., R.G., T.T.M.N., and N.R. performed all the experiments except for the following: S.M. performed the yeast experiments. H.-J.O. and B.S. performed Trichoplax DNA/RNA extractions and cDNA library preparations. J.D.S. expressed and purified recombinant proteins, performed affinity measurements, and grew crystals. S.B. performed affinity measurements, collected, and analyzed crystallographic data. A.A.-E.-S. expressed and purified recombinant proteins. M.K. analyzed affinity measurements and built the crystallographic model. N.P., M.G.H., H.-J.O., B.S., P.O.H., R.R., G.G., and M.K. participated in the study design and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All coordinate files have been deposited to the Protein Data Bank under the accession codes 6YLD and 6YLI. All raw diffraction images were deposited to the SBGrid Data Bank (53) using their PDB accession codes 6YLD and 6YLI. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/40/eabc4149/DC1
REFERENCES AND NOTES
- 1.Zmasek C. M., Godzik A., Evolution of the animal apoptosis network. Cold Spring Harb. Perspect. Biol. 5, a008649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs Y., Steller H., Live to die another way: Modes of programmed cell death and the signals emanating from dying cells. Nat. Rev. Mol. Cell Biol. 16, 329–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banjara S., Suraweera C. D., Hinds M. G., Kvansakul M., The Bcl-2 family: Ancient origins, conserved structures, and divergent mechanisms. Biomolecules 10, 128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengartner M. O., Ellis R. E., Horvitz H. R., Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356, 494–499 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Quinn L., Coombe M., Mills K., Daish T., Colussi P., Kumar S., Richardson H., Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 22, 3568–3579 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lasi M., Pauly B., Schmidt N., Cikala M., Stiening B., Käsbauer T., Zenner G., Popp T., Wagner A., Knapp R. T., Huber A. H., Grunert M., Söding J., David C. N., Böttger A., The molecular cell death machinery in the simple cnidarian Hydra includes an expanded caspase family and pro- and anti-apoptotic Bcl-2 proteins. Cell Res. 20, 812–825 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Kratz E., Eimon P. M., Mukhyala K., Stern H., Zha J., Strasser A., Hart R., Ashkenazi A., Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ. 13, 1631–1640 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Lee E. F., Clarke O. B., Evangelista M., Feng Z., Speed T. P., Tchoubrieva E. B., Strasser A., Kalinna B. H., Colman P. M., Fairlie W. D., Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes. Proc. Natl. Acad. Sci. U.S.A. 108, 6999–7003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh R., Letai A., Sarosiek K., Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20, 175–193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kvansakul M., Hinds M. G., The structural biology of BH3-only proteins. Methods Enzymol. 544, 49–74 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Strasser A., Vaux D. L., Viewing BCL2 and cell death control from an evolutionary perspective. Cell Death Differ. 25, 13–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kale J., Osterlund E. J., Andrews D. W., BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 25, 65–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tait S. W. G., Oberst A., Quarato G., Milasta S., Haller M., Wang R., Karvela M., Ichim G., Yatim N., Albert M. L., Kidd G., Wakefield R., Frase S., Krautwald S., Linkermann A., Green D. R., Widespread mitochondrial depletion via mitophagy does not compromise necroptosis. Cell Rep. 5, 878–885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moldoveanu T., Follis A. V., Kriwacki R. W., Green D. R., Many players in BCL-2 family affairs. Trends Biochem. Sci. 39, 101–111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caria S., Hinds M. G., Kvansakul M., Structural insight into an evolutionarily ancient programmed cell death regulator – the crystal structure of marine sponge BHP2 bound to LB-Bak-2. Cell Death Dis. 8, e2543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galasso C., D’Aniello S., Sansone C., Ianora A., Romano G., Identification of cell death genes in sea urchin Paracentrotus lividus and their expression patterns during embryonic development. Genome Biol. Evol. 11, 586–596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moya A., Sakamaki K., Mason B. M., Huisman L., Forêt S., Weiss Y., Bull T. E., Tomii K., Imai K., Hayward D. C., Ball E. E., Miller D. J., Functional conservation of the apoptotic machinery from coral to man: The diverse and complex Bcl-2 and caspase repertoires of Acropora millepora. BMC Genomics 17, 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill K. L., Huang K., Zhang J., Chen Y., Luo X., Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 30, 973 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang K., O’Neill K. L., Li J., Zhou W., Han N., Pang X., Wu W., Struble L., Borgstahl G., Liu Z., Zhang L., Luo X., BH3-only proteins target BCL-xL/MCL-1, not BAX/BAK, to initiate apoptosis. Cell Res. 29, 942–952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu D., Isozaki Y., Zhang X., Han J., Maruyama S., Birth and early evolution of metazoans. Gondwana Res. 25, 884–895 (2014). [Google Scholar]
- 21.Osigus H.-J., Rolfes S., Herzog R., Kamm K., Schierwater B., Polyplacotoma mediterranea is a new ramified placozoan species. Curr. Biol. 29, R148–R149 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Syed T., Schierwater B., Trichoplax adhaerens: Discovered as a missing link, forgotten as a hydrozoan, re-discovered as a key to metazoan evolution. Vie Milieu 52, 177–187 (2002). [Google Scholar]
- 23.Schierwater B., My favorite animal, Trichoplax adhaerens. BioEssays 27, 1294–1302 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Srivastava M., Begovic E., Chapman J., Putnam N. H., Hellsten U., Kawashima T., Kuo A., Mitros T., Salamov A., Carpenter M. L., Signorovitch A. Y., Moreno M. A., Kamm K., Grimwood J., Schmutz J., Shapiro H., Grigoriev I. V., Buss L. W., Schierwater B., Dellaporta S. L., Rokhsar D. S., The Trichoplax genome and the nature of placozoans. Nature 454, 955–960 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Green D. R., Fitzgerald P., Just so stories about the evolution of apoptosis. Curr. Biol. 26, R620–R627 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popgeorgiev N., Jabbour L., Gillet G., Subcellular localization and dynamics of the Bcl-2 family of proteins. Front. Cell. Dev. Biol. 6, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartron P.-F., Arokium H., Oliver L., Meflah K., Manon S., Vallette F. M., Distinct domains control the addressing and the insertion of Bax into mitochondria. J. Biol. Chem. 280, 10587–10598 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Edlich F., Banerjee S., Suzuki M., Cleland M. M., Arnoult D., Wang C., Neutzner A., Tjandra N., Youle R. J., Bcl-xL retrotranslocates Bax from the mitochondria into the cytosol. Cell 145, 104–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schinzel A., Kaufmann T., Schuler M., Martinalbo J., Grubb D., Borner C., Conformational control of Bax localization and apoptotic activity by Pro168. J. Cell Biol. 164, 1021–1032 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simonyan L., Légiot A., Lascu I., Durand G., Giraud M.-F., Gonzalez C., Manon S., The substitution of Proline 168 favors Bax oligomerization and stimulates its interaction with LUVs and mitochondria. Biochim. Biophys. Acta Biomembr. 1859, 1144–1155 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Popgeorgiev N., Bonneau B., Ferri K. F., Prudent J., Thibaut J., Gillet G., The apoptotic regulator Nrz controls cytoskeletal dynamics via the regulation of Ca2+ trafficking in the zebrafish blastula. Dev. Cell 20, 663–676 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Aouacheria A., Brunet F., Gouy M., Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-Only, and BNip families of apoptotic regulators. Mol. Biol. Evol. 22, 2395–2416 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Aouacheria A., Rech de Laval V., Combet C., Hardwick J. M., Evolution of Bcl-2 homology motifs: Homology versus homoplasy. Trends Cell Biol. 23, 103–111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou J. J., Li H., Salvesen G. S., Yuan J., Wagner G., Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell 96, 615–624 (1999). [DOI] [PubMed] [Google Scholar]
- 35.McDonnell J. M., Fushman D., Milliman C. L., Korsmeyer S. J., Cowburn D., Solution structure of the proapoptotic molecule BID: A structural basis for apoptotic agonists and antagonists. Cell 96, 625–634 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Thijssen R., Roberts A. W., Venetoclax in lymphoid malignancies: New insights, more to learn. Cancer Cell 36, 341–343 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., McHenry K. T., Pinchback R. M., Ligon A. H., Cho Y.-J., Haery L., Greulich H., Reich M., Winckler W., Lawrence M. S., Weir B. A., Tanaka K. E., Chiang D. Y., Bass A. J., Loo A., Hoffman C., Prensner J., Liefeld T., Gao Q., Yecies D., Signoretti S., Maher E., Kaye F. J., Sasaki H., Tepper J. E., Fletcher J. A., Tabernero J., Baselga J., Tsao M.-S., Demichelis F., Rubin M. A., Janne P. A., Daly M. J., Nucera C., Levine R. L., Ebert B. L., Gabriel S., Rustgi A. K., Antonescu C. R., Ladanyi M., Letai A., Garraway L. A., Loda M., Beer D. G., True L. D., Okamoto A., Pomeroy S. L., Singer S., Golub T. R., Lander E. S., Getz G., Sellers W. R., Meyerson M., The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell K. J., Dhayade S., Ferrari N., Sims A. H., Johnson E., Mason S. M., Dickson A., Ryan K. M., Kalna G., Edwards J., Tait S. W. G., Blyth K., MCL-1 is a prognostic indicator and drug target in breast cancer. Cell Death Dis. 9, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang Z., Luo H., Payton J. E., Cain J., Ley T. J., Opferman J. T., Tomasson M. H., Mcl1 haploinsufficiency protects mice from Myc-induced acute myeloid leukemia. J. Clin. Invest. 120, 2109–2118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zervantonakis I. K., Iavarone C., Chen H.-Y., Selfors L. M., Palakurthi S., Liu J. F., Drapkin R., Matulonis U., Leverson J. D., Sampath D., Mills G. B., Brugge J. S., Systems analysis of apoptotic priming in ovarian cancer identifies vulnerabilities and predictors of drug response. Nat. Commun. 8, 365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotschy A., Szlavik Z., Murray J., Davidson J., Maragno A. L., Le Toumelin-Braizat G., Chanrion M., Kelly G. L., Gong J.-N., Moujalled D. M., Bruno A., Csekei M., Paczal A., Szabo Z. B., Sipos S., Radics G., Proszenyak A., Balint B., Ondi L., Blasko G., Robertson A., Surgenor A., Dokurno P., Chen I., Matassova N., Smith J., Pedder C., Graham C., Studeny A., Lysiak-Auvity G., Girard A.-M., Gravé F., Segal D., Riffkin C. D., Pomilio G., Galbraith L. C. A., Aubrey B. J., Brennan M. S., Herold M. J., Chang C., Guasconi G., Cauquil N., Melchiore F., Guigal-Stephan N., Lockhart B., Colland F., Hickman J. A., Roberts A. W., Huang D. C. S., Wei A. H., Strasser A., Lessene G., Geneste O., The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Eitel M., Schierwater B., The phylogeography of the Placozoa suggests a taxon-rich phylum in tropical and subtropical waters. Mol. Ecol. 19, 2315–2327 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Arnaud E., Ferri K. F., Thibaut J., Haftek-Terreau Z., Aouacheria A., Le Guellec D., Lorca T., Gillet G., The zebrafish bcl-2 homologue Nrz controls development during somitogenesis and gastrulation via apoptosis-dependent and -independent mechanisms. Cell Death Differ. 13, 1128–1137 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Rong Y.-P., Aromolaran A. S., Bultynck G., Zhong F., Li X., Coll K. M., Matsuyama S., Herlitze S., Roderick H. L., Bootman M. D., Mignery G. A., Parys J. B., De Smedt H., Distelhorst C. W., Targeting Bcl-2-IP3 receptor interaction to reverse Bcl-2’s inhibition of apoptotic calcium signals. Mol. Cell 31, 255–265 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Marshall B., Puthalakath H., Caria S., Chugh S., Doerflinger M., Colman P. M., Kvansakul M., Variola virus F1L is a Bcl-2-like protein that unlike its vaccinia virus counterpart inhibits apoptosis independent of Bim. Cell Death Dis. 6, e1680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy A. J., Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X., Dai S., Zhu Y., Marrack P., Kappler J. W., The structure of a Bcl-xL/Bim fragment complex: Implications for Bim function. Immunity 19, 341–352 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Echols N., Grosse-Kunstleve R. W., Afonine P. V., Bunkóczi G., Chen V. B., Headd J. J., McCoy A. J., Moriarty N. W., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Adams P. D., Graphical tools for macromolecular crystallography in PHENIX. J. Appl. Crystallogr. 45, 581–586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., Sliz P., Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer P. A., Socias S., Key J., Ransey E., Tjon E. C., Buschiazzo A., Lei M., Botka C., Withrow J., Neau D., Rajashankar K., Anderson K. S., Baxter R. H., Blacklow S. C., Boggon T. J., Bonvin A. M. J. J., Borek D., Brett T. J., Caflisch A., Chang C.-I., Chazin W. J., Corbett K. D., Cosgrove M. S., Crosson S., Dhe-Paganon S., Cera E. D., Drennan C. L., Eck M. J., Eichman B. F., Fan Q. R., Ferré-D’Amaré A. R., Fromme J. C., Garcia K. C., Gaudet R., Gong P., Harrison S. C., Heldwein E. E., Jia Z., Keenan R. J., Kruse A. C., Kvansakul M., McLellan J. S., Modis Y., Nam Y., Otwinowski Z., Pai E. F., Pereira P. J. B., Petosa C., Raman C. S., Rapoport T. A., Roll-Mecak A., Rosen M. K., Rudenko G., Schlessinger J., Schwartz T. U., Shamoo Y., Sondermann H., Tao Y. J., Tolia N. H., Tsodikov O. V., Westover K. D., Wu H., Foster I., Fraser J. S., Maia F. R. N. C., Gonen T., Kirchhausen T., Diederichs K., Crosas M., Sliz P., Data publication with the structural biology data grid supports live analysis. Nat. Commun. 7, 10882 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Westerfield, A Guide for the Laboratory Use of Zebrafish Danio (Brachydanio) Rerio (University of Oregon Press, ed. 3, 1995). [Google Scholar]
- 55.Arokium H., Camougrand N., Vallette F. M., Manon S., Studies of the interaction of substituted mutants of BAX with yeast mitochondria reveal that the C-terminal hydrophobic α-helix is a second ART sequence and plays a role in the interaction with anti-apoptotic BCL-xL. J. Biol. Chem. 279, 52566 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Renault T. T., Teijido O., Missire F., Ganesan Y. T., Velours G., Arokium H., Beaumatin F., Llanos R., Athané A., Camougrand N., Priault M., Antonsson B., Dejean L. M., Manon S., Bcl-xL stimulates Bax relocation to mitochondria and primes cells to ABT-737. Int. J. Biochem. Cell Biol. 64, 136 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/40/eabc4149/DC1


