Thermofluidics actuates genetic patterning in artificial tissue.
Abstract
Spatial patterns of gene expression in living organisms orchestrate cell decisions in development, homeostasis, and disease. However, most methods for reconstructing gene patterning in 3D cell culture and artificial tissues are restricted by patterning depth and scale. We introduce a depth- and scale-flexible method to direct volumetric gene expression patterning in 3D artificial tissues, which we call “heat exchangers for actuation of transcription” (HEAT). This approach leverages fluid-based heat transfer from printed networks in the tissues to activate heat-inducible transgenes expressed by embedded cells. We show that gene expression patterning can be tuned both spatially and dynamically by varying channel network architecture, fluid temperature, fluid flow direction, and stimulation timing in a user-defined manner and maintained in vivo. We apply this approach to activate the 3D positional expression of Wnt ligands and Wnt/β-catenin pathway regulators, which are major regulators of development, homeostasis, regeneration, and cancer throughout the animal kingdom.
INTRODUCTION
Cells transform noisy environmental signals into spatial and dynamic gene expression patterns that guide biological form and function. Information describing how these transcriptional networks are patterned is exploding because of revolutions in single-cell RNA sequencing and spatial transcriptomics. Recapitulating this spatiotemporal information transfer in three-dimensional (3D) tissue settings remains a pivotal yet elusive goal of diverse fields, such as tissue engineering (1), synthetic biology (2, 3), and developmental biology (4, 5).
To control gene expression, biologists have developed diverse technologies to rewire cells at the genetic level, such as gene knockout, inhibition, overexpression, and editing (6–8). To further enable spatial and dynamic control of gene expression, several of these tools have been adapted to be triggered by exogenous stimuli such as light (e.g., optogenetic transcriptional control) (9, 10). Light-based actuation of gene expression patterning has been especially useful in 2D culture or optically transparent settings. However, the inherently poor penetration of light in densely populated tissues (11), long exposure times needed to activate molecular switches, and corresponding challenges in patterning light delivery have limited widespread adoption of light-based patterning of gene expression in 3D settings (12).
We hypothesized that we could overcome these challenges by exploiting more penetrant forms of energy to drive gene patterning. In particular, mild heating is an attractive option for 3D patterning across length scales, as heat can be targeted locally and penetrate tissues at depth. Furthermore, heat can diffuse through tissues to establish thermal gradients in predictable and controllable patterns that are dictated by established rules of heat transfer (13). Last, advances in molecular engineering have led to proliferation of thermal molecular bioswitches to regulate gene expression (14, 15), with mammalian systems activating in the mild hyperthermia range (~38° to 45°C).
Heat transfer has a long industrial history, as heat is often added, removed, or moved between processes using heat exchangers, which transfer heat between fluidic networks. Recently, heat exchanger fabrication has undergone a radical shift due to developments in advanced manufacturing (e.g., 3D printing). Predating its history in industry, biological organisms have also long used heat exchanger design principles for thermoregulation. We reasoned that instead of building heat exchangers from hard materials, developing methods to build heat exchangers in materials compatible with living cells could facilitate volumetric heat patterning in artificial tissues.
RESULTS
Heat exchangers for actuation of transcription
We introduce a thermofluidic method for mesoscale spatiotemporal control of gene expression in artificial tissues that exploits volumetric fluid-based heat transfer, which we call “heat exchangers for actuation of transcription” (HEAT; Fig. 1A). HEAT leverages our open-source projection stereolithography bioprinting technology (16) to fabricate topologically complex fluidic channels of user-defined geometries in hydrogels (Fig. 1B, top and middle). 3D printed hydrogels are laden with genetically engineered heat-inducible cells during the printing process (Fig. 1A). Encased channel networks are perfused with precisely heated fluid from a power-supplied heating element. During perfusion, tissue temperature is monitored in real-time using an infrared camera (Fig. 1A). We find that thermofluidic perfusion facilitates heat transfer from the channels into the bulk hydrogel and enables architectural heat patterning in hydrogels (Fig. 1B, bottom).
Fig. 1. Thermofluidic heating in 3D bioprinted hydrogels.
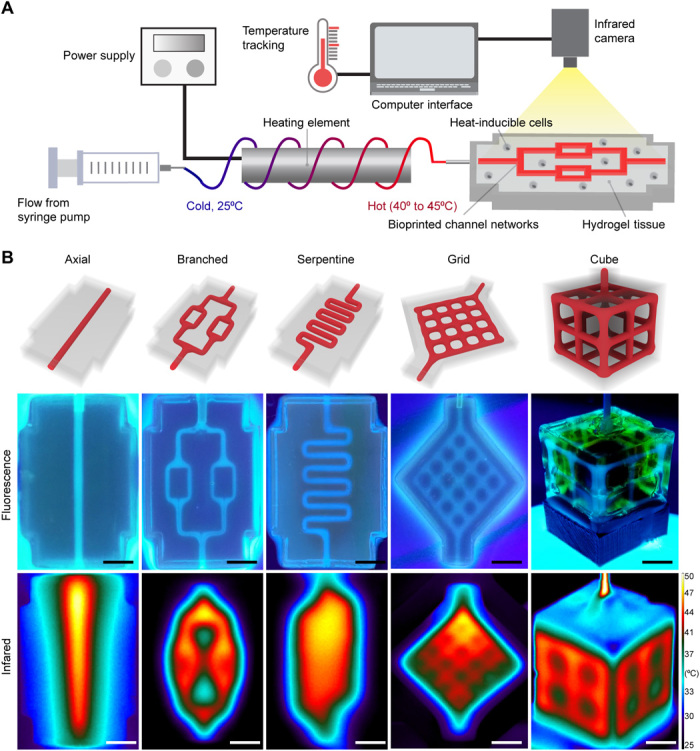
(A) Schematic of thermofluidic workflow. A biocompatible fluid flows around a power supplied heating element to preheat the fluid before entry in perfusable channel networks within hydrogel tissue constructs laden with heat-sensitive cells. During perfusive heating, hydrogel temperature is continuously monitored using an infrared camera. (B) Perfusable channel networks of varying spatial geometries can be bioprinted within biocompatible 3D hydrogels. Top: 3D rendering of network architectures. Middle: Hydrogel channels infused with tonic water fluoresce when imaged under ultraviolet backlight. Bottom: Infrared thermography of heat-perfused hydrogels demonstrates that during perfusion, heat traces the path of fluid flow and dissipates into the bulk hydrogel. Scale bars, 5 mm.
Thermal characterization in single-channel hydrogels
Most mammalian thermally inducible gene switches require exposure to mild hyperthermia (39° to 45°C) for prolonged periods of ~15 to 60 min to activate transcription (15, 17). We therefore tested whether this approach could precisely regulate tissue temperature over prolonged periods of time by maintaining steady-state thermal profiles in perfused hydrogels. To do this, we first printed hydrogels that contained a single channel (Fig. 2A). We then perfused precisely heated fluid through this channel while tracking hydrogel temperature in real-time using infrared thermography (Fig. 2B). Upon initiating perfusion, we observed that hydrogel temperature underwent an initial ramp-up phase (~5 min) followed by a steady-state plateau in which temperature deviated by <±0.4°C/min at three separate regions measured across the hydrogel (Fig. 2B, right).
Fig. 2. Thermal profile characterization in perfused hydrogels.
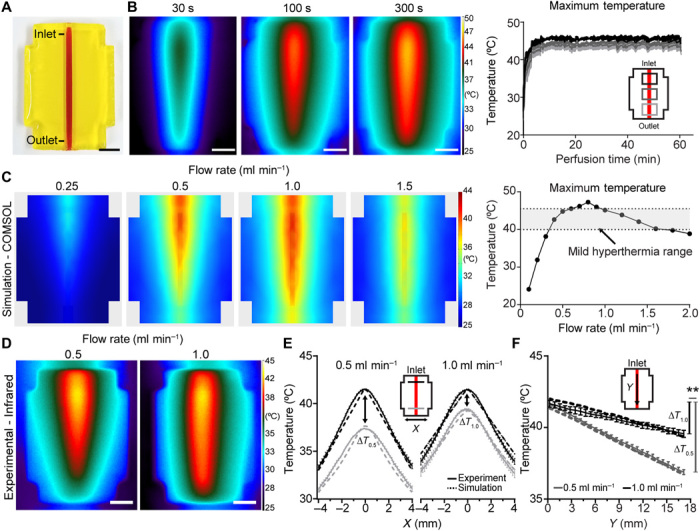
(A) Photograph of a single-channel bioprinted hydrogel used for initial thermal characterization. Scale bar, 5 mm. (B) Representative infrared images from controlled perfusion of heated fluid through the channel over time (left). Scale bars, 5 mm. (C) Representative finite-element modeling images depicting steady-state predictions on the surface of perfused hydrogels at varying flow rates and constant heater power (left; full dataset in fig. S1B). Computational modeling predicts that flow rate can achieve maximal hydrogel temperatures in the mild hyperthermia temperature range (right, gray shading denotes mild hyperthermia range). (D) Hydrogels were experimentally perfused at flow rates of 0.5 and 1.0 ml min−1 and imaged using infrared thermography. Scale bars, 5 mm. (E) Hydrogel temperature plotted orthogonal (x) to the flow direction at inlet and outlet positions show agreement between thermal gradients in computational and experimental measurements (computational, dashed lines; experimental, solid lines). (F) Hydrogel temperature plotted parallel (y) to flow direction demonstrates a larger temperature drop from inlet to outlet (y) during flow at 0.5 ml min−1 (∆T0.5) compared to flow at 1.0 ml min−1 (∆T1.0) in computational and experimental models (computational, dashed lines; experimental, solid lines; n = 5, data are mean temperature ± standard error, **P < 0.01 by Student’s t test). Photo credit: Daniel Corbett, University of Washington.
During perfusion, heat is transferred from fluidic channels to the bulk through convection and conduction, resulting in thermal gradients throughout the bulk volume (18). The perfusate input temperature is known to govern the rate and magnitude of heat transfer, while fluid flow rate influences the thermal profile (18). To determine the relative effects of perfusate temperature and flow rate on hydrogel heating at biologically relevant temperatures, we sought to develop a finite element model of heated hydrogel perfusion for mild hyperthermia that incorporated thermal and flow parameters from our heating system. To derive these parameters, we first incrementally increased flow rate over a range of heating element powers and measured fluid temperature at the point of heater outflow (i.e., hydrogel inlet; fig. S1). We then implemented perfusate temperature values observed from each flow rate at 13.5-W heater power into a computational model of single-channel hydrogel heating (Fig. 2C and fig. S1B). Computational simulations predicted that hydrogel temperatures in the range for mild hyperthermia were achievable using flow rates from 0.4 to 1.6 ml min−1, but not for slower or faster flow rates (Fig. 2C and fig. S1B). Within this window, we observed that flow rates of 0.5 and 1.0 ml min−1 produced subtle differences in the shape of thermal profiles, despite roughly equivalent input temperatures (Fig. 2C and fig. S1B). Thus, these flow rates provided a set of conditions to further examine the effects of flow rate on heat transfer.
We therefore performed experimental validation studies of perfused single-channel hydrogels at 0.5 or 1.0 ml min−1 and analyzed the steady-state thermal profiles from infrared images (Fig. 2D). Experimental temperature measurements (solid lines) and computational simulation predictions (dashed lines) showed agreement when measured both orthogonal (Fig. 2E) and parallel (Fig. 2F) to channel flow. Both physical measurements and simulations demonstrated thermal gradients in the hydrogel. Temperature along the channel was better maintained under flow at 1.0 ml min−1 compared to flow at 0.5 ml min−1 (**P < 0.01; Fig. 2, E and F), and flow at 0.5 ml min−1 promoted more heat transfer at the channel inlet (fig. S2A). Addition of cells to single-channel hydrogels did not affect temperature profile after thermofluidic perfusion (fig. S2B) nor did differences in hydrogel weight percent in ranges commonly used for 3D printing of cellularized hydrogels [i.e., 10 to 20 weight % (wt %); fig. S2C] (16). Stiffer hydrogel formulations (i.e., 25 wt %) did exhibit different temperatures at the hydrogel edge, although these formulations are less commonly used for bioprinting due to their limited support of cell viability (16).
These findings led us to further computationally explore the potential spatial design space for a single-channel system. To do this, we assessed how varying channel length and ambient temperature affect the thermal profile in our model. Predictions showed that single channels up to 30 mm long achieved hyperthermic temperatures (40° to 45°C) along their entire length, with outlet temperatures falling out of the hyperthermic range at greater lengths (fig. S3A). Spatial heat distribution was only marginally affected within the ambient temperature range used in our studies here (20° to 22°C; fig. S3B), but more substantive increases in ambient temperature (e.g., to 30, 37°C) produced wider spatial gradients in hyperthermic range (fig. S3B). Together, these studies showed that the rules of heat transfer could be leveraged to predict thermal spatial profiles in perfused hydrogels and that these profiles could be finely tuned by varying parameters such as flow rate, channel length, and input and ambient temperature.
Generation and characterization of heat-inducible cells
We next aimed to genetically engineer heat-inducible cells that activate gene expression upon exposure to mild hyperthermia. To do this, we implemented a temperature-responsive gene switch-based on the human heat shock protein 6A (HSPA6) promoter, which exhibits a low level of basal activity and a high degree of up-regulation in response to mild heating (19). This promoter activates heat-regulated transcription through consensus pentanucleotide sequences (5′-NGAAN-3′) called heat shock elements, which are binding sites for heat shock transcription factors (19). We transduced human embryonic kidney (HEK) 293T cells with a lentiviral construct in which a 476–base pair (bp) region of the HSPA6 promoter containing eight canonical heat shock elements was placed upstream of a firefly luciferase (fLuc) reporter gene (Fig. 3A). Initial characterization of temperature-sensitive promoter activity in engineered cells in 2D tissue culture demonstrated a temperature-dose dependent up-regulation of luciferase activity in the range of mild hyperthermia (fig. S4A). Statistically significant up-regulation was observed in heated cells compared to nonheated controls after hyperthermia for 30 min at 45°C or 60 min from 43° to 45°C, while peak bioluminescence occurred after 60 min at 44°C (292 ± 26-fold increase in bioluminescence relative to 37°C controls). Bioluminescent signal was first detected 8 hours after heat shock, peaked at 16 hours (110 ± 30-fold increase), and fell back to baseline by 2 days (fig. S4B). Administration of a second heat shock stimulus 3 days later reinduced bioluminescent signal (fig. S4C). Thus, gene activation with this promoter system is transient but can be reactivated with pulsing.
Fig. 3. Fluidic heating induces gene expression in 3D artificial tissues.
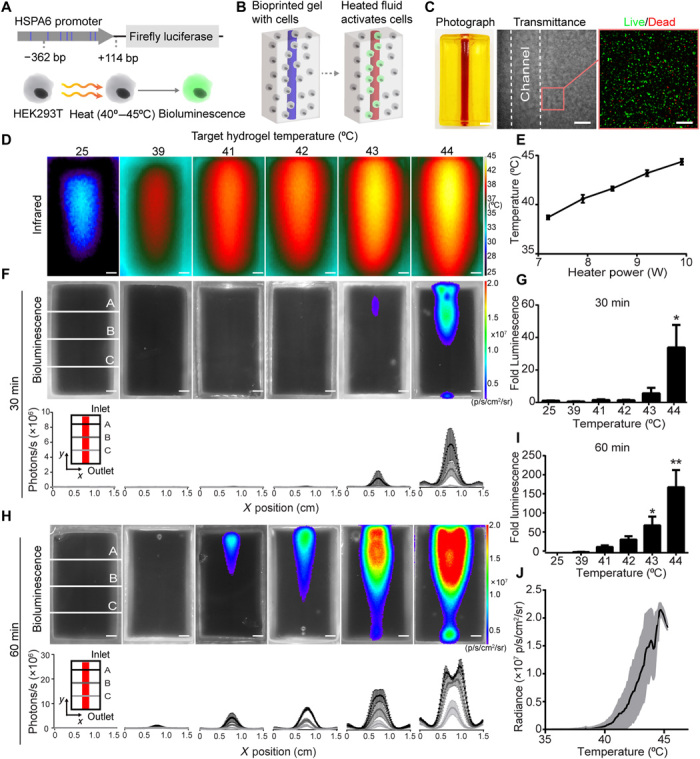
(A) HEK293T cells were engineered to express fLuc under the HSPA6 promoter. (B) Schematic of thermofluidic activation of encapsulated cells. (C) Single-channel tissue used for 3D heat activation (left). Scale bar, 3 mm. Transmittance image of cellularized hydrogel after printing (middle). Scale bar, 500 μm. HEK293T cells in bioprinted tissues stained with calcein-AM (“live,” green) and ethidium homodimer (“dead,” red; right). Scale bars, 200 μm. (D) Representative infrared images of thermofluidic perfusion in single-channel hydrogels. Scale bars, 2 mm. (E) Hydrogel temperatures are tuned by changing heater power at constant flow rate (n = 3, mean temperature ± standard error). (F) Representative bioluminescence images of hydrogels (top; scale bars, 2 mm) and intensity traces at three positions (A to C) across the width (x) of the hydrogel after 30 min of perfused heating. (G) Fold change in bioluminescence after 30 min of heating relative to 25°C controls. (H) Representative bioluminescence images of hydrogels (top; scale bars, 2 mm) and intensity traces after 60 min of perfused heating (bottom; scale bars, 2 mm). (I) Fold change in bioluminescence after 60 min of heating demonstrates a temperature-dependent dosage response in gene expression [(G and I); n = 3, mean fold luminescence ± standard error; *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Dunnett’s multiple comparison test]. (J) Temperature-expression response curve (black) shows mean bioluminescent radiance across temperature; shaded regions (gray) indicate ± SD. n = 3. Photo credit: Daniel Corbett, University of Washington.
We observed that our highest heat exposure (45°C for 60 min) led to a tradeoff between bioluminescence and cell integrity, as indicated by reduced cell metabolic activity and substrate detachment (fig. S5A). These findings suggested that fine control of heat would be needed for thermofluidics to be useful in cellularized applications. We therefore rigorously characterized the effect of heating on HEK293T cells embedded in the hydrogel formulation used for our thermofluidic studies. Similar to 2D studies, cell viability fell significantly only after exposure to our highest temperature, 45°C (fig. S5B). Together, these studies demonstrate engineering of human cells with a heat-sensitive gene switch and identification of a tight window of thermal exposure parameters that both differentially up-regulate gene bioluminescence and maintain cell integrity.
Thermofluidic activation of gene expression in artificial tissues
We sought to determine whether thermofluidic heating could be used to induce gene expression in heat-inducible cells encased within 3D artificial tissues (Fig. 3B). To do this, we encapsulated heat-inducible cells in the bulk of bioprinted constructs that contained a single perfusable channel (Fig. 3, B and C). Since tissue constructs were printed from biocompatible materials without ultraviolet light cross-linking, most cells remained viable upon encapsulation, similar to our previous studies (16) (Fig. 3C). To determine whether our heat-inducible cells could be activated using thermofluidics, we perfused channels at 0.5 ml min−1 using thermal exposure parameters identified in 2D culture (Fig. 3, D and E). Similar to 2D, we observed that thermal dose-dependent luciferase up-regulation (Fig. 3, F to J) was statistically significant after 30 min of heating to a target hydrogel temperature of 44°C or after 60 min of heating to temperatures of 43° and 44°C by whole-gel bioluminescent output (71 ± 22-fold and 169 ± 44-fold increase relative to controls, respectively; Fig. 3, H and I). To more finely characterize how bioluminescent intensity correlates with temperature, infrared and bioluminescence images were overlaid to map individual pixels and generate temperature-bioluminescence response curves. The shape of temperature-response curves appeared similar in shape across various target temperatures (Fig. 3J, all data overlaid; fig. S6, individual response curves). Similar to whole-gel analyses, greater target temperatures generated the most robust activation (Fig. 3J and fig. S6). In initial studies, we noted that leakage at the hydrogel inlet or outlet could activate cells. Subsequent improvements to fluidic connectivity with a custom-printed perfusion apparatus led to higher precision thermal patterning (fig. S7; see link to open source perfusion apparatus design in Methods). Last, multiperspective imaging and bioluminescence quantification of single-channel perfused hydrogels from both “top-down” and “cross-sectional” perspectives demonstrated that reporter gene activation had a 3D radial gradient topology around each channel (fig. S8). Together, these results illustrate that thermofluidics can be used to activate varying levels of gene expression in 3D artificial tissues.
Heat exchangers for spatial and dynamic control of gene expression patterning
Spatial patterns of gene expression within native tissues vary widely in magnitude, scale, and spatial complexity. While we achieved variation in magnitude in our signal-channel studies, the expression profile geometry across the hydrogel remained similar at various perfusion temperatures. This raised the question of how to design heat delivery schemes that enable more spatially complex expression patterns across the hydrogel. Our thermal characterization (Fig. 2) revealed flow rate as one parameter that we could use, but changing flow rate alone imparted only subtle differences to the spatial thermal profile (Fig. 2, D to F). To identify a more perturbative and user-defined means of affecting heat distribution across the hydrogel, we turned to industrial heat transfer applications, in which heat exchangers are optimized to transfer heat between fluids by controlling parameters such as channel placement and flow pattern.
We mimicked a double pipe heat exchanger design within cellularized hydrogels by printing two channels at varying distances from one another (Fig. 4A, narrow versus wide). We then perfused hydrogels under different conditions for flow direction (concurrent versus countercurrent) and fluid temperature [hot (44°C) versus cold (25°C)]. Similar to our single-channel characterization, double-channel tissues showed close matching between thermal and bioluminescence profiles (Fig. 4A). Concurrent flow in narrow spaced channels created elongated spatial plateaus of heat and bioluminescence between the channels. Conversely, widely spaced hot channels generated mirror-imaged thermal and bioluminescent profiles, with distinct spatial separation between channels. Countercurrent flow patterns generated parallelogrammic thermal and bioluminescent profiles in both channel spacings. Substituting a hot channel for a cold channel attenuated bioluminescence in a manner that depended on channel spacing (Fig. 4A). Computational models of a similar bifurcating channel geometry further demonstrated how simple changes to parameters such as channel spacing can alter spatial thermal profile (fig. S9).
Fig. 4. Heated perfusion of complex network architectures localizes gene expression in space and time.
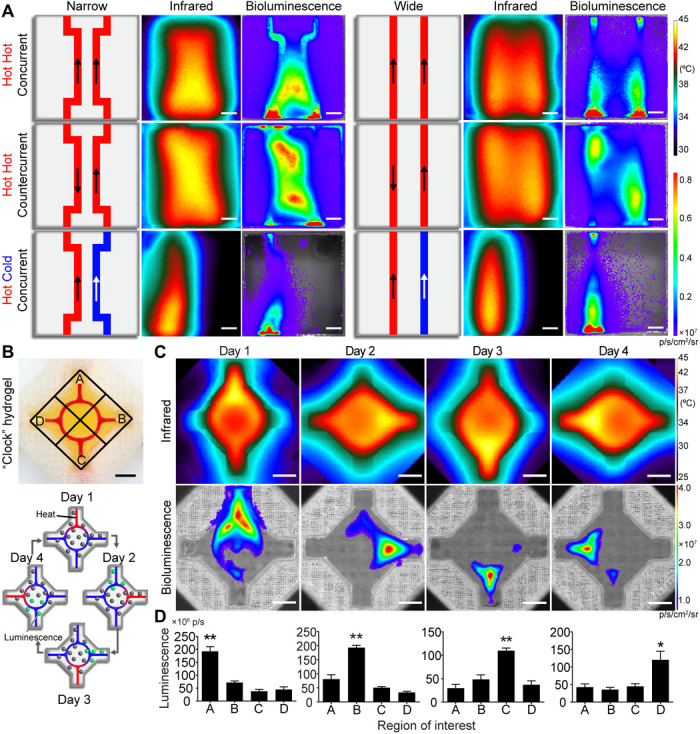
(A) Heat exchanger inspired designs for various flow directions, fluid temperatures, and channel architectures (schematics; left and center). Representative thermal (middle) and bioluminescent (right) images demonstrate spatial tunability of thermal and gene expression patterning. Scale bars, 5 mm. (B) Photographic image of four-armed clock-inspired hydrogel used for dynamic activation (top; channel filled with red dye). Each inlet is assigned to a local region (A to D). Schematic shows the spatial and dynamic heating pattern for the 4-day study (bottom). (C) Representative infrared (top) and bioluminescence expression (bottom) images for dynamic hydrogel activation at each day during the time course. (D) Quantification of local bioluminescent signals from regions of interest corresponding to each day of heating. Across all 4 days, regions corresponding to perfused arms had higher bioluminescent signals than nonperfused arms (n = 5, data are mean luminescence ± standard error; *P < 0.05 and **P < 0.01 by one-way ANOVA followed by Tukey’s post hoc test).
As biological gene expression patterns are transient and fluctuating, we next tested whether thermofluidics could dynamically localize regions of gene expression over time. To do this, we printed clock-inspired constructs, in which four separate inlets converged on a circular channel (Fig. 4B, top). We then perfused heated fluid through each inlet over four consecutive days (Fig. 4B, bottom) and imaged tissues for bioluminescence. Bioluminescent images demonstrated statistically significant luciferase up-regulation for regions surrounding heated inlets compared to nonheated inlet regions on all 4 days (Fig. 4, C and D.) Together, our results illustrate that by exploiting heat transfer design principles, thermofluidics enables user-defined spatial and dynamic patterning of mesoscale gene expression patterns in 3D artificial tissues.
Maintenance of gene patterning in vivo
To test whether gene patterning could be maintained after engraftment of artificial tissues in vivo, we stimulated tissues with HEAT and implanted these tissues into athymic mice. All tissues contained HEK293T cells expressing fLuc under the control of the heat-inducible HSPA6 promoter. All tissue constructs contained a single channel and were stimulated in one of three ways: (i) thermofluidic perfusion at 44°C for 60 min, (ii) bulk heating in a cell culture incubator at 44°C for 60 min, or (iii) bulk exposure in a cell culture incubator to 37°C. Tissues were implanted into mice immediately after heating, and bioluminescence imaging was performed 24 hours later. We found that thermofluidic spatial control of gene expression was maintained after in vivo tissue engraftment (Fig. 5A and movie S1).
Fig. 5. HEAT gene patterning is maintained after tissue implant in vivo.
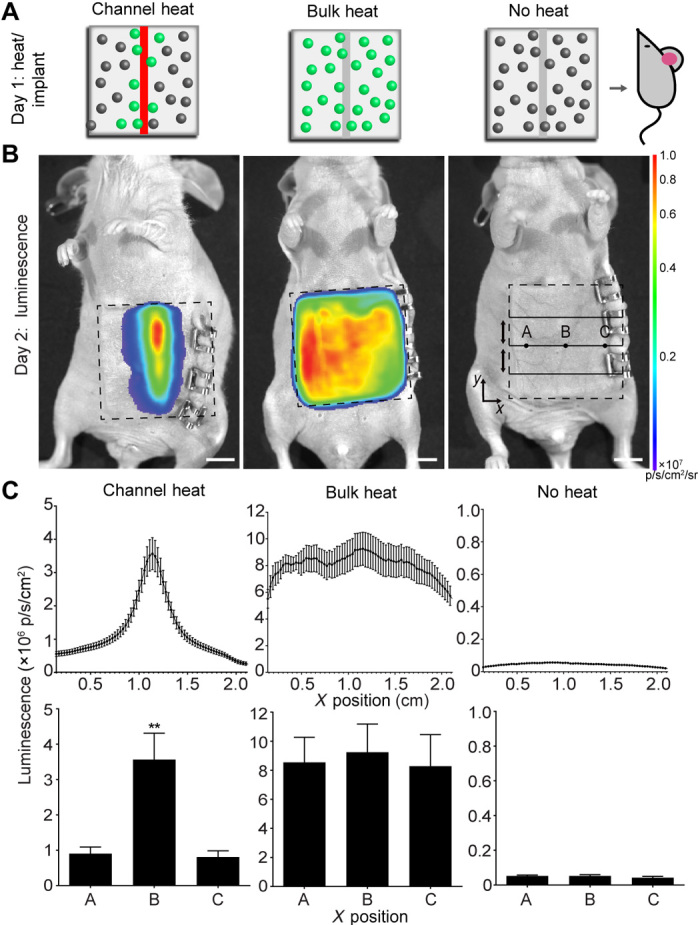
(A) Artificial tissues with embedded heat-inducible fLuc HEK293T cells received 44°C thermofluidic heating (channel heat, n = 5), 44°C global heating (bulk heat, n = 3), or remained at 37°C (no heat, n = 3) for 1 hour before immediate implantation into athymic mice. (B) Bioluminescence from implanted hydrogels (dashed lines) showed region specific signal only in channel heated hydrogels. (C) Average line profiles (top) across the width (x) of the hydrogel for inlet, middle, and outlet positions show that only channel heated gels induced a spatially coordinated response that was statistically significant (bottom) between the center (position B) and edges of the hydrogel (position A and C; channel heat, n = 5; bulk heat, n = 3; no heat, n = 3; data are mean luminescence ± standard error; **P < 0.01, by one-way ANOVA.
Spatial control of Wnt/β-catenin signaling pathway
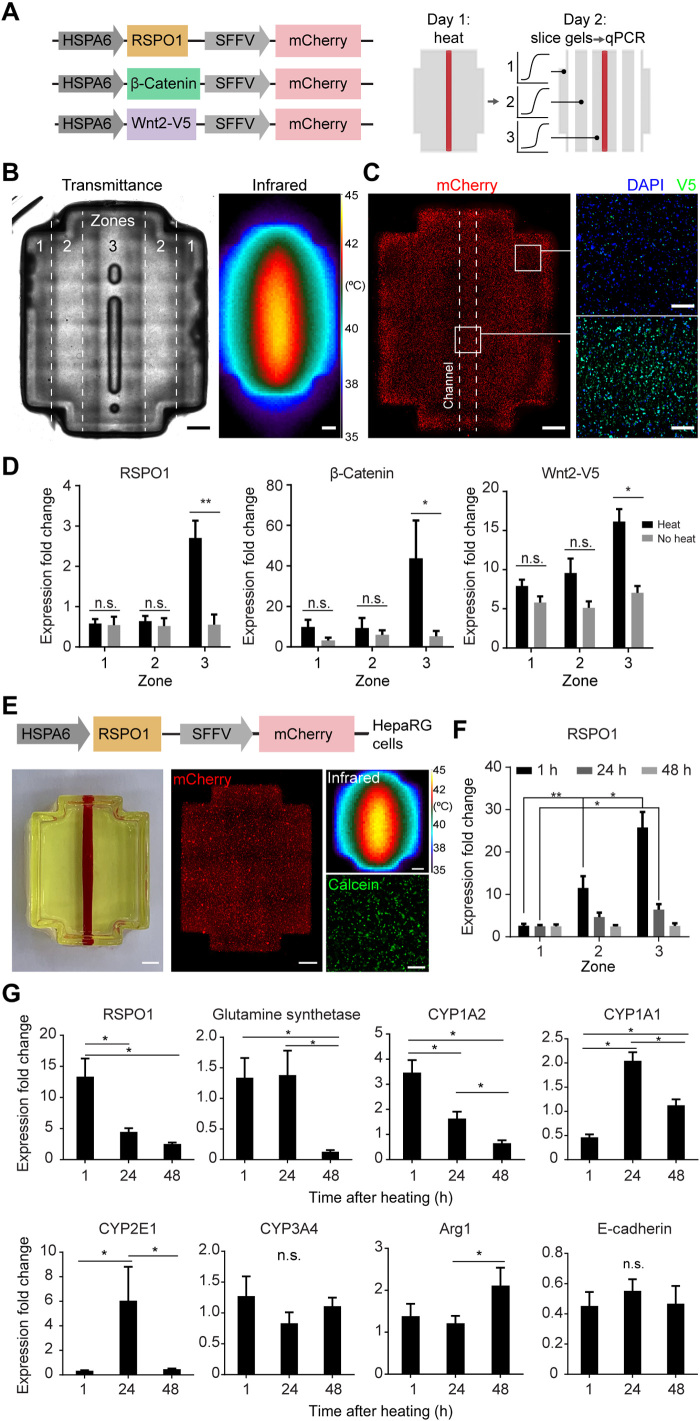
We next sought to demonstrate the modularity of our system for spatially regulating expression of the Wnt/β-catenin signaling pathway, which directs diverse aspects of embryonic development, tissue homeostasis, regeneration, and disease (20). We engineered heat-inducible constructs to drive expression of three genes in the Wnt/β-catenin signaling pathway: (i) R-spondin-1 (RSPO1), a potent positive regulator of Wnt/β-catenin signaling (21); (ii) β-catenin, a critical transcriptional coregulator that translates to the nucleus upon canonical Wnt signaling (22); and (iii) Wnt-2, a ligand that binds to membrane-bound receptors to activate the Wnt/β-catenin signaling pathway. The Wnt-2 gene was also tagged with V5 (23). We engineered lentiviral constructs in which RSPO1, β-catenin, or Wnt2-V5 is driven by the heat-inducible HSPA6 promoter, and mCherry is driven by a constitutive promoter [spleen focus-forming virus (SFFV); Fig. 6A]. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis of each engineered cell line for mCherry expression relative to GAPDH expression suggested lentiviral integration (fig. S10A). We then printed artificial tissues containing heat-inducible β-catenin, RSPO1, or Wnt2 HEK293T cells and a single fluidic channel (Fig. 6B). Constructs were heated fluidically and then sliced into longitudinal zones (Fig. 6, A and B) to analyze expression of the Wnt family gene expression by RT-qPCR. Representative artificial tissues contained mCherry+ cells across the tissue (Fig. 6C). Immunostaining for the V5 tag fused to Wnt2 appeared higher near the heated channel compared to the gel periphery (Fig. 6C). RSPO1, β-catenin, or Wnt2 expression was highest in the zone surrounding the heated channel (Fig. 6D). These results show that HEAT can be leveraged to activate expression of various family members of the Wnt/β-catenin signaling pathway.
Fig. 6. Thermofluidic Wnt regulation in engineered HEK293T and HepaRG cells.
(A) Schematics of lentiviral constructs (left) and thermofluidic HEK293T tissue experiments (right). (B) Transmittance image of cellularized construct after printing (left; zones indicated by dashed lines). Infrared image of construct during heating (right). Scale bars, 1 mm. (C) mCherry+ HEK293T cells in printed tissues (left). Scale bars, 1 mm. Images of thermofluidically heated Wnt2 constructs after immunostaining for V5 tag (coexpressed with Wnt2; right; images taken near the tissue’s channel and periphery as indicated by insets). Scale bars, 200 μm. (D) Wnt family genes were up-regulated in zone 3 of thermofluidically perfused gels compared to controls (n = 4, mean fold change ± standard error; *P < 0.05 and **P < 0.01 by two-way ANOVA followed by Tukey’s multiple comparison test). (E) Differentiated HepaRG cells were engineered with a heat-inducible RSPO1 construct (schematic, top) and printed in single-channel hydrogels (photograph, left). Scale bars, 1 mm. After heating (infrared), HepaRGs remained viable in printed constructs (calcein). Scale bar, 200 μm. (F) Thermofluidically heated RSPO-1 HepaRG hydrogels were dissected into zones 1 to 3 based on distance from the heat channel for RT-qPCR analysis at 1, 24, and 48 hours after heating. Expression fold change was normalized to no heat control samples. qPCR analysis of RSPO-1 across dissected zones (n = 5 to 10, data are mean fold change ± standard error; *P < 0.05 by one-way ANOVA followed by Tukey’s multiple comparison test). (G) RT-qPCR analysis of pooled RNA across all zones at each time point for pericentral associated genes, glutamine synthetase, CYP1A2, CYP1A1, CYP2E1, and CYP3A4, and periportal/midzonal genes, Arg1 and E-cadherin (n = 15 to 30, data are mean fold change ± standard error, **P < 0.01 and *P < 0.05 by one-way ANOVA followed by Tukey’s multiple comparison test). Photo credit: Daniel Corbett, University of Washington. n.s., not significant.
Effect of thermofluidic activation of RSPO1 on the expression of key metabolic liver enzymes
We reasoned that the ability to activate expression of Wnt/β-catenin signaling pathway members could be useful for the emerging human “organ-on-a-chip” field by affecting functional cellular phenotypes in vitro. To test this, we turned to the liver, which performs hundreds of metabolic functions essential for life, including central roles in drug metabolism. To carry out these functions, hepatocytes divide the labor, with hepatocytes in different spatial locations performing different functions, a phenomenon called liver zonation. Recent studies have shown that liver zonation is regulated at the molecular level by Wnt/β-catenin signaling (22), with higher Wnt activity associated with a pericentral vein phenotype and lower Wnt activity characteristic of a periportal phenotype. However, the extent to which different members of this pathway affect human zonated hepatic phenotypes remains unclear. A better understanding of this process would accelerate development of zonated human liver models for hepatotoxicity and drug metabolism studies.
We hypothesized that thermofluidic activation of RSPO1 in human hepatic cells would be sufficient to activate zonated hepatic gene expression profiles, as ectopic expression of RSPO1 in mouse liver has recently been shown to induce a pericentral zonation phenotype in vivo (24). To test this hypothesis, we transduced human HepaRG cells, an immortalized human hepatic cell line that retains characteristics of primary human hepatocytes, with our lentiviral construct in which HSPA6 drives RSPO1, and SFFV drives mCherry (Fig. 6E). Transduced human hepatic cells were then printed in artificial tissues with a single fluidic channel, to mimic central lobular placement of the central vein (Fig. 6E). Constructs were heated fluidically and then sliced into zones (Fig. 6A), and gene expression was measured by RT-qPCR (Fig. 6F). Fold up-regulation values were normalized to identically fabricated control artificial tissues maintained at 37°C. We found that RSPO1 expression increased in a dose-dependent and spatially defined manner, with expression in zone 3 nearest the channel (“central vein”) 10-fold higher than in zone 1 by 1 hour after heating. RSPO1 expression was transient, falling with each day after heating, similar to our luciferase studies (Fig. 4C and fig. S5C). Thermofluidic activation of RSPO1 induced expression of key pericentral marker genes, including glutamine synthetase, an enzyme involved in nitrogen metabolism, and the cytochrome P450 (CYP) drug-metabolizing enzymes CYP1A2, CYP1A1, and CYP2E1 relative to control tissues that were not heated, although with varied timing and without spatial localization in this study (Fig. 6G and fig. S10). Expression of pericentral drug-metabolizing enzyme CYP3A4 was not induced with heating, consistent with other studies in which adding Wnt3a ligand to primary human hepatocyte cultures did not alter CYP3A4 expression (25). Periportal marker E-cadherin was not induced, but periportal/midzonal gene Arg1 increased at 48 hours, especially in the zone 2 midzonal region (fig. S10). Together, these studies contribute a fundamental understanding of how various liver zonation genes are induced by RSPO1 activation in human hepatic cells.
DISCUSSION
In this study, we demonstrate that thermal patterning via bioprinted fluidics can directly pattern gene expression in 3D artificial tissues. A key advantage of the HEAT method is that it leverages the recent explosion in accessible additive manufacturing tools (16, 26, 27) by using open-source bioprinting methods that are readily available to the broader community. Furthermore, the entire patterned network is stimulated nearly simultaneously (as opposed to sequentially by time-intensive rastering), and this parallel stimulation can be sustained for exposure times required to trigger gene expression. Together, the sheer rapidity and highly parallel nature of this process enable spatial and dynamic genetic patterning at length scales and depths not previously possible in 3D artificial tissues.
Most previous methods to elicit cellular signaling in artificial tissues have focused on tethering extracellular cues to hydrogels (28, 29). Innovations in stimuli-responsive or “smart” biomaterials enabled activation of these chemistries by exogenous physical stimuli, such as light, to control the spatial position and timing of extracellular cues (30, 31). Although useful, these material-focused methods are unlikely to provide complete control even in fully defined starting environments because cells rapidly remodel their microenvironments (32). Moreover, these technologies offer an imprecise means to control downstream transcription because many, often unknown, intermediary steps modify intracellular signal transduction before gene activation. Our thermofluidic approach provides a complementary new technology to these methods that target extracellular signals by facilitating spatiotemporal control at the intracellular genetic level.
While our studies here reveal the potential power of HEAT for gene patterning, the first-generation system presented here does have limitations in its ability to fully control heat transfer both spatially and temporally. In our studies here, we found that channels up to 30 mm long (but no longer) could achieve hyperthermic temperature ranges along the entire channel length. Furthermore, the effect of heat-mediated stimulation on gene expression was transient. These limits could be overcome through a variety of design modifications. For example, the hydrogel or perfusate’s thermal conductivity could be increased by materials engineering to extend patterning area or length, such as by cross-linking metal nanoparticles into the polymer backbone as has been done before for other applications (33). To achieve different activation temperatures or dynamics, further genetic engineering of the heat shock promoter or other heat-activatable gene switches could be used (14). Thus, we envision that our initial system here will establish an important foundation that leads to a new family of studies that will ultimately describe a far greater “design space” for thermofluidic patterning.
To fully realize the vision of precision-controlled 3D artificial tissues, a diverse toolkit of orthogonal physical delivery and molecular remote control agents will likely be needed (34, 35). Thermofluidics could be coupled with other tissue engineering strategies that program extracellular (3, 29–31) or intracellular (10, 14) signal presentation, cell patterning (36), or tissue curvature (37). Thermofluidics could also be used orthogonally with other remote control agents, such as those leveraging small-molecule (12), ultrasound (38), radio wave (39), magnetic (40), or light-based activation (41). Coupled with rapid advances in gene editing (10), synthetic morphogenesis (2, 3), and stem cell technology (4, 5), thermofluidics could be useful for spatially and temporally activating genes across tissues to drive cell proliferation, fate, or assembly decisions. While we demonstrate utility for activating Wnt/β-catenin signaling pathway genes here, this approach could be rapidly adapted to activate any gene of interest. In our studies, we demonstrate one application of this approach by driving human hepatic cells toward a more pericentral liver phenotype in 3D artificial tissues. In doing so, we gain fundamental insights into how activation of Wnt agonist RSPO1 regulates expression of various metabolic zonation genes. These findings have important implications for developing both organ-on-chip systems for pharmacology and hepatotoxicity, as well as artificial tissues for human therapy. By blurring the interface between the advanced fabrication and biological realms, thermofluidics creates a new avenue for bioactive tissues with applications in both basic and translational biomedicine.
MATERIALS AND METHODS
Materials and photopolymer synthesis
Poly(ethylene glycol) diacrylate (PEGDA; 6000 Da) and lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) were prepared as previously described (16, 42). Gelatin methacrylate (GelMA) was synthesized as previously described, with slight modifications (43). Methacrylic anhydride was added dropwise to gelatin dissolved in carbonate-bicarbonate buffer at 50°C for 3 hours, followed by precipitation in ethanol. The precipitate was allowed to dry, dissolved in phosphate-buffered saline (PBS), frozen at −80°C, and then lyophilized for up to 1 week. GelMA was stored at −20°C until use. Tartrazine (Sigma-Aldrich T0388, St. Louis, MO, USA) was added to prepolymer solutions as a photoabsorber to increase print resolution as previously described (16). Prepolymer mixtures for all cellular studies contained 7.5 wt % 6 K PEGDA, 7.5 wt % GelMa with 17 mM LAP, and 1.591 mM tartrazine. For characterization of heat transfer with respect to gel density, the overall polymer weight percent was varied while holding the ratio of 6 K PEGDA to GelMA constant at 50:50 (for example, 20 wt % = 10 wt % 6 K PEGA + 10 wt % GelMa).
Model design
Hydrogels with perfusable channel networks were designed in an open-source 3D computer graphics software Blender 2.7 (Blender Foundation, Amsterdam, Netherlands) or in SolidWorks (Dassault Systemes SolidWorks Corp., Waltham, MA).
3D printing
Our stereolithography apparatus for tissue engineering bioprinting system was used in this study (16). Briefly, the system contains three major components: (i) a Z-axis with stepper motor linear drive, (ii) an open-source RepRap Arduino Mega Board (UltiMachine, South Pittsburg, TN) microcontroller for Z-axis control of the build platform, and (iii) a projection system consisting of a DLP4500 Optical Engine with a 405-nm light-emitting diode output (Wintech, Carlsbad, CA) connected to a laptop for photomask projection and motor control. The projector is placed in front of the Z-axis, and a mirror is positioned at 45° to the projection light path to reflect projected images onto the build platform. A sequence of photomasks based on a 3D model is prepared using Creation Workshop software (www.envisionlabs.net/), which also controls the Z-axis movement of the build platform. Printing is achieved by curing sequential model layers of the photosensitive prepolymer. All printing was conducted in a sterile tissue culture hood. For visualization of channel networks, we perfused open channels with ultraviolet fluorescent tonic water or India ink dyes (P. Martin’s, Oceanside, CA).
In-line fluid heating system
To control temperature distribution in perfused hydrogels, an in-line fluid heater was developed to prewarm perfusate solutions before infusion in hydrogel channel networks. The fluid heater consists of four components: (i) an adjustable dc Power Supply (Yescom USA Inc., City of Industry, CA), (ii) a cylindrical cartridge heater (Uxcell, Hong Kong), (iii) perfusate tubing (peroxide-cured silicone tubing, Cole Parmer, Vernon Hills, IL), and (iv) a syringe pump (Harvard Apparatus, Holliston, MA). To construct the in-line fluid heater, perfusate tubing was connected to the syringe pump for flow rate control, while the cartridge heater was connected to the power supply for heating control. Perfusate tubing was then wounded around the cylindrical cartridge heater, allowing for heat transfer from the heater into the flowing perfusate. The temperature of the fluid was then controlled by changing the flow rate or heater power. In all studies, we used PBS (Thermo Fisher Scientific, Hampton, NH) for the perfusate solution.
Hydrogel fluidic connections
To establish a fluidic connection between the heating system and hydrogel channel networks, we used custom-designed 3D printed perfusion chips printed on a MakerGear M2 3D printer (MakerGear, Beachwood, OH) in consumer-grade poly(lactic acid) plastic filament. Perfusion chips were fabricated with (i) an open cavity to insert 3D bioprinted hydrogels and (ii) attachment ports for fluid-dispensing nozzles. The outflow of the fluid heater was fitted with a male luer hose barb (Cole Parmer) connected to a flexible tip, polypropylene nozzle (Nordson EFD, East Providence, RI) and inserted into 3D printed attachment ports. Hydrogels were then inserted to perfusion chips, and proper fluidic connections were ensured before beginning perfusion. Model files for 3D printed perfusion holders are provided in the open repository data of our previously published work (16).
Infrared thermography
Fluid temperature and heat distribution were measured in perfused hydrogels by infrared thermography. Images were acquired by an uncooled microbolometer-type infrared camera (FLIR A655sc, Wilsonville, OR) that detects a 7.5- to 14.0-μm spectral response with a thermal sensitivity of <0.05°C and analyzed for temperature values using the FLIR ResearchIR software (Wilsonville, OR).
Computational models
We built finite element models of perfused hydrogels in COMSOL 4.4 software (COMSOL AB, Burlington, MA). Simulations were run under transient conditions using the “Conjugate heat transfer” module and 3D printed hydrogel and housing geometries to predict the temperature distribution. The model was based on (i) forced convective heat transfer from the perfusion channel to the hydrogel volume and (ii) conductive heat transfer within the hydrogel volume.
Equation for (i): Heat transfer in a fluid
Where ρ is the fluid density, T is the temperature, Cρ is the heat capacity at constant pressure, u is the velocity field, α is the thermal expansion coefficient, pA is the absolute pressure, τ is the viscous stress tensor, S is the strain rate tensor, k is the fluid thermal conductivity, and Q is the heat content.
Equation for (ii)
Where ρ is the hydrogel density, T is the temperature, k is the hydrogel thermal conductivity, and Q is the heat content.
Material properties of both the hydrogel and perfusate were modeled as water. Heat flux boundary conditions were included to model heat loss to the ambient environment, heat transfer coefficients of 5 and 30 W/(m2 * K) were applied to the sides and upper boundaries of the hydrogel, respectively, with an infinite temperature condition of 22.0°C applied for all boundaries. Boundary temperature and fluid inflow conditions at the channel inlet were used to simulate the effect of changing perfusate temperature and flow rate, respectively. Model geometry was manipulated for studies on channel length and channel branching. Prescribed external temperature was varied for ambient temperature studies.
Cell culture
HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning, NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco) and 1% (v/v) penicillin-streptomycin (GE Healthcare Life Sciences, WA, USA). Differentiated HepaRG cells (Fisher Scientific) were maintained at confluence in six-well plates at a density of 2 × 106 cells per well in Williams E media (Lonza, MD, USA) supplemented with 5× HepaRG Thaw, Plate & General Purpose Medium Supplement (Fisher), and 1% (v/v) Glutamax (Fisher).
Construction of heat-sensitive reporter gene cells
A vector containing a 476-bp version of the human HSPA6 promoter driving expression of fLuc reporter gene (gift of R. Schez Shouval from the Weizmann Institute of Science) was packaged into lentivirus using helper plasmids pMDLg/pRRE (Addgene no. 12251), pMD2.G (Addgene no. 12259), and pRSV-Rev (Addgene no. 12253) by cotransfection into HEK293T cells. Crude viral particles were harvested after 48 hours of transfection. For viral transduction, crude lentivirus was diluted 1:20 in DMEM containing polybrene (6 μg/ml; Invitrogen), added to competent HEK293T cells in six-well tissue culture plates, and incubated overnight (Corning). The next day, virus-containing media was removed and replaced with fresh DMEM containing 10% FBS. After transduction, cells were heat-activated (see below) and flow-sorted to obtain a pure cell population.
Heat treatment
To activate transgene expression under the HSPA6 promoter, engineered HEK293T cells were exposed to varying levels of hyperthermia in 2D and 3D. For 2D heat treatment studies, cells were seeded at 8 × 104 cells/cm2 in tissue culture plates 1 day before heat treatment. The next day, tissue culture plates were exposed to indicated heat treatments in thermostatically controlled cell culture incubators. Temperature was verified with a secondary method by a thermocouple placed inside the incubator. Upon completion of heat treatment, cells were returned to a 37°C environment and sorted or analyzed at later time points. For the luminescent transient studies in fig. S4B, cells were lysed in TE buffer [100 mM tris and 4 mM EDTA (pH 7.5)] and stored at 4°C until imaging. For the pulsed activation studies in fig. S4C, cells received two heat shocks as described previously at days 0 and 3. Luminescence was quantified across days 1 to 4 and normalized to cell counts from tissue culture plates that were processed in parallel according to each experimental temperature. For 3D heat shock studies, cells were encapsulated and printed in 3D perfusable hydrogels (see below) 1 day before heating. 3D hydrogels were then heat-perfused in a room temperature environment. Hydrogel temperature was monitored continuously with the infrared camera, and small adjustments to heater power were made as necessary to maintain a stable temperature profile. During perfused heating, outlet medium was continuously discarded. Upon completion of perfused heating, hydrogels were dismounted from the perfusion chips and returned to a cell culture incubator.
Cell encapsulation and printing of cell-laden hydrogels
Cultured HEK293T cells were detached from tissue culture plates with 0.25% trypsin solution (Corning), counted, centrifuged at 1000 rpm for 5 min, and resuspended in liquid prepolymer (7.5 wt % 6 K PEGDA, 7.5 wt % GelMA, 17 mM LAP, and 1.591 mM tartrazine). For characterization of heat transfer with respect to cell density, cells were encapsulated in prepolymer mixtures at final densities from 0 to 24 × 106 cells ml−1 before printing. For HEK293T expression studies, cells were encapsulated at a final density of 6 × 106 cells ml−1. For HepaRG studies, cells were encapsulated at a final density of 2.5 × 106 cells ml−1. Printing was performed as previously described under DLP light intensities ranging from 17 to 24.5 mW cm−2, with bottom layer exposure times from 30 to 35 s and remaining layer exposure times from 12 to 17.5 s. Upon print completion, fabricated hydrogels were removed from the platform with a sterile razor blade and allowed to swell in cell culture media. Hydrogels were changed to fresh media 15 min after swelling and allowed to incubate overnight. Media was replaced the following morning. We tested the viability of both HEK293T and HepaRG cells following 3D printing by incubating cell-laden hydrogels with Live/Dead viability/cytotoxicity kit reagents (Life Technologies, Carlsbad, CA) according to manufacturer’s instructions. Fluorescence imaging was performed on a Nikon Eclipse Ti inverted epifluorescent microscope, and images were quantified using ImageJ’s built-in particle analyzer tool [National Institutes of Health, Bethesda, Maryland].
Bioluminescent imaging
To visualize the magnitude and spatial localization of heat-induced luciferase expression, bioluminescence imaging was performed on heated cells and hydrogels using the in vivo imaging system (IVIS) Spectrum imaging system (PerkinElmer, Waltham, MA). Immediately before bioluminescence imaging, cell culture media was changed to media containing d-luciferin (0.15 mg/ml; PerkinElmer), and images were taken every 2 min until a bioluminescent maximum was reached. Images were analyzed using Living Image software (PerkinElmer). Luminescent imaging was performed from a “top-down” view (perspective orthogonal to hydrogel channel axis) for most studies. For cross-sectional images in fig. S8, hydrogels were manually sliced, incubated in luciferin containing media, and imaged under cross-section view (perspective parallel to hydrogel channel axis).
Pixel-to-pixel temperature-to-expression correlation
Data for the expression versus temperature plot was obtained by aligning thermal and bioluminescent images using MATLAB. To align the images, four reference points corresponding to the corners of the hydrogel were manually selected on both thermal and bioluminescence images. Then, an orthogonal transformation was performed on each image to align the corners of the hydrogel, after which the areas outside the selection were cropped. Pixel values from each image were then plotted against each other to produce the expression versus temperature plot.
In vivo implantation of HEAT-modulated artificial tissues
Heat-inducible cells were generated as previously described and embedded into 3D-printed artificial tissues with single channels before being placed at 37°C overnight. The next day, artificial tissues received either thermofluidic heat stimulation via flow of 44°C biocompatible fluid at 1.0 ml min−1 for 60 min (n = 5), global heat stimulus by being placed in a 44°C tissue culture incubator for 60 min (n = 3), or were maintained in a 37°C tissue culture incubator (n = 3). The artificial tissues were then immediately implanted subcutaneously on the ventral side of female NCr nude mice aged 8 to 12 weeks old (Taconic). Twenty-four hours after implantation, mice were anesthetized and injected with luciferin (15 mg/ml; PerkinElmer, Waltham, MA). Bioluminescence was then recorded via the IVIS Spectrum Imaging System (PerkinElmer). For 3D images, a custom 3D imaging unit developed by A. D. Klose and N. Paragas (44) (InVivo Analytics, New York, NY) was used. Briefly, anesthetized mice were placed into body-fitting animal shuttles and secured into the custom 3D imaging unit that uses a mirror gantry for multiview bioluminescent imaging. Collected images were then compiled and overlaid onto a standard mouse skeleton for perspective.
Spatial analysis of in vivo IVIS images
Line profiles in the x-direction across the inlet, middle, and outlet of 2D IVIS projection images from artificial gels were generated using Living Systems software (PerkinElmer, Waltham, MA). The three line profiles (inlet, middle, and outlet) from each artificial tissue were then averaged together with the average line profiles from the other artificial gels within each respective group (experimental group, n = 5; positive control group, n = 3; negative control group, n = 3). The average line profile of each group was then plotted, and average radiance values from positions 0.75 cm from the center of the channel (denoted positions A and C) were then statistically compared to the average radiance value at the center of the channel (position B) within each group by one-way analysis of variance (ANOVA).
Generation of Wnt constructs and cells
Lentiviral constructs in which the HSPA6 promoter drives a Wnt family gene were subcloned using Gibson assembly by the UW BioFab facility. Human β-catenin pcDNA3 was a gift from E. Fearon (Addgene plasmid no. 16828; http://n2t.net/addgene:16828; RRID: Addgene_16828) (45). Active Wnt2-V5 was a gift from X. He (Addgene plasmid no. 43809; http://n2t.net/addgene:43809; RRID:Addgene_43809) (46). RSPO1 was subcloned using a complementary DNA (cDNA) clone plasmid. (Sino Biological, Beijing, China). All plasmids contained a downstream cassette in which a constitutive promoter (SFFV) drives the reporter gene mCherry (gift from G. A. Kwong, Georgia Institute of Technology). Lentivirus was generated by cotransfection of HEK293Ts with HSPA6➔Wnt transfer plasmids with third-generation packaging plasmids (pMDLg/pRRE, pMD2.G, pRSV-REV) in DMEM supplemented with 0.3% Xtreme Gene Mix (Sigma-Aldrich). Crude virus was harvested starting the day after initial transfection for four consecutive days. For viral transduction, HEK293Ts at 70% confluency and HepaRGs at 100% confluency were treated with crude virus containing polybrene (8 μg/ml; Sigma-Aldrich) for 24 hours. Five days following viral transduction, mCherry+ HEK293Ts were sorted from the bulk population by flow cytometry at the UW Flow analysis facility. HepaRGs were not sorted by flow cytometry. mCherry expression in positive HEK293T cell populations was performed using RT-qPCR.
Wnt up-regulation in HEAT-induced constructs
To quantify Wnt regulator levels in HEAT-treated gels, HEK293Ts and HepaRGs for a given construct were encapsulated and heated in 3D hydrogels as previously described. “No heat control” samples remained at 37°C in tissue culture incubators until RNA isolation. One to 48 hours following heat treatment, hydrogels were manually sliced into corresponding zones (1 to 3) and RNA was isolated using phenol-chloroform extraction (47). cDNA was synthesized using the Superscript III First-Strand Synthesis Kit (Thermo Fisher Scientific), and qPCR was performed using iTaq Universal SYBR Green Supermix (Biorad, Hercules, CA) on the 7900HT Real Time PCR System (Applied Biosystems, Waltham, MA). Primers for Wnt and housekeeping genes were designed and synthesized by Integrated DNA Technologies (Coraville, IA). Relative gene expression was normalized against the housekeeping gene 18S RNA calculated using the ∆∆Ct method. Data are presented as the mean relative expression ± SEM. Data for HEK293T studies were normalized to relative expression of the Wnt target in 2D culture at 37°C. Data for HEK293T mCherry expression were normalized to 18 s RNA and compared to GAPDH (also normalized to 18S RNA) expression levels. Data for HepaRG studies were normalized by relative expression of the Wnt target or pericentral/periportal gene marker to no heat control samples.
V5 staining/clearing in Wnt2 HEAT gel
HSPA6➔Wnt2/V5 gels were fixed in 4% paraformaldehyde 24 hours postheating. For staining, samples are blocked overnight at room temperature in 1% bovine serum albumin, 1% normal donkey serum, 0.1 M tris, and 0.3% Triton X-100 with agitation. After blocking, samples are incubated in Anti-V5 tag antibody (Abcam, ab27671) diluted 1:100 in fresh blocking buffer and 5% dimethyl sulfoxide for 24 hours at 37°C and agitation. Samples are washed and then incubated in secondary antibody diluted 1:500 in fresh blocking buffer and 5% dimethyl sulfoxide overnight at 37°C and agitation. After incubation, samples are washed in PBS + 0.2% Triton X-100 + 0.5% 1-thioglycerol three times at room temperature and agitation, changing fresh buffer every 2 hours. To begin clearing, samples are incubated in clearing enhanced 3D (Ce3D) (48) solution at room temperature overnight with agitation protected from light. 4′,6-Diamidino-2-phenylindole is diluted 1:500 in the Ce3D solution to counter stain for nuclei. To 3D image the cleared samples, the gels are placed on glass-bottom dishes and imaged overnight on an SP8 Resonant Scanning Confocal Microscope.
Statistics
Data in graphs are expressed as the SE or SEM ± SD, as denoted in figure legends. Statistical significance was determined using two-tailed Student’s t test for two-way comparisons or one-way ANOVA or two-way ANOVA followed by Dunnett’s, Sidak’s, or Tukey’s multiple comparison test.
Supplementary Material
Acknowledgments
Funding: This research was funded by NIH NHLBI grant DP2HL137188 (K.R.S.), the NIH NIBIB Cardiovascular Training Grant (D.C.C., T32EB001650), the NIH Environmental Pathology and Toxicology Training (W.B.F., T32ES007032), the NSF Graduate Research Fellowship (B.G., 1450681), the Ford Foundation Predoctoral Fellowship (C.E.O.), the Washington Research Foundation postdoctoral fellowship (M.C.R.), the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (J.S.M.), and the Gree Foundation (I.B., K.R.S., and C.A.D.). We thank the large number of open-source and related projects that critically facilitated this work, including Arduino.cc, RepRap.org, UltiMachine.com, Ultimaker.com, Blender.org, Python.org, ImageMagick.org, Git, NIH ImageJ, Fiji.sc, and the NIH 3D Print Exchange. We thank J. Jang, H. Reincke, P. Grosjean, Y. Zheng, B. Ratner, and A. Emery for helpful discussions. We thank D. Hailey from the UW Garvey Imaging Core for assistance with imaging, S. Halabiya, and C. Cordray for assistance with subcloning, and N. Paragas for assistance with the IVIS Imaging System. Author contributions: D.C.C. and K.R.S conceived and initiated the project. All authors contributed to experimental design, experimental planning, experimental execution, data analysis, and to the writing of the manuscript. Competing interests: J.S.M. and B.G. are co-founders of and hold an equity stake in Volumetric Inc., which is a startup company. D.C.C., K.R.S., B.G., and J.S.M. are listed as co-inventors in a pending U.S. Patent application 62/756,106. Remaining co-authors report that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data and hydrogel design files for 3D printing are available in Zenodo (https://zenodo.org/record/4014310#.X1Fxz8hKhPY). Computer code generated to analyze data sets is available in Zenodo. Data, hydrogel design files, and computer code are available in Zenodo. Additional data related to this paper may be requested from the authors. Computer code generated to analyze datasets is available in Zenodo. Data, hydrogel design files, and computer code are available in Zenodo.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/40/eabb9062/DC1
REFERENCES AND NOTES
- 1.Green J. J., Elisseeff J. H., Mimicking biological functionality with polymers for biomedical applications. Nature 540, 386–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogt N., Customizing cell-cell communication. Nat. Methods 13, 285 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Morsut L., et al. , Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell 118, 6072–6078 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warmflash A., Sorre B., Etoc F., Siggia E. D., Brivanlou A. H., A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martyn I., Kanno T. Y., Ruzo A., Siggia E. D., Brivanlou A. H., Self-organization of a human organizer by combined Wnt and Nodal signaling. Nature 558, 132–135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J.-W., Xi J. J., Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Cell Res. 23, 465–472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrangou R., Doudna J. A., Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Ye H., Baba M. D., Peng R., Fussenegger M., A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 332, 1565–1569 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Polstein L. R., Juhas M., Hanna G., Bursac N., Gersbach C. A., An engineered optogenetic switch for spatiotemporal control of gene expression, cell differentiation, and tissue morphogenesis. ACS Synth. Biol. 6, 2003–2013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ntziachristos V., Going deeper than microscopy : The optical imaging frontier in biology. Nat. Methods 7, 603–614 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Deans T. L., Singh A., Gibson M., Elisseeff J. H., Regulating synthetic gene networks in 3D materials. Proc. Natl. Acad. Sci. 109, 15217–15222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipkin M., Hardy D., Measurement of some thermal properties of human tissues. J. Appl. Physiol. 7, 212–217 (1954). [DOI] [PubMed] [Google Scholar]
- 14.Piraner D. I., Abedi M. H., Moser B. A., Lee-Gosselin A., Shapiro M. G., Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat. Chem. Biol. 13, 75–80 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Miller I. C., Gamboa Castro M., Maenza J., Weis J. P., Kwong G. A., Remote control of mammalian cells with heat-triggered gene switches and photothermal pulse trains. ACS Synth. Biol. 7, 1167–1173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigoryan B., Paulsen S. J., Corbett D. C., Sazer D. W., Fortin C. L., Zaita A. J., Greenfield P. T., Calafat N. J., Gounley J. P., Ta A. H., Johansson F., Randles A., Rosenkrantz J. E., Louis-Rosenberg J. D., Galie P. A., Stevens K. R., Miller J. S., Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 364, 458–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith R. C., Machluf M., Bromley P., Atala A., Walsh K., Spatial and temporal control of transgene expression through ultrasound-mediated induction of the heat shock protein 70B Promoter In Vivo. 706, 697–706 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Fedorov A. G., Viskanta R., Three-dimensional conjugate heat transfer in the microchannel heat sink for electronic packaging. Heat Mass Transf. 43, 399–415 (2000). [Google Scholar]
- 19.Dreano M., Brochot J., Myers A., Cheng-Meyer C., Rungger D., Voellmy R., Bromley P., High-level, heat-regulated synthesis of proteins in eukaryotic cells. Gene 49, 1–8 (1986). [DOI] [PubMed] [Google Scholar]
- 20.Nusse R., Clevers H., Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 21.de Lau W., Peng W. C., Gros P., Clevers H., The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 28, 305–316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kretzschmar K., Clevers H., Wnt/β-catenin signaling in adult mammalian epithelial stem cells. Dev. Biol. 428, 273–282 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Si-Tayeb K., Lemaigre F. P., Duncan S. A., Organogenesis and development of the liver. Dev. Cell 18, 175–189 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Rocha A. S., Vidal V., Mertz M., Kendall T. J., Charlet A., Okamoto H., Schedl A., The angiocrine factor Rspondin3 is a key determinant of liver xonation. Cell Rep. 13, 1757–1764 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Gerbal-Chaloin S., Dumé A.-S., Briolotti P., Klieber S., Raulet E., Duret C., Fabre J.-M., Ramos J., Maurel P., Daujat-Chavanieu M., The WNT/ β-catenin pathway is a transcriptional regulator of CYP2E1, CYP1A2, and Aryl hydrocarbon receptor gene expression in primary human hepatocytes. Mol. Pharmacol. 86, 624–634 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Lee A., Hudson A. R., Shiwarski D. J., Tashman J. W., Hinton T. J., Yerneni S., Bliley J. M., Campbell P. G., Feinberg A. W., 3D bioprinting of collagen to rebuild components of the human heart. Science 365, 482–487 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Miller J. S., Stevens K. R., Yang M. T., Baker B. M., Nguyen D.-H. T., Cohen D. M., Toro E., Chen A. A., Galie P. A., Yu X., Chaturvedi R., Bhatia S. N., Chen C. S., Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 11, 768–774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutolf M. P., Hubbell J. A., Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 23, 47–55 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Martino M. M., Briquez P. S., Güç E., Tortelli F., Kilarski W. W., Metzger S., Rice J. J., Kuhn G. A., Müller R., Swartz M. A., Hubbell J. A., Growth factors engineered for super-affinity to the extracellular matrix enhance tissue healing. Science 343, 885–889 (2014). [DOI] [PubMed] [Google Scholar]
- 30.DeForest C. A., Tirrell D. A., A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat. Mater. 14, 523–531 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Lee T. T., García J. R., Paez J. I., Singh A., Phelps E. A., Weis S., Shafiq Z., Shekaran A., del Campo A., García A. J., Light-triggered in vivo activation of adhesive peptides regulates cell adhesion, inflammation and vascularization of biomaterials. Nat. Mater. 14, 352–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loebel C., Mauck R. L., Burdick J. A., Local nascent protein deposition and remodelling guide mesenchymal stromal cell mechanosensing and fate in three-dimensional hydrogels. Nat. Mater. 18, 883–891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schexnailder P., Schmidt G., Nanocomposite polymer hydrogels. Colloid Polym. Sci. 287, 1–11 (2009). [Google Scholar]
- 34.Wu C.-Y., Roybal K. T., Puchner E. M., Onuffer J., Lim W. A., Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toettcher J. E., Voigt C. A., Weiner O. D., Lim W. A., The promise of optogenetics in cell biology: Interrogating molecular circuits in space and time. Nat. Methods 8, 35–38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens K. R., Ungrin M. D., Schwartz R. E., Ng S., Carvalho B., Christine K. S., Chaturvedi R. R., Li C. Y., Zandstra P. W., Chen C. S., Bhatia S. N., InVERT molding for scalable control of tissue microarchitecture. Nat. Commun. 4, 1847 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes A. J., Miyazaki H., Coyle M. C., Zhang J., Laurie M. T., Chu D., Vavrušova Z., Schneider R. A., Klein O. D., Gartner Z. J., Engineered tissue folding by mechanical compaction of the mesenchyme. Dev. Cell 44, 165–178.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan Y., Yoon S., Sun J., Huang Z., Lee C., Allen M., Wu Y., Chang Y.-J., Sadelain M., Shung K. K., Chien S., Wang Y., Mechanogenetics for the remote and noninvasive control of cancer immunotherapy. Proc. Natl. Acad. Sci. 115, 992–997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanley S. A., Sauer J., Kane R. S., Dordick J. S., Friedman J. M., Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat. Med. 21, 92–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long X., Ye J., Zhao D., Zhang S. J., Magnetogenetics: Remote non-invasive magnetic activation of neuronal activity with a magnetoreceptor. Science bulletin, 60, 2107–2119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Q., Wang X., Tibbitt M. W., Anseth K. S., Montell D. J., Elisseeff J. H., Light activated cell migration in synthetic extracellular matrices. Biomaterials 33, 8040–8046 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fairbanks B. D., Schwartz M. P., Bowman C. N., Anseth K. S., Photoinitiated Polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: Polymerization rate and cytocompatibility. Biomaterials 30, 6702–6707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shirahama H., Lee B. H., Tan L. P., Cho N., Precise tuning of facile one-pot gelatin methacryloyl (GelMA) synthesis. Nat. Publ. Gr. , 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klose A. D., Paragas N., Automated quantification of bioluminescence images. Nat. Commun. 9, 4262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolligs F. T., Hu G., Dang C. V., Fearon E. R., Neoplastic transformation of RK3E by mutant β-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol. Cell. Biol. 19, 5696–5706 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacDonald B. T., Hien A., Zhang X., Iranloye O., Virshup D. M., Waterman M. L., He X., Disulfide bond requirements for active Wnt ligands. J. Biol. Chem. 289, 18122–18136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toni L. S., Garcia A. M., Jeffrey D. A., Jiang X., Stauffer B. L., Miyamoto S. D., Sucharov C. C., Optimization of phenol-chloroform RNA extraction. MethodsX 5, 599–608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W., Germain R. N., Gerner M. Y., Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (C e 3D). Proc. Natl. Acad. Sci. 114, E7321–E7330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/40/eabb9062/DC1