Summary
Background
The number of cases of gonorrhoea in the USA and worldwide caused by Neisseria gonorrhoeae is increasing (555 608 reported US cases in 2017, and 87 million cases worldwide in 2016). Many countries report declining in vitro susceptibility of azithromycin, which is a concern because azithromycin and ceftriaxone are the recommended dual treatment in many countries. We aimed to identify strain types associated with decreased susceptibility to azithromycin.
Methods
We did a genomic analysis of N gonorrhoeae isolates obtained by the US Gonococcal Isolate Surveillance Project. Isolates were whole-genome sequenced based on decreased susceptibility to azithromycin (minimal inhibitory concentration [MIC] ≥2 μg/mL, using agar dilution antibiotic susceptibility testing) and geographical representation. Bioinformatic analyses established genomic diversity, strain population dynamics, and antimicrobial resistance profiles.
Findings
410 isolates were sorted into more than 20 unique phylogenetic clades. One predominant persistent clade (consisting of 97 isolates) included the most isolates with azithromycin MICs of 2 μg/mL or higher (61 of 97 [63%] vs 59 of 311 [19%]; p<0·0001) and carried a mosaic mtr (multiple transferable resistance) locus (68 of 97 [70%] vs two of 313 [1%]; p<0·0001). Of the remaining 313 isolates, 57 (18%) had decreased susceptibility to azithromycin (MIC ≥4 μg/mL), which was attributed to 23S rRNA variants (56 of 57 [98%]) and formed phylogenetically diverse clades, showing various levels of clonal expansion.
Interpretation
Reduced azithromycin susceptibility was associated with expanding and persistent clades harbouring two well described resistance mechanisms, mosaic mtr locus and 23S rRNA variants. Understanding the role of recombination, particularly within the mtr locus, on the fitness and expansion of strains with decreased susceptibility has important implications for the public health response to minimise gonorrhoea transmission.
Funding
US Centers for Disease Control and Prevention (CDC), CDC Combating Antibiotic Resistant Bacteria initiative, Oak Ridge Institute for Science Education, US Department of Energy/CDC/Emory University, National Institutes of Health, and Biomedical Laboratory Research and Development Service of the US Department of Veterans Affairs.
Introduction
Neisseria gonorrhoeae, a Gram-negative diplococcus, is the causative agent of gonorrhoea. The rates of reported cases of gonorrhoea in the USA have increased by 75·2% between 2009 and 2017, from a historic low of 98·1 cases per 100 000 population in 2009 to 170·6 cases per 100 000 population reported in the USA in 2017.1 Global prevalence and incidence of gonorrhoea is high,2 with an estimated 87 million cases worldwide in 2016.3 N gonorrhoeae was identified as an urgent-level antibiotic resistance threat by the US Centers for Disease Control and Prevention (CDC) in 2013,4 and again in 2019.5 The CDC currently recommends dual therapy with azithromycin and ceftriaxone for first-line treatment of uncomplicated gonorrhoea (urogenital infections in adults).6
Since its development in the 1980s, azithromycin has proven effective against gonorrhoea, chlamydia, and other non-sexually transmitted bacterial infections,7 and this antibiotic has been widely used. However, the long half-life of azithromycin (serum t1/2 68 h) might increase the risk of selection for resistance through prolonged subinhibitory intracellular and extracellular concentrations. 7 Reduced susceptibility of gonococci to azithromycin was reported in the 1990s, and although azithromycin has not been recommended as a monotherapy, growing reports of decreased azithromycin susceptibility are being monitored worldwide.8–13 Azithromycin was recommended to be used in dual therapy with ceftriaxone in 2015,6 to function as a shield to potentially delay the emergence of resistance to ceftriaxone, the last remaining antimicrobial option. This recommendation was modelled after combination antimicrobial therapy for prevention of resistance used for other infectious diseases such as tuberculosis.14
The Clinical and Laboratory Standards Institute set interpretive susceptibility criteria for azithromycin at the discrete, in vitro minimum inhibitory concentration (MIC) of 1 μg/mL or lower.15 They did not define resistance, since the level at which in vitro susceptibility isolates would be associated with azithromycin monotherapy treatment failure is unknown; thus, an MIC greater than 1 μg/mL was defined as decreased susceptibility. Similarly, the European Committee on Antimicrobial Susceptibility Testing defined an epidemiological cutoff of 1 μg/mL.15
In the USA, the Gonococcal Isolate Surveillance Project (GISP) showed that the percentage of isolates with decreased susceptibility to azithromycin (MIC ≥2 μg/mL) remained low for decades, until a sharp rise from 0·6% in 2013 to 2·5% in 2014 and to 4·4% by 2017.1 The predominant reason for this increase is the growth (from 2013 to 2017) in the proportion of isolates with low-level decreased susceptibility to azithromycin (MIC 2–4 μg/mL).1 WHO9 and other international organisations10–12 reported an increase in the percentage of isolates with reduced susceptibility to azithromycin across multiple countries, approaching the level at which WHO recommends the review, modification, or discontinuation of an antimicrobial in empirical treatment.9 Several cases of decreased susceptibility to both azithromycin and ceftriaxone have been recorded internationally.16 To date, no known treatment failures with this susceptibility pattern have been identified in the USA;15 however, CDC monitors resistance levels because of its mandate for preparedness.
N gonorrhoeae antibiotic resistance variants emerge through either plasmid-mediated or chromosomal-mediated mechanisms, including single nucleotide polymorphisms (SNPs) or recombination events.17 Antibiotic resistance genes for azithromycin include the 23S rRNA genes, which result in intermediate-level or high-level resistance, and the mtr (multiple transferable resistance) efflux pump locus (mtr regulatory and mtr pump).17–19 Multiple variations within the mtr locus, which affect either mtr regulation, overexpression of the pump, or elevated efflux of the pump,20 might work in concert to elevate MIC to levels of decreased susceptibility (MIC 2–4 μg/mL).
We aimed to extend our understanding of N gonorrhoeae genetic diversity, variation in strain prevalence, and antibiotic resistance determinants beyond previously published data for surveillance isolates obtained by GISP in 2000–1318 and 2014–16.21 We did a surveillance-based genome analysis of GISP isolates from 2017 to identify strains and antibiotic resistance profiles associated with the overall increase in proportion of isolates with decreased susceptibility to azithromycin (MIC ≥2 μg/mL) in the USA.
Methods
GISP protocols
Isolates were selected from the national sentinel surveillance project GISP.22 GISP guidelines in 2017 included collection of isolates from the first 25 males who presented with gonococcal urethritis every month at one of 27 clinical sites across the USA. Isolates underwent antimicrobial susceptibility testing for seven drugs, using standardised agar-dilution methods.22 Confirmation antimicrobial susceptibility testing was done at the CDC (Atlanta, GA, USA). Collection of clinical specimens, culturing of isolates, and antimicrobial susceptibility testing procedures are described in the GISP protocol.22
Isolate selection
From GISP isolates obtained in 2017, 5061 met all GISP qualifications,22 of which, 410 were sequenced (appendix 1 pp 3–4). First, a stratified random sample of 13 isolates from every sentinel site was selected without replacement for sequencing (n=283). Next, from the remaining GISP specimens, all isolates meeting the following phenotypic criteria were selected, exclusive of previous subsets (appendix 1 p 3): azithromycin MIC 4 μg/mL or higher (n=74); cefixime MIC 0·25 μg/mL or higher (n=12); or ceftriaxone MIC 0·125 μg/mL or higher (n=6). Finally, isolates with an azithromycin MIC of 2 μg/mL were selected at random (appendix 1 p 4), to achieve a 30% sampling frame (n=37). Only 30% of isolates with an MIC of 2 μg/mL were sequenced because of an increase in isolates with decreased susceptibility to azithromycin, limited capacity for sequencing, and prioritisation to sequence isolates with intermediate-level and high-level decreased susceptibility to azithromycin (MIC ≥4 μg/mL). All sequences had to pass quality cutoffs. GISP whole-genome sequencing (WGS) datasets from 2000–1318 and 2014–1621 included isolate selection, as previously described. Appendix 2 presents the 410 isolates from GISP sequenced in 2017, with accession numbers, MICs for azithromycin, ceftriaxone, and cefixime, multilocus sequence typing, N gonorrhoeae multiantigen sequence typing, antimicrobial resistance determinant calls, and demographic data (sexual orientation, HIV status, and geographical distribution according to Health and Human Services [HHS]-defined US region;23 appendix 1 p 5). The representation of sequenced isolates per HHS region is provided in appendix 1 (pp 6–7).
WGS of N gonorrhoeae isolates
WGS was done at the Antibiotic Resistance Laboratory Network (AR Lab Network) and Hawaii Department of Health, using PulseNet protocols (appendix 1 p 8)24 and MiSeq with V2/V3 reagents (Illumina, San Diego, CA, USA), or at the CDC Biotechnology Core Facility Branch (Atlanta, GA, USA) using HiSeq2500 and V2 reagents (Illumina). Read data are deposited at the National Center for Biotechnology Information Sequence Read Archive (project accession PRJNA317462 and PRJNA329501; appendix 2).
Bioinformatics analyses
The WGS analysis pipeline21 is described in appendix 1 (pp 9–10) and included the following processes: quality assessment; trimming; assembly; multilocus sequence typing; N gonorrhoeae multiantigen sequence typing; and maximum-likelihood core-genome SNPs, removal of recombination, phylogenetic alignment (1 785 354 nucleotides, 16 396 SNPs), and interactive tree of life (iTOL) visualisation. The N gonorrhoeae strain FA19 (GenBank accession CP012026.1) served as the reference genome. AMR-profiler (CDC) is a customised script for antibiotic resistance determinant identification (alleles or SNPs) from raw reads and genomic assemblies.21 Maximum-likelihood core-genome SNPs, removal of recombination, phylogenetic alignment (including 410 isolates from 2017 and 646 isolates from 2014 to 2016) was generated for review (1 583 698 nucleotides, 20 758 SNPs).
Statistical analysis
Fisher’s exact test was used to calculate associations between pairwise categorical variables such as increased MIC, resistance determinants, sequence type, and clade (appendix 1 pp 11–13).
Role of the funding source
The funder had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Results
The 410 isolates that were sequenced represented 70 unique sequence types defined by multilocus sequence typing, including 11 novel multilocus sequence types (MLSTs). 22 unique sequence types were represented by five or more isolates (figure 1); ST9363 represented 81 (20%) of 410 isolates. Among isolates with decreased susceptibility to azithromycin (n=120), the most common sequence types were ST9363 (42%), ST7371 (16%), ST1584 (13%), and ST1579 (7%). The MLST ST9363 was recorded in the USA in previous years (2014–16) as a predominant strain that included nearly half the isolates with decreased susceptibility to azithromycin (96 of 198 [48%]).21
Figure 1: Distribution of multilocus sequence types for 410 isolates from the USA in 2017.
The 22 most numerous sequence types, which include five or more isolates, are shown according to susceptibility to azithromycin, ceftriaxone, and cefixime. Decreased susceptibility to azithromycin is defined as MIC ≥2 μg/mL. Decreased susceptibility to cefixime is defined as MIC ≥0·250 μg/mL. Decreased susceptibility to ceftriaxone is defined as MIC ≥0·125 μg/mL. MIC=minimum inhibitory concentration.
Sequence types and associated phenotypic antibiotic susceptibilities for azithromycin, ceftriaxone, and cefixime are shown in figure 1. Differences were noted in the percentage of isolates within each sequence type that had decreased susceptibility to azithromycin. In ST9363, 51 (63%) of 81 isolates had decreased susceptibility to azithromycin; 46 (57%) had low-level decreased susceptibility (MIC 2–4 μg/mL) and five (6%) had intermediate-level to high-level decreased susceptibility (MIC 8–16 μg/mL). By comparison, 20 (87%) of 23 isolates in ST7371 had intermediate-level decreased susceptibility to azithromycin (MIC 8–16 μg/mL); moreover in ST7371, 22 (95%) of 23 isolates had decreased susceptibility to ciprofloxacin (MIC 32 μg/mL). ST1584 contained 16 (47%) of 34 isolates with decreased susceptibility to azithromycin (MIC 4 μg/mL). In ST1579, nine (75%) of 12 isolates not only had decreased susceptibility to azithromycin (MIC 8 μg/mL) but also to ciprofloxacin (MIC 4 μg/mL).
Figure 2 shows the maximum-likelihood core-genome phylogenetic tree based on 410 isolates, with more than 19 clades. Clades typically consisted of one predominant MLST or contained related MLSTs (eg, ST9363 and ST11422 share six out of seven alleles). The table presents clade descriptors, including predominant MLSTs, number of isolates per clade, and antimicrobial resistance profiles. Variants are described in appendix 1 (pp 19–20).
Figure 2: Maximum-likelihood core-genome phylogenetic tree of 410 Neisseria gonorrhoeae isolates from the USA in 2017.
The reference genome was FA19 (GenBank accession CP012026.1). 19 clades (labelled A–S) are shown, with susceptibility profiles and relevant antibiotic resistance determinants. Variants are described in appendix 1 (pp 19–20). Isolates susceptible to azithromycin are defined as MIC ≤1 μg/mL; decreased susceptibility to azithromycin is categorised with MICs of 2–4 μg/mL, 8 μg/mL, and ≥16 μg/mL. Isolates susceptible to cefixime are defined as MIC ≤0·125 μg/mL, and those with decreased susceptibility are defined as MIC ≥0·250 μg/mL. Isolates susceptible to ceftriaxone are defined as MIC ≤0·060 μg/mL, and those with decreased susceptibility are defined as MIC ≥0·125 μg/mL. Isolates susceptible to ciprofloxacin are defined as MIC ≤0·5 μg/mL, and those with decreased susceptibility are defined as MIC ≥1·0 μg/mL. The antimicrobial resistance determinants 23S rRNA 2611C→T and 2059A→G are categorised by cumulative number of variant rRNA copies present (one to four copies). penA identifies mosaic and non-mosaic penA alleles. Categorisation of isolates by sexual orientation of the individual from whom the isolate was obtained is also shown. MIC=minimum inhibitory concentration. MSW=men who have sex with women. MSM=men who have sex with men. MSMW=men who have sex with men and women.
Clade A, the largest in this dataset, consisted of 97 isolates, including those both susceptible to azithromycin and with decreased susceptibility. 61 (63%) of 97 isolates had decreased susceptibility to azithromycin (MIC ≥2 μg/mL), compared with 59 (19%) of the remaining 311 isolates (p<0·0001; appendix 1 p 11). Clade A was predominantly represented by MLST ST9363, in 81 (84%) of 97 isolates versus none of 313 isolates (p<0·0001; appendix 1 p 13), and to a lesser extent by MLST ST11422 (four of 97 [4%]), ST11417 (two of 97 [2%]), and ST11428 (two of 97 [2%]; figure 2, table). Clade A (ST9363) was persistent throughout the year and represented diverse demographic groups. Isolates from clade A (ST9363) were from eight of ten HHS geographical regions in the USA,23 with two-thirds (67%) of isolates from two HHS geographical areas (region 5 [IL, IN, MI, MN, OH, and WI] and region 9 [AR, CA, HI, NV, GU, American Samoa, Northern Mariana Islands, Federated States of Micronesia, Marshall Islands, and Palau]) and were equally represented by men who have sex with men (45%) and men who have sex with women (45%). Clade A consisted of multiple hierarchical subclades; figure 3 shows clade A (ST9363; 32 isolates, average SNP distance 147 [SD 96]) and subclade sc9363.1 (65 isolates, average SNP distance 44 [23]). 61 (63%) of 97 isolates in clade A had decreased susceptibility to azithromycin (MIC ≥2 μg/mL) and were predominantly clustered in sc9363.1 (57 of 65 [88%] vs four of 32 [13%]; p<0·0001; appendix 1 p 13). Six of the remaining eight isolates in sc9363.1 had an azithromycin MIC of 1 μg/mL, which is within one dilution of the decreased susceptibility designation.
Table:
Summary of genotypic and phenotypic properties of phylogenetic clades containing isolates with decreased susceptibility to azithromycin
| Year | MLST | NG-MAST | SNP distance, mean (SD) | Total, n | AZMds, n | CIPds, n | HHS region | AZM MIC, μg/mL | 2611C→T, n(N)*† | 2059A→G, n(N)*† | mtrR mosaic* | MtrR H105Y* | mtrR premature stop* | mtrR promoter*‡ | MtrD*§ | penA mosaic*¶ | PBP2 D345 insertion* | PBP1 L421P* | PorB* ∥ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade A, ST9363 | 2014–17 | 9363, 11428 | 654, 2992, 3935 | 147 (96) | 32 | 4 | 1 | 5, 9 | 0.25–16 | 2 (4), 1 (2) | .. | .. | .. | .. | .. | ✓ | 2.001 | ✓ | .. | .. |
| Subclade sc9363.1 | 2014–17 | 9363, 11422 | 3935, 8241, 12302 | 44 (23) | 65 | 57 | 10 | 5, 9 | 2–16 | 2 (3), 1 (1) | 2 (4) | ✓ | .. | .. | A→C | ✓ | 2.001 | ✓ | .. | KN |
| Clade D, ST7371 | 2016–17 | 7371 | 3169 | 8 (5) | 22 | 20 | 21 | 3 | 8–16 | 21 (4) | .. | .. | .. | .. | DelA | .. | 18.001 | ✓ | ✓ | KD |
| Clade Q, ST1584 | 2014–17 | 1584 | 865, 8532, 4248 | 118 (85) | 13 | .. | .. | 2, 5, 6 | .. | .. | .. | .. | .. | ✓ | .. | .. | 14.001 | ✓ | .. | .. |
| Subclade scl584.1 | 2015–17 | 1584 | 7638 | 9 (4) | 16 | 16 | .. | 9 | 4 | 16 (4) | .. | .. | .. | ✓ | .. | .. | 14.002 | ✓ | .. | .. |
| Subclade scl584.2 | 2015–17 | 10317 | Multiple | 39 (13) | 5 | .. | .. | 5 | .. | .. | .. | .. | .. | ✓ | .. | .. | 14.001 | ✓ | .. | .. |
| Clade B, ST1579 | 2016–17 | 1579 | 17645, 18497 | 8 (3) | 9 | 9 | 9 | 5, 9 | 8 | 9 (4) | .. | .. | ✓ | .. | DelA | .. | 2.002 | ✓ | ✓ | |
| Clade 0, ST10932 | 2016–17 | 10932 | Multiple | 149 (122) | 4 | .. | .. | 9, 7 | .. | .. | .. | .. | .. | .. | .. | .. | 22.001 | ✓ | .. | .. |
| Subclade scl0932.1 | 2015–17 | 10932 | 17636 | 13 (8) | 4 | 4 | .. | 9, 10 | 4–8 | 4 (4) | .. | .. | .. | .. | .. | .. | 22.001 | ✓ | .. | ND |
| Clade L, ST12093 | 2017 | 12093 | 18435, 4990 | 87 (67) | 4 | 3 | .. | 9 | 2–16 | 3 (4) | .. | .. | ✓ | .. | DelA | .. | 9.001 | ✓ | ✓ | .. |
| Clade E, ST8156 | 2017 | 8156, 1583 | 5441, 16009 | 186 (90) | 11 | .. | .. | 9, 3 | .. | .. | .. | .. | ✓ | .. | DelA | .. | 2.002 | ✓ | ✓ | .. |
| Subclade sc8156.1 | 2017 | 8156 | 16099, 18487 | .. | .. | 2 | .. | 3, 5 | 8 | 2 (4) | .. | .. | ✓ | .. | DelA | .. | 2.001 | ✓ | ✓ | .. |
Clades defined within the phylogenetic tree (figure 2) are labelled A–S. Every clade is defined by the most common MLST of the isolates within the clade (eg, ST9363). For any category, if the variable represents >50% of isolates, the number is bold; if >90% the number is bold and underlined. Year=isolate collection year in which the isolates of that strain were present (2014–16;22 2017 [current dataset]). MLST=multilocus sequence type. NG-MAST=Neisseria gonorrhoeae multiantigen sequence type. SNP=single nucleotide polymorphism. Total=number of isolates defined in the clade or the part of the clade not represented in the subclade. AZMds=number of isolates in the clade with decreased susceptibility to azithromycin. CIPds= number of isolates in the clade with decreased susceptibility to ciprofloxacin. HHS=US Department of Health and Human Services; HHS regions are numbered 1–10.23 AZM MIC=minimum inhibitory concentration of azithromycin.
Antibiotic resistance determinants are recorded by clade, and only variants from wild-type are listed. Descriptions and accession numbers of variants are available in appendix 1 (pp 19–20). One letter amino acid codes are shown in headings, for ease of reading.
Number of isolates that have the 23S rRNA variant (number of variant alleles present in the isolate).
Either adenine to cytosine variant (A→C) or adenine nucleotide deletion (DelA).
Ser821Ala and Lys823Glu variations.
Numbers refer to the most common penA allele identified for the isolates in the cluster.
KN denotes Gly120Lys and Gly121Asn variations. KD denotes Gly120Lys and Gly121Asp variations. ND denotes Gly121Asn and Gly121Asp variations.
Figure 3: Region of the maximum-likelihood core-genome phylogenetic tree representing clade A.
Variants are described in appendix 1 (pp 19–20). Aligned antimicrobial determinants emphasise the separation of the parental clade (ST9363) and subclade 9363.1. Isolates susceptible to azithromycin are defined as MIC ≤1 μg/mL; decreased susceptibility to azithromycin is categorised with MICs of 2–4 μg/mL, 8 μg/mL, and ≥16 μg/mL. The antimicrobial resistance determinants 23S rRNA 2611C→T and 2059A→G are categorised by cumulative number of variant rRNA copies present (one to four copies). mtrR locus is described by mtrR mosaic (wild-type non-mosaic or mosaic) and mtrR promoter (C substitution or A deletion in the 13-bp inverted repeat). Amino acid substitutions in MtrR (Ala39Thr, Gly45Asp, and His105Tyr), MtrD (Lys823Glu, representing mtrD mosaicity), and PorB (Gly120Lys and Gly121Asp or Asn) are also shown. mtr operon and porB and penA are defined antibiotic resistance genes for cefixime, ceftriaxone, and penicillin.19,20 penA identifies mosaic and non-mosaic penA alleles. MIC=minimum inhibitory concentration.
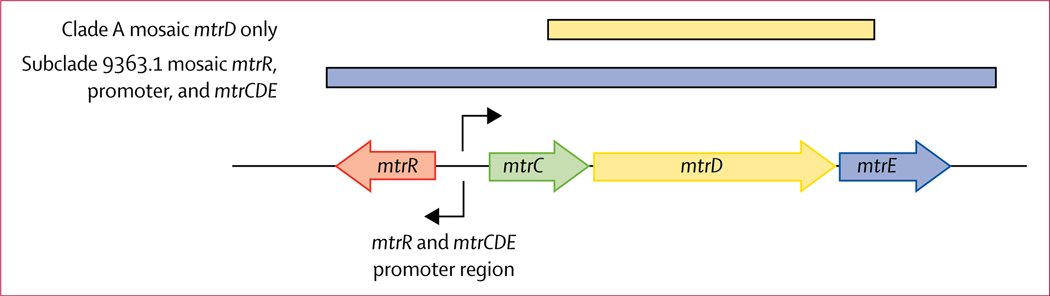
68 (70%) of 97 isolates in clade A carried a mosaic mtr locus (68 of 97 [70%] vs two of 313 [1%]; p<0·0001; figure 3; appendix 1 p 12). 65 (100%) of 65 isolates in subclade sc9363.1 carried multiple variants in the mtr locus (figure 4), including mosaic mtrR (gene encoding the repressor), an A→C substitution in the mtrR and mtrCDE promoter region, and mosaic mtrD (gene encoding the cytoplasmic membrane protein of the MtrCDE efflux pump) identified by protein variant Lys823Glu (figure 3; appendix 1 pp 19–20). These variants contributed to decreased susceptibility to azithromycin.20 The adenosine to cytosine nucleotide change in the mtrR and mtrCDE promoter region might affect mtrR and mtrCDE expression, whereas a mosaic mtrR could decrease functionality of the repressor and in turn affect mtrCDE expression.20 Variations in mtrD affect antimicrobial efflux by the pump. The remainder of isolates in clade A (ST9363, 32 of 97 isolates) did not carry mosaic mtrR (32 of 32 [100%]) or mtrR promoter variants (28 of 32 [88%]), but did carry the mosaic mtrD (variant Lys823Glu; 32 of 32 [100%]; figures 3, 4) and MtrR Ala39Thr (28 of 32 [88%]), which could result in increased mtrCDE expression and a change in MtrD functionality and efflux; however, these mutations were associated with susceptibility to azithromycin (MIC <2 μg/mL). In previous studies from the USA, strain ST9363 with decreased susceptibility to azithromycin and mosaic mtr locus was reported (appendix 1 pp 14–15).18,21 The mosaic mtr locus was initially identified associated with decreased susceptibility to azithromycin in the USA in 2012,18 but only represented a minority of isolates studied with decreased susceptibility to azithromycin in 2012–13 (22% [11 of 50]). In 2014–16, strain ST9363 with decreased susceptibility to azithromycin and mosaic mtr locus represented 57% (113 of 198) of isolates with decreased susceptibility to azithromycin;21 in 2017, the clade with mosaic mtr represented 69% (155 of 222, with estimation to 100% of 2017 isolates with decreased susceptibility to azithromycin [MIC 2 μg/mL]) of isolates with decreased susceptibility to azithromycin (appendix 1 pp 16–17), suggesting persistence and expansion of the strain. Strain ST9363 with mosaic mtrD only was initially identified in 2009 and shows strain persistence to 2017.
Figure 4: Schematic of mtr locus of Neisseria gonorrhoeae.
Arrows indicate the direction of transcription. The coloured lines above the mtr locus indicate the mosaic region in clade A isolates with mosaic mtrD only (upper line) and in subclade 9363.1 with mosaic mtrR, promoter, and mosaic mtrCDE (lower line).
Dispersed within clade A (both ST9363 and sc9363.1), eight isolates carried 23S rRNA mutations: six carried 2611C→T (one, two, three, or four copies), and two carried four copies of 2059A→G (appendix 1 pp 19–20). These variants resulted in azithromycin MICs of 2–16 μg/mL (appendix 1 p 18). Moreover, ten isolates within subclade sc9363.1 (N gonorrhoeae multiantigen sequence type [NG-MAST] 12302, predominantly in men who have sex with men [eight of ten]) carried ciprofloxacin resistance (MIC ≥1 μg/mL) attributed to mutations in GyrA and ParC (appendix 1 pp 19–20), suggesting continued evolution of this clade.
Isolates with decreased susceptibility to azithromycin in clades other than clade A (57 of 313) had intermediate-level (MIC 4–16 μg/mL) or high-level (MIC >16 μg/mL) decreased susceptibility and carried 23S rRNA variants. These isolates were phylogenetically dispersed and predominantly found in clade Q (represented by ST1584), clade D (represented by ST7371), and clade B (represented by ST1579; figure 2; table).
Clade Q, represented by ST1584, contained 34 isolates and included multiple subclades (table). 13 isolates of ST1584 represented multiple HHS geographical areas (region 5, region 6 [AR, LA, NM, OK, and TX], and region 2 [NJ, NY, PR, and VI]), multiple NG-MASTs (865, 8532, and 8194), had an average SNP distance of 118 (SD 85), and were predominantly found in men who have sex with women (12 of 13). The 13 isolates carried an mtrR premature stop sequence (truncated MtrR protein; appendix 1 pp 19–20) without 23S rRNA variants, contributing to their relatively low azithromycin MIC (≤1 μg/mL). Within clade Q was a subclade (sc1584.1) of 16 phylogenetically homogeneous isolates (average SNP distance 9 [SD 5]). These isolates were NG-MAST 7638, solely from HHS region 9, and predominantly found in men who have sex with men (13 of 16). 16 (100%) of 16 sc1584.1 isolates had decreased susceptibility to azithromycin (MIC 4 μg/mL) and contained four copies of 23S rRNA 2611C→T and the mtrR premature stop sequence. Although clade Q (represented by ST1584) was present in 2014–16,21 the homogeneous subclade sc1584.1 with the 23S rRNA mutation 2611C→T (MIC 4–8 μg/mL) was first identified in 2015–16, showing persistence to 2017 (appendix 1 pp 14–15).
Clade D, represented by ST7371, contained 20 of 22 isolates with intermediate-level decreased susceptibility to azithromycin (MIC 8–16 μg/mL). Clade D (ST7371) was highly homogenous (NG-MAST 3169 [19 of 22]), had an average SNP distance of 8 (SD 5), and was predominantly found in HHS region 3 (DE, DC, MD, PA, VA, and WV) and in men who have sex with women (21 of 22). 21 of 22 isolates carried four copies of 23S rRNA 2611C→T and variants in the mtr promoter associated with decreased susceptibility to azithromycin. Furthermore, variants were recorded in PBP2, encoded by the penA gene (Ala501Thr), PBP1, encoded by the ponA gene (Leu421Pro), PorB (Gly120Lys and Gly121Asp or Gly121Asn), which are associated with reduced susceptibility to other antibiotics, and GyrA (Ser91Phe and Asp95Asn) and ParC (Ser87Ile), associated with ciprofloxacin resistance (MIC 32 μg/mL; appendix 1 pp 19–20). Clade D (ST7371) isolates with decreased susceptibility to azithromycin were previously recorded in 201621 showing strain persistence to 2017 (appendix 1 pp 14–15).
Clade B, represented by ST1579, contained nine (100%) of nine isolates with decreased susceptibility to azithromycin (MIC 8 μg/mL). Isolates in this clade carried the 23S rRNA mutation 2611C→T, were highly homogenous (average SNP distance 8 [SD 3]), and were predominantly found in men who have sex with men (eight of nine) and in the HHS geographical region 5 (six of nine). The clade carried variants associated with decreased susceptibility to azithromycin (adenosine deletion in the mtr promoter, and MtrR His105Tyr) and a variant associated with decreased susceptibility to cefixime (Leu421Pro in PBP1; appendix 1 pp 19–20). Isolates were collected throughout 2017, suggesting persistence of the ST1579 strain, with 23S rRNA variants seen in 2016 (appendix 1 pp 14–15).21 All isolates were resistant to ciprofloxacin (MIC 4 μg/mL).
Other isolates with decreased susceptibility to azithromycin that carried the 23S rRNA mutation 2611C→T were distributed among phylogenetically diverse clades and with low representation, implying limited expansion. These isolates were present in clade O (represented by ST10932; four of eight isolates with MIC 4–8 μg/mL), clade L (represented by ST12093; three of four isolates with MIC 2–16 μg/mL), clade E (represented by ST8156 and ST1583; two of 13 isolates with MIC 8 μg/mL), clade G (represented by ST7827; one of seven isolates with MIC 8 μg/mL), and clade C (represented by ST1901; one of 22 isolates with MIC 4 μg/mL). Clades represented by ST8156 and ST10932 were present in 2014–16 and 2015–16, respectively, and carried the 2611C→T mutation. Although present earlier,21 neither ST12093 (2014) nor ST7827 (2014–16) carried 23S rRNA variants before 2017 (appendix 1 pp 14–15), implying introduction of antimicrobial resistance into this strain. Only two isolates contained the 23S rRNA 2059A→G vari ant (appendix 1 pp 19–20); these isolates were in clade A (sc9363.1), were predominantly found in the HHS geographical region 5, and had high-level decreased susceptibility to azithromycin (MIC >16 μg/mL). The 23S rRNA 2059A→G variant seen in previous years was recorded in separate clades represented by ST1901 (n=4) in 2016 and ST7822 (n=3) in 2014–15 (appendix 1 pp 14–15).21
23S rRNA variants have been identified in GISP isolates every year (2000–17), albeit sporadically. Grad and colleagues18 reported 56% of isolates with decreased susceptibility to azithromycin were associated with variants of the ribosome, including 23S rRNA variants, whereas Thomas and colleagues21 reported 20% of isolates with decreased susceptibility to azithromycin could by accounted for by 23S rRNA variants.21 In this analysis, with estimation to 100% of 2017 isolates with decreased susceptibility to azithromycin (MIC 2 μg/mL), 66 (30%) of 222 isolates with decreased susceptibility to azithromycin were associated with 23S rRNA variants (appendix 1 pp 16–17).
Discussion
The findings of our genomic analysis provide an in-depth description of N gonorrhoeae strain populations and antibiotic resistance determinants among isolates obtained from the USA in 2017, and a comparison with populations in 2000–16. Our findings offer an explanation for the increasing number of patients with decreased susceptibility to azithromycin because of persistence and expansion of a predominant strain, ST9363. Our genomic characterisation shows the continued persistence of strain ST9363, with mosaic mtr locus associated with the phenotype of low-level decreased susceptibility to azithromycin (MIC 2–4 μg/mL), from its first observation in the USA in 2012 to the current analysis in 2017. Although resistance criteria have not been established for azithromycin, and insufficient clinical data are available about treatment failures associated with low-level resistance to azithromycin (MIC 2–4 μg/mL), isolates with these MICs fall outside defined susceptibility criteria15 and are, therefore, concerning for potential treatment failures.
Subclade sc9363.1 carries various mutations of the mtr operon, encoding a multiple transferable resistance efflux pump (MtrCDE), which exports a diverse array of substances, including antimicrobials and antibiotics. Mutations in mosaic mtrR, the mtrR promoter, and mosaic mtrD augment expression of the efflux pump (MtrCDE), provide increased pump efficacy, and contribute to improved biological fitness and survival of N gonorrhoeae in vivo.19,25,26 Since 2014, this subclade showed pervasiveness in most HHS-defined US regions and in populations of men who have sex with men and men who have sex with women. Isolates in ST9363 carrying mosaic mtr were identified internationally in Australia in 2012; these isolates accounted for 85% of those with decreased susceptibility to azithromycin (MIC ≥1·0 μg/mL) in Australia by 2017.27 Similar isolates were identified in Canada in 2014,28 suggesting mosaic mtr locus operon could be contributing to the worldwide rise of low-level azithromycin resistance (MIC 2–4 μg/mL). Highly functional DNA repair mechanisms provide N gonorrhoeae with the capability for gene transfer recombination events to improve infectivity and survival against host defences and to produce gain-of-resistance determinants (mosaic mtr). We need to understand these mechanisms to minimise future recombination events that achieve resistance. Similarly, if bacteria are induced to recombine or select for resistance because of prolonged exposure to subinhibitory concentrations of azithromycin, based on the long half-life of the antibiotic after treatment,7 then minimisation of exposure (reduction in clinical treatment) could be one approach to solve this problem.
In addition to a mosaic mtr locus that increases azithromycin MICs just above the susceptibility level,20,25 multiple copies of 23S rRNA variants result in decreased susceptibility to azithromycin (MICs ≥4 μg/mL).17 Our genomic analysis identified diverse populations of isolates carrying the 23S rRNA variants 2611C→T and 2059A→G, which are associated, respectively, with intermediate-level (MIC 4–16 μg/mL) and high-level (MIC >16 μg/mL) decreased susceptibility to azithromycin. Persistence and loss of these clades, along with introduction of new strains with 23S rRNA variants, maintained the proportion (0·2–0·9%) of isolates with decreased susceptibility to azithromycin (MIC 8–16 μg/mL) reported in 2013–17.1 The 23S rRNA variant 2611C→T was phylogenetically distributed within multiple clades (represented, for example, by ST9363, ST7371, ST1584, and ST1579). These persistent populations belonged either to highly clonal clades (eg, ST7371 in men who have sex with women or ST1584 in men who have sex with men) represented in one HHS-defined US region or to genetically similar, geographically dispersed populations (eg, ST1579 in men who have sex with men). Other sequence types (eg, ST12093 with the 2611C→T variant, or ST9363 with the 2059A→G variant) suggest novel introduction of or potential losses of 23S rRNA variants within a strain (eg, ST7822 with the 2611C→T variant, or ST1901 with the 2059A→G variant). Previous studies suggest biological fitness and stability of strains with 23S rRNA variants (eg, 2059A→G)29,30 or mosaic mtr locus,19,25,26 which continue to be of concern for the potential clinical failure of monotherapy.15,16 However, strains with 23S variants in our study represented homogeneous less expansive networks than presumed for the ST9363 strain. Understanding the reasons behind the low expansion of 23S rRNA variants, with respect to the expansive transmission of clade ST9363, in addition to the role of DNA repair and recombination in biological fitness and transmissibility, could be invaluable to slow the spread of drug-resistant N gonorrhoeae. Continued genomic monitoring of strains is crucial,31 and increased understanding of the networks of transmission and spread within these networks might provide further direction for public health response in the control of drug-resistant N gonorrhoeae.
A strength of our study was the origin of sequenced isolates from robust sentinel surveillance in GISP,22 whereas many other surveillance systems include more passively collected strains, sometimes emphasising cases with clinical complications. Our study also had several limitations. GISP collection includes only specimens from men. GISP does not discriminate from repeat individuals or networked partners; both conditions can introduce bias. Although there is no way to quantify the degree of overlap, the occurrence is expected to be low. We assume that all isolates are independent. The number of isolates studied (n=410) could be insufficient for complete accurate representation of low-prevalence strains or low-frequency antibiotic resistance determinants, but the number was sufficient for full geographical representation across the major clades. Selection of 30% of isolates with low-level resistance to azithromycin (MIC 2 μg/mL), although balanced in representation across most HHS geographical regions, required estimation to 100% of isolates with MIC of 2 μg/mL, which could result in unknown biases. An additional limitation is that the selection criteria and number of isolates included in our analysis differed from those in previous studies, and direct comparison of counts or prevalence has limitations. 2017 was the first year when GISP laboratory work was mostly done in public health laboratories from the regional AR Lab Network. Since isolates were submitted for sequencing before finalisation of the reconciliated year-end GISP list, 4% of isolates in our bioinformatic analyses were later removed from GISP accounting. It is unknown how this minor discrepancy in isolate selection affects the analyses; a potential minor and unknown bias is acknowledged. Some isolates selected were not available for sequencing because of unforeseen circumstances (appendix 1 pp 3–4). Going forward, CDC is building a laboratory network for sustainable, future, molecular surveillance efforts.31
Supplementary Material
Research in context.
Evidence before this study
For molecular analyses of Neisseria gonorrhoeae, including whole-genome sequence analyses, with target surveillance dates of 2013–17, we searched PubMed between 2014 and 2019 with the keywords “Neisseria gonorrhoeae”, “antibiotic susceptibility”, “resistance”, “resistance determinants”, “genomic”, “surveillance”, “mtrR”, “mtr operon”, and “ST 9363”. For 23S rRNA variants, we searched PubMed with no date restriction with the keywords “Neisseria gonorrhoeae”, “rRNA variant”, and “23S rRNA”. For drug studies, we searched PubMed between 1990 and 2019 with the keywords “Neisseria gonorrhoeae”, “gonorrhea”, “treatment”, “therapy”, and “azithromycin”. Reports of molecular surveillance from Europe, Canada, China, Japan, and the USA were reviewed, and decreasing susceptibility to azithromycin was reported. For example, in the USA, the Gonococcal Isolate Surveillance Project (GISP) showed that the percentage of N gonorrhoeae isolates with reduced azithromycin susceptibility increased from 0·6% in 2013 to 4·4% in 2017. The WHO Gonococcal Antimicrobial Surveillance Programme showed decreasing susceptibility to azithromycin across multiple WHO regions during 2009–14. Reports provided descriptions of prevalent strain types, common antimicrobial-resistant determinants, and initial observations of novel resistant mechanisms, but they did not address whether reduced susceptibility of azithromycin to N gonorrhoeae strains was associated with a particular resistance mechanism. This paucity of information raises the question as to which resistance mechanisms are associated with the declining susceptibility to azithromycin and whether there is spread of strains harbouring select resistance mutations.
Added value of this study
We did a genomic analysis of N gonorrhoeae isolates obtained by GISP. We identified one predominant phylogenetic cluster with decreased susceptibility to azithromycin (minimum inhibitory concentration 2–4 μg/mL) carrying a mosaic mtr efflux pump locus sequence. In 2017, this clade in the USA represented 69% of isolates with decreased susceptibility to azithromycin, whereas in 2012 it represented a minority of isolates with reduced susceptibility. The increase in prevalence of this clade could account for the rise in isolates with decreased susceptibility to azithromycin recorded in GISP and emphasises the importance of recombination as an antimicrobial-resistance determinant and the biological advantage and stability of this strain. Additionally, persistence of 23S rRNA resistance determinants in multiple strains suggests stability of isolates with this mechanism of decreased susceptibility to azithromycin.
Implications of all the available evidence
Our data provide genetic characterisation of and a genomic mechanism for the decreasing susceptibility to azithromycin noted in the USA between 2013 and 2017. One strain with a mosaic mtr locus shows a wide demographic distribution and persistence and increased representation over multiple years. Continued transmission of this clade is likely and might support prolonged rises in the prevalence of isolates with decreased azithromycin susceptibility in the USA. The importance of this finding is emphasised by the concern of potential global spread of this strain, with initial observations of a similar strain in Australia (2012) and Canada (2014). By 2017, the Australian strain accounted for 85% of isolates with decreased susceptibility to azithromycin. Use of ongoing global surveillance assists in guiding decisions about recommendations for gonorrhoea treatment.
Acknowledgments
This work was supported by the US Centers for Disease Control and Prevention (CDC) and in part made possible through support from CDC’s Combating Antibiotic Resistant Bacteria programmes and Advanced Molecular Detection (to KMG, SS, MWS, JCT, CDP, BHR, and ENK). This project was also supported in part by an appointment to the Research Participation Program at the US CDC administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US CDC (to SS and JCT). This work was also supported by funds from the Division of STD Prevention at the US CDC (to SSC and HW); an Intergovernmental Personnel Act (to WMS); a National Institutes of Health grant (number R37AI21150–34, awarded to WMS); and the Biomedical Laboratory Research and Development Service of the US Department of Veterans Affairs (Senior Research Career Scientist Award to WMS). We thank the Gonococcal Isolate Surveillance Project (GISP), US CDC GISP contributors, and all US GISP sites for submitting gonococcal isolates and epidemiological data to the US CDC; Elizabeth A Torrone for dedication to and management of the GISP project; Robert Kirkcaldy for critical review of the report; A J A McLean and J Cartee (CDC) for isolate propagation and transfer; and Samera Sharpe (CDC) for confirmatory antimicrobial susceptibility testing measurements. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US CDC.
Footnotes
Antimicrobial-Resistant Neisseria gonorrhoeae Working Group
Sopheay Hun, Chi Hua, Ryan Ruiz (Antibiotic Resistance Laboratory Network [AR Lab Network], Washington State Department of Health, WA, USA); Olusegun O Soge (Department of Global Health and Medicine, University of Washington, Seattle, WA, USA); Catherine Dominguez, Jillian Loomis, Ami Patel (AR Lab Network, Maryland Department of Health, MD, USA); Jenny Zhang, Tamara Baldwin, Chun Wang, John Leavitt (AR Lab Network, Texas Department of State Health Services, TX, USA); Christina Moore (AR Lab Network, Tennessee Department of Health, TN, USA); Christian Whelen, Pamela O’Brien (Hawaii Department of Health State Laboratories Division, HI, USA); and Alesia Harvey (Centers for Disease Control and Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD and TB Prevention, Division of STD Prevention, Surveillance and Data Management Branch, Atlanta, GA, USA).
Declaration of interests
We declare no competing interests.
For PulseNet protocols see https://www.cdc.gov/pulsenet
For the Sequence Read Archive see https://www.ncbi.nlm.nih.gov/sra
References
- 1.US Centers for Disease Control and Prevention. Sexually transmitted disease surveillance 2017. September, 2018. https://www.cdc.gov/std/stats17/2017-STD-Surveillance-Report_CDC-clearance-9.10.18.pdf (accessed June 2, 2020). [Google Scholar]
- 2.Rowley J, Vander Hoorn S, Korenromp E, et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull World Health Organ 2019; 97: 548–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Sexually transmitted infections (STIs): key facts. June 14, 2019. https://www.who.int/news-room/fact-sheets/detail/sexually-transmitted-infections-(stis) (accessed June 2, 2020). [Google Scholar]
- 4.US Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013. April 23, 2013. https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed June 2, 2020). [Google Scholar]
- 5.US Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. December, 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threatsreport-508.pdf (accessed June 2, 2020). [Google Scholar]
- 6.Workowski KA, Bolan GA. Sexually transmitted diseases treatment guidelines, 2015. June 5, 2015. https://www.cdc.gov/std/tg2015/tg-2015-print.pdf (accessed June 2, 2020). [Google Scholar]
- 7.Kong FYS, Horner P, Unemo M, Hocking JS. Pharmacokinetic consideration regarding the treatment of bacterial sexually transmitted infections with azithromycin: a review. J Antimicrob Chemother 2019; 74: 1157–66. [DOI] [PubMed] [Google Scholar]
- 8.Whittles LK, White PJ, Paul J, Didelot X. Epidemiological trends of antibiotic resistant gonorrhoea in the United Kingdom. Antibiotics 2018; 7: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wi T, Lahra MM, Ndowa F, et al. Antimicrobial resistance in Neisseria gonorrhoeae: global surveillance and a call for international collaborative action. PLoS Med 2017; 14: e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George CRR, Enriquez RP, Gafus BJ, et al. Systematic review and survey of Neisseria gonorrhoeae ceftriaxone and azithromycin susceptibility data in the Asian Pacific, 2011 to 2016. PLoS One 2019; 14: e0213312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin I, Sawatzky P, Allen V, et al. Multidrug-resistant and extensively drug-resistant Neisseria gonorrhoeae in Canada, 2012–2016. Can Commun Dis Rep 2019; 45: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuda M, Hatazaki K, Ito S, et al. Antimicrobial susceptibility of Neisseria gonorrhoeae in Japan from 2000 to 2015. Sex Transm Dis 2017; 44: 149–53. [DOI] [PubMed] [Google Scholar]
- 13.Public Health England. Surveillance of antimicrobial resistance in Neisseria gonorrhoeae in England and Wales: key findings from the Gonococcal Resistance to Antimicrobials Surveillance Programme (GRASP)—data to May 2018. October, 2018. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/746261/GRASP_2017_report.pdf (accessed June 2, 2020).
- 14.Tamma PD, Cosgrove SE, Maragakis LL. Combination therapy for treatment of infections with Gram-negative bacteria. Clin Microbiol Rev 2010; 25: 450–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kersh EN, Allen V, Ransom E, et al. Rationale for a Neisseria gonorrhoeae susceptible-only interpretive breakpoint for azithromycin. Clin Infect Dis 2020; 70: 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyre DW, Sanderson ND, Lord E, et al. Gonorrhoea treatment failure caused by a Neisseria gonorrhoeae strain with combined ceftriaxone and high-level azithromycin resistance, England, February 2018. Euro Surveill 2018; 23: 1800323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 2014; 27: 587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grad YH, Harris SR, Kirkcaldy RD, et al. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis 2016; 214: 1579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadsworth CB, Arnold BJ, Sater MRA, Grad Y. Azithromycin resistance through interspecific acquisition of an epistasis-dependent efflux pump component and transcriptional regulator in Neisseria gonorrhoeae. mBio 2018; 9: e01419–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouquette-Loughlin CE, Reimche JL, Balthazar JT, et al. Mechanistic basis for decreased antimicrobial susceptibility in a clinical isolate of Neisseria gonorrhoeae possessing a mosaic-like mtr efflux pump locus. mBio 2018; 9: e02281–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas JC, Seby S, Abrams AJ, et al. Evidence of recent genomic evolution in gonococcal strains with decreased susceptibility to cephalosporins or azithromycin in the United States, 2014–2016. J Infect Dis 2019; 220: 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.US Centers for Disease Control and Prevention. Gonococcal Isolate Surveillance Project (GISP): protocol. May, 2016. https://www.cdc.gov/std/gisp/GISP-Protocol-May-2016.pdf (accessed June 2, 2020). [Google Scholar]
- 23.US Department of Health and Human Services. Regional offices. April 15, 2014. https://www.hhs.gov/about/agencies/iea/regionaloffices/index.html (accessed June 2, 2020). [Google Scholar]
- 24.Ribot EM, Freeman M, Hise KB, et al. PulseNet: entering the age of next-generation sequencing. Foodborne Pathog Dis 2019; 16: 451–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shafer W. Mosaic drug efflux gene sequences from commensal Neisseria can lead to low-level azithromycin resistance expressed by Neisseria gonorrhoeae clinical isolates. mBio 2018; 9: e01747–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handing JW, Ragland SA, Bharathan UV, Criss AK. The MtrCDE efflux pump contributes to survival of Neisseria gonorrhoeae from human neutrophils and their antimicrobial components. Front Microbiol 2018; 9: 2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williamson DA, Chow EPF, Gorrie CL, et al. Bridging of Neisseria gonorrhoeae lineages across sexual networks in the HIV pre-exposure prophylaxis era. Nat Commun 2019; 10: 3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demczuk W, Martin I, Peterson S, et al. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J Clin Microbiol 2016; 54: 1304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, van der Veen S. Neisseria gonorrhoeae 23S rRNA A2059G mutation is the only determinant necessary for high-level azithromycin resistance and improves in vivo biological fitness. J Antimicrob Chemother 2019; 74: 407–15. [DOI] [PubMed] [Google Scholar]
- 30.Jacobsson S, Golparian D, Cole M, et al. WGS analysis and molecular resistance mechanisms of azithromycin-resistant (MIC >2 mg/L) Neisseria gonorrhoeae isolates in Europe from 2009 to 2014. J Antimicrob Chemother 2016; 71: 3109–16. [DOI] [PubMed] [Google Scholar]
- 31.Kersh EN, Pham CD, Papp JR, et al. Expanding U.S. laboratory capacity for Neisseria gonorrhoeae antimicrobial susceptibility testing and whole-genome sequencing through the CDC’s Antibiotic Resistance Laboratory Network. J Clin Microbiol 2020; 58: e01461–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.