Abstract
Two recurrent mutations, K27M and G34R/V, in H3F3A, encoding non-canonical histone H3.3, are reported in pediatric and young adult gliomas, whereas G34W mutation is prevalent in bone tumors. In contrast to K27M mutation, it remains elusive how G34 mutations affect the epigenome. Here we performed whole-genome bisulfite sequencing of four G34R-mutated gliomas and the G34V-mutated glioma cell line KNS-42 for comparison with gliomas harboring K27M and no mutations in H3F3A and with G34W-mutated bone tumors. G34R-mutated gliomas exhibited lower global methylation levels, similar CpG island (CGI) methylation levels, and compromised hypermethylation of telomere-proximal CGIs, compared to the other two glioma subgroups. Hypermethylated regions specific to G34R-mutated gliomas were enriched for CGIs, including those of OLIG1, OLIG2, and canonical histone genes in the HIST1 cluster. They were notably hypermethylated in osteosarcomas with, but not without, G34W mutation. Independent component analysis revealed that G34 mutation-specific components shared a significant similarity between glioma and osteosarcoma, suggesting that G34 mutations exert characteristic methylomic effects regardless of the tumor tissue-of-origin. CRISPR/Cas9-mediated disruption of G34V-allele in KNS-42 cells led to demethylation of a subset of CGIs hypermethylated in G34R-mutated gliomas. These findings will provide a basis for elucidating epigenomic roles of G34 oncohistone in tumorigenesis.
Subject terms: Cancer, Epigenomics, DNA methylation
Introduction
DNA methylation is a major epigenetic modification involved in a wide variety of biological processes and hence various diseases, especially, cancers. Global CpG hypomethylation and focal hypermethylation at CpG islands (CGIs) are well-known hallmarks of cancers1–3 and contribute to carcinogenesis via distinct mechanisms: global hypomethylation causes chromosome instability leading to various structural variations, whereas hypermethylation of promoter CGIs suppresses the expression of downstream genes, especially, tumor suppressor genes.
Genetic alterations involved in gliomagenesis have been extensively investigated4–7. These alterations include IDH1/2 mutations and 1p/19q co-deletion, which have served as the bases for the new nomenclature of central nervous system tumors8. The association of epigenetic alterations with glioma has also been reported. For instance, IDH1/2-mutated gliomas harbor a characteristic methylation pattern defined as the glioma-CGI methylator phenotype and associate with favorable prognosis. Methylation of the promoter of MGMT, encoding O6-methylguanine-DNA methyltransferase, was demonstrated to be a predictive factor for chemosensitivity to alkylating agents9. A recent study demonstrated the utility of methylation profiling in auxiliary diagnosis of central nervous system tumors10.
The H3F3A gene is one of the two genes encoding histone H3.3, a non-canonical histone variant, and has been established as a major driver gene of malignant gliomas, particularly pediatric and young adult gliomas11–13. Histone H3.3 is loaded to transcriptionally active regions in a replication-independent manner and is conserved among various species, ranging from yeast to human14. Several cancers bear missense mutations that affect the N-terminal tail of histone H3.3. Two recurrent mutations of H3F3A, K27M and G34R/V, have been identified in glioma, whereas G34W/R/L mutations of H3F3A are prevalent in giant cell tumor of bone (GCTB) and K36M mutation of H3F3B recurs in chondrosarcoma15–17.
Previous studies on gliomas revealed that K27M and G34R/V subgroups correlate with distinct clinicopathological features such as disease prevalence, age, tumor location, prognosis, MRI findings, and histopathological diagnosis11,18. Distinct patterns of gene expression and DNA methylation, which likely affect disease phenotypes, have also been reported for these gliomas primarily by microarray analyses12. Epigenetic mechanisms of tumor development have been well elucidated for the K27M subgroup, which is defined as "diffuse midline glioma, H3 K27M-mutant" in the latest WHO classification10. Histone H3.3 bearing K27M substitution traps the Polycomb Repressive Complex 2 (PRC2) to inhibit its histone methyltransferase activity, thereby leading to a global decrease and a local increase of histone H3 trimethylated at its K27 residue19,20. Accordingly, EZH2, the catalytic component of PRC2, is a promising target of new molecular targeted therapy21,22.
On the other hand, the mechanism behind the contribution of G34 mutations to tumorigenesis remains elusive. As an approach to address the epigenetic mechanisms of action for G34 variants, we conducted whole-genome bisulfite sequencing (WGBS) by post-bisulfite adaptor tagging (PBAT)23,24 to delineate single base-resolution methylomes of four G34-mutated glioma cases characterized in our previous study25. Comparison of the WGBS data with those of other types of gliomas and bone tumors highlighted a unique methylomic signature shared with bone tumors bearing G34 mutations.
Results
Global hypomethylation and exacerbated variation of regional methylation in G34-mutated gliomas
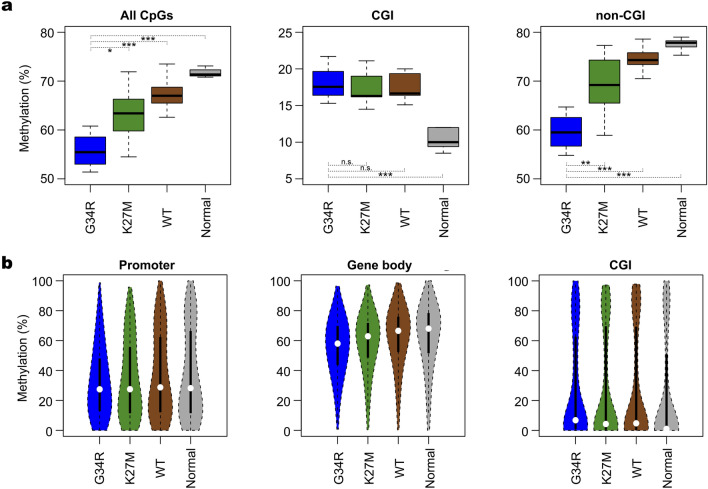
We used the PBAT method to conduct PCR-free WGBS of four, seven, and three cases of primary gliomas harboring G34R mutation (G34 subgroup), K27M mutation (K27 subgroup), and no mutation in the H3F3A gene (WT subgroup), respectively, as well as the G34V-mutated glioma cell line KNS-42 (Tables S1 and S2). Combining these WGBS datasets with those obtained from the ICGC database, we compared the mean CpG methylation levels among the three subgroups. G34 subgroup exhibited the lowest mean methylation level (55.8%), which was followed by those of K27 (63.6%) and WT (67.3%) subgroups (Fig. 1). This order was maintained in the promoters and gene bodies but not in CGIs; mean CGI methylation levels were comparable among the three subgroups (Fig. 1). All the three subgroups exhibited lower global and higher CGI methylation than the normal brain tissues.
Figure 1.
Global methylation levels of gliomas and normal brain tissue. (a) Box plots of mean CpG methylation levels. Data on all CpGs, those in CGIs, and those in non-CGI regions are shown for the three glioma subgroups and the normal brain tissue (*p < 0.05; **p < 0.01; ***p < 0.001; n.s., not significant). G34R, N = 4; K27M, N = 13; WT, N = 8; Normal, N = 8. (b) Violin plots of CpG methylation levels in promoters, gene bodies, and CGIs. Mean methylation levels are calculated for non-overlapping 1-Mb sliding windows.
To further evaluate the methylation pattern among the three subgroups, we divided the genome into non-overlapping 1-Mb sliding windows, calculated mean methylation levels of individual windows, and examined their distribution (Figure S1a). While the methylation means of the windows exhibited a unimodal distribution in each subgroup, the peak of G34 subgroup was relatively the most left-shifted (i.e., hypomethylated) and broadest among the three subgroups. When normalized to each genome-wide methylation mean, the G34 subgroup showed a wider distribution in both directions (i.e., relative hypermethylation and hypomethylation) compared to the other two subgroups, indicating a larger variance in local methylation levels (Figure S1b).
We also analyzed partially methylated domains (PMDs), which are large genomic regions with intermediate methylation levels and often overlap with the lamina-associated domains (LADs)26. Almost all PMDs common to the four G34R-mutated gliomas overlapped with those identified in at least one case of the other two subgroups (Figure S2). Thus, there were likely very few, if any, qualitative differences of PMDs between G34 and the other two subgroups.
Mitigation of telomere-proximal CGI hypermethylation in G34-mutated glioma
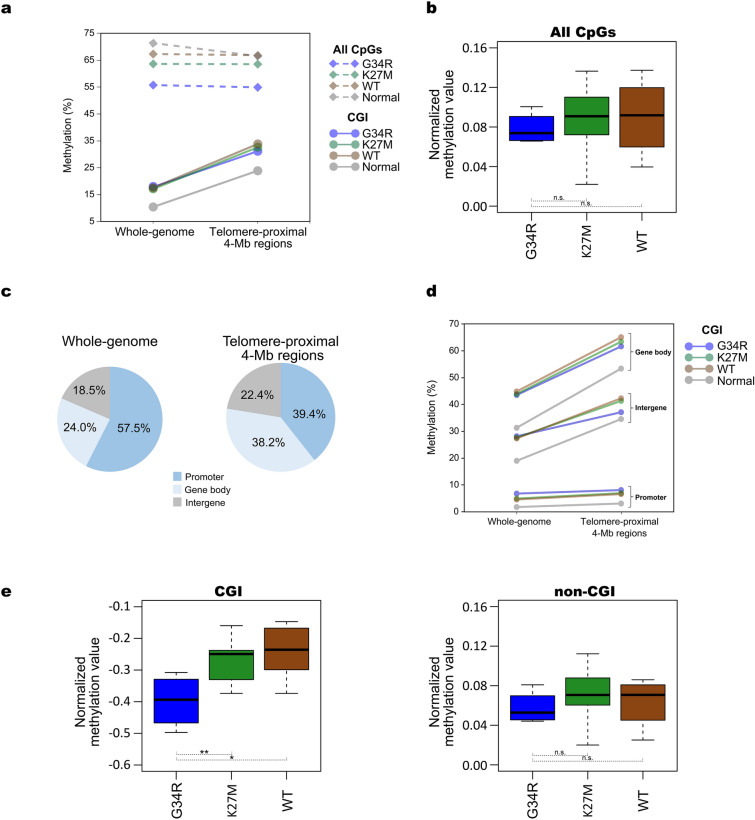
A previous methylation array study reported “enhanced hypomethylation” at the chromosomal ends (i.e., telomere-proximal 4-Mb regions) as a characteristic of G34-mutated glioma12. However, current WGBS data indicated that mean methylation levels of these regions are comparable to the genome-wide mean in all the three subgroups (Fig. 2a, broken lines). In the normal brain tissues, methylation of these regions was lower than the genome-wide mean (Fig. 2a). Even after normalization to the normal brain tissue, telomere-proximal 4-Mb regions failed to show a significant difference among the three subgroups in contrast to the previous study12 (Fig. 2b).
Figure 2.
Methylation trends in telomere-proximal regions. (a) Mean methylation levels of whole-genome and telomere-proximal 4-Mb regions. Mean methylation levels are separately plotted for all CpG sites and those within CGIs. Only mean values of individual sample categories are indicated for clarity. G34R, N = 4; K27M, N = 13; WT, N = 8; Normal, N = 8. (b) Normalized mean methylation value of telomere-proximal 4-Mb regions. The methylation value was calculated using all CpG sites as follows. Mean methylation level of telomere-proximal 4-Mb regions of each sample category was first normalized by its whole-genome mean methylation level. The normalized value of each tumor subgroup was further normalized to that of the normal brain tissue. Log2-values are indicated. (c) Compositional difference of CGIs between whole-genome and telomere-proximal regions. CGIs are divided into those at promoters, gene bodies, and the other regions (intergenic regions). (d) Mean methylation levels of three distinct CGI classes in whole-genome and telomere-proximal 4-Mb regions. (e) Normalized mean methylation values of CGIs and non-CGI regions in telomere-proximal regions. Each methylation value was calculated as in (b).
To reconcile this apparent discrepancy, we separately analyzed CGIs and non-CGI portions of the genome. Notably, the mean CGI methylation level in telomere-proximal 4-Mb regions was higher than the genome-wide mean in both normal and tumor samples (Fig. 2a, solid lines). To address the reason for this elevated methylation, we classified CGIs into those at promoters, gene bodies, and intergenic regions. Intriguingly, CGIs at gene bodies and promoters were over- and underrepresented, respectively, in telomere-proximal 4-Mb regions, compared to the genome-wide mean (Fig. 2c). Gene body CGIs showed the highest mean methylation level, which was further elevated near the telomeres (Fig. 2d). By contrast, promoter CGIs showed the lowest mean methylation level with little, if any, sign of the telomere-associated elevation (Fig. 2d). These differences likely contributed to the observed elevation of CGI methylation levels at the chromosomal ends.
We next compared the extent of elevation in telomere-proximal CGI methylation among the three subgroups. Compared to K27 and WT subgroups, G34 subgroup showed a sign of mitigation in the elevation of methylation (Fig. 2a, solid lines), which was most evident in the intergenic CGIs (Fig. 2d). Indeed, when normalized to the normal brain tissue, G34 subgroup showed a significantly lower normalized methylation value than the other two subgroups (Fig. 2e, left). Importantly, however, non-CGI portions of telomere-proximal regions failed to show any significant difference in the normalized methylation values among the three subgroups (Fig. 2e, right). In the unbiased sampling by WGBS, the number of interrogated CpG sites from non-CGI portions overwhelms those from CGIs. Hence, current WGBS data likely masked the methylation changes at CGIs by those at non-CGI portions, leading to the apparently comparable methylation values among the three subgroups (Fig. 2b).
Taken together, while all gliomas exhibited the elevation of CGI methylation level in telomere-proximal regions, its extent was less prominent in the G34 subgroup compared to the other two subgroups.
Methylation changes specific to G34-mutated gliomas
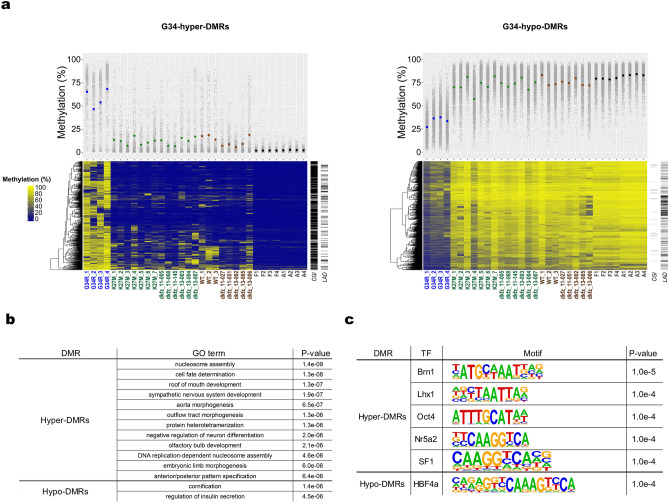
To identify the most characteristic methylomic changes, we aimed to identify differentially methylated regions (DMRs) between G34 and the other two subgroups (i.e., G34 subgroup-specific DMRs). For this purpose, we first identified DMRs between the G34 and K27 subgroups, between the G34 and WT subgroups, and between the G34 subgroup and the normal brain tissue. Next, we determined the intersection of these three DMR sets to define 687 G34 subgroup-specific hyper-DMRs (G34-hyper-DMRs; 0.2 Mb in total) and 2,064 G34 subgroup-specific hypo-DMRs (G34-hypo-DMRs; 1 Mb in total) (Figure S3a; Table S3). Similarly, we identified K27-specific DMRs, WT-specific DMRs, and DMRs common to all the three subgroups (Figures S3b–S3f).
G34-hyper-DMRs and G34-hypo-DMRs were enriched for CGIs and LADs, respectively (Fig. 3a). Genes proximal to G34-hyper-DMRs were enriched for those associated with the GO term “nucleosome assembly”, including canonical histone genes, especially, those in the HIST1 cluster (Fig. 3b; Table S3). They were also enriched for genes involved in neural differentiation, including those encoding oligodendrocyte-specific transcription factors OLIG1 and OLIG2, consistent with a previous study12. The binding motif of BRN1/POU3F3, a POU family transcription factor involved in neural development, was overrepresented in G34-hyper-DMRs (Fig. 3c). On the other hand, G34-hypo-DMRs failed to enrich notable proximal genes and transcription factor-binding motifs (Fig. 3b,c; Table S3).
Figure 3.
Hypermethylated and hypomethylated regions specific to the G34 subgroup. (a) G34-hyper-DMRs and G34-hypo-DMRs. Raw methylation levels are used for hierarchical clustering of DMRs. Rows and columns of the heatmap indicate DMRs and samples, respectively. Ladders on the right side of each panel indicate CGIs and LADs. G34R, N = 4; K27M, N = 13; WT, N = 8; Normal, N = 8. (b) GO enrichment analysis of DMRs. See Table S3 for details. (c) Transcription factor-binding motifs enriched in DMRs.
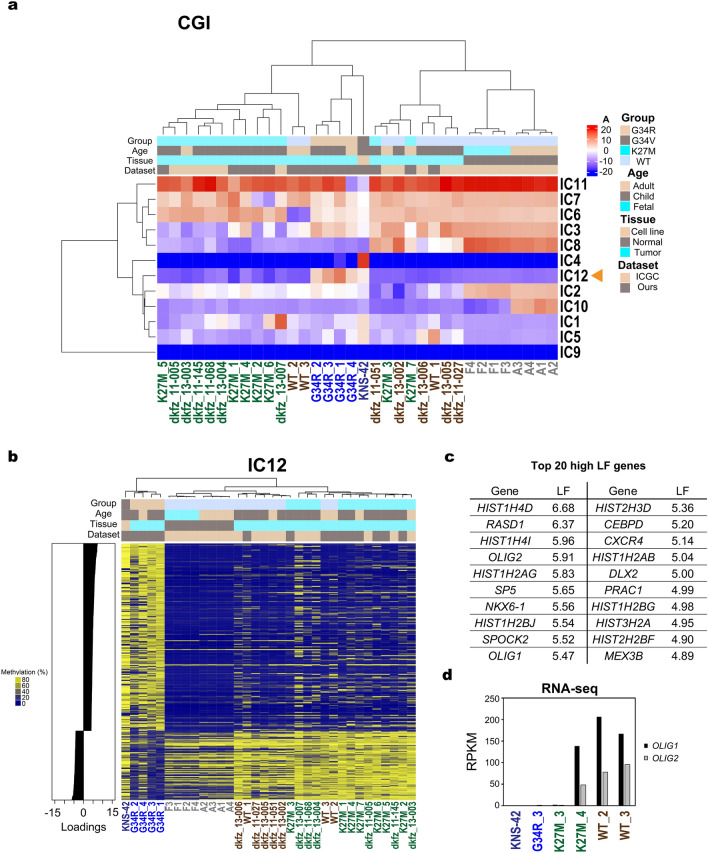
As an alternative approach to identifying G34 subgroup-specific alterations, we applied independent component analysis (ICA) to the WGBS data. ICA is a method for blind source separation to decompose a data matrix into the product of a mixing matrix and independent components (ICs), the latter of which represent hidden source signals27. The elements of the mixing matrix can be used for hierarchical clustering of samples, based on the rationale that biologically similar samples should share a similar pattern in weighting to individual source signals. The mixing matrix also helps identify subgroup-specific ICs, which are weighted either positively or negatively in certain subgroups, but not in the others. Major contributors showing large, either positive or negative, loading factors (LFs) to such a subgroup-specific IC should serve as good methylation markers for the subgroup.
We separately applied ICA to the methylation data for promoters, gene bodies, and CGIs (Figs. 4a and S4a). In hierarchical clustering based on the mixing matrix, all G34R-mutated gliomas and KNS-42 formed a monophyletic cluster excluding the others when CGIs and gene bodies were used in ICA (Figs. 4a and S4a). On the other hand, two K27M-mutated gliomas were co-clustered with the G34R-mutated gliomas in the ICA of promoters (Figure S4A). Note that IC12 (CGIs), IC3 (gene bodies), and IC3 (promoters) exhibited significantly different weighting patterns between G34 and the other two subgroups, thus serving as G34-specific ICs (Figs. 4b and S4).
Figure 4.
Independent component analysis (ICA). (a) Unsupervised hierarchical clustering of samples and ICs by mixing matrix coefficients. Results are shown for ICA of CGIs. Arrowhead indicates IC12 showing distinct weighting patterns between the G34 subgroup and the others. G34R/V, N = 5; K27M, N = 13; WT, N = 8; Normal, N = 8. (b) Unsupervised hierarchical clustering of samples by raw methylation levels of CGIs with high loading factors (LFs) for IC12 of ICA using CGIs. CGIs with LFs larger than 3 or smaller than − 3 are selected for the hierarchical clustering. (c) List of the top 20 genes with high LFs for G34 subgroup-specific IC of ICA using gene bodies. (d) Expression levels of OLIG1 and OLIG2. RPKM values calculated from RNA-seq data are shown for the indicated samples.
Genes with large-positive LFs to these G34 subgroup-specific ICs were enriched for canonical histone genes in the HIST1 cluster and the GO term “nucleosome assembly” (Fig. 4c; Table S4), consistent with the DMR analysis (Fig. 3; Table S3). The large positive LF genes also included OLIG1 and OLIG2, silencing of which was indicated by RNA-seq in the G34 subgroup (Fig. 4d). On the other hand, genes with large negative LFs failed to enrich any notable GO terms (Table S4).
G34 variant-specific methylomic signature shared between gliomas and bone tumors
Mutations affecting the G34 residue of histone H3.3 are prevalent in both gliomas and GCTBs. The G34W mutation occurs in most, if not all, GCTBs and a fraction of osteosarcomas. A study of GCTBs and osteosarcomas using methylation array attracted our attention, because it identified HIST1H2BB as the most differentially methylated gene between osteosarcomas with and without G34W mutation15 and because this gene was found within G34-hyper-DMRs and among the large-positive LF genes for G34 subgroup-specific ICs in our study (Tables S3 and S4). We thus wondered whether the G34W mutation in bone tumors induces methylomic changes resembling those induced by G34R mutation in gliomas.
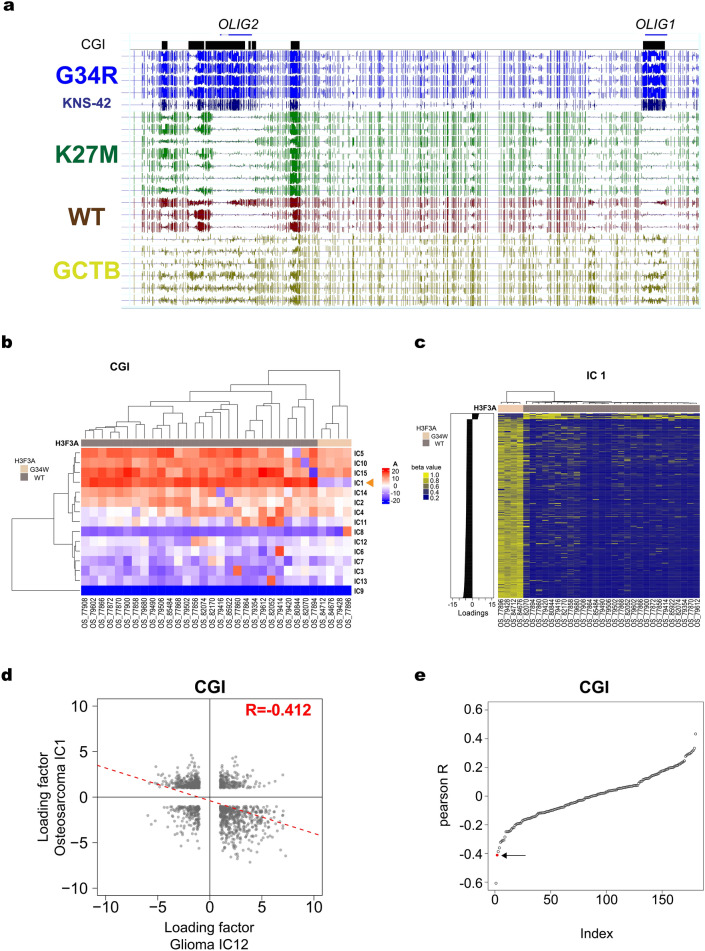
To directly address this possibility, we performed shallow WGBS on six GCTB cases, all of which were confirmed to harbor the G34W mutation16, and found that the HIST1 cluster, OLIG1, and OLIG2 exhibited modest levels of hypermethylation (Fig. 5a). Judging from the mutant allele frequency revealed by exome sequencing (0.09–0.24)16, we attributed the modest levels to the relatively low tumor cell content in these specimens. However, it remains possible that the G34W mutation was not related to the observed methylation changes because no data was available for GCTB cases lacking H3F3A mutations.
Figure 5.
Methylomic signature shared by gliomas and bone tumors bearing G34 mutations. (a) Genome browser shot of OLIG1/OLIG2 locus in gliomas and GCTBs. (b) Unsupervised hierarchical clustering of osteosarcoma samples and ICs by mixing matrix coefficients. Results are shown for ICA of CGIs based on the methylation array data (N = 32)15. Arrowhead indicates IC1, which shows distinct weighting patterns between tumors with and without G34W mutation. (c) Unsupervised hierarchical clustering of osteosarcoma samples by raw methylation levels of CGIs. Genes with LFs larger than 3 or smaller than − 3 are used for clustering. (d) Correlation plot of LFs between glioma IC12 and osteosarcoma IC1, both specific to G34 subgroup. ICA was performed using CGIs. (e) Pearson’s correlation coefficients of all possible pairs between ICs identified in ICA of glioma and osteosarcoma. ICA uses CGIs and the coefficients are sorted by their values. Red circle pointed by the arrow indicates the pair of G34-specific ICs (i.e., glioma IC12 and osteosarcoma IC1).
We thus applied ICA to the methylation array data on osteosarcomas including both tumors with and without G34W mutation15. Interestingly, ICA readily identified G34W-specific ICs. For instance, ICA of CGIs identified IC1 to be G34W-specific (Fig. 5b,c). Similarly, ICA of promoters and gene bodies identified IC9 and IC12, respectively, as G34W-specific ICs, major contributors to which were notably enriched for genes associated with the GO term “nucleosome assembly” or noncanonical histone genes in the HIST1 cluster (Figure S5a, Table S5). In addition, OLIG1 and OLIG2 showed large negative LFs to G34W-specific IC12 (OLIG1−5.71, OLIG2−4.28, Table S5).
We reasoned that if G34 mutations have similar methylome-wide effects in gliomas and osteosarcomas, G34 mutation-specific ICs identified in these two tumors should show an overall similarity. Indeed, LFs of highly contributing CGIs in glioma IC12 and osteosarcoma IC1 showed a good correlation (Fig. 5d). Note that the correlation was negative, because positive and negative LFs corresponded to hypermethylation in the glioma and osteosarcoma ICA, respectively (Figs. 4b, 5c). To evaluate the correlation further, we calculated Pearson’s correlation coefficients (PCCs) in all possible combinations between glioma and osteosarcoma ICs identified in ICA of CGIs. Notably, G34 mutation-specific ICs of glioma and osteosarcoma showed the mutual top-hit relation: glioma IC12 showed the negatively largest PCC with IC1 among 15 osteosarcoma ICs, whereas osteosarcoma IC1 showed the negatively largest PCC with IC12 among 12 glioma ICs. The pair of G34 mutation-specific ICs was in those displaying the highest PCCs among the 180 (= 12 × 15) possible combinations (Fig. 5e). We confirmed similar findings using the methylation array data of G34-mutated gliomas12 instead of the WGBS data (Figures S5b and S5c).
Taken together, these results suggested that G34 mutations exert a characteristic methylome-wide effect regardless of the tumor tissue of origin, presumably through a common underlying mechanism.
Possible role of G34-substituted histone H3.3 in CGI methylation
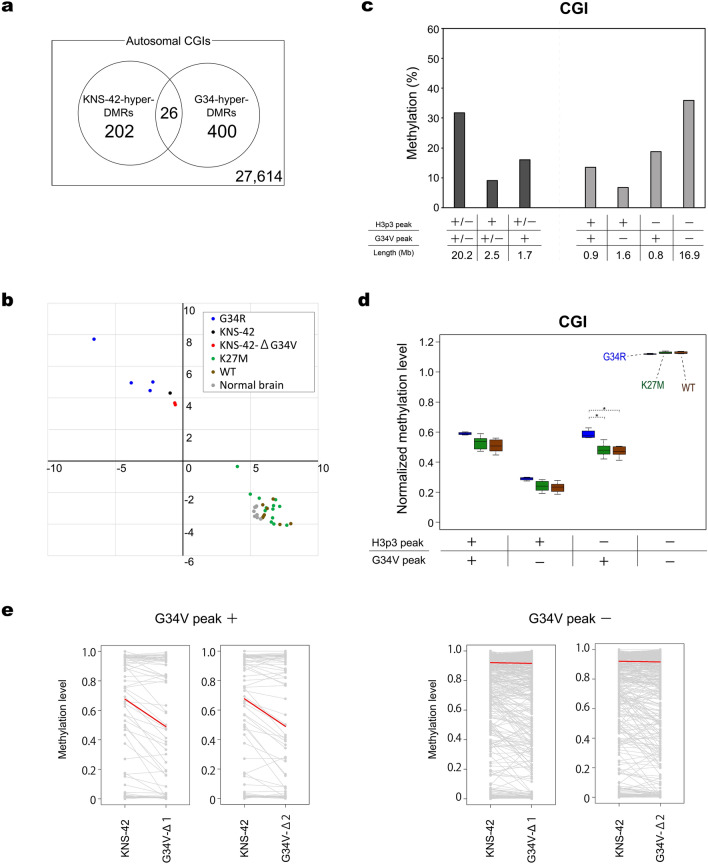
As an approach to address the mechanism underlying the G34-specific methylomic signature, we examined the effects of CRISPR/Cas9-mediated disruption of G34V-allele in KNS-42 cells. We selected two disruptants (Figures S6a–S6c) and identified DMRs between them and their parental KNS-42 cells by WGBS (Table S6). Although the overall effects of G34V-allele disruption were rather limited, CGIs in KNS-42-hyper-DMRs (i.e., genomic regions demethylated in the disruptants compared to KNS-42 cells) showed a significant overlap with CGIs in G34-hyper-DMRs (Fig. 6a; p = 2.4 × 10−15, odds ratio = 8.7). In ICA, the two disruptants had lower weightings on G34-specific ICs than their parental KNS-42 cells (Figs. 6b and S6d). It thus seemed that the contribution of G34-specific methylomic signature was diminished in the absence of G34V-substituted histone H3.3. These results suggested that G34V mutation contributes, at least, partly to maintain the G34-specific methylome signature.
Figure 6.
Effect of G34-substituted histone H3.3 on CGI hypermethylation. (a) Overlap between CGIs in G34-hyper-DMRs and those in KNS-42-hyper-DMRs. p value = 2.4 × 10−15; odds ratio = 8.7 (Fisher’s exact test). (b) Effect of G34V-allele disruption on G34-specific methylomic signature in KNS-42 cells. ICA including the two disruptants identified two IC’s to be G34-specific, one from ICA using CGIs and the other from ICA using promoters (Figure S6d). For each sample, weightings to the former and latter ICs are plotted to horizontal and vertical axes, respectively. Note that both disruptants depart from their parental KNS-42 cells toward the other 13 samples without G34 mutations, or in the opposite direction to four G34R gliomas. (c) Methylation levels of CGIs with differential histone H3.3 deposition patterns in KNS-42 cells. CGIs are classified into four groups based on the presence (+) or absence (−) of overlapping H3p3 and G34V peaks in KNS-42 cells. Each bar indicates mean methylation level of each CGI group. Length indicates the total length of genomic regions exhibiting the indicated ChIP-seq peak patterns. (d) Normalized methylation levels of the four CGI groups in clinical glioma samples. CGIs are classified as in c, and their methylation levels in glioma subgroups are displayed by box plots. Methylation levels are normalized to the genome-wide mean of individual tumor samples. G34, N = 4; K27M, N = 13; WT, N = 8. (e) Effect of G34V-allele disruption on G34-hyper-DMRs. Ladder charts are shown for methylation levels of CGIs in G34-hyper-DMRs in KNS-42 and each disruptant. CGIs in G34-hyper-DMRs are divided into those overlapping with G34V peaks (N = 92) and those without them (N = 334). Red lines indicate the median methylation change.
We also performed ChIP-seq using two monoclonal antibodies, one specific to histone H3.3 and the other specific to histone H3.3-G34V. We hereinafter refer to ChIP-seq peaks obtained by the former and latter antibodies as H3p3 and G34V peaks, respectively. Both H3p3 and G34V peaks were enriched in CGIs and promoters in KNS-42 cells, sharing a largely similar distribution pattern (Figures S7a and S7b). We examined the overlap between ChIP-seq peaks and G34-hyper/hypo-DMRs. G34V peaks overlapped with 13% and 0.9% of G34-hyper-DMRs and G34-hypo-DMRs, respectively (Figure S7c). Note that G34-hyper-DMRs were enriched for CGIs (Fig. 3a; p < 2.2 × 10−16, odds ratio = 97.0) and that CGIs were enriched for H3p3 and G34V peaks (Figure S7a, p < 2.2 × 10−16; odds ratio = 7.72 for H3p3, 3.61 for G34V).
To determine the relationship between histone H3.3 deposition and CGI methylation, we first classified CGIs into four distinct groups based on the presence and absence of overlapping ChIP-seq peaks in KNS-42 cells and then compared methylation levels among the four groups. CGIs with histone H3.3 were less methylated than those without it, irrespective of whether the 34th residue was Gly or Val (Fig. 6c). Among histone H3.3-bearing CGIs, those with G34V peaks showed higher methylation levels than those without them (Fig. 6c). A similar trend was observed in clinical tumor samples, even in those without G34R mutation (Fig. 6d). It was thus possible that these CGIs were intrinsically prone to hypermethylation and that the G34 variants were preferentially incorporated to and/or stably retained in the hypermethylated nucleosomes. Intriguingly, however, the G34 subgroup exhibited a larger extent of hypermethylation than the other two subgroups (Fig. 6d). These results suggest an involvement of G34R variants in further elevating, or counteracting to decrease, CGI methylation levels. In this context, it is intriguing to note that CGIs in G34-hyper-DMRs with G34V peaks, but not those without them, were efficiently demethylated upon G34V-allele disruption in KNS-42 cells (Fig. 6e). Collectively, these results suggested an involvement of G34-substituted histone H3.3 in hypermethylation of a subset of CGIs.
Discussion
We used the enhanced PBAT protocol24 to generate WGBS data on 14 gliomas, including the first four WGBS data on cases with histone H3.3-G34R. These data not only consolidated the findings in a previous study using the methylation array12 but extend the characteristics of G34-mutated gliomas, including a specific signature shared by G34-mutated bone tumors.
The previous study reported that G34-mutated gliomas exhibit two features, namely, global CpG hypomethylation and enhanced hypomethylation at the chromosomal ends12. In contrast to global hypomethylation (Fig. 1), enhanced hypomethylation at the chromosomal ends was not readily apparent in the WGBS data (Fig. 2). We demonstrated that the enhanced hypomethylation reported previously was based on the less prominent elevation of CGI methylation levels in telomere-proximal 4-Mb regions in the G34 subgroup compared to that in the other two subgroups (Fig. 2). It remains to be seen in future studies how G34 mutations, or coincident loss of ATRX, mitigates the elevation of CGI methylation levels in the telomere-proximal regions, a common feature shared between normal brain tissue and glioma.
We also identified methylomic changes specific to G34 subgroup using both conventional DMR search and ICA (Figs. 3, 4). We confirmed the previously reported G34 subgroup-specific hypermethylation and downregulation of OLIG1 and OLIG212. The requirement of OLIG2 in gliomagenesis has been shown to be context-dependent28 and the pathogenic significance of its suppression in G34 subgroup remains an open question. We also found an enrichment of the binding motif of BRN1/POU3F3 in G34-hyper-DMRs. Hypermethylation around these sites may prevent BRN1/POU3F3 from binding to its target sites, thereby exerting an effect similar to its downregulation. In this context, it is interesting to note that the overexpression of Linc-POU3F3, a non-coding RNA that suppresses the expression of POU3F3 mRNA, promotes cell viability and proliferation of glioma cells29. In addition, we revealed coordinated hypermethylation of canonical histone genes in the HIST1 cluster, pathophysiological significance of which remains elusive. Since histone genes form a specific subnuclear structure called the histone locus body30, coordinated methylation changes may reflect alterations of the structure. A recent study on hypermethylation of histone-related genes in lung cancer proposed HIST1H4F hypermethylation as a potential pan-cancer biomarker, based on the analysis of 17 tumor types, not including glioma, from the TCGA database31. Our data, however, showed that hypermethylation of HIST1H4F occurs in G34 subgroup but not in the others. It is also intriguing to note that although the G34 subgroup seemed to show more unified methylomic changes than the K27 subgroup, tumor phenotypes such as the MRI findings and pathological diagnosis were more variable in the G34 subgroup than in the K27 subgroup32,33.
Furthermore, we applied ICA to the methylation array data on osteosarcomas to identify a G34W-specific signature and revealed its similarity to the G34-specific signature of gliomas (Fig. 5). Although GCTB seemed to share the signature, its clear identification was hampered by the unavailability of rare GCTB cases lacking G34 mutation and by the low tumor cell content in the samples. The latter issue may be overcome by single-cell methylome sequencing in future. In anyway, these results suggest that G34-mutated histone H3.3 exerts a characteristic methylomic effect regardless of the tumor tissue of origin, presumably through a common mechanism. Hence G34-specific DMRs identified in this study would provide good reporters to experimentally pursue the mechanism. These results also indicate a potential of ICA to uncover hidden methylomic signals.
How do G34 variants of histone H3.3 induce epigenomic changes including those of DNA methylation? Previous studies reported that G34R/V and G34W/L mutations locally affect di-/tri-methylation of K36 of histone H3 (H3K36me2/3) in the same nucleosome particle, when overexpressed in 293 T and HeLa cells, respectively19,34. Indeed, these substitutions were shown to inhibit SETD2, which is responsible for H3K36me2/3, in vitro35. On the other hand, a study using mouse embryonic stem cells showed that a knocked-in G34R allele induces widespread changes in H3K9me3 and H3K36me3 through inhibition of the KDM4 family of K9/K36 demethylases36. Although the effects of G34 mutations on H3K36me2/3 remain controversial, the well-established link between H3K36me3 and DNA methylation likely plays a role in methylomic alterations.
In this study, we used KNS-42 cells to examine the effects of G34V-allele disruption on the methylome and to reveal genomic distribution of G34V-substituted histone H3.3. These approaches are straightforward but have inevitably limitations. First, considering the context-dependent and feedback-rich nature of epigenetic regulation, we should note that simple disruption experiments allow us to examine only immediate effects on the maintenance, but not the generation, of methylome patterns. Molecular mechanisms for de novo formation of a methylome pattern can be dispensable for its maintenance, and the well-known recurrence of G34 mutations in distinct tumor types may be a reflection of developmental context-dependence of their epigenetic effects. Second, ChIP-seq results could be confounded if wild-type and mutant proteins are located at the same loci, because no antibody was available to distinguish wild-type histone H3.3 from G34-substituted proteins. Despite these limitations, our results collectively suggested a role for the deposition of G34-substituted histone H3.3 in elevating methylation levels of a subset of CGIs (Fig. 6). There should obviously be additional mechanisms that generate methylome patterns in genomic regions other than these CGIs. We expect that appropriate transgenic models for induced expression of G34 oncohistones and antibodies specific to wild-type histone H3.3 would help future studies to fully elucidate the mechanistic aspects and tumorigenic roles of the characteristic methylome patterns revealed in this study.
Methods
Glioma samples
From our previous comprehensive study analyzing H3F3A mutations in gliomas25, we included 11 H3F3A-mutated primary gliomas comprised of four G34R and seven K27M cases. As a control, we included three cases of age-matched glioblastomas, in which genetic analyses confirmed the absence of driver mutations, such as those in IDH1/2, H3F3A, and BRAF (V600E), in addition to 1p/19q codeletion37. These cases are summarized in Table S1. This investigation was approved by the Ethics Committee of Kyushu University.
CRISPR/Cas9-mediated targeting of G34V allele in KNS-42 cells
The G34V-mutated glioma cell line KNS-42 was obtained from Japanese Collection of Research Bioresources Cell Bank. For CRISPR/Cas9-mediated disruption of G34V allele, a target DNA sequence (5′- TTC TTC ACC ACT CCA GTA GA-3′) was selected from the second exon of H3F3A with CHOPCHOP (New England Biolabs). A template oligonucleotide was designed with EnGen sgRNA Template Oligo Designer (New England Biolabs), synthesized by Eurofins genomics, and used to prepare the single guide RNA (sgRNA). A ribonucleoprotein complex formed between the sgRNA and EnGen Cas9 NLS, S. pyogenes (New England Biolabs) was electroporated into KNS-42 cells using NEON Transfection System (Thermo Fisher Scientific). Following the cloning by limiting dilution, the targeted region was amplified by PCR, cloned with Topo TA cloning kit (Thermo Fisher Scientific), and sequenced using BigDye Terminator v1.1 cycle sequencing kit and SeqStudio genetic analyzer (Thermo Fisher Scientific). Loss of G34V-substituted histone H3.3 was confirmed by Western blotting with anti-histone H3.3 G34V rabbit monoclonal antibody (RevMAb Biosciences, 31-1193-00).
WGBS
We used an improved PBAT protocol based on a highly efficient method for single-stranded DNA ligation24 to prepare PCR-free WGBS libraries from 100 ng of genomic DNAs and subjected them to paired-end sequencing (150 nt × 2) using the Illumina HiSeq X Ten platform at Macrogen Japan Corp. (Kyoto, Japan). WGBS reads were processed as described previously24. The statistics are summarized in Table S2.
Other methylome data
We obtained WGBS data on six and five high-grade gliomas bearing K27M and no mutation in H3F3A (EGAS00001000578), respectively, and eight normal cerebellum samples (EGAS00001000561) from the International Cancer Genome Consortium (ICGC). Methylation array data on GCTBs and osteosarcomas15 were obtained from the authors.
Methylome bioinformatics
Autosomal CpG sites covered at least 10 times were used for downstream analyses. DMRs were identified using the program metilene with default parameters38. For clinical samples, DMRs were filtered to retain those containing at least 20 CpGs, with P-values less than 0.01 and with a methylation difference larger than 30%. For the comparison between KNS-42 with and without G34V allele, we used DMRs with P-values less than 10−5. ICA was performed using the fastICA package in R as described previously39. Outputs of ICA were used to generate Euclidian distance matrices for hierarchical clustering by Ward’s method. All heatmaps were generated using the ComplexHeatmap library40. Motif enrichment analysis was carried out using the findMotifsGenome.pl of HOMER41. All functional enrichment analyses with the Gene Ontology (GO) biological process terms were performed using the topGO package in R.
RNA-seq
Total RNAs were prepared from frozen tissues of five glioma cases and KNS-42 cells using the RNeasy Mini Kit (Qiagen) with DNase treatment. RNA-seq libraries were prepared using the TruSeq stranded mRNA Kit (Illumina) and sequenced using NovaSeq6000 at Macrogen Japan Corp. RNA-seq reads were processed using the CLC Genomics Workbench (Qiagen).
ChIP-seq
KNS-42 cells were fixed with 1% formaldehyde and incubated at room temperature for 15 min. The cells were collected with centrifugation and re-suspended in ChIP-lysis buffer (50 mM HEPES-KOH, pH7.5, 150 mM NaCl, 1 mM EDTA, 0.5% (v/v) sodium deoxycholate, 1% (v/v) NP-40, 0.1% (w/v) sodium dodecyl sulfate) containing 1 × protease inhibitor cocktail (Nacalai Tesque). The chromatin was sheared by sonication with Focused-ultrasonicator S220 (Covaris) using milliTUBE 1-ml AFA Fiber. For ChIP, anti-histone H3.3 rabbit monoclonal antibody (Abcam, ab176840) and anti-histone H3.3 G34V rabbit monoclonal antibody were used as primary antibodies with pre-washed Dynabeads Protein A (Dynal). The epitope of the former antibody lies between the 50th and C-terminal residues of human histone H3.3. The ChIP-ed DNA was purified using the DNeasy Blood and Tissue kit (Qiagen) with some modifications, and 10 ng of the DNA was used for library preparation with the SMARTer ThruPLEX DNA-seq Kit (Takara Bio) according to the manufacturer's instruction. Sequencing was performed using the HiSeq X Ten or NovaSeq 6000 at Macrogen Japan Corp. ChIP-seq reads were mapped to the reference human genome GRCh37 (hg19) and peak calling was performed using MACS242.
Supplementary information
Acknowledgements
We thank Christian Koelsche for the methylation array data of bone tumors and Yoshinao Oda for useful comments on GCTB. This work was supported by AMED under Grant Numbers JP16am0101059 and JP19am0101103 and by JSPS KAKENHI 17H06305.
Author contributions
Y.S., F.M., K.Y., K.I., and T.I. conceived the project; Y.S., F.M., and H.A. performed the experiments and data analyses; Y.S., M.M., N.H., D.K., R.H., Yo.A., T.A., Y.F., Ya.A., A.Y., T.S., K.Y., and K.I. provided the tumor samples; Y.S. and T.I. wrote the paper.
Data availability
WGBS, RNA-seq, and ChIP-seq data have been deposited in the Japanese Genotype–phenotype Archive (JGA) under the accession number JGAS00000000197 and in the DDBJ Sequence Read Archive (DRA) under the accession numbers DRA010212 and DRA010335.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-73116-x.
References
- 1.Klutstein M, Nejman D, Greenfield R, Ceder H. DNA Methylation in cancer and aging. Cancer Res. 2016;76:3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 2.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckel-Passow JE, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N. Engl. J. Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceccarelli M, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis DN, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 9.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 10.Capper D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 12.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi L, Wen H, Shi X. The histone variant H3.3 in transcriptional regulation and human disease. J. Mol. Biol. 2017;429:1934–1945. doi: 10.1016/j.jmb.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koelsche C, et al. Histone 3.3 hotspot mutations in conventional osteosarcomas: A comprehensive clinical and molecular characterization of six H3F3A mutated cases. Clin. Sarcoma Res. 2017;7:9. doi: 10.1186/s13569-017-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogura K, et al. Highly recurrent H3F3A mutations with additional epigenetic regulator alterations in giant cell tumor of bone. Genes Chromosomes Cancer. 2017;56:711–718. doi: 10.1002/gcc.22469. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto H, et al. Diagnostic utility of histone H3.3 G34W, G34R, and G34V mutant-specific antibodies for giant cell tumors of bone. Hum. Pathol. 2018;73:41–50. doi: 10.1016/j.humpath.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 18.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammad F, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 2017;23:483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 21.Piunti A, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med. 2017;23:493–500. doi: 10.1038/nm.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjerke L, et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 2013;3:512–519. doi: 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miura F, Enomoto Y, Dairiki R, Ito T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012;40:e136. doi: 10.1093/nar/gks454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura F, et al. Highly efficient single-stranded DNA ligation technique improves low-input whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2019;47:e85. doi: 10.1093/nar/gkz435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimoto K, et al. Prevalence and clinicopathological features of H3.3 G34-mutant high-grade gliomas: A retrospective study of 411 consecutive glioma cases in a single institution. Brain Tumor Pathol. 2017;34:103–112. doi: 10.1007/s10014-017-0287-7. [DOI] [PubMed] [Google Scholar]
- 26.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyvarinen A, Oja E. Independent component analysis: Algorithms and applications. Neural Netw. 2000;13:411–430. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 28.Kosty J, Lu F, Kupp R, Mehta S, Lu QR. Harnessing OLIG2 function in tumorigenicity and plasticity to target malignant gliomas. Cell Cycle. 2017;16:1654–1660. doi: 10.1080/15384101.2017.1361062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, Wu L, Yang Q, Ye M, Zhu X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene. 2015;554:114–119. doi: 10.1016/j.gene.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 30.Marzluff WF, Koreski KP. Birth and death of histone mRNAs. Trends Genet. 2017;33:745–759. doi: 10.1016/j.tig.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong S, et al. Histone-related genes are hypermethylated in lung cancer and hypermethylated HIST1H4F could serve as a pan-cancer biomarker. Cancer Res. 2019;79:6101–6112. doi: 10.1158/0008-5472.CAN-19-1019. [DOI] [PubMed] [Google Scholar]
- 32.Broniscer A, et al. Gliomatosis cerebri in children shares molecular characteristics with other pediatric gliomas. Acta Neuropathol. 2016;131:299–307. doi: 10.1007/s00401-015-1532-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korshunov A, et al. Histologically distinct neuroepithelial tumors with histone 3 G34 mutation are molecularly similar and comprise a single nosologic entity. Acta Neuropathol. 2016;131:137–146. doi: 10.1007/s00401-015-1493-1. [DOI] [PubMed] [Google Scholar]
- 34.Shi L, Shi J, Shi X, Li W, Wen H. Histone H3.3 G34 mutations alter histone H3K36 and H3K27 methylation in Cis. J. Mol. Biol. 2018;430:1562–1565. doi: 10.1016/j.jmb.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang J, et al. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSalpha interaction. Proc. Natl. Acad. Sci. USA. 2018;115:9598–9603. doi: 10.1073/pnas.1806355115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voon, H. P. J. et al. Inhibition of a K9/K36 demethylase by an H3.3 point mutation found in paediatric glioblastoma. Nat. Commun.9, 3142 (2018). [DOI] [PMC free article] [PubMed]
- 37.Hatae R, et al. Precise detection of IDH1/2 and BRAF hotspot mutations in clinical glioma tissues by a differential calculus analysis of high-resolution melting data. PLoS ONE. 2016;11:e0160489. doi: 10.1371/journal.pone.0160489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jühling F, et al. metilene: Fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;26:256–262. doi: 10.1101/gr.196394.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Araki H, et al. Base-resolution methylome of retinal pigment epithelial cells used in the first trial of human induced pluripotent stem cell-based autologous transplantation. Stem Cell Rep. 2019;13:761–774. doi: 10.1016/j.stemcr.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 41.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGBS, RNA-seq, and ChIP-seq data have been deposited in the Japanese Genotype–phenotype Archive (JGA) under the accession number JGAS00000000197 and in the DDBJ Sequence Read Archive (DRA) under the accession numbers DRA010212 and DRA010335.