Abstract
Co-occurrence of two devastating foliar-fungal diseases of peanut, viz., late leaf spot (LLS), and rust may cause heavy yield loss besides adversely affecting the quality of kernel and fodder. This study reports the mapping of seven novel stress-related candidate EST-SSRs in a region having major QTLs for LLS and rust diseases using an F2 mapping population (GJG17 × GPBD4) consisting of 328 individuals. The parental polymorphism using 1311 SSRs revealed 84 SSRs (6.4%) as polymorphic and of these 70 SSRs could be mapped on 14 linkage groups (LG). QTL analysis has identified a common QTL (LLSQTL1/RustQTL) for LLS and rust diseases in the map interval of 1.41 cM on A03 chromosome, explaining 47.45% and 70.52% phenotypic variations, respectively. Another major QTL for LLS (LLSQTL1), explaining a 29.06% phenotypic variation was also found on LG_A03. A major rust QTL has been validated which was found harboring R-gene and resistance-related genes having a role in inducing hypersensitive response (HR). Further, 23 linked SSRs including seven novel EST-SSRs were also validated in 177 diverse Indian groundnut genotypes. Twelve genotypes resistant to both LLS and rust were found carrying the common (rust and LLS) QTL region, LLS QTL region, and surrounding regions. These identified and validated candidate EST-SSR markers would be of great use for the peanut breeding groups working for the improvement of foliar-fungal disease resistance.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02446-4) contains supplementary material, which is available to authorized users.
Keywords: Expressed sequence tag, Foliar-fungal disease, Groundnut, LLS, QTL mapping
Introduction
Globally, peanut (Arachis hypogaea L.) is one of the most important oilseed crops, having multiple economic uses like vegetable oil, confectionery, and feed (Nawade et al. 2019; Bhalani et al. 2019). It is widely cultivated as a grain-legume in Asia, Africa, and America, especially in the arid and semi-arid regions (Nawade et al. 2018). Besides the rich source of oil (40–60%), protein (25–30%), and carbohydrate (10–20%), it also contains various cardio-protective and anti-carcinogenic compounds (Aggarwal et al. 2004; Ko et al. 2017; Pandey et al. 2012). Globally, the peanut is cultivated in 28.52 Mha with 45.95 Mt of yield (FAOSTAT 2018). Though, India is the second-largest peanut producer (6.70 Mt), but its productivity is very low (1.36 t/ha), when equated with other major producers like China (3.75 t/ha) and USA (4.47 t/ha) (FAOSTAT 2018).
Among biotic constraints, rust, leaf spots, stem rot, and collar rot are the major ones restricting the optimum expression of yield potential (Varshney et al. 2014; Dodia et al. 2019; Bosamia et al. 2020). Late leaf spot [Phaseoisariopsis personata (Berk. and Curt) Deighton] and rust (Puccinia arachidis Speg) are the two most prevalent and devastating foliar-fungal disease of peanut which mostly occur together and may result in yield loss to the tune of 50–70% (Subrahmanyam et al. 1984; Dwivedi et al. 2002). Furthermore, these diseases also badly affect the quality of both kernel and fodder (Dwivedi et al. 2002). The strong linkage between fungal disease resistance with low-productivity and various undesirable pod related traits poses a challenge for the breeding of foliar disease-resistant varieties (Shirasawa et al. 2018). Also, due to the non-availability of high yielding foliar disease-resistant cultivars, fungicide application seems the only option for disease control. However, chemical fungicides are not only costly but their application results in enhanced environmental pollutions (Monyo et al. 2009). Thus, the development of high yielding cultivars with resistance to these fungal diseases is a more eco-friendly approach for sustainable agriculture.
Although, conventional breeding approaches have been attempted, limited success has been achieved for the imposition of foliar-fungal disease resistance in peanut. The conventional disease screening methods are facing a serious challenge due to the defoliating behavior and co-occurrence of these two foliar diseases (Sujay et al. 2012). Further, the tetraploid nature, cross-incompatibility with wild relatives, and low genetic variability in the cultivated gene pool also prevents the sharing of desired alleles from wild and other sources (Pandey et al. 2012).
During the last two decades, significant efforts have been made in groundnut breeding for the improvement of foliar-fungal disease resistance (Mishra et al. 2015). Promising cultivar such as GPBD4 (Gowda et al. 2002) and breeding line VG9514 (Varman et al. 1999) was developed using wild species (Arachis cardenasii), through conventional breeding, and was subsequently used for the development of mapping populations for resistance to foliar-fungal diseases. Substantial efforts have been made for the construction of high-density genetic linkage maps using various DNA markers (Sujay et al. 2012; Varshney et al. 2009; Khedikar et al. 2010; Gautami et al. 2012; Kolekar et al. 2016). The first major genomic region (QTLrust01) contributing up to 55.20% phenotypic variation for rust was identified by Khedikar et al. (2010). Subsequently, this genomic region was saturated and two QTLs each for LLS and rust resistance were mapped (Sujay et al. 2012). Afterward, a few SSR markers linked with these diseases could be validated in certain genetic backgrounds (Khedikar et al. 2010; Kolekar et al. 2016; Gajjar et al. 2014; Sukruth et al. 2015; Yeri et al. 2014; Yol et al. 2016). Recently, using the QTL-seq approach, candidate genes imparting resistance to LLS and rust diseases were also identified (Shirasawa et al. 2018; Pandey et al. 2017; Clevenger et al. 2018) and a major QTL for rust resistance on A03 chromosome was mapped (Mondal and Badigannavar 2018).
Although, major QTLs for rust have been fine mapped, these need to be validated in a large and diverse set of peanut genotypes. Also, a fine map for QTL(s) controlling LLS resistance is not yet available. Thus, scope is there for the identification of more polymorphic markers in this QTL region for its wider use in different genetic backgrounds for marker-assisted selection. In this backdrop, the objectives of this investigation were to saturate both LLS and rust QTLs using novel EST-SSR markers having functional relevance to biotic stresses; and to validate the linked markers in a large set of Indian groundnut genotypes including released varieties.
Materials and methods
Development of mapping population
An F2 mapping population, consisting of 328 individuals, derived from a cross between two Indian varieties (GJG17 × GPBD4) was used for the mapping of rust and LLS disease resistance loci. GJG17 is high a yielding Virginia type, foliar disease susceptible variety developed from the cross between JSSP11 × GG6 at Junagadh Agricultural University, Junagadh, Gujarat (Rathnakumar et al. 2013). Whereas, GPBD4 is a Spanish bunch variety that is highly resistant to foliar fungal diseases such as LLS and rust. This variety was the second cycle interspecific derivative which is derived from the cross between KRG1 × CS16 (ICGV86855) (Gowda et al. 2002).
Phenotyping and disease scoring for LLS and rust
Phenotyping of 328 F2 individuals for LLS and rust reaction was carried out at ICAR-Directorate of Groundnut Research, Junagadh during the year 2014 by the creation of artificial disease epiphytotic conditions in the field, using ‘spreader row technique’. The F2 plants along with their parents were raised in 5 m rows with 10 cm intra-row and 45 cm inter-row spacing. Forty-five days after sowing (DAS), the plants were inoculated uniformly in the evening with LLS/rust for ten days (Sujay et al. 2012). Further, to increase the disease pressure, sprinkler irrigation was given in the evening to maintain sufficient humidity. Disease scoring was done at 90 DAS using a modified 9-point scale (Subbarao et al. 1990). For comparison of the disease reaction, the susceptible or the spreader row genotype (TMV2) and the resistant (GPBD4) genotypes were used.
Parental polymorphism and genotyping of mapping population
The genomic DNA was extracted from the tender leaves of parents and the F2 individuals using the modified CTAB method (Nawade et al. 2016); its integrity was checked on 0.8% agarose gel and quantified using Nanodrop ND-1000 spectrophotometer (Thermo Scientific, UK). A total of 1311 SSR markers were used for the parental polymorphism survey. Of these 900 were developed previously by our group (Bosamia et al. 2015), while 411 were selected from the previous reports (Sujay et al. 2012; Khedikar et al. 2010; He et al. 2003; Mondal and Badigannavar 2010).
The polymerase chain reaction (PCR) was set with a reaction mixture volume of 10 µL including, 1.0 µL (20 ng) template DNA, 2.0 μL of 5 × PCR buffer (Promega, USA), 2.5 mM MgCl2 (Promega, USA), 0.2 μM dNTP (Thermo Fisher Scientific, USA), 20 μM of each forward and reverse primer, 1.0 U of Taq DNA polymerase (Promega, USA) and sterile double distilled water to make up the final volume. The touchdown PCR program was set with initial denaturation for 3 min at 94 ºC, five cycles of 30 s denaturation at 94 ºC, 30 s annealing at 65 ºC (with 1 ºC decrement per cycle for remaining four cycles), and extension for 1 min at 72 ºC. The remaining 30 cycles were performed to amplify only specific DNA fragments with denaturation for 30 s at 94 ºC, annealing for 30 s at 60 ºC and extension for 1 min at 72 ºC. In the end, the final extension was done at 72 ºC for 5 min. The amplified products were resolved on 6% non-denaturing polyacrylamide gel electrophoresis (PAGE) gels which were stained and visualized using an automated gel documentation system (Fujifilm FLA-5000) (Bosamia et al. 2015).
Bulked segregant analysis (BSA)
The BSA was performed to find the EST-SSRs putatively linked to LLS and rust resistance gene(s)/QTLs. As the F2 population size was quite large (328 plants), so we could easily identify ten plants each which showed extreme phenotypes (susceptible and resistance) for both LLS and rust. These were used to prepare bulks viz. bulk-1 (susceptible bulk) and bulk-2 (resistant bulk) by pooling the equal quantity (50 ng/µL) of genomic DNA from the ten selected F2 plants each. All the polymorphic EST-SSR markers were used to check the bulks along with the parents.
Construction of the genetic map
The genotypic data were used for the Chi square (χ2) analysis to test the null hypothesis of 1:2:1 expected polymorphic marker ratio. The linkage analysis was carried out using QTL IciMapping ver 4.1 (Wang et al. 2016) with LOD (logarithm of odds) cutoff value as 3.0 and Kosambi map function was used to express the map distance in centiMorgan (cM) (Kosambi 1944). The novel EST-SSR markers were also integrated into the existing linkage map of Sujay et al. (2012).
QTL analysis for LLS and Rust
The LLS and rust score and genotyping data of polymorphic SSR markers were generated for the F2 population and QTL analysis was performed using the Inclusive Composite Interval Mapping (ICIM) algorithm (Li et al. 2007). QTL IciMapping software ver. 4.1 (Wang et al. 2016) was used to detect the significant main effect QTLs. Moreover, for additive QTLs, the scanning speed of 1.0 cM and threshold LOD value of 2.5 was set and a significant LOD threshold was determined by 1000 permutations with a probability of type I error as 0.01 (Doerge 2002).
Physical mapping of linked markers and its collinearity assessment
EST sequences of all the markers on A03 chromosome were used as query sequences and using nucleotide BLAST (https://peanutbase.org/blast/nucleotide/nucleotide) against cultivated peanut genome database, the physical position of each SSR was retrieved and the linkage map was integrated to the physical map. The order and positions of the markers mapped on the A03 chromosome containing major QTL for LLS and rust were compared with the previous maps using Strudel 1.15.08.25 (Bayer et al. 2011).
Validation of identified markers
A total of 177 genotypes, consisting of 171 Indian varieties and 06 advanced breeding lines (Table S1) were phenotyped for rust and LLS disease reactions using a modified 9-point scale (Subbarao et al. 1990) during kharif seasons of the year 2015 and 2016. A set of 23 SSR markers located between the marker interval GM2009 and GM1954 on the A03 chromosome (Table S2) were selected for the validation in these diverse genotypes.
Results
Phenotypic evaluation, BSA and construction of genetic map
The phenotyping of 328 F2 individuals and parental lines for LLS and rust diseases under artificial epiphytotic conditions revealed significant variations (Fig. S1), and frequency distribution of the disease revealed normal distribution (Fig. S2). Among 1311 SSR markers screened, 84 (6.4%) showed polymorphism in the parental genotypes (GJG17 and GPBD4); of which, 43 (51%) were novel EST-SSRs developed by our lab (Table S3). The BSA identified 04 novel SSRs namely, DGR329 (Genbank accession No. XM_025836111.1), DGR508 (XM_025784199.1), DRG800 (XM_025835763.1), and DGR2409 (XM_025836145.1) which clearly distinguished the bulks (Fig. S3). Seventy SSRs could be mapped on 14 linkage groups (LGs) covering a total map distance of 797.55 cM with an average inter-marker distance of 11.39 cM (Fig. S4; Table S4), while 14 remained unlinked.
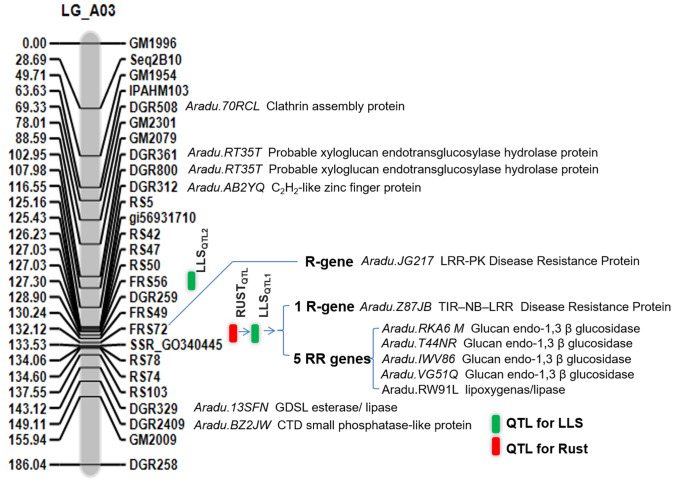
The length of the largest LG was 186.04 cM (LG_A03), while the smallest was 5.25 cM (LG_A07). The number of loci mapped per LG ranged from 2.0 (LG_A06, LG_A07, LG_B05, LG_B06, and LG_B08) to 27 (LG_A03). The LGs were assigned to the polymorphic markers using the details of Sujay et al. (2012) and the physical positions of the respective ESTs. Seven novel EST-SSRs viz., DGR259 (XM_025835922.2), DGR312 (XM_025835733.2), DGR329, DGR361 (XM_025784168.1), DGR508, DGR800, and DGR2409 could be mapped on the LG_A03; which is known to harbor major LLS and rust resistance QTLs (Sujay et al. 2012; Khedikar et al. 2010; Mondal and Badigannavar 2018).
QTL(s) for LLS and rust resistance
The genotypic data of 70 polymorphic SSRs and phenotypic data of 328 F2 individuals were used for the identification of QTLs imparting disease resistance. Two major QTLs viz., LLSQTL1, and LLSQTL2 are identified for LLS resistance on LG_A03 explaining 47.45% and 29.06% PVE (percentage of variance explained), respectively (Table 1). The LLSQTL1 was found located in the map interval of 1.41 cM and is flanked by the markers SSR_GO340445 and FRS72 (Fig. 1). Whereas, LLSQTL2 was found located between the markers DGR259 and FRS59. Incidentally, the LLSQTL2 was found located very close to LLSQTL1 at a distance of 3.22 cM from the marker FRS72 (Fig. S5). Also, a major QTL for rust resistance (RustQTL) with 70.52% PVE (Fig. S5) was validated which is flanked by the markers SSR_GO340445 and FRS72; and is also located in the region harboring LLSQTL1 on LG_A03.
Table 1.
Summary of major QTLs identified for late leaf spot and rust resistance on A03 chromosome
| Trait | QTL name | Position | Left marker | Right marker | PVE (%) | LOD | Add |
|---|---|---|---|---|---|---|---|
| LLS | LLSQTL1 | 133 | FRS72 | SSR_GO340445 | 47.45 | 50.39 | 1.24 |
| LLSQTL2 | 128 | FRS56 | DGR259 | 29.06 | 34.53 | 0.96 | |
| Rust | RustQTL | 133 | FRS72 | SSR_GO340445 | 70.52 | 87.81 | 2.24 |
Where, PVE percentage of variance explained; LOD logarithm of odds; Add additive variance
Fig. 1.
Chromosome A03 carrying major QTLs for rust and late leaf spot disease resistance and functional annotation of the genes in the identified QTL region
The common QTL region lying between markers GO340445 and FRS72 spanning 332.7 kb was reported harboring one R-gene (Aradu.Z87JB; TIR–NB–LRR) and four PR-genes (Aradu.RKA6M, Aradu.T44NR, Aradu.1WV86, and Aradu.NG5IQ) having vital roles in imparting resistance to foliar fungal disease (Mondal and Badigannavar 2018). In addition to the five genes identified by Mondal and Badigannavar (2018), we identified one lipooxygenase/lipase gene (Aradu.RW91L) having resistance function (Table 2).
Table 2.
Summary of EST-SSR markers and genes in the targeted genomic region of A03 chromosome
| SSR marker (Gene ID) | Position (cM) | Functional annotation |
|---|---|---|
| GM1954 | 49.71 | – |
| IPAHM103 | 63.63 | – |
| DGR508* (Aradu.70RCL) | 69.33 | Putative clathrin assembly protein |
| GM2301 | 78.01 | – |
| GM2079 | 88.59 | – |
| DGR800* (Aradu.RT35T) | 102.95 | Xyloglucan endotransglucosylase |
| DGR361* (Aradu.RT35T) | 107.98 | Xyloglucan endotransglucosylase |
| DGR312* (Aradu.AB2YQ) | 116.55 | C2H2-like zinc finger protein |
| RS5 | 125.16 | – |
| gi56931710 | 125.43 | – |
| RS42 | 126.23 | – |
| RS47 | 127.03 | – |
| RS50 | 127.03 | – |
| FRS56 | 127.3 | – |
| DGR259* (Aradu.UG0RV) | 128.9 | MADS-box transcription factor |
| FRS49 | 130.24 | – |
| (Aradu.JG217) | – | LRR-Protein kinase superfamily protein |
| FRS72 | 132.12 | – |
| (Aradu.Z87JB) | – | Disease resistance protein (TIR-NBS-LRR class) |
| (Aradu.RKA6M) | – | Glucan endo-1,3 beta-glucosidase-like |
| (Aradu.T44NR) | – | Glucan endo-1,3 beta-glucosidase-like |
| (Aradu.1WV86) | – | Glucan endo-1,3 beta-glucosidase-like |
| (Aradu.RW91L) | – | Lipase/lipooxygenase PLAT/LH2 family |
| (Aradu.NG5IQ) | – | Glucan endo-1,3 beta-glucosidase-like |
| SSR_GO340445 | 133.53 | – |
| RS78 | 134.06 | – |
| RS74 | 134.6 | – |
| RS103 | 137.55 | – |
| DGR329* (Aradu.13SFN) | 143.12 | GDSL esterase/lipase |
| DGR2409* (Aradu.BZ2JW) | 149.11 | CTD small phosphatase-like protein |
| GM2009 | 155.94 | – |
Where, *Indicate the identified EST-SSR markers in the present study between marker locus GM2009 and GM1954, a QTL hot spot region for LLS and rust; (–) details not available
Collinearity between linkage map and physical map
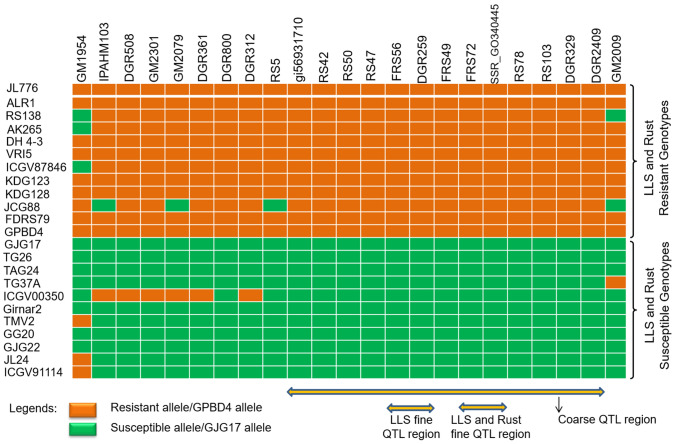
Twenty-seven SSRs were mapped on A03 chromosome, including 23 SSRs in the LLS and rust QTL region spanning between markers GM1954 and GM2009 (Table S1). Moreover, good collinearity was also observed for these SSRs on the A03 chromosome for their order and positions (Fig. 2). A small part of this region (between gi56931710 and DGR2409) was identified as a coarse QTL region imparting LLS and rust resistance. Further, a refined region (between markers DGR259 and FRS56) imparting LLS resistance and a common QTL region (between SSR_GO340445 and FRS72) for both LLS and rust resistance were identified as fine QTL regions (Fig. 3).
Fig. 2.
Collinearity between linkage map and physical map for the SSR markers linked with the rust and LLS diseases on A03 chromosome
Fig. 3.
Allelic pattern of all the markers flanking major rust and LLS QTL region
Validation studies
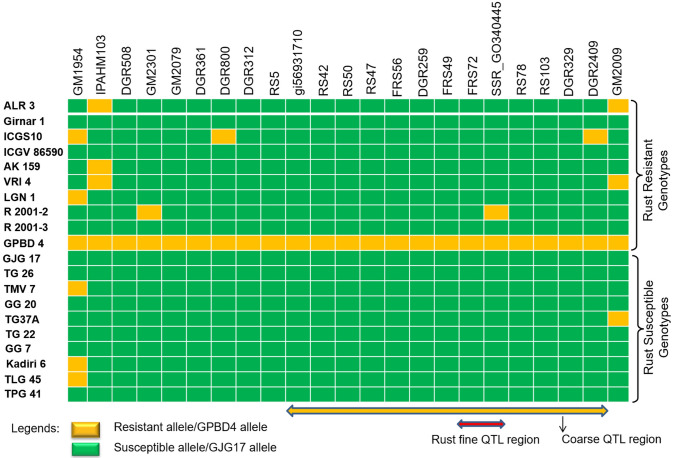
Twenty-three SSR markers between marker locus GM1954 and GM2009 were used for validation in 177 diverse genotypes (Table S1; Table S5). Twelve genotypes resistant to both LLS and rust were found harboring resistant alleles of 13 markers viz., DGR2409, DGR329, RS103, RS78, SSR_GO340445, FRS72, FRS49, DGR259, FRS56, RS50, RS42, gi56931710, and DGR800 (Fig. 3). However, eight markers (RS5, DGR312, DGR361, GM2079, GM2301, DGR508, IPAHM103, and GM1954) present on the left end of the coarse QTL region and one marker (GM2009) present on the right end of LLS-rust QTL region could not discriminate these genotypes. The LLS and rust-resistant genotypes (RS138, AK 265, and ICGV87846) showed susceptible allele amplification by GM1954 marker; whereas, the genotype RS138 showed susceptible allele amplification by GM2009 marker. Similarly, JCG88 a resistant genotype to both the diseases also amplified susceptible allele from four markers viz., IPAHM103, GM2079, RS5, and GM2009 (Fig. 3).
Moreover, almost all the LLS and rust susceptible genotypes (except ICGV00350) showed susceptible allele amplification for all the tested marker loci, except for GM1954 and GM2009 markers, which are situated at the terminal end of the region. However, ICGV00350, a susceptible genotype to both the diseases showed resistant allele amplification by markers such as IPAHM103, DGR508, GM2301, GM2079, DGR361, and DGR312. These markers are found located on the left side of the coarse QTL region. In addition to the marker SSR_GO340445 and FRS72 which are found carrying fine QTL region, other markers like DGR2409, DGR329, RS103, RS78, FRS49, DGR259, FRS56, RS50, RS42, and gi56931710 also showed very tight association with both the diseases (Fig. S3). Two markers namely, GM1954 and GM2009 which are positioned at the border of the rust and LLS QTL region, showed less correspondence with the susceptible and resistant genotypes.
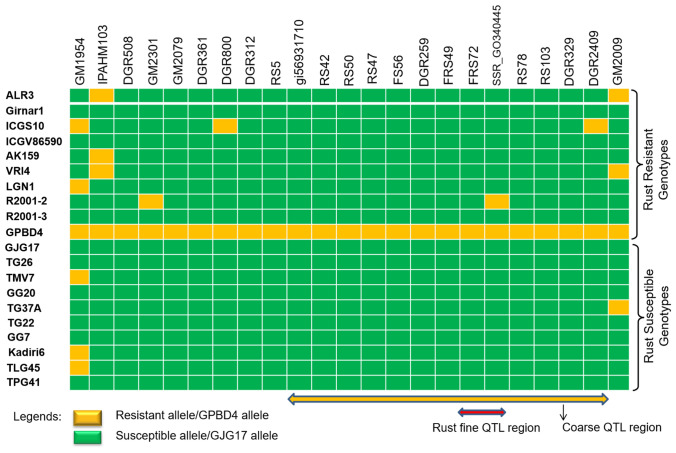
Surprisingly, none of the nine genotypes resistant to only rust disease were found having the marker alleles associated with coarse QTL region except markers loci SSR_GO340445 and DGR2409 in the resistant genotypes R2001-2 and ICG10, respectively. The marker validation details for all the genotypes are presented in Table S1 and Figs. 2, 3, 4.
Fig. 4.
Validation of all the markers flanking the major rust QTL region in a set of genotypes
Discussion
Simultaneous screening of an F2 population for rust and LLS diseases under open field conditions through the spreader row technique has resulted in reliable phenotyping for these diseases. On a similar note, in wheat Alahmad et al. (2018) have also screened an F2 population for two fungal diseases i.e. crown rot and leaf rust for the identification of linked QTLs. A set of 1311 SSR markers when tested between the parents (GJG17 and GPBD4) of the mapping population, resulted in a very low (6.04%) level of polymorphism, which was in concurrence with the previous reports of 6.07–6.65% polymorphism (Sujay et al. 2012; Khedikar et al. 2010; Mondal et al. 2012a). The low genetic diversity in cultivated peanut is mainly due to its recent origin from a single polyploidization event (Young et al. 1996). Of 900 novel EST-SSR markers studied, 43 (4.77%) are found polymorphic in the parents which were a little bit more than the previous reports (Hong et al. 2010; Mondal et al. 2012b). The very low polymorphism by the EST-SSRs could be due to the amplification of conserved genic regions. Four EST-SSR markers viz., DGR329, DGR508, DGR800, and DGR2409 were able to discriminate against the resistant and susceptible bulks for both LLS and rust diseases in BSA. However, an SSR marker PM384100 (Shoba et al. 2012), and a RAPD marker J71350 (Mondal et al. 2008) did not show complete correspondence with resistance when validated in a set of genotypes.
Out of 84 polymorphic SSRs, 23 (27.38%) showed segregation distortion (SD), which is quite similar to the earlier reports (Sujay et al. 2012; Khedikar et al. 2010). However, a few studies have also recorded a relatively low level of SD for the studied markers (Mondal et al. 2012a; Hong et al. 2010, 2008). Out of the seven novel EST-SSR markers identified, four markers viz., DGR329, DGR508, DGR2409, and DGR800 could distinguish the bulks in BSA, while three (DGR259, DGR312, and DGR361) could not differentiate the bulks. Interestingly all the seven markers were fine mapped on A03 chromosome, in the region which is known to harbor QTLs for LLS and rust resistance. Gajjar et al. (2014) in their validation studies also reported the presence of maximum numbers of linked-markers on LG_03 (now chromosome A03).
The identification of QTL(s) is a prerequisite in the marker-assisted improvement of any quantitative trait. Khedikar et al. (2010) first mapped a major QTL conferring rust resistance having 55.20% PVE on LG_6 (later identified as A03) using the IPAHM103 marker (Fig. S6). Afterward, Mondal et al. (2012a) identified a more tightly linked SSR_GO340445 (1.9 cM) to the rust resistance gene. Further, Sujay et al. (2012) have identified two common regions each carrying a major QTL for rust and LLS between marker GM1954 and GM2009.
Several studies have shown the presence of QTLs imparting LLS and/or rust disease resistance between the markers GM1954 and GM2009 on A03 chromosome (Sujay et al. 2012; Khedikar et al. 2010; Gajjar et al. 2014). Further, the QTL-seq analysis has also revealed the presence of SNPs in the candidate genes affecting LLS and rust resistance (Pandey et al. 2017; Shirasawa et al. 2018). Mondal et al. (2012a) have reported SSR_GO340445 marker is tightly liked to rust resistance QTL. Recently, Mondal and Badigannavar (2018) have identified the fine-region of QTL (1.25 cM, 96.3% PVE) for rust resistance between the markers SSR_GO340445 and FRS72. Thus, the region between the markers GM1954 and GM2009 was targeted for its validation and further saturation with newly developed EST- SSR markers.
Our analysis has validated the presence of 1.25 cM QTL region for rust resistance (Mondal and Badigannavar 2018). Interestingly, the same QTL region (1.41 cM) located between the markers SSR_GO340445 and FRS72 was also found harboring a common QTL for LLS resistance (47.45% PVE). Further, we have also identified a major QTL for LLS resistance (LLSQTL2; 29.06% PVE) which is situated at 3.22 cM distance from FRS72 marker. Thus, the resistance to both LLS and rust diseases in the peanut was found governed by a common genomic region on chromosome A03.
We could also validate one R-gene (Aradu.Z87JB) and four resistance-related genes (Aradu.RKA6M, Aradu.T44NR, Aradu.1WV86, and Aradu.NG5IQ) in the LLS-Rust common QTL region on the A03 chromosome (Table 2) as reported by Mondal and Badigannavar (2018). R-gene resistance is due to the incompatible reaction, where elicitor is recognized by the receptor and causes hypersensitive response (HR) (Eitas and Dangl 2010; Bernoux et al. 2014). The resistance-related genes were also known to impart host plant resistance by encoding various antifungal proteins which can degrade the cell-wall polysaccharides of the pathogen (Xu et al. 1992; Serba et al. 2015). The overexpression of tobacco β-1,3-glucanase imparted resistance to Cercospora arachidicola and Aspergillus flavus (Sundaresha et al. 2010); while radish and fenugreek fusion defensin gene showed improved resistance to Cercospora arachidicola and Phaeoisariopsis personata in transgenic peanut (Bala et al. 2016).
Lipoxygenase (Aradu.RW91L) ‒ a defense-related gene was found located between SSR_GO340445 and FRS72 markers. Lipoxygenase has irreversible membrane damage function by peroxidation of membrane lipids, which causes cell death and thereby checks the pathogen spread (Keppler and Novacky 1986). LRR-PK (Aradu.JG217) which is equivalent to the RHG4 gene in soybean, was found flanked by two major QTLs viz., RUSTQTL/LLSQTL1 and LLSQTL2. This R‒gene might be involved in the initiation of HR by helping TIR-NBS-LRR (Aradu.Z87JB) in resistant peanut plants (Mondal and Badigannavar 2018).
EST-SSRs have great potential to tag and map QTLs since they are derived from the expressed regions of the genome (Oliveira et al. 2007). An EST-SSR marker (DGR259) was found close to the fine QTL region, while six more markers viz. DGR329, DGR2409, DGR312, DGR361, DGR800, and DGR508 could be integrated into the vicinity of a major QTL imparting disease resistance. The marker DGR329 was derived from the EST of Aradu.13SFN gene which encodes for the GDSL esterase/lipase enzyme having a defense role as reported in Arabidopsis (Kwon et al. 2009; Lee et al. 2009) and wheat (Schweiger et al. 2016). Similarly, a gene Aradu.AB2YQ, the source of DGR312 marker, encodes for the C2H2 zinc finger domain of transcription factors, and are known to impart defense in pepper, capsicum, and potato against Pseudomonas syringae (Oh et al. 2005), Xanthomonas campestris (Kim et al. 2004), and Phytophthora infestans (Tian et al. 2010), respectively.
The sequence analysis of QTL region on the A03 chromosome identified the SNPs imparting resistance against LLS and rust diseases (Shirasawa et al. 2018; Pandey et al. 2017). Also, the R-gene (Aradu.Z87JB) encoding disease resistance protein (TIR-NBS-LRR class) and PR genes (Aradu.1WV86, Aradu.NG5IQ, and Aradu.RW91L) encoding glucan endo-1,3-beta-glucosidase-2 like protein, glucan endo-1, 3 beta-glucosidase-4 like protein and lipase/lipooxygenase, respectively have been identified in the 1.41 cM fine QTL region imparting LLS and rust resistance. Similar results were also obtained for rust by Mondal and Badigannavar (2018).
Two genes viz., Aradu.RT35T and Aradu.AB2YQ encoding for xyloglucan endotransglucosylase/ hydrolases (XTHs) enzyme and C2H2-like zinc finger protein, respectively, have been identified, which is very close to the fine QTL region imparting resistance. These genes were found harboring non-synonymous SNPs when compared among the resistant and susceptible genotypes (Shirasawa et al. 2018; Pandey et al. 2017), which could be the possible reason for alteration of gene function and making the genotypes as resistant or susceptible.
It is interesting to note that, two markers (DGR361 and DGR800) which are derived from the ESTs of Aradu.RT35T gene were mapped at different positions on the A03 chromosome. A detailed analysis has revealed that the markers DGR361 and DGR800 have not only amplified different SSR motifs viz. (CT)7(TCT)5 (DGR361) and CT(14) (DGR800) but were also found located on different positions viz. Aradu03:29,051,583 and Aradu03:131,814,096, respectively. Thus, the primes have targeted the tandemly repeated sequences of the paralogous genes located on A03 chromosome (Bosamia et al. 2015).
Detailed expression studies of the identified genes in the QTL region will help in elucidation of the comprehensive resistance mechanism operating in peanut for rust and LLS diseases. The collinearity analysis has shown the similarity between the marker order when the linkage map (LG_A03) was compared with the physical map of A03 chromosome. A few markers did not follow the order compared to physical map and previous linkage maps (Sujay et al. 2012; Khedikar et al. 2010; Mondal and Badigannavar 2018) as linkage maps depict only relative positions of markers to each other (Sourdille et al. 2003). An improved and saturated map of the targeted genomic region has been developed by integrating seven novel EST-SSR markers. For MABC, MAS, and also for the QTL pyramiding, there is a need to identify very tightly linked and polymorphic markers flanking LLS and rust QTL region in different genotypic background.
The effective use of linked SSR markers in any marker-assisted breeding program demands its validation in varied genetic backgrounds (Mondal et al. 2008). Validation of 23 SSR markers in a set of 177 diverse Indian groundnut genotypes revealed 12 as resistant to both LLS and rusts and carrying coarse QTL region. Thirteen markers in the coarse QTL region (DGR2409, DGR329, RS103, RS78, SSR_GO340445, FRS72, FRS49, DGR259, FRS56, RS50, RS42, gi56931710, and DGR800) showed correspondence with LLS and rust resistance reaction of the genotypes studied. Likewise, three markers viz., SSR_GO340445, FRS72, and FRS49 in the rust fine QTL region have also shown correspondence for rust reaction in 95 diverse groundnut genotypes (Mondal and Badigannavar 2018). Similarly, a marker (gi56931710) in the coarse QTL region, differentiated the rust-resistant and susceptible genotypes (Mondal and Badigannavar 2018; Mondal et al. 2012a), while five markers (GM2009, GM2301, GM2079, GM1954, and IPAHM103) in the major QTL region have shown significant association with rust resistance in different mapping populations (Sukruth et al. 2015).
Interestingly, three markers namely, GM2009, GM1954, and RS5, which flanked the coarse QTL region, did not show any correspondence with the foliar disease resistance in our population. On the contrary, Sujay et al. (2012) observed a moderate phenotypic effect with marker GM1954; while Yol et al. (2016) found a significant association of GM1954 marker to rust resistance when tested in 256 groundnut genotypes.
A set of nine genotypes resistant to only rust disease are found lacking the specific marker alleles linked with the coarse QTL region governing the rust resistance (Table S1). This means that either the specific linked marker alleles are absent in these genotypes or these genotypes could be carrying the QTL(s) other than those present in the coarse QTL region of A03 chromosome for rust resistance. Thus, there is a need to focus on these genotypes to find the actual reason for the same. However, two genotypes viz., R2001-2, and ICG10 showed resistant allele of marker SSR_GO0340445 and DGR2409, respectively.
It is important to note that the majority of these genotypes are derived from NcAcs (North Carolina Accessions) such as ALR3, Girnar1, ICGS10, VRI4, and LGN1. Mondal et al. (2012b) upon validation also found that one SSR marker gi56931710 amplified a resistant allele in all the non-NcAcs derived rust-resistant genotypes, but not in NcAcs derived resistant genotype (NcAc343). Further, of five NcAcs derived lines, two genotypes viz., Girnar1 and ICGS10 did not amplify the IPAHM103 marker loci. Also, the genotype ICGV86590 remained unclassified as resistant by the IPAHM103 allele, as also reported by Khedikar et al. (2010). It seems that the NcAcs derived lines may carry a different genomic region controlling the rust resistance in peanuts.
Overall, 21 genotypes were found resistant to foliar diseases, and of these 12 (57%) were found resistant to both LLS and rust, while 09 (43%) were resistant to only rust. Further, 16 (76%) genotypes are of ssp. fastigiata type; while, 05 (24%) are of ssp. hypogaea type (Table S5). Similarly, Subrahmanyam et al. (1989) reported 87% of the resistant genotypes as ssp. fastigiata type; while, 13% as ssp. hypogaea type. Also, various sources of LLS and rust resistance have been identified in the cultivated gene pool (Varman 1999; Subrahmanyam et al. 1989; Mehan et al. 1996; Singh et al. 1997) and majority of them are of subspecies fastigiata type (Subrahmanyam et al. 1995; Liao 2003). Since primitive peanut subspecies fastigiata has its probable origin in Peru, which is a secondary gene center and also the source of foliar-fungal disease resistance (Subrahmanyam et al. 1995), therefore, this subspecies was found dominating as a source of resistance to these diseases. Furthermore, when we looked at the market type, all the resistant genotypes of ssp. fastigiata are of Spanish bunch type (Table S5). Spanish types mostly have superior agronomic traits (Mehan et al. 1996) and thus are frequently used as a resistance source in various peanut breeding programs.
Conclusions
This study reports the validation of QTLs for rust and the identification of novel QTLs for LLS resistance. Both the QTLs have been mapped as a common major QTL region imparting LLS and rust resistance on A03 chromosome. Also, this region was further saturated by integrating a greater number of novel markers that can be used to introgress these fine QTL regions to improve LLS and rust resistance more efficiently by minimizing the risk of linkage drag. Since the novel EST-SSR markers are derived from the genes known to impart biotic stress resistance, therefore the linked marker associated genes may further be studied for its exact mode of action in imparting the resistance to LLS and rust diseases. The marker validation has identified the genotypes having varied genomic backgrounds which may be used as a potential source of LLS and rust resistance in the groundnut improvement as per the need of the breeder.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgment
Financial support received from the Indian Council of Agricultural Research (ICAR), New Delhi, India, and Maulana Azad National Fellowship of UGC to the first author is gratefully acknowledged. The technical assistance in maintaining the plant population rendered by M.B. Sheikh and M.B. Kandoliya is also thankfully acknowledged.
Author contributions
Conceptualized, Investigation and Supervision: GPM, RT, HPG; Data curation and analysis: SA, BN, CS, TCB; Project administration, methodology, and resources: NK, JRD; Writing—original draft: SA, GPM; Writing—review and editing: GPM, RT.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical statements
The corresponding author on behalf of all the co-authors testifies that this manuscript has not currently being considered for publication or published in whole or in part in any other journal. Further, we also affirm that there are no other ethical issues of any sort are involved while performing any experiment mentioned in this manuscript.
Contributor Information
Gyan P. Mishra, Email: gyan.gene@gmail.com
Radhakrishnan T., Email: radhakrishnan.nrcg@gmail.com.
References
- Aggarwal BB, Bhardwaj A, Aggarwal RS, Seeram NP, Shishodia S, Takada Y. Role of resveratrol in prevention and therapy of cancer: preclinical and clinical studies. Anticancer Res. 2004;24:2783–2840. [PubMed] [Google Scholar]
- Alahmad S, Dinglasan E, Leung KM, Riaz A, Derbal N, Voss-Fels KP, et al. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods. 2018;14:36. doi: 10.1186/s13007-018-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala M, Radhakrishnan T, Kumar A, Mishra GP, Dobaria JR, Kirti PB. Over-expression of a fusion gene of radish and fenugreek defensins improves the resistance to leaf spot diseases caused by Cercospora arachidicola and Phaeoisariopsis personata in transgenic peanut. Turkish J Biol. 2016;40:139–149. doi: 10.3906/biy-1412-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M, Milne I, Stephen G, Shaw P, Cardle L, Wright F, et al. Comparative visualization of genetic and physical maps with Strudel. Bioinformatics. 2011;27:1307–1308. doi: 10.1093/bioinformatics/btr111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Moncuquet P, Kroj T, Dodds PN. A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front Plant Sci. 2014;5:606. doi: 10.3389/fpls.2014.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalani H, Thankappan R, Mishra GP, Sarkar T, Bosamia TC, Dobaria JR. Regulation of antioxidant mechanisms by AtDREB1A improves soil-moisture deficit stress tolerance in transgenic peanut (Arachis hypogaea L) PLoS ONE. 2019;14(5):e0216706. doi: 10.1371/journal.pone.0216706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosamia TC, Mishra GP, Thankappan R, Dobaria JR. Novel and stress relevant EST-derived SSR markers developed and validated in peanut. PLoS ONE. 2015;10(7):e0133537. doi: 10.1371/journal.pone.0129127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosamia TC, Dodia SM, Mishra GP, Ahmad S, Joshi B, Thirumalaisamy PP, et al. Unraveling the mechanisms of resistance to Sclerotium rolfsii in peanut (Arachis hypogaea L.) using comparative RNA-Seq analysis of resistant and susceptible genotypes. PLoS ONE. 2020;15(8):e0236823. doi: 10.1371/journal.pone.0236823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger J, Chu Y, Chavarro C, Botton S, Culbreath A, Isleib TG, et al. Mapping late leaf spot resistance in peanut (Arachis hypogaea) using QTL-seq reveals markers for marker-assisted selection. Front Plant Sci. 2018;9:83. doi: 10.3389/fpls.2018.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodia SM, Joshi B, Gangurde SG, Thirumalaisamy PP, Mishra GP, Kumar N, et al. Genotyping-by-sequencing based genetic mapping reveals a large number of epistatic interactions for stem rot resistance in groundnut. Theor Appl Genet. 2019;132:1001–1016. doi: 10.1007/s00122-018-3255-7. [DOI] [PubMed] [Google Scholar]
- Doerge RW. Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet. 2002;3:43–52. doi: 10.1038/nrg703. [DOI] [PubMed] [Google Scholar]
- Dwivedi SL, Pande S, Rao JN, Nigam SN. Components of resistance to late leaf spot and rust among interspecific derivatives and their significance in a foliar disease resistance breeding in groundnut (Arachis hypogaea L.) Euphytica. 2002;125:81–88. doi: 10.1023/A:1015707301659. [DOI] [Google Scholar]
- Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT (2018) Food and agricultural organization of the United Nations. https://www.fao.org/faostat/en/#data/QC. Accessed 7 Aug 2020
- Gajjar K, Mishra GP, Radhakrishnan T, Dodia S, Rathnakumar A, Kumar N et al (2014) Validation of SSR markers linked to the rust and late leaf spot diseases resistance in diverse peanut genotypes. Aust J Crop Sci 86:927–936. https://www.cropj.com/thankapan_8_6_2014_927_936.pdf. Accessed 1 Mar 2020
- Gautami B, Foncéka D, Pandey MK, Moretzsohn MC, Sujay V, Qin H, et al. An international reference consensus genetic map with 897 marker loci based on 11 mapping populations for tetraploid groundnut (Arachis hypogaea L) PLoS ONE. 2012;7:e41213. doi: 10.1371/journal.pone.0041213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda MV, Motagi BN, Naidu GK, Diddimani SB, Sheshagiri R. GPBD 4: a Spanish bunch groundnut genotype resistant to rust and late leaf spot. Intl Arachis Newslett. 2002;22:29–32. [Google Scholar]
- He G, Meng R, Newman M, Gao G, Pittman RN, Prakash CS. Microsatellites as DNA markers in cultivated peanut (Arachis hypogaea L) BMC Plant Biol. 2003;3:3. doi: 10.1186/1471-2229-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YB, Liang XQ, Chen XP, Liu HY, Zhou GY, Li SX, et al. Construction of genetic linkage map based on SSR markers in peanut (Arachis hypogaea L.) Agric Sci China. 2008;7:915–921. doi: 10.1016/S1671-2927(08)60130-3. [DOI] [Google Scholar]
- Hong Y, Chen X, Liang X, Liu H, Zhou G, Li S, et al. A SSR based composite genetic linkage map for the cultivated peanut Arachis hypogaea L. genome. BMC Plant Biol. 2010;10:17–30. doi: 10.1186/1471-2229-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler LD, Novacky A. Involvement of membrane lipid peroxidation in the development of a bacterially induced hypersensitive reaction. Phytopathol. 1986;76:104–108. doi: 10.1094/Phyto-76-104. [DOI] [Google Scholar]
- Khedikar YP, Gowda MV, Sarvamangala C, Patgar KV, Upadhyaya HD, Varshney RK. A QTL study on late leaf spot and rust revealed one major QTL for molecular breeding for rust resistance in groundnut (Arachis hypogaea L.) Theor Appl Genet. 2010;121:971–984. doi: 10.1007/s00122-010-1366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Hong JK, Lee SC, Sohn KH, Jung HW, Hyang BK. CAZFP1, Cys2/His2-type zinc-finger transcription factor gene functions as a pathogen-induced early-defense gene in Capsicum annuum. Plant Mol Biol. 2004;55:883–904. doi: 10.1007/s11103-004-2151-5. [DOI] [PubMed] [Google Scholar]
- Ko JH, Sethi G, Um JY, Shanmugam MK, Arfuso F, Kumar AP, et al. The role of resveratrol in cancer therapy. Int J Mol Sci. 2017;18(12):2589. doi: 10.3390/ijms18122589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolekar RM, Sujay V, Shirasawa K, Sukruth M, Khedikar YP, Gowda MV, et al. QTL mapping for late leaf spot and rust resistance using an improved genetic map and extensive phenotypic data on a recombinant inbred line population in peanut (Arachis hypogaea L.) Euphytica. 2016;209:147–156. doi: 10.1007/s10681-016-1651-0. [DOI] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- Kwon SJ, Jin HC, Lee S, Nam MH, Chung JH, Kwon SI, et al. GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J. 2009;58:235–245. doi: 10.1111/j.1365-313X.2008.03772.x. [DOI] [PubMed] [Google Scholar]
- Lee DS, Kim BK, Kwon SJ, Jin HC, Park OK. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem Biophys Res Commun. 2009;379:1038–1042. doi: 10.1016/j.bbrc.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Li H, Ye G, Wang J. A modified algorithm for the improvement of composite interval mapping. Genetics. 2007;175:361–374. doi: 10.1534/genetics.106.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BS (2003) The groundnut, Hubei Press for Science and Technology, Wuhan, China, https://www.peanutscience.com/doi/pdf/10.3146/AT07-004.1. Accessed 1 Mar 2020
- Mehan VK, Reddy PM, Subrahmanyam P, McDonald D, Singh AK. Identification of new sources of resistance to rust and late leaf spot in peanut. Int J Pest Manag. 1996;42:267–271. doi: 10.1080/09670879609372004. [DOI] [Google Scholar]
- Mishra GP, Radhakrishnan T, Kumar A, Thirumalaisamy PP, Kumar N, Bosamia TC, et al. Advancements in molecular marker development and their applications in the management of biotic stresses in peanuts. Crop Prot. 2015;77:74–86. doi: 10.1016/j.cropro.2015.07.019. [DOI] [Google Scholar]
- Mondal S, Badigannavar AM. Molecular diversity and association of SSR markers to rust and late leaf spot resistance in cultivated groundnut (Arachis hypogaea L) Plant Breed. 2010;129(1):68–71. doi: 10.1111/j.1439-0523.2009.01635.x. [DOI] [Google Scholar]
- Mondal S, Badigannavar AM. Mapping of a dominant rust resistance gene revealed two R genes around the major Rust_QTL in cultivated peanut (Arachis hypogaea L.) Theor Appl Genet. 2018;131:1671–1681. doi: 10.1007/s00122-018-3106-6. [DOI] [PubMed] [Google Scholar]
- Mondal S, Badigannavar AM, Murty GSS. RAPD markers linked to a rust resistance gene in cultivated groundnut (Arachis hypogaea L.) Euphytica. 2008;159:233–239. doi: 10.1007/s10681-007-9482-7. [DOI] [Google Scholar]
- Mondal S, Badigannavar AM, D’Souza SF. Development of genic molecular markers linked to a rust resistance gene in cultivated groundnut (Arachis hypogaea L.) Euphytica. 2012;188:163–173. doi: 10.1007/s10681-011-0619-3. [DOI] [Google Scholar]
- Mondal S, Badigannavar AM, D’Souza SF. Molecular tagging of a rust resistance gene in cultivated groundnut Arachis hypogaea L. introgressed from Arachis cardenasii. Mol Breed. 2012;29:467–476. doi: 10.1007/s11032-011-9564-z. [DOI] [Google Scholar]
- Monyo ES, Osiru MO, Kadyampakeni D, Mponda O, Chinyamunyamu B (2009) Improving food security and nutrition in Malawi and Tanzania through research on edible legumes. Proceedings of stakeholder workshops on groundnut production in Malawi and Tanzania held 1–2 March and 13 April 2007, Lilongwe Malawi and Mtwara Tanzania. Patancheru 502 324, Andhra Pradesh, India: ICRISAT, 96. (ISBN:978–92–9066–515–1)
- Nawade B, Bosamia TC, Thankappan R, Rathnakumar AL, Kumar A, Dobaria JR, et al. Insights into the Indian peanut genotypes for ahFAD2 gene polymorphism regulating its oleic and linoleic acid fluxes. Front Plant Sci. 2016;7:1271. doi: 10.3389/fpls.2016.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawade B, Mishra GP, Radhakrishnan T, Dodia SM, Ahmad S, Kumar A, et al. High oleic peanut breeding: achievements perspectives and prospects. Trends Food Sci Technol. 2018;78:107–119. doi: 10.1016/j.tifs.2018.05.022. [DOI] [Google Scholar]
- Nawade B, Mishra GP, Radhakrishnan T, Sangh C, Dobariya JR, Kundu R. Development of high oleic peanut lines through marker-assisted introgression of mutant ahFAD2 alleles and its fatty acid profiles under open-field and controlled conditions. Biotech. 2019;9:243. doi: 10.1007/s13205-019-1774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Park JM, Joung YH, Lee S, Chung E, Kim S, et al. A plant EPF-type zinc-finger protein, CaPIF1, involved in defence against pathogens. Mol Plant Pathol. 2005;63:269–285. doi: 10.1111/j.1364-3703.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- Oliveira KM, Pinto LR, Marconi TG, Margarido GR, Pastina MM, Teixeira LH, et al. Functional integrated genetic linkage map based on EST-markers for a sugarcane (Saccharum spp.) commercial cross. Mol Breed. 2007;20:189–208. doi: 10.1007/s11032-007-9082-1. [DOI] [Google Scholar]
- Pandey MK, Monyo E, Ozias-Akins P, Liang X, Guimarães P, Nigam SN, et al. Advances in Arachis genomics for peanut improvement. Biotechnol Adv. 2012;30:639–651. doi: 10.1016/j.biotechadv.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Pandey MK, Khan AW, Singh VK, Vishwakarma MK, Shasidhar Y, Kumar V, et al. QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.) Plant Biotechnol J. 2017;15:927–941. doi: 10.1111/pbi.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnakumar AL, Singh R, Parmar DL, Misra JB (2013) A crop profile and compendium of varieties notified in India, Directorate of Groundnut research, PB No. 5 Junagadh-362001 Gujarat India, 118. https://www.dgr.org.in/wp-content/uploads/2015/07/Groundnut-a-crop-profile-and-compendium-of-notified-varieties-of-India.pdf. Accessed 1 Mar 2020
- Schweiger W, Steiner B, Vautrin S, Nussbaumer T, Siegwart G, Zamini M, et al. Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor Appl Genet. 2016;129:1607–1623. doi: 10.1007/s00122-016-2727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serba DD, Uppalapati SR, Mukherjee S, Krom N, Tang Y, Mysore KS, et al. Transcriptome profiling of rust resistance in switchgrass using RNA-Seq analysis. Plant Genome. 2015;82:1–12. doi: 10.3835/plantgenome2014.10.0075. [DOI] [PubMed] [Google Scholar]
- Shirasawa K, Bhat RS, Khedikar YP, Sujay V, Kolekar RM, Yeri SB, et al. Sequencing analysis of genetic loci for resistance for late leaf spot and rust in peanut (Arachis hypogaea L) Front Plant Sci. 2018;9:1727. doi: 10.3389/fpls.2018.01727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoba D, Manivannan N, Vindhiyavarman P, Nigam SN. SSR markers associated for late leaf spot disease resistance by bulked segregant analysis in groundnut (Arachis hypogaea L.) Euphytica. 2012;188:265–272. doi: 10.1007/s10681-012-0718-9. [DOI] [Google Scholar]
- Singh AK, Mehan VK, Nigam SN (1997) Sources of resistance to groundnut fungal and bacterial disease: an update and appraisal. Technical Report, Information bulletin, No. 50, International Crops Research Institute for the Semi-Arid Tropics, Patencheru 502324, Andhra Pradesh, India. https://oar.icrisat.org/id/eprint/6676. Accessed 1 Mar 2020
- Sourdille P, Cadalen T, Guyomarch H, Snape J, Perretant M, Charmet G, et al. An update of the Courtot 9 Chinese spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor Appl Genet. 2003;106:530–538. doi: 10.1007/s00122-002-1044-8. [DOI] [PubMed] [Google Scholar]
- Subbarao PV, Subrahmanyam P, Reddy PM. A modified nine point disease scale for assessment of rust and late leaf spot of groundnut. Montpellier, France: Second International Congress of French Phytopathological Society; 1990. [Google Scholar]
- Subrahmanyam P, Williams JH, McDonald D, Gibbons RW. The influence of foliar diseases and their control by selective fungicides on a range of groundnut (Arachis hypogaea L.) genotypes. Ann Appl Bio. 1984;1043:467–476. doi: 10.1111/j.1744-7348.1984.tb03029.x. [DOI] [Google Scholar]
- Subrahmanyam P, Rao VR, McDonald D, Moss JP, Gibbons RW. Origins of resistances to rust and late leaf spot in peanut (Arachis hypogaea, Fabaceae) Econ Bot. 1989;43:444. doi: 10.1007/BF02935917. [DOI] [Google Scholar]
- Subrahmanyam P, McDonald D, Waliyar F, Reddy LJ, Nigam SN, Gibbons RW (1995) Screening methods and sources of resistance to rust and late leaf spot of groundnut. In: Information bulletin, No 47, ICRISAT, Patancheru, India, https://oar.icrisat.org/id/eprint/3477. Accessed 1 Mar 2020
- Sujay V, Gowda MV, Pandey MK, Bhat RS, Khedikar YP, Nadaf HL, et al. QTL analysis and construction of consensus genetic map for foliar diseases resistance based on two RIL populations in cultivated groundnut (Arachis hypogaea L.) Mol Breed. 2012;32:773–788. doi: 10.1007/s11032-011-9661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukruth M, Paratwagh SA, Sujay V, Kumari V, Gowda MV, Nadaf HL, et al. Validation of markers linked to late leaf spot and rust resistance, and selection of superior genotypes among diverse recombinant inbred lines and backcross lines in peanut (Arachis hypogaea L.) Euphytica. 2015;204:343–351. doi: 10.1007/s10681-014-1339-2. [DOI] [Google Scholar]
- Sundaresha S, Kumar AM, Rohini S, Math S, Keshamma E, Chandrashekar S, et al. Enhanced protection against two major fungal pathogens of groundnut, Cercospora arachidicola and Aspergillus flavus in transgenic groundnut over expressing a tobacco β 1–3 glucanase. Eur J Plant Pathol. 2010;126:497–508. doi: 10.1007/s10658-009-9556-6. [DOI] [Google Scholar]
- Tian ZD, Zhang Y, Liu J, Xie CH. Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays role in salt tolerance. Plant Biol. 2010;12:689–697. doi: 10.1111/j.1438-8677.2009.00276.x. [DOI] [PubMed] [Google Scholar]
- Varman PA. Foliar disease resistant line developed through interspecific hybridization in groundnut (Arachis hypogaea) Indian J Agr Sci. 1999;69:67–68. [Google Scholar]
- Varshney RK, Bertioli DJ, Moretzsohn MD, Vadez V, Krishnamurthy L, Aruna R, et al. The first SSR-based genetic linkage map for cultivated groundnut (Arachis hypogaea L.) Theor Appl Genet. 2009;118:729–739. doi: 10.1007/s00122-008-0933-x. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Pandey MK, Janila P, Nigam SN, Sudini H, Gowda MV, et al. Marker-assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.) Theor Appl Genet. 2014;127:1771–1781. doi: 10.1007/2Fs00122-014-2338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li H, Zhang L, Meng L. User’s Manual of QTL Genetic Resources IciMapping Version 4.0, The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences CAAS), Beijing 100081, China, and Genetic Program. Mexico, Mexico: International Maize and Wheat Improvement Center CIMMYT; 2016. [Google Scholar]
- Xu PL, Wang J, Fincher GB. Evolution and differential expression of the 1→3-beta-glucan endohydrolase-encoding gene family in barley, Hordeum vulgare. Gene. 1992;120:157–165. doi: 10.1016/0378-1119(92)90089-8. [DOI] [PubMed] [Google Scholar]
- Yeri SB, Shirasawa K, Pandey MK, Gowda MV, Sujay V, Shriswathi M, et al. Development of NILs from heterogeneous inbred families for validating the rust resistance QTL in peanut (Arachis hypogaea L.) Plant Breed. 2014;133:80–85. doi: 10.1111/pbr.12130. [DOI] [Google Scholar]
- Yol E, Upadhyaya HD, Uzun B. Identification of rust resistance in groundnut using a validated SSR marker. Euphytica. 2016;210:405–411. doi: 10.1007/s10681-016-1705-3. [DOI] [Google Scholar]
- Young ND, Weeden NF, Kochert G. Genome mapping in legumes. In: Austin A, Paterson AH, editors. Genome mapping in plants. USA: Landes Company; 1996. pp. 211–227. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.