Abstract
Tuberculosis (TB) is one of the major causes of death related to antimicrobial resistance worldwide because of the spread of Mycobacterium tuberculosis multi- and extensively drug resistant (multi-drug resistant (MDR) and extensively drug-resistant (XDR), respectively) clinical isolates. To fight MDR and XDR tuberculosis, three new antitubercular drugs, bedaquiline (BDQ), delamanid, and pretomanid were approved for use in clinical setting. Unfortunately, BDQ quickly acquired two main mechanisms of resistance, consisting in mutations in either atpE gene, encoding the target, or in Rv0678, coding for the repressor of the MmpS5-MmpL5 efflux pump. To better understand the spreading of BDQ resistance in MDR- and XDR-TB, in vitro studies could be a valuable tool. To this aim, in this work an in vitro generation of M. tuberculosis mutants resistant to BDQ was performed starting from two MDR clinical isolates as parental cultures. The two M. tuberculosis MDR clinical isolates were firstly characterized by whole genome sequencing, finding the main mutations responsible for their MDR phenotype. Furthermore, several M. tuberculosis BDQ resistant mutants were isolated by both MDR strains, harboring mutations in both atpE and Rv0678 genes. These BDQ resistant mutants were further characterized by studying their growth rate that could be related to their spreading in clinical settings. Finally, we also constructed a data sheet including the mutations associated with BDQ resistance that could be useful for the early detection of BDQ-resistance in MDR/XDR patients with the purpose of a better management of antibiotic resistance in clinical settings.
Keywords: Mycobacterium tuberculosis, bedaquiline, multi-drug resistance, Rv0678, MmpL5, AtpE
Introduction
According to the World Health Organization (WHO) report, in 2018, tuberculosis (TB), caused by Mycobacterium tuberculosis, was one of the major causes of death related to antimicrobial resistance (World Health Organization [WHO], 2019a). Globally, in 2018 about half a million TB infections were rifampicin-resistant, of which 78% were multi-drug resistant (MDR)-TB (World Health Organization [WHO], 2019a). Among these cases, 6.2% were estimated to have extensively drug-resistant (XDR)-TB (World Health Organization [WHO], 2019a). Even if it is a relatively small percentage of all MDR-TB cases, these infections are more complicated to treat and to manage and are a challenge for the health systems worldwide.
Recently, three new antitubercular drugs, bedaquiline (BDQ) (Janssen, Beerse, Belgium), delamanid (Otsuka, Tokyo, Japan), and pretomanid (TB Alliance) were approved for the treatment of MDR-TB (Li et al., 2019; Nieto Ramirez et al., 2020). Interestingly, several studies demonstrated that patients treated with a BDQ-containing regimen showed a high culture conversion rate (65–100%) (Li et al., 2019; Pontali et al., 2019).
BDQ is a diarylquinoline that targets atpE gene, coding for the subunit c of the ATP synthase complex (Andries et al., 2005). Its use reduces the mortality when added to treatment for MDR- and XDR-TB (Li et al., 2019; Conradie et al., 2020). The potential risk of BDQ of prolonging the QT interval has occurred in only 0.6% of treated patients; consequently, the advantage in its use is uncontested, even if it is still under investigation. In fact, many clinical studies are testing the effectiveness of new drug combinations, which include BDQ, to design the next generation regimens (Sharma et al., 2020).
In this context, WHO has recently updated the treatment for MDR-TB, by recommending two possible regimens (the longer regimen and the shorter one), both including BDQ and other drugs (Caminero et al., 2019; World Health Organization [WHO], 2019b). Interestingly, in a recent study, NIX-TB trial, a three-drug regimen including linezolid, BDQ and pretomanid was tested with XDR- and MDR-TB patients; the therapy was successful for 90% of patients (Conradie et al., 2020). As evident, BDQ use is rapidly spreading, and 90 countries reported having imported or started using BDQ by the end of 2018 (World Health Organization [WHO], 2019a).
In spite of its recent use in clinical practice, primary BDQ resistance appeared among M. tuberculosis clinical isolates (Veziris et al., 2017; Villellas et al., 2017; Zimenkov et al., 2017; Ismail et al., 2018). BDQ resistance is especially associated with mutations in atpE and Rv0678 genes.
The most common mutations linked to low-level of BDQ resistance are present in Rv0678 gene coding for the M. tuberculosis repressor of MmpS5-MmpL5 efflux system. This transporter pumps out of the cells also clofazimine and azoles (Milano et al., 2009; Hartkoorn et al., 2014; Smith et al., 2017). In some cases, Rv0678 mutations occurred together with polymorphisms in other genes encoding the uncharacterized transporter Rv1979c and the cytoplasmic peptidase PepQ (Rv2535c), both associated with cross-resistance to clofazimine (CFZ) (Nieto Ramirez et al., 2020). Furthermore, a report demonstrated that mutations in pepQ gene confer low-level of BDQ resistance in mice (Almeida et al., 2016).
As expected, high BDQ resistance levels are caused by mutations in atpE gene, even if their frequency is extremely low among TB patients (Nieto Ramirez et al., 2020).
The surveillance of drug resistance during clinical management is mandatory in order to prevent the occurrence of BDQ resistance among TB patients. To this aim, in vitro studies could be a valuable tool for understanding the reasons linked to the spreading of BDQ resistance in particular amongst M. tuberculosis MDR and XDR clinical isolates. While acquiring resistance to first-line drugs such as rifampicin (RIF) and isoniazid (INH) is linked to a perturbance in the M. tuberculosis fitness (Kodio et al., 2019), mutations in Rv0678 and atpE have not been yet demonstrated to have this behavior (Andries et al., 2014; Nieto Ramirez et al., 2020). On the other hand, the low frequency of atpE mutants in the clinical setting in comparison to Rv0678 mutations could suggest a possible reduced fitness cost linked to some atpE mutations (Nieto Ramirez et al., 2020).
To better understand the spreading of BDQ resistance in MDR- and XDR-TB, we reported an in vitro generation of M. tuberculosis mutants resistant to BDQ starting from MDR clinical isolates as parental cultures, since BDQ is used to treat patients affected by MDR-TB. Moreover, we performed growth curves of both obtained BDQ resistant mutants and original MDR isolates to detect possible differences in strains harboring either Rv0678 or atpE mutations. Furthermore, we compared these mutations to a compiled data sheet of previously published SNPs, deriving from in vitro, in vivo and clinically resistant strains, thus providing additional information for rapid and efficient detection of all known BDQ-resistance associated mutations to ensure an optimal treatment monitoring.
Materials and Methods
Bacterial Strains, Growth Conditions and Drugs
Mycobacterium tuberculosis H37Rv and clinical isolates as well as their mutants were grown at 37°C in Middlebrook 7H9 broth (Becton Dickinson), supplemented with 0.05% w/v Tween 80 or on Middlebrook 7H11, both supplemented with 0.2% w/v glycerol, and 10% v/v Middlebrook OADC enrichment (oleic acid, albumin, D-glucose, catalase; Becton Dickinson). M. tuberculosis MDR clinical isolates were collected and characterized at the Sondalo Division of the Valtellina and Valchiavenna, Italy, hospital authority in 2012 (Menendez et al., 2013). Bedaquiline (D.B.A. Italia s.r.l.) was dissolved in DMSO (Sigma Aldrich).
All the experiments with M. tuberculosis were performed in Biosafety level 3 laboratory by authorized and trained researchers.
Genomic DNA Preparation and Whole-Genome Sequencing of M. tuberculosis Clinical Isolates
Genomic DNA of M. tuberculosis MDR clinical isolates (hereafter named IC1 and IC2) was extracted as previously described (Belisle and Sonnenberg, 1998). Genomic DNA samples were sequenced by using an Illumina HiSeq2000 technology at Fisabio (Valencia, Spain). Illumina reads were aligned to the annotated genome sequence of the wild-type H37Rv (Cole et al., 1998) (NC_000962.3) to identify SNPs. For the bioinformatic analysis of Illumina data, repetitive PE and PPE gene families were discarded as well as SNPs and indels with less than 50% probability. The possible polymorphisms associated to the resistance to the following drugs were investigated: streptomycin (rrs, rpsL, gidB), INH (katG, inhA, ndh, nat), RIF (rpoB), ethambutol (embA, embB, embC, embR), ethionamide (ethA, inhA, ethR), pyrazinamide (pncA, rpsA, panD), capreomycin (tlyA, rrs), and BDQ (Rv0678, atpE, pepQ).
Determination of Minimal Inhibitory Concentration (MIC)
The drug susceptibility of M. tuberculosis strains was determined using the resazurin microtiter assay (REMA), as previously described (Palomino et al., 2002). Briefly, log-phase bacterial cultures were diluted to a theoretical OD600 = 0.0005 and grown in a 96-well black plate (Fluoronunc, Thermo Fisher) in the presence of serial compound dilution. A growth control containing no compound and a sterile control without inoculum were also included. After 7 days of incubation at 37°C, 10 μl of resazurin (0.025% w/v) were added and fluorescence was measured after 24 h further incubation using a FluoroskanTM Microplate Fluorometer (Thermo Fisher Scientific; excitation = 544 nm, emission = 590 nm). Bacterial viability was calculated as a percentage of resazurin turnover in the absence of compound.
Isolation and Characterization of M. tuberculosis Spontaneous Mutants Resistant to BDQ
Mycobacterium tuberculosis BDQ resistant mutants were isolated by plating approximately 108 and 109 CFU from exponential growth phase cultures of IC1 and IC2 clinical isolates onto solid medium containing drug at concentrations exceeding the MIC (5X, 10X, 20X MIC). Following 6–8 weeks of incubation, BDQ resistant colonies were streaked onto 7H11 medium. At the same time, these colonies were streaked also onto 7H11 medium plus the same BDQ concentration used for mutant isolation to confirm the resistant phenotype. BDQ MIC values were also assessed by REMA. Genomic DNA was extracted from each mutant and Rv0678, atpE, and pepQ genes were amplified by PCR (oligonucleotides in Supplementary Table S1), purified using Wizard® SV Gel and PCR Clean-Up System (Promega) and analyzed by conventional Sanger sequencing (Eurofins Genomics, Italy).
Growth Curves of M. tuberculosis BDQ Resistant Mutants and MDR Clinical Isolates
The cultures of M. tuberculosis mutants, as well as their corresponding parental strain, were inoculated in 7H9 medium in round bottom tubes at 37°C to reach an early exponential phase. Then, each strain was reinoculated in new 7H9 medium at final OD600 = 0.06. The cultures were incubated in standing at 37°C for 8 days. After 24, 48, 96, 168, 192 h, the optical densities at 600 nm were recorded to plot growth curves. The H37Rv strain was also included as control.
Data Sheet Creation
The Medical Subject Headings vocabulary of biomedical terms (MeSH) search builder was used to construct the query for the terms “Bedaquiline,” “Mycobacterium,” and “Mutation”, with which the Pubmed and Pubmed Central databases were skimmed, then all the abstracts were downloaded in a MEDLINE format. The information gathered was then manually filtered in two categories: relevant papers (i.e., original works, case studies, and clinical studies) and papers not pertinent to our purpose. The filtered-as-relevant papers were downloaded as full text, thoroughly analyzed and the type of mutations linked to BDQ resistance were annotated to set-up the data sheet.
Results
Characterization of M. tuberculosis MDR Clinical Isolates
IC1 and IC2 strains are two M. tuberculosis MDR clinical isolates previously characterized (Menendez et al., 2013). In detail, IC1 is resistant to streptomycin (SM), INH, RIF, ethambutol (EMB), ethionamide (ETH); IC2 is resistant not only to the previously mentioned drugs, but also to pyrazinamide (PYR), and capreomycin (CM) (Menendez et al., 2013).
REMA was used to determine the BDQ MIC values of both isolates (MIC = 0.06 μg/ml, as for the H37Rv wild-type strain). This MIC value (0.06 μg/ml) for M. tuberculosis BDQ sensitive strains is in agreement with that proposed in 7H9 medium by both EUCAST and previously (Kaniga et al., 2016; EUCAST, 2020).
In order to pinpoint the SNPs responsible for their drug-resistance profile, the M. tuberculosis clinical strains were subjected to whole-genome sequencing (WGS) analysis. Using the obtained Illumina data, the genes involved in the resistance to the following drugs were checked: SM (rrs, rpsL, gidB), INH (katG, inhA, ndh, nat), RIF (rpoB), EMB (embA, embB, embC, embR), ETH (ethA, inhA, ethR), PYR (pncA, rpsA, panD), CAP (tlyA, rrs), and BDQ (Rv0678, atpE, pepQ).
For both M. tuberculosis isolates, the non-synonymous mutations found to be associated to their drug-resistance phenotype are enlisted in Table 1. As expected, no mutation was found in Rv0678, atpE, and pepQ genes according to their BDQ sensitivity.
TABLE 1.
Phenotypic and genotypic characteristics of M. tuberculosis IC1 and IC2 clinical isolates.
| Resistance | Genome position (bp) | Gene | Mutation | Amino acid substitution |
| Clinical isolate IC1 | ||||
| STR | 4407880 | gidB | T323G | L108R |
| INH | 2155168 | katG | G944C | S315T |
| RIF | 761155 | rpoB | C1349T | S450L |
| EMB | 4242803 | embC | G2941C | V981L |
| EMB | 4247429 | embB | A916G | M306V |
| ETH | 4327058 | ethA | G416A | G139D |
| Clinical isolate IC2 | ||||
| STR | 781687 | rpsL | A128G | K43R |
| INH | 2155168 | katG | G944C | S315T |
| RIF | 761155 | rpoB | C1349T | S450L |
| EMB | 4242803 | embC | G2941C | V981L |
| EMB | 4247429 | embB | A916G | M306V |
| PYR | 2288807 | pncA | C435G | D145E |
| PYR | 2289106 | pncA | G136A | A46T |
| ETH | 4327058 | ethA | G416A | G139D |
| CM | 1918707 | tlyA | insAG | Frameshift |
Mycobacterium tuberculosis IC1 and IC2 clinical strains were used for further experiments because of their drug-resistance phenotype as well as their BDQ sensitivity.
Isolation and Phenotypic Characterization of Spontaneous M. tuberculosis Mutants Resistant to BDQ
Once shown their BDQ sensitivity, M. tuberculosis IC1 and IC2 clinical isolates were used to select and to isolate BDQ-resistant spontaneous mutants, since patients affected by MDR-TB are likely to receive BDQ as part of their therapy.
Mutants were selected onto solid medium containing high BDQ concentrations (0.3, 0.6, 1.2 μg/ml, corresponding to 5, 10, 20-fold MIC, respectively). Mycobacterium tuberculosis BDQ-resistant mutants were isolated at a frequency of about 1.8 × 10–8 for IC1 and 6 × 10–9 for IC2.
All the 12 isolated mutants showed to be BDQ resistant and their MIC value was confirmed by REMA, ranging from 0.25 μg/ml (4X MIC of sensitive strain) to 8 μg/ml (128X MIC of sensitive strain) (Table 2). The different levels of drug-resistance could be linked to different associated mutations.
TABLE 2.
Phenotypic and genotypic characteristics of mutants resistant to BDQ obtained from M. tuberculosis clinical isolates IC1 and IC2.
| M. tuberculosis strains | MIC (μg/ml) | Mutation | Amino acid substitution |
| H37Rv | 0.06 | ||
| IC1 | 0.06 | ||
| IC2 | 0.06 | ||
| IC1 B | 8 | atpE: g187c | A63P |
| IC1 C | 4 | atpE: g187c | A63P |
| IC1 D | 4 | atpE: g187c | A63P |
| IC1 F | 4 | atpE: g187c | A63P |
| IC1 G | 4 | atpE: g187c | A63P |
| IC1 H | 2 | atpE: a83c | D28A |
| IC2 Q | 0.5 | atpE: a83g | D28G |
| IC1 L | 0.5 | rv0678: c400t | R124Stop |
| IC1 M | 0.5 | rv0678: g120c | L40F |
| IC1 N | 0.5 | rv0678: a271c | T91P |
| IC1 O | 0.5 | rv0678: g61t | E21Stop |
| IC2 P | 0.25 | rv0678: g197a | G66E |
In order to investigate this possibility, Rv0678, atpE, and pepQ genes were amplified by PCR from the genomic DNA of M. tuberculosis BDQ resistant mutants and sequenced by Sanger method.
None of 12 M. tuberculosis resistant mutants had mutation in pepQ gene, while polymorphisms were found either in atpE or Rv0678 genes (Table 2 and Supplementary Data Sheet S1).
In particular, seven strains carried a mutation in AtpE, the cellular BDQ target. Among them, five mutants harbored the same replacement of alanine at position 63 by a proline (A63P). IC2Q mutant had a substitution of the aspartic acid at position 28 with a glycine (D28G), while IC1H mutant presented at the same position a substitution with an alanine (D28A), (Table 2 and Supplementary Data Sheet S1). Furthermore, these mutants were characterized by a high level of BDQ resistance (2–8 μg/ml) ranging from 32 to 128X MIC of the wild-type strain.
The other five isolated M. tuberculosis mutants harbored mutations in Rv0678, encoding the MmpR transcriptional repressor of the efflux pump MmpS5-MmpL5. These mutants were characterized by a low level of BDQ resistance (0.25–0.5 μg/ml) corresponding to 4–8X MIC the wild-type strain. Three mutants, IC1M, IC1N, and IC2P, presented an amino acid change: respectively, the leucine at position 40 was replaced by a phenylalanine, the tyrosine at position 91 by a proline, and, finally, the glycine at position 66 by a glutamate (Table 2 and Supplementary Data Sheet S1). The MmpR of the other two mutants, IC1L and IC1O, was truncated by a stop codon at position 400 and at position 61, respectively. Additional experiments to demonstrate the role of Rv0678 in BDQ resistance were not performed. None of these Rv0678 polymorphisms was already published, at the best of our knowledge.
Evaluation of the Possible Influence of atpE and Rv0678 Mutations to the Growth Rate of M. tuberculosis BDQ Resistant Mutants
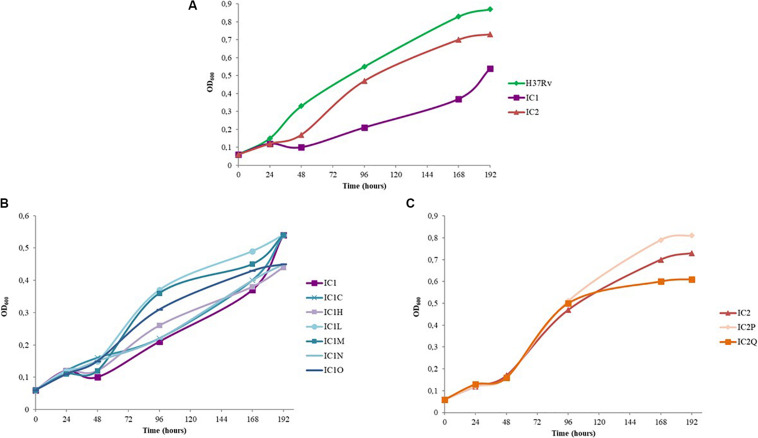
The growth curves of BDQ resistant mutants with respect to that of the M. tuberculosis H37Rv strain and the two parental MDR isolates were also evaluated (Figure 1).
FIGURE 1.
Growth curves of standing cultures of M. tuberculosis strains (H37Rv, clinical isolates and mutant strains). The experiment was repeated three times giving comparable results. The figure shows one representative experiment. (A) Growth curves of M. tuberculosis H37Rv strain and IC1 and IC2 clinical isolates. (B) Growth curves of IC1 clinical isolate and the following BDQ resistant mutants: IC1 C (representative mutant strain with A63P mutation in AtpE), IC1 H, IC1 L, IC1 M, IC1 N, IC1 O. (C) Growth curves of IC2 clinical isolate and the BDQ resistant mutants IC2 P and IC2 Q.
As expected, IC1 and IC2 isolates presented a longer lag phase with respect to that of the wild-type strain. This is also in agreement with their lower growth rate (Figure 1A).
Interestingly, the rate of growth of IC2Q (atpE mutant) was lower than that of IC2 strain and of IC2P mutant (Rv0678 mutant); on the other hand, IC2P grew faster than the other two strains (Figure 1C).
In the case of IC1-derived mutants, the lag phase length is similar between the mutants and the parental strain, while the rate of growth of IC1L, IC1M, IC1N, and IC1O (Rv0678 mutants) was faster than that of IC1 (Figure 1B). These latter harbor mutations in Rv0678 which do not perturb M. tuberculosis essential functions, whilst IC1C and IC1N (both atpE mutants) displayed a rate of growth similar to the parental one.
Overall, our data highlight that the Rv0678 mutations do not affect growth rate of our parental strains, but could actually give an advantage in the growth rate.
Collection of All Known Polymorphisms Causing BDQ Resistance in M. tuberculosis and Other Mycobacteria
The previously published mutations associated with BDQ-resistance as well as the new ones found in this work were included in Supplementary Data Sheet S1.
BDQ is also active against non-tuberculous mycobacteria (NTM) belonging to the Mycobacterium avium-intracellulare complex (MAC) and the Mycobacterium abscessus complex (MABSC) (Philley et al., 2015). Its possible use against these other mycobacterial species is under investigation. In NTM species BDQ presents the same mechanisms of resistance found in M. tuberculosis; consequently, the evaluation of the BDQ resistance associated polymorphisms could be useful also in these species. For this reason, in this collection, all the mycobacterial species in which an actual clinical use or possible use is under evaluation were included. Moreover, both in vitro isolated mutants and clinical isolates were added. At the end, all the mutations linked to BDQ resistance in mycobacteria have been considered in this data sheet that could be useful for the better understanding of BDQ resistance.
Discussion
The current use of BDQ in the treatment for MDR- and XDR-TB reduces the mortality and it is highly effective (Ahmad et al., 2018; Conradie et al., 2020).
Nevertheless, the two main mechanisms of BDQ resistance, which are already widespread, are: modification of target (mutations in atpE, coding for the target) and over-expression of an efflux pump (mutations in Rv0678 gene, coding for the repressor of MmpS5-MmpL5 efflux system). Several reports showed that the most spreading mechanism of BDQ resistance in clinical setting is represented by mutations in Rv0678 gene even if with a low level of BDQ-resistance (Villellas et al., 2017; Nieto Ramirez et al., 2020). To fight BDQ-resistance caused by Rv0678 mutations and to allow its use for the largest possible part of patients, verapamil, an efflux inhibitor, could be the keystone, since it has been demonstrated to increase the efficacy of BDQ against both M. tuberculosis and Mycobacterium abscessus (Ghajavand et al., 2019; Viljoen et al., 2019).
In this study, an in vitro generation of M. tuberculosis mutants resistant to BDQ was performed starting from two MDR clinical isolates as parental cultures since patients affected by MDR-TB are eligible to receive BDQ as part of their therapy.
Polymorphisms were identified in both Rv0678 and atpE genes. Our results confirm that these genes represent the main genetic drivers for the onset of BDQ-resistance, as previously pointed out.
Most in vitro isolated mutants harbored mutations in the BDQ target, AtpE at positions 28 and 63, according to previous multiple reports (Huitric et al., 2010; Segala et al., 2012; Zimenkov et al., 2017; Ismail et al., 2018). In particular, the mutation A63P was found in the first report regarding BDQ discovery (Andries et al., 2005). The 28 and 63 amino acid positions are considered mutation hotspots, as described in Supplementary Data Sheet S1. In fact, the AtpE D28 and A63 are not directly involved in the BDQ binding, but the disruption of the non-covalent bonds they form causes resistance (Preiss et al., 2015). Thanks to the previously published structure of complex crystals obtained by the co-crystallization of the Mycobacterium phlei c-ring with BDQ, it is well-known that BDQ forms an extensive amount of van derWaals interactions with a stretch of nine residues (in M. phlei: G62, L63, E65, A66, A67, Y68, F69, I70, and L72) provided by two adjacent c-subunits (Preiss et al., 2015). The mutations (D28A/G, A63P) harbored by BDQ-resistant mutants isolated in this study are positioned close to BDQ-binding site causing indirect structural interference with BDQ binding (Preiss et al., 2015), as evident by the higher MIC showed.
Different mutations could be linked to different levels of drug-resistance, as previously shown (Andries et al., 2005; Hartkoorn et al., 2014; Almeida et al., 2016). In general, atpE gene associated variants lead to high level of BDQ-resistance, but the most troublesome polymorphisms are linked to Rv0678 gene, that are also the most represented ones found in clinical isolates even if the Rv0678 mutations are linked to a lower level of BDQ resistance (Villellas et al., 2017; Nieto Ramirez et al., 2020), as typical for efflux pump mechanism. When over-expressed, MmpS5-MmpL5 efflux system can extrude different classes of drugs, for example CFZ and azoles, further limiting the therapeutic options of patients affected by M/XDR-TB. As well exemplified also in this present study, different mutations were identified in Rv0678 and were disseminated across the gene (Table 2 and Supplementary Data Sheet S1). Both the missense mutations and the nonsense mutations are reported to abolish the repressor activity of Rv0678, causing an over-expression of the MmpL5 efflux pump leading to drug extrusion (Zhang et al., 2015). It is worth noting that a G66 missense mutation was found not only in this study (G66E), but also in CFZ-resistant M. tuberculosis mutant isolated in vitro (G66V) (Zhang et al., 2015).
As responsible for BDQ resistance, mutations in the intergenic region between Rv0678 and MmpS5 as well as mutations in the genes encoding the efflux pump MmpS5/MmpL5 were also reported (Ghajavand et al., 2019).
Furthermore, several reports showed that Rv0678 mutations could be present prior the BDQ treatment both in vitro and in vivo (Pang et al., 2017; Veziris et al., 2017; Villellas et al., 2017; Xu et al., 2017; Chawla et al., 2018; Martinez et al., 2018). Consequently, it could be hypothesized that these mutations are adaptative or could improve the growth rate of the MDR mutants, representing an advantage for them. In this work, we evaluated the growth rate of our M. tuberculosis BDQ-resistant mutants in comparison with that of the parental strains (two MDR clinical isolates). The atpE mutants presented a growth rate similar or lower than that of the parental strains, since atpE is essential for M. tuberculosis growth, while Rv0678 mutants showed either a similar growth rate as parental strains or better. Noteworthy, Rv0678 gene is not essential for M. tuberculosis growth (Radhakrishnan et al., 2014). Finally, from our work the relative fitness of the mutants could be speculated. In fact, fitness cost determines in part the fate of resistance mutations (Melnyk et al., 2015). In vitro the atpE mutants seem to show a little decrease in fitness relative to that of the respective parental strain. On the opposite hand, Rv0678 mutants seem to have the same fitness in comparison to the corresponding isolate or even a little advantage.
Previous studies showed that Rv0678 repressor controls the expression of MmpS5-MmpL5 efflux system as well as of other transporters such as IniAB and DrrA (Milano et al., 2009; Andries et al., 2014). Among the regulated proteins, there were also some essential enzymes and proteins important for the virulence (e.g., PimA, an antitoxin VapB1, etc.) (Andries et al., 2014), that could play a role in M. tuberculosis growth and/or infection. Apart from a genomic approach, proteomics-based approaches coupled with bioinformatics could be useful for the characterization of novel proteins which might be related to drug resistance, especially when no related mutations could explain it (Sharma et al., 2018).
Conclusion
The presented in vitro growth rate data could explain the spreading of Rv0678 naturally occurring mutations in clinical settings also prior BDQ treatment, even if we cannot exclude the presence of other compensatory mutations that alleviate the cost of resistance without altering it. This evidence also suggests a role for fitness in BDQ-resistance spread, even if further investigations are needed to clearly elucidate this mechanism.
Overall, due to an increasing BDQ usage, it is urgent to implement a more extensive surveillance for such resistance in order to prevent the emergence of resistance in clinical settings. Our collection of polymorphisms responsible for BDQ resistance could be used as theranostics targets, such as in the development of a diagnostic kit for the early detection of BDQ resistant isolates to better manage the available therapeutic options.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
GD and MP designed this study and interpreted the data. GD, JS, and MP wrote the manuscript. GD, JS, VS, PM, and AU performed experiments. All authors approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. GD is funded by a fellowship from the University of Pavia (FRG – Fondo Ricerca and Giovani: “Assegno di ricerca di tipo A”). This research was supported by the Italian Ministry of Education, University and Research (MIUR): Dipartimenti di Eccellenza Program (2018–2022) – Department of Biology and Biotechnology “Lazzaro Spallanzani”, University of Pavia.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.559469/full#supplementary-material
References
- Ahmad N., Ahuja S. D., Akkerman O. W., Alffenaar J. C., Anderson L. F., Baghaei P., et al. (2018). Treatment correlates of successful outcomes in pulmonary multidrug-resistant tuberculosis: an individual patient data meta-analysis. Lancet 392 821–834. 10.1016/S0140-6736(18)31644-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida D., Ioerger T., Tyagi S., Li S. Y., Mdluli K., Andries K., et al. (2016). Mutations in pepQ Confer Low-level resistance to Bedaquiline and Clofazimine in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 60 4590–4599. 10.1128/AAC.00753-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries K., Verhasselt P., Guillemont J., Göhlmann H. W., Neefs J. M., Winkler H., et al. (2005). A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307 223–227. 10.1126/science.1106753 [DOI] [PubMed] [Google Scholar]
- Andries K., Villellas C., Coeck N., Thys K., Gevers T., Vranckx L., et al. (2014). Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 9:e102135. 10.1371/journal.pone.0102135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle J. T., Sonnenberg M. G. (1998). Isolation of genomic DNA from Mycobacteria. Methods Mol. Biol. 101 31–44. 10.1385/0-89603-471-2:31 [DOI] [PubMed] [Google Scholar]
- Caminero J. A., García-Basteiro A. L., Rendon A., Piubello A., Pontali E., Migliori G. B. (2019). The future of drug-resistant tuberculosis treatment: learning from the past and the 2019 World Health Organization consolidated guidelines. Eur. Respir. J. 54:1901272. 10.1183/13993003.01272-2019 [DOI] [PubMed] [Google Scholar]
- Chawla K., Martinez E., Kumar A., Shenoy V. P., Sintchenko V. (2018). Whole-genome sequencing reveals genetic signature of bedaquiline resistance in a clinical isolate of Mycobacterium tuberculosis. J. Glob. Antimicrob. Resist. 15 103–104. 10.1016/j.jgar.2018.09.006 [DOI] [PubMed] [Google Scholar]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393 537–544. [DOI] [PubMed] [Google Scholar]
- Conradie F., Diacon A. H., Ngubane N., Howell P., Everitt D., Crook A. M., et al. (2020). Treatment of highly drug-resistant pulmonary tuberculosis. N. Engl. J. Med. 382 893–902. 10.1056/NEJMoa1901814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2020). Bedaquiline/Mycobacterium tuberculosis 7H9 International MIC Distribution - Reference Database 2020-08-09. Available online at: https://mic.eucast.org/Eucast2/regShow.jsp?Id=44655 (accessed August 9, 2020). [Google Scholar]
- Ghajavand H., Kargarpour Kamakoli M., Khanipour S., Pourazar Dizaji S., Masoumi M., Rahimi Jamnani F., et al. (2019). High prevalence of bedaquiline resistance in treatment-naive tuberculosis patients and verapamil effectiveness. Antimicrob. Agents Chemother. 63:e02530-18. 10.1128/AAC.02530-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkoorn R. C., Uplekar S., Cole S. T. (2014). Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58 2979–2981. 10.1128/AAC.00037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitric E., Verhasselt P., Koul A., Andries K., Hoffner S., Andersson D. I. (2010). Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 54 1022–1028. 10.1128/AAC.01611-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N. A., Omar S. V., Joseph L., Govender N., Blows L., Ismail F., et al. (2018). Defining Bedaquiline susceptibility, resistance, cross-resistance and associated genetic determinants: a retrospective cohort study. Ebiomedicine 28 136–142. 10.1016/j.ebiom.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K., Cirillo D. M., Hoffner S., Ismail N. A., Kaur D., Lounis N., et al. (2016). A multilaboratory, multicountry study to determine bedaquiline MIC quality control ranges for phenotypic drug susceptibility testing. J. Clin. Microbiol. 54 2956–2962. 10.1128/JCM.01123-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodio O., Georges Togo A. C., Sadio Sarro Y. D., Fane B., Diallo F., Somboro A., et al. (2019). Competitive fitness of Mycobacterium tuberculosis in vitro. Int. J. Mycobacteriol. 8 287–291. 10.4103/ijmy.ijmy_97_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Sun F., Zhang W. (2019). Bedaquiline and delamanid in the treatment of multidrug-resistant tuberculosis: Promising but challenging. Drug Dev Res. 80 98–105. 10.1002/ddr.21498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E., Hennessy D., Jelfs P., Crighton T., Chen S. C., Sintchenko V. (2018). Mutations associated with in vitro resistance to bedaquiline in Mycobacterium tuberculosis isolates in Australia. Tuberculosis 111 31–34. 10.1016/j.tube.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Melnyk A. H., Wong A., Kassen R. (2015). The fitness costs of antibiotic resistance mutations. Evol. Appl. 8 273–283. 10.1111/eva.12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez C., Rodriguez F., Ribeiro A. L., Zara F., Frongia C., Lobjois V., et al. (2013). Synthesis and evaluation of α-ketotriazoles and α,β-diketotriazoles as inhibitors of Mycobacterium tuberculosis. Eur. J. Med. Chem. 69 167–173. 10.1016/j.ejmech.2013.06.042 [DOI] [PubMed] [Google Scholar]
- Milano A., Pasca M. R., Provvedi R., Lucarelli A. P., Manina G., Ribeiro A. L., et al. (2009). Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis 89 84–90. 10.1016/j.tube.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Nieto Ramirez L. M., Quintero Vargas K., Diaz G. (2020). Whole genome sequencing for the analysis of drug resistant strains of Mycobacterium tuberculosis : a systematic review for Bedaquiline and Delamanid. Antibiotics 9:E133. 10.3390/antibiotics9030133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino J. C., Martin A., Camacho M., Guerra H., Swings J., Portaels F. (2002). Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 46:2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Zong Z., Huo F., Jing W., Ma Y., Dong L., et al. (2017). In vitro drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing, China. Antimicrob. Agents Chemother. 61:e0900-17. 10.1128/AAC.00900-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philley J. V., Wallace R. J., Jr., Benwill J. L., Taskar V., Brown-Elliott B. A., Thakkar F. (2015). Preliminary Results of Bedaquiline as salvage therapy for patients with nontuberculous mycobacterial lung disease. Chest 148 499–506. 10.1378/chest.14-2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontali E., Raviglione M. C., Migliori G. B. The writing group members of the Global TB Network Clinical Trials Committee (2019). Regimens to treat multidrug-resistant tuberculosis: past, present and future perspectives. Eur. Respir. Rev. 28:190035. 10.1183/16000617.0035-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss L., Langer J. D., Yildiz O., Eckhardt-Strelau L., Guillemont J. E., Koul A., et al. (2015). Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. Sci. Adv. 1:e1500106. 10.1126/sciadv.1500106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A., Kumar N., Wright C. C., Chou T. H., Tringides M. L., Bolla J. R., et al. (2014). Crystal structure of the transcriptional regulator Rv0678 of Mycobacterium tuberculosis. J. Biol. Chem. 289 16526–16540. 10.1074/jbc.M113.538959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segala E., Sougakoff W., Nevejans-Chauffour A., Jarlier V., Petrella S. (2012). New mutations in the mycobacterial ATP synthase: new insights into the binding of the diarylquinoline TMC207 to the ATP synthase C-ring structure. Antimicrob. Agents Chemother. 56 2326–2334. 10.1128/AAC.06154-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Bisht D., Khan A. U. (2018). Potential alternative strategy against drug resistant tuberculosis: a proteomics prospect. Proteomes 6:26. 10.3390/proteomes6020026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Sharma S., Sharma J. (2020). Potential strategies for the management of drug-resistant tuberculosis. J. Glob. Antimicrob. Resist. 22 210–214. 10.1016/j.jgar.2020.02.029 [DOI] [PubMed] [Google Scholar]
- Smith C. S., Aerts A., Saunderson P., Kawuma J., Kita E., Virmond M. (2017). Multidrug therapy for leprosy: a game changer on the path to elimination. Lancet Infect. Dis. 17:e00293-97. 10.1016/S1473-3099(17)30418-8 [DOI] [PubMed] [Google Scholar]
- Veziris N., Bernard C., Guglielmetti L., Le Du D., Marigot-Outtandy D., Jaspard M., et al. (2017). Rapid emergence of Mycobacterium tuberculosis bedaquiline resistance: lessons to avoid repeating past errors. Eur. Respir. J. 49:1601719. 10.1183/13993003.01719-2016 [DOI] [PubMed] [Google Scholar]
- Viljoen A., Raynaud C., Johansen M. D., Roquet-Banères F., Herrmann J. L., Daher W., et al. (2019). Improved activity of bedaquiline by verapamil against Mycobacterium abscessus in vitro and in macrophages. Antimicrob. Agents Chemother. 63:e0705-19. 10.1128/AAC.00705-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villellas C., Coeck N., Meehan C. J., Lounis N., de Jong B., Rigouts L., et al. (2017). Unexpected high prevalence of resistance-associated Rv0678 variants in MDR-TB patients without documented prior use of clofazimine or bedaquiline. J. Antimicrob. Chemother. 72 684–690. 10.1093/jac/dkw502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2019a). Global Tuberculosis Report. Available online at: https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf?ua=1 (accessed October 17, 2019). [Google Scholar]
- World Health Organization [WHO] (2019b). World Health Organization [WHO] Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment. Available at: https://www.who.int/tb/publications/2019/consolidated-guidelines-drug-resistant-TB-treatment/en/ (accessed October 17, 2019). [PubMed] [Google Scholar]
- Xu J., Wang B., Hu M., Huo F., Guo S., Jing W., et al. (2017). Primary clofazimine and bedaquiline resistance among isolates from patients with multidrug-resistant tuberculosis. Antimicrob. Agents Chemother. 61:e0239-17. 10.1128/AAC.00239-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Chen J., Cui P., Shi W., Zhang W., Zhang Y. (2015). Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 70 2507–2510. 10.1093/jac/dkv150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimenkov D. V., Nosova E. Y., Kulagina E. V., Antonova O. V., Arslanbaeva L. R., Isakova A. I., et al. (2017). Examination of bedaquiline- and linezolid-resistant Mycobacterium tuberculosis isolates from the Moscow region. J. Antimicrob. Chemother. 72 1901–1906. 10.1093/jac/dkx094 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.