Abstract
We first reported a phenomenon of cross-resistance to vancomycin (VCM) and daptomycin (DAP) in methicillin-resistant Staphylococcus aureus (MRSA) in 2006, but mechanisms underlying the cross-resistance remain incompletely understood. Here, we present a follow-up study aimed to investigate genetic determinants associated with the cross-resistance. Using 12 sets of paired DAP susceptible (DAPS) and DAP non-susceptible (DAPR) MRSA isolates from 12 patients who had DAP therapy, we (i) assessed susceptibility to DAP and VCM, (ii) compared whole-genome sequences, (iii) identified mutations associated with cross-resistance to DAP and VCM, and (iv) investigated the impact of altered gene expression and metabolic pathway relevant to the cross-resistance. We found that all 12 DAPR strains exhibiting cross-resistance to DAP and VCM carried mutations in mprF, while one DAPR strain with reduced susceptibility to only DAP carried a lacF mutation. On the other hand, among the 32 vancomycin-intermediate S. aureus (VISA) strains isolated from patients treated with VCM, five out of the 18 strains showing cross-resistance to DAP and VCM carried a mprF mutation, while 14 strains resistant to only VCM had no mprF mutation. Moreover, substitution of mprF in a DAPS strain with mutated mprF resulted in cross-resistance and vice versa. The elevated lysyl-phosphatidylglycerol (L-PG) production, increased positive bacterial surface charges and activated cell wall (CW) synthetic pathways were commonly found in both clinical isolates and laboratory-developed mutants that carry mprF mutations. We conclude that mprF mutation is responsible for the cross-resistance of MRSA to DAP and VCM, and treatment with DAP is more likely to select for mprF-mediated cross-resistance than is with VCM.
Subject terms: Genetics, Microbiology, Pathogenesis
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) infections are serious clinical problems causing high morbidity and mortality worldwide. MRSA is resistant to not only β-lactam antibiotics but also other classes of antibiotics such as aminoglycosides, tetracyclines, or fluoroquinolones, restricting the available antibacterial agents for MRSA treatment1–4. Vancomycin (VCM), a glycopeptide antibiotic exerting bactericidal activity by binding to D-ala-D-ala residues of peptidoglycan to inhibit bacterial cell wall (CW) synthesis, is the first-line antibiotic against MRSA infections5. Emergence of MRSA with reduced susceptibility to VCM therefore further limits the scarcely available treatment options3,4,6–8.
Daptomycin (DAP), a cyclic lipopeptide antibiotic with potent bactericidal activity, is frequently used as salvage therapy after VCM treatment failure9. In the presence of calcium, the anionic DAP molecule attains its active cationic peptide form, which will then insert its lipophilic tail into the negative-charged cell membrane (CM)10,11. The interaction between DAP and the CM causes potassium leakage and membrane depolarization that ultimately contribute to cell death12. This means that DAP and VCM differ in not only chemical structure but also in their bactericidal mechanism7,13. Nevertheless, MRSA strains with cross-resistance to DAP and VCM, which was first reported by our group in 200614, have been frequently isolated from patients treated with either DAP or VCM8,15,16.
Multiple peptide resistance factor (MprF) is reported to mediate DAP non-susceptibility in S. aureus17. Mutation of mprF is associated with gain-of-function, in which lysinylation of phosphatidylglycerol (PG) is enhanced, thus increasing membrane lysyl-phosphatidylglycerol (L-PG) production17,18. This positively charged L-PG will then be translocated from the inner membrane to the outer leaflet of the CM by the flippase domain of the MprF protein, causing an increased net positive charge on the CM19. Eventually, the more positively charged CM surface may serve as a protective barrier against DAP binding20,21. However, this remains controversial since only some DAP non-susceptible (DAPR) strains displayed enhanced L-PG concentration in outer leaflet although most DAPR strains carrying mprF mutation showed increased L-PG production19,22–25. Aside from changes in CM properties, increased thickness of the CW is also proposed to cause ineffective binding of DAP to the CM14,26. Some DAPR strains are accompanied by an increased expression of genes involved in CW synthesis, such as murAB or pbp2, a response similar to those induced by VCM and the other CW-targeting agents27,28. As a salient feature of vancomycin-intermediate S. aureus (VISA), CW thickening was reported one of the contributing factors to VCM resistance in some DAPR strains. In fact, mutations in either walK, encoding the sensor protein kinase of a two-component regulatory system, or vraSR, involved in cell envelope homeostasis, both of which resulted in CW thickening, were related to the DAP/VCM cross-resistance29,30. However, phenotypic changes in CW thickness were not consistently observed in all DAPR strains26,31. Consequently, the mechanism(s) conferring resistance of S. aureus to the two different classes of antibacterial agents remain largely unknown.
This study aimed to investigate genetic determinants of the cross-reduced susceptibility to DAP and VCM in clinically isolated MRSA. A total of 12 sets of DAP susceptible (DAPS) and DAPR MRSA isolates collected from different hospitals in Japan were whole-genome sequenced, and gene mutations associated with the phenotype of cross-reduced susceptibility were identified and functionally characterized. Our results indicated that reduced susceptibility to both DAP and VCM was regulated by mprF mutation via increased L-PG production, subsequent alteration of bacterial surface charge, and CW biosynthetic pathways.
Results
Reassessment of VCM and DAP susceptibilities
This study began with analysis and validation of VCM and DAP susceptibilities for all paired isolates collected from 12 patients from whom DAPR MRSA strains were generated during DAP therapy (Supplemental Table 1). DAP and VCM susceptibility tests on the 13 DAPR strains could classify the DAPR strains into two different resistance groups, judged by minimum inhibitory concentrations (MICs) and population analysis profiles, namely, cross-reduced susceptibility to DAP and VCM (termed reduced DAP/VCM susceptibility) and reduced susceptibility to only DAP (Table 1). Among the 13 DAPR strains, 12 strains that belonged to the cross-reduced DAP/VCM susceptibility group showed DAP and VCM MICs of 1.5 to 3 mg/L. There is a 3.0- to 8.0-fold increase in DAP MIC and a 1.3- to 1.5-fold increase in VCM MIC of DAPR strains when compared to their corresponding DAPS parent strains, except for strain F-2 which showed an exceptional twofold increase in VCM MIC. One strain, K-2, showed reduced susceptibility to only DAP with a MIC of 2 mg/L but had no change in VCM susceptibility compared to its parent strain K-1 (Table 1). These susceptibility patterns were also confirmed by analysis of a resistant subpopulation against DAP and VCM (Supplemental Fig. 1) and determination of MICs with a different method (Supplemental Table 4). In addition, almost all DAPR strains had increased doubling time compared to their DAPS counterparts, but there were two exceptions (D-2 and G-2) (Table 1).
Table 1.
Summary of MIC, gene mutation, MLST, doubling time, cell-wall thickness, cytochrome c uptake and L-PG content on the isolates from DAP treatment patients.
| Patient | Strain | DAP MIC | VCM MIC | MLST | Mutation in | Doubling time (min) | Cell wall thickness (nm) | Cytochrome c uptake (%)g | L-PG content (%)h | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mg/L | Ratioa | mg/L | Ratioa | mprF | Others | |||||||
| Cross-reduced susceptibility group (to DAP and VCM) | ||||||||||||
| A | A-1 | 0.38 | 1.00 | 1.5 | 1.00 | 764 | –b | – | 26.6 | 23.18 ± 2.50 | 100 | 100 |
| A-2 | 1.5 | 3.95 | 2 | 1.33 | 764 | T345I | – | 27.0 | 24.40 ± 1.72 | 73.27 ± 6.79* | 177.20 ± 18.47* | |
| B | B-1 | 0.19 | 1.00 | 1.5 | 1.00 | 764 | – | – | 30.0 | 23.28 ± 2.73 | 100 | 100 |
| B-2 | 1.5 | 7.89 | 2 | 1.33 | 764 | L776S | B1_1709 (N31_fs)c | 30.7 | 22.41 ± 1.58 | 78.75 ± 6.83** | 128.86 ± 10.58* | |
| C | C-1 | 0.5 | 1.00 | 2 | 1.00 | 764 | – | – | 29.5 | 24.28 ± 2.00 | 100 | 100 |
| C-3 | 1.5 | 3.00 | 3 | 1.50 | 764 | A475P | – | 30.2 | 22.17 ± 1.86 | 71.45 ± 4.51** | 147.52 ± 18.09* | |
| C-4 | 3 | 6.00 | 3 | 1.50 | 764 | L459_H466 del | – | 30.9 | 23.17 ± 2.32 | 70.65 ± 11.63* | 140.48 ± 11.44* | |
| D | D-1 | 0.25 | 1.00 | 1.5 | 1.00 | 1 | – | – | 32.8 | 20.60 ± 2.77 | 100 | 100 |
| D-2 | 2 | 8.00 | 2 | 1.33 | 1 | L826F | ir-1, ir-2d | 27.6 | 19.86 ± 1.80 | 52.37 ± 15.09* | 144.14 ± 12.20* | |
| E | E-1 | 0.25 | 1.00 | 1 | 1.00 | 764 | – | – | 32.6 | 25.30 ± 2.46 | 100 | 100 |
| E-2 | 1.5 | 6.00 | 1.5 | 1.50 | 764 | L826F | – | 36.2 | 23.64 ± 2.67 | 61.79 ± 3.30** | 166.21 ± 24.86* | |
| F | F-1 | 0.25 | 1.00 | 0.75 | 1.00 | 764 | – | – | 33.3 | 24.89 ± 2.39 | 100 | 100 |
| F-2 | 2 | 8.00 | 1.5 | 2.00 | 764 | L826F | agrA(T210I), F1_0943(A363T) | 36.5 | 25.86 ± 2.15 | 71.98 ± 4.81* | 124.00 ± 12.49* | |
| G | G-1 | 0.5 | 1.00 | 1 | 1.00 | 2809 | – | – | 32.2 | 30.02 ± 2.32 | 100 | 100 |
| G-2 | 1.5 | 3.00 | 1.5 | 1.50 | 2809 | T345A | ir | 31.4 | 29.72 ± 2.15 | 45.99 ± 2.14** | 158.88 ± 26.37* | |
| H | H-1 | 0.75 | 1.00 | 2 | 1.00 | 5 | – | – | 27.1 | 23.31 ± 1.84 | 100 | 100 |
| H-3 | 0.5 | 0.67 | 2 | 1.00 | 5 | – | hisF(G207_del), H1_0704(C241Y) | 25.9 | NDe | 92.05 ± 4.73 | 89.62 ± 7.58 | |
| H-5 | 3 | 4.00 | 3 | 1.50 | 5 | L291I | hisF(G207_del), H1_0704(C241Y) | 26.6 | 22.92 ± 2.35 | 78.46 ± 0.77** | 155.3 ± 13.74* | |
| H-5(mprF_H-1) | 0.5 | 1.00 | 2 | 1.00 | 5 | – | hisF(G207_del), H1_0704(C241Y) | ND | ND | 100 | 100 | |
| H-3(mprF_H-5) | 3 | 6.00 | 3 | 1.50 | 5 | L291I | hisF(G207_del), H1_0704(C241Y) | ND | ND | 69.26 ± 7.81* | 149.26 ± 21.91* | |
| I | I-2 | 0.75 | 1.00 | 2 | 1.00 | NT | – | – | 27.0 | 22.11 ± 1.83 | 100 | 100 |
| I-3 | 3 | 4.00 | 3 | 1.50 | NT | W424R | – | 33.8 | 25.55 ± 2.92** | 68.88 ± 5.90* | 165.51 ± 12.12* | |
| J | J-1 | 0.38 | 1.00 | 1.5 | 1.00 | 764 | – | – | 27.1 | 23.44 ± 1.37 | 100 | 100 |
| J-3 | 1.5 | 3.95 | 2 | 1.33 | 764 | L341S | – | 30.1 | 23.40 ± 1.63 | 73.32 ± 3.19** | 119.66 ± 7.16* | |
| L | L-1 | 0.25 | 1.00 | 1.5 | 1.00 | 380 | – | – | 26.5 | 22.12 ± 1.80 | 100 | 100 |
| L-2 | 2 | 8.00 | 2 | 1.33 | 380 | S337L | L1_0548(T134I) | 29.9 | 22.24 ± 1.29 | 69.99 ± 9.15* | 116.96 ± 4.30* | |
| Reduced susceptibility to only DAP group | ||||||||||||
| K | K-1 | 0.38 | 1 | 1.5 | 1 | 764 | – | lacF(trunc)f | 24.7 | 21.50 ± 1.82 | 100 | 100 |
| K-2 | 2 | 5.26 | 1.5 | 1.00 | 764 | – | lacF(H41) | 27.1 | 21.15 ± 1.75 | 122.16 ± 1.00* | 55.01 ± 15.16** | |
a) MIC ratio of DAPR strain to its parent DAPS strain; b) no mutation; c) fs: frameshift; d) ir: intergenic region; e) not determined; f) truncated at position 42; g & h) relative values compared to corresponding parent strains (*p < 0.05; **p < 0.01).
Comprehensive mutation identification
To determine genomic alterations associated with reduced susceptibilities to DAP and VCM, whole-genome sequences of 12 pairs of DAPS and DAPR MRSA strains from 12 patients were determined. Comparative genome analysis found that all DAPR strains with reduced susceptibility to both DAP and VCM, carried at least one non-synonymous mutation. All identified mutations were validated using PCR-based sequencing and are listed in Table 1. Interestingly, these strains unanimously carried mutations on mprF gene encoding an L-PG synthetase, which is known to synthesize positively charged lipid L-PG. On the other hand, the DAPR strain K-2 with reduced susceptibility to only DAP had an insertion mutation in lacF, which encodes a conserved ATP-binding domain homologous to ABC transporters known in bacteriocin immunity systems32. The lacF of K-1 differed from that of K-2 for the presence of one thymine deletion at position 125 that generated a premature stop codon (Supplemental Fig. 2A) and resulted in LacF truncation at position 42 (Supplemental Fig. 2B), indicating that the restoration of LacF function is responsible for reduced susceptibility of K-2 to DAP.
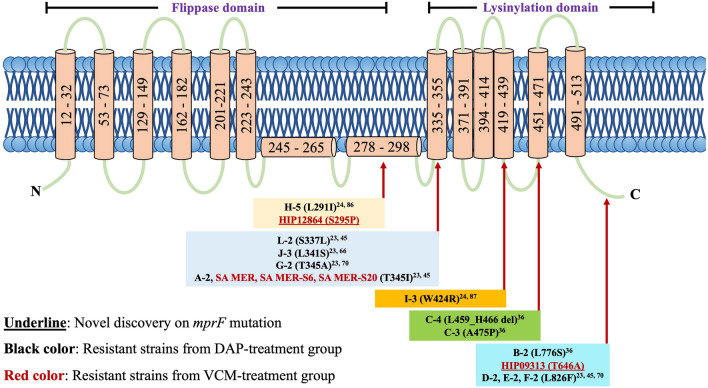
For the mprF mutation, 10 types of point mutations were identified in this study, most of which were located on the lysinylation domain of MprF (Fig. 1). In addition, as shown in Table 1, DAPR strains D-2 and G-2 carried intergenic region mutations besides the mprF mutation. Four out of 11 cross-reduced susceptibility strains had additional mutations that resulted in amino acid substitutions of B1_1709(N31_fs) in DAPR strain B-2, agrA(T210I) and F1_0943(A363T) in DAPR strain F-2, L1_0548(T134I) in DAPR strain L-2, and hisF(G207_del) and H1_0704(C241Y) in DAPR strains H-5. The mutations of hisF(G207_del) and H1_0704(C241Y) could also be found in DAPS strain H-3, indicating that these mutations seem to not be directly involved in the mechanism of reduced DAP/VCM susceptibility. In summary, mprF mutations were commonly found in the MRSA isolates with cross-reduced susceptibility to DAP and VCM that were isolated from patients who had DAP therapy.
Figure 1.
The location of mprF mutations in DAPR strains and VISA strains. Most DAPR (black text) and VISA (red text) isolates in this experiment carried mprF mutations on the lysinylation domain. The underlined mutations refer to newly discovered mprF mutations. The MprF structure is modified from previous studies21.
Detection of genes reported to be associated with decreased susceptibility to VCM or DAP in S. aureus
Many genes have been reported to be associated with conversion of vancomycin-susceptible S. aureus to VISA, including walK, clpP, graSR, vraSR, msrR, and rpoB30,33,34. Some of these VISA-related genes were also reported to reduce DAP susceptibility in MRSA33,35. Therefore, we examined the sequences of these genes for all strains in DAP treatment group, but no differences were found between any pair of DAPS and DAPR strains. The phenomenon of DAP and VCM cross-resistance was first reported in VISA strains in 200614, and afterward, it became recognized in clinical settings during the VCM therapy. Therefore, we further investigated and characterized the pattern of DAP and VCM susceptibilities for 32 VISA strains isolated from patients worldwide during years of 1996 – 2004 when the DAP was not available in clinical setting34. We found that among the 32 VISA strains, 18 showed both intermediate VCM resistance and reduced susceptibility to DAP (cross-reduced susceptibility) (Table 2). All 32 strains were examined for single nucleotide polymorphisms (SNPs) associated with VCM resistance (Table 2). We also determined full mprF sequences for all 32 VISA strains using PCR and Sanger sequencing methods, and the results were combined with our previous results on the SNPs of walK, clpP, graSR, vraSR, msrR, and rpoB in Table 2. We found point mutations in mprF in five out of 18 VISA strains with cross-reduced susceptibility to DAP and VCM, but not in any of the 14 strains with only intermediate VCM resistance. No other mutations found in genes or intergenic region of cross-reduced susceptible DAPR strains from DAP-treated patients were identified in the VISA isolates. These results suggested that there is a high prevalence of reduced DAP/VCM susceptibility among the VISA strains and that mprF mutations play a role in conferring the cross-reduced susceptibility to VCM and DAP in some VISA strains that were generated during the prolonged VCM chemotherapy.
Table 2.
Summary of MIC and gene mutation of VISA strains.
| Strain name | Etest MIC (mg/L) | Gene Mutationsa | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DAP | VCM | mprF | walK | clpP | graS | graR | vraS | vraR | msrR | rpoB | |
| Cross-reduced susceptibility group (to DAP and VCM) | |||||||||||
| MI (HIP5827) | 1.5 | 6 | – | V494L30 | – | – | – | – | – | – | R140S34 |
| SA MER | 2 | 3 | T345I | – | ND | ND | ND | ND | ND | ND | ND |
| SA MER-S6 | 4 | 3 | T345I | – | ND | ND | – | – | ND | – | ND |
| SA MER-S20 | 4 | 6 | T345I | – | ND | ND | – | – | ND | – | – |
| HIP06297 (PC) | 2 | 4 | – | A567D30 | ND | ND | – | – | ND | – | Q468L34 |
| HIP08926 | 2 | 3 | – | R222I, T492K30 | ND | L26F, I59L, T224I30 | D148Q30 | – | ND | – | ND |
| HIP09143 | 1.5 | 3 | – | – | ND | ND | – | – | ND | – | ND |
| HIP12864 | 2 | 4 | S295P | – | ND | ND | – | – | ND | – | P519L34 |
| HIP13057 | 1.5 | 4 | – | R282C30 | ND | ND | E15K30 | – | ND | – | H481Y34 |
| HIP13036 | 1.5 | 6 | – | – | ND | ND | – | T104A26 | ND | – | – |
| Mu50 | 1.5 | 6 | – | – | – | – | N197S30 | I5N26 | – | E146K34 | H481Y34 |
| HIP06854 | 1.5 | 6 | – | T492K30 | ND | ND | ND | ND | ND | ND | ND |
| HIP09313 | 2 | 4 | T646A | L10F, S437T30 | R152H26 | ND | ND | P327S26 | E59D30 | ND | ND |
| HIP09662 | 1.5 | 3 | – | Ins 433N, Ins 434D30 | ND | L26F, I59L, T224I30 | D148Q30 | – | E59D30 | – | D471N, S486L34 |
| LY-1999–01 | 1.5 | 4 | – | N48K, R222K, A468T30 | ND | L26F, I59L, T224I30 | D148Q30 | ND | E59D30 | K321R34 | R406S34 |
| 99/3700-W | 1.5 | 3 | – | R222K, V366M, A468T30 | ND | L26F, I59L, T224I30 | D148Q30 | – | – | – | – |
| 28,160 | 1.5 | 3 | – | ND | ND | L26F, I59L, T224I30 | D148Q30 | ND | E59D30 | ND | S529L34 |
| BR5 | 1.5 | 3 | – | R222K, V366M, A468T30 | ND | L26F, I59L, T224I30 | D148Q30 | – | E59D30 | ND | I527M30 |
| Intermediate resistance to only VCM group | |||||||||||
| NJ (HIP5836) | 0.75 | 4 | – | I28T, 1341V30 | – | – | S79F30 | A260V30 | – | – | H481Y34 |
| HIP07256 | 1 | 3 | – | ND | ND | ND | ND | ND | ND | ND | ND |
| LIM2 | 1 | 3 | – | ND | ND | L26F, I59L, T224I30 | D148Q30 | – | E59D30 | ND | H481N, S529L34 |
| HIP09740 | 0.75 | 3 | – | V380I30 | ND | ND | – | – | ND | – | H481D34 |
| BR15 | 1 | 3 | – | ND | ND | ND | ND | ND | ND | ND | ND |
| HIP10540 | 1 | 4 | – | – | ND | L26F, I59L, T224I30 | D148Q30 | – | ND | – | A477V34 |
| P1V44 | 0.75 | 3 | – | – | ND | L26F, I59L, T224I30 | D148Q30 | – | ND | – | H481N, S529L34 |
| 99/3759-V | 0.75 | 3 | – | V156Q30 | M1V26 | L26F, I59L, T224I30 | D148Q30 | ND | E59D, H481N, S539L30 | ND | H481N, S529L34 |
| AMC11094 | 0.75 | 3 | – | ND | ND | ND | – | – | A113V30 | – | – |
| LY-1999–03 | 1 | 4 | – | N48K, R222K, A468T30 | ND | L26F, I59L, T224I30 | D148Q30 | – | E59D30 | K312R30 | – |
| C2000001227 | 1 | 4 | – | A243T30 | ND | ND | – | A314V30 | ND | – | – |
| NRS118 | 1 | 4 | – | F330S30 | ND | ND | – | – | ND | – | H481N, S529L34 |
| NRS126 | 0.5 | 3 | – | – | ND | ND | – | – | ND | – | H481N34 |
| 98,141 | 0.75 | 3 | – | – | ND | L26F, I59L, T224I30 | D148Q30 | ND | E59D30 | ND | H481N, S529L34 |
a) mprF mutation was determined in this study, and the other gene mutations were detected in the previous studies (references were indicated); -: no mutation; ND: Not determined.
Substitution of mprF in DAPS strain with mutated mprF identified in the DAPR strain caused reduced DAP and VCM susceptibility
To confirm the role of mprF mutations in reduced DAP and VCM susceptibility, an identified mprF mutation (mprF(L291I)) of DAPR strain H-5 was cloned and introduced into its corresponding DAPS strain H-3 to replace mprF of H-3. The H-3 strain (an isogenic strain of H-1 and H-5), isolated in between H-1 and H-5 during DAP therapy, was chosen for mprF substitution to eliminate the confounding effect of other gene mutations (hisF and H1_0704), which cannot be found in DAPS strain H-1 (Table 1). The MIC test showed that the H-3 strain carrying mprF(L291I) had increased MICs of both DAP and VCM, from 0.5 and 2 respectively, to 3 mg/L, showing reduced DAP/VCM susceptibility to the same levels of the H-5 strain; vice versa, replacement of mprF(L291I) of H-5 with mprF of H-3 resulted in decreased MICs of both DAP and VCM from 3 to 0.5 and 2 mg/L, respectively, for the H-5 strain (Table 1). These results demonstrated that mprF mutations cause reduced susceptibility to both DAP and VCM in the MRSA H-5 strain.
Reduced DAP and VCM susceptibility associated with mprF mutation was found in in vitro selected mutants
The clinical DAPR strains isolated from patients who had DAP therapy exhibited reduced susceptibility to both DAP and VCM due to mprF mutations. The mutation position in mprF varied among strains from different patients, as shown in the above results (Table 1, Fig. 1) and the findings of Kanesaka et al.36. To understand whether this is also the case for in vitro selected DAPR mutants with cross-reduced DAP/VCM susceptibilities, we generated DAPR strains with reduced DAP/VCM susceptibility in vitro by exposing DAPS MRSA to DAP of gradually increasing concentrations and examined mutations for mprF and lacF. Two DAPR mutants were obtained from DAPS strain C-1 (DAP MIC, 0.5 mg/L) by stepwise selection on Mueller Hinton (MH) agar containing increasing DAP concentrations from 0.5 to 4 mg/L. These mutants could grow in the presence of 4 mg/L DAP. We found that these two mutants had reduced susceptibility to both DAP and VCM, increasing the MICs of DAP from 0.5 to 3 and 6 mg/L and VCM from 2 to 3 and 4 mg/L, and are accompanied by mprF mutations, mprF(T472K) for the mutant C-1_DAPR#1 and mprF(R50L) for mutant C-1_DAPR#2, respectively (Table 3).
Table 3.
Summary of MIC, doubling time and mutations in mprF and lacF on in vitro derivatives of the C-1 and K-1 strains.
| Strain | Etest MIC (mg/L) | Doubling Time (min) | Mutation in | ||
|---|---|---|---|---|---|
| DAP | VCM | mprF | lacF | ||
| Clinical isolates from Patient C | |||||
| C-1 | 0.5 | 2 | 29.5 | – | – |
| C-3 | 1.5 | 3 | 30.2 | A475P | – |
| C-4 | 3 | 3 | 30.9 | L459_H466 del | – |
| In vitro derivatives of C-1 strain | |||||
| C-1_DAPR#1 | 3 | 3 | 30.8 | T472K | – |
| C-1_DAPR#2 | 6 | 4 | 28.0 | R50L | – |
| Clinical isolates from Patient K | |||||
| K-1 | 0.38 | 1.5 | 24.7 | – | trunc* |
| K-2 | 2 | 1.5 | 27.1 | - | H41 |
| In vitro derivatives of K-1 strain | |||||
| K-1_DAPR#1 | 6 | 3 | 54.6 | T472K | H41A |
| K-1_DAPR#2 | 6 | 3 | 35.4 | T472K | H41A |
–: no mutation; DAPR: Daptomycin non-susceptible strain; *: truncated at position 42.
The DAPR strain K-2 with reduced susceptibility to only DAP carried a lacF mutation that has not been previously reported (Table 1). To valuate this mutation, a similar stepwise DAP selection was performed on its DAPS counterpart strain K-1, and two DAP-resistant mutants (K-1_DAPR#1 and K-1_DAPR#2) were generated. Interestingly, these two in vitro selected mutants showed decreased susceptibility to both DAP and VCM, increasing the MICs of DAP from 0.38 to 6 mg/L and VCM from 1.5 to 3 mg/L; it was also accompanied by a mprF mutation in addition to the restoration of LacF as seen in K-2 strain (Table 3 and Supplemental Fig. 2B). These results, together with the above study on clinical DAPR strains, demonstrated that the phenomenon of reduced susceptibility to DAP and VCM in MRSA is strongly associated with mprF mutations.
Reduced DAP/VCM susceptibilities and CW thickness
Thickened CW is known as a phenotypic determinant for VCM resistance in VISA7,8,37. Although not consistently reported, alteration of CW structure and/or changes in expression of genes involved in CW metabolic pathways have also been found in some DAPR strains26,31. Therefore, alteration of CW structure might be one of the factors involved in reduced DAP and VCM susceptibility. In order to test this hypothesis, the CW thickness of 30 cells from each DAPS/DAPR strain were measured by transmission electron microscopy (TEM). The results showed that only one strain in the group of cross-reduced DAP and VCM susceptibility (I-3) carrying a mprF mutation (mprF(W424R)) displayed significantly increased CW thickness (25.55 ± 2.92 nm) compared with its susceptible counterpart I-2 (22.11 ± 1.83 nm) (Table 1). In contrast, the other 11 DAPR strains in reduced DAP/VCM susceptibility group did not exhibit significant CW thickening compared with their corresponding parental strains (Table 1).
There was also no difference in CW thickness between DAPS isolate K-1 (21.50 ± 1.82 nm) and DAPR isolate K-2 (21.15 ± 1.75 nm) exhibiting resistance to only DAP due to a lacF mutation (Table 1). These results did not clearly support the association of CW thickening with the phenomenon of reduced susceptibility to DAP in the DAPR strains with mprF or lacF mutations.
mprF mutation and bacterial surface charge
The mprF mutation had been previously reported in MRSA with reduced DAP susceptibility. MprF is a membrane-bound enzyme that adds lysine to phosphatidylglycerol in the cytoplasmic membrane. This modification is reported to change the electrostatic repulsive forces of the bacterial CM, which then conferred reduced susceptibility to cationic antimicrobial peptides19,23. To determine whether mprF mutations identified in this study resulted in such alterations, we carried out a cytochrome c binding assay on all DAPR strains to examine the alteration of bacterial surface charges. A cationic cytochrome c can bind a negatively-charged bacterial cell surface and, hence, has been widely employed to determine the relative surface charges of the cell envelope38,39. Our results showed that all strains carrying a mprF mutation had significantly reduced cytochrome c binding when compared to their parental strains, indicating that all mprF mutations identified in this study caused increased positive surface charge (Table 1). Similarly, increased positive charge on bacterial surface was observed when the mprF of DAPS strain H-3 was replaced with mutated mprF, while replacement of mutated mprF in DAPR strain H-5 with that of its wild-type counterpart reduced the positive surface charge (Table 1). The DAPR strain with reduces susceptibility to only DAP carrying a lacF mutation did not exhibit reduced negative charges (Table 1). These results suggested that alteration of bacterial surface charge is associated with mprF mutation-mediated reduced DAP/VCM susceptibility in MRSA.
mprF mutation and L-PG production
The increased cationic phospholipid L-PG production in cytoplasmic membranes has been reported to decrease DAP susceptibility in MRSA40. To understand whether the mprF mutations found in this study are implicated in L-PG production, we set out to determine membrane L-PG levels for all DAPR and DAPS strains using the thin-layer chromatography (TLC) assay. Altered L-PG production of DAPR strains over corresponding DAPS strains was calculated in relative values (percentages) and is summarized in Table 1. As shown in Table 1, all DAPR strains with cross-reduced DAP/VCM susceptibility (carrying a mprF mutation) showed increased L-PG production. Although a more than 50% increase in L-PG production can be found in most DAPR strains, four strains (B-2, F-2, J-3, and L-2) harboring mprF mutations at different locations displayed only a marginal increase (10% or 20%) (Table 1). These results suggested that increased L-PG production regulated by mprF mutation may contribute to cross-reduced DAP/VCM susceptibility. In addition, we found that the K-2 strain with reduced susceptibility to only DAP (carries lacF mutation) had decreased L-PG production compared to its DAPS counterpart. This indicated that lacF mutation may raise reduced susceptibility to DAP through a different metabolic pathway from what has been reported so far.
Transcriptional analysis on representative DAPR strains carrying mprF or lacF mutation and their DAPS counterparts
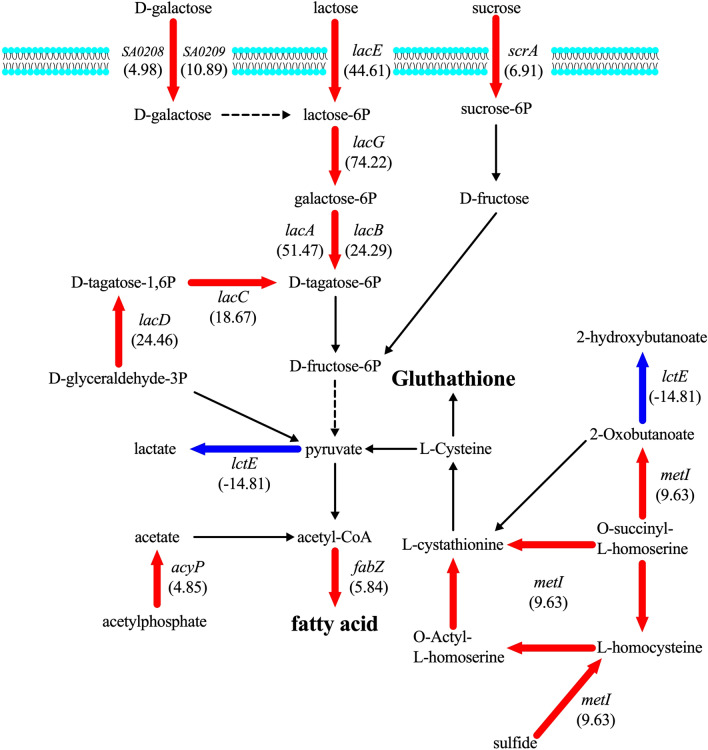
In the above results, the association of mprF mutation and altered membrane metabolic pathways with cross-reduced DAP/VCM susceptibility was clearly demonstrated; however, the impact of the mprF mutations found in this study on metabolic regulations toward the cross-reduced susceptibility remains to be clarified. To this end, a representative pair of DAPS and DAPR strains, H-3 and H-5, isolated from patient H were selected for a whole-genome-scale gene expression profiling by RNA-sequencing. The mprF(L291I) mutation identified in H-5 is the only genomic alteration found between H-5 and H-3 and is considered to be responsible for cross-reduced susceptibility to DAP and VCM, as verified by gene substitution experiments (see elsewhere above). A total of 103 genes differentially expressed by more than fourfold between H-3 and H-5 were found (Supplemental Table 5). Among them, 61 genes were upregulated (59.22%) and 42 genes were downregulated (40.78%). These genes could be roughly classified into four functional categories: metabolism (27.18%), information storage and processing (13.59%), cellular process and signaling (6.80%), and the others (52.43%). As shown in Supplemental Table 5, a number of genes directly or indirectly involved in metabolism of fatty acid and peptidoglycan are found to be upregulated in DAPR strain H-5. These include genes responsible for fructose-6-phosphate (F-6P) synthesis such as gatA (5.2-fold), tal (8.4-fold), and pmi (4.8-fold); or fatty acid synthesis such as hlb-2 (113.8-fold), fadA (4.3-fold) and plsY (4.6-fold), all of which were highlighted on the map of fatty acid metabolic pathway (Fig. 2). It was also noted that seven out of 61 upregulated genes in DAPR strain H-5 were involved in CW metabolism. Upregulation of nagD (6.6-fold), pyrR (4.1-fold) and pmi (4.8-fold) genes and downregulation of psuG (-5.1-fold) collectively affect the intracellular pool of uridine diphosphate-N-acetylglucosamine (UDP-NAG) and UDP-N-aceylmuramic acid (UDP-NAM), which serves as backbone for peptidoglycan (Fig. 2)41. In addition, upregulation of the genes associated with staphylococcal “cell wall stimulon”28,42,43, such as spsA (4.3-fold), ssaA (5.9-fold), relP (5.8-fold) and sasA (4.3-fold) was also found. Thus, mechanism of reduced DAP/VCM susceptibility by mprF mutation may be resulted from changes in CW/CM metabolism.
Figure 2.
Gene expression in contribution to cross-reduced susceptibility in DAPR strain H-5. Increased gene expression in fatty acid and peptidoglycan via carbohydrate metabolism (galactitol, ribose, or mannose) was observed. The red arrows refer to gene upregulation. The blue arrows refer to gene downregulation.
Unlike the association between mprF mutations and reduced DAP/VCM susceptibility, which can be deduced from our current study, the regulatory function of a lacF mutation on DAP single resistance in the K-2 strain is still unclear, prompting us to perform RNA-Seq analysis. The gene expression profiles of K-1 and K-2 strains are shown in Supplemental Table 6. There are 37 (68.52%) upregulated and 17 (31.48%) downregulated genes with a fourfold change between the DAPS (K-1) and DAPR (K-2) strains. Fifty percent of the differentially expressed genes are involved in amino acid or carbohydrate transport and metabolism, and energy production and conversion. This is followed by 11.11% of genes associated with defense mechanism and 7.4% with CW, CM, and envelop metabolisms. Among those upregulated genes, six lac operon genes (lacABCDEG) that comprise tagatose 6-phosphate pathway and lactose- and galactose- metabolizing enzymes overexpressed by 18- to 74-fold. In addition, altered expression in genes associated with CM metabolism were identified, including upregulation of acyP (4.9-fold), fabZ (5.8-fold), and metI (9.6-fold), and downregulation of lctE (-14.8-fold). Alteration of cellular defense associated genes was also found in the K-2, such as overexpression of gpxA2 (6.8-fold) and downregulation of opp-4D (-4.1-fold).
Discussion
The current study was conducted to investigate genetic determinants of cross-reduced susceptibility to DAP and VCM in MRSA. DAP and VCM are two different classes of antibiotics exhibiting distinct modes of bactericidal actions, thus triggering different resistance mechanisms in bacterial strains. Nevertheless, MRSA strains with reduced susceptibility to both DAP and VCM, a phenomenon known as cross-resistance to DAP and VCM, have been reported14–16. Owing to the fact that DAP and VCM are primary treatment options for MRSA infections, understanding the regulatory pathways leading to cross-resistance is crucial to facilitate the identification of novel target sites and the development of new therapeutic agents, contributing to the management of difficult-to-treat bacterial infections.
MprF is known to play a role in protecting bacteria against cationic antimicrobial peptides (CAMPs), including DAP, by altering bacterial membrane surface charges. Principally, MprF regulates the transition of phospholipid PG to L-PG by the addition of a lysine residue, causing an increased positive charge in CM, which is repulsive toward cationic antibiotics13,16,38. Accordingly, cells lacking the mprF gene showed increased susceptibility toward many positively charged antibiotics, including CAMPs, DAP, or VCM16,44. Previous studies frequently attributed reduced DAP susceptibility to mprF mutation, but a few mentioned about its association with alteration of VCM susceptibility25,45,46. Our current study demonstrated that mprF mutations are major determinants of cross-reduced susceptibility to DAP and VCM in MRSA during DAP therapy but have only partial contribution during the course of VCM chemotherapy (Table 1 and 2). The cross-reduced susceptibility to DAP and VCM mediated via mprF mutation was confirmed by a gene replacement assay whereby introduction of mprF mutation from DAPR strain H-5 to DAPS strain H-3 resulted in increased DAP and VCM MICs of DAPS strain H-3 (Table 1). This cross-reduced susceptibility by mprF mutation might partially be explained with the change of bacterial surface charges by increment of lysyl-PG (L-PG) in the mutant strains (see later). Moreover, in our study, we showed that mprF mutations contribute to increase in MICs of DAP and VCM located on the lysinylation domain (Fig. 1). The mutation in the lysinylation domain of mprF is not just limited to clinical isolates, since most laboratory-derived reduced DAP/VCM susceptibility isolates obtained by stepwise DAP selection on DAPS strains from both cross-reduced susceptibility group (strain C-1) and single-reduced susceptibility group (strain K-1) also carried mprF mutations in the lysinylation domain (Table 3).
Despite having a pronounced association with DAP-selected cross-reduced susceptibility strains, involvement of mprF mutations in the reduced DAP/VCM susceptibility during VCM exposure is less evident. Regardless, glycopeptide-resistant bacterial isolates exhibiting reduced DAP/VCM susceptibility phenotype can be observed in previous14,47 and current studies (Table 2). VISA strains display thickened CW to allow increased binding of VCM to false targets in peptidoglycan (affinity trapping), thereby contributing to their reduced VCM susceptibility8,14. Similar to VCM, the target site of DAP is located in the CM. Moreover, DAP is bigger than VCM in molecular sizes (1,620.67 for DAP and 1,485.7 for VCM). DAP molecules need to penetrate through the CW, the primary barrier of bacterial defense mechanism, before reaching their lethal targets. Therefore, one possible pathway leading to DAP resistance in VISA strains may be increased CW thickness7,37. CW thickening could also explain the reduced DAP susceptibility in DAP-selected reduced DAP/VCM susceptible strains, as reduced DAP binding at CM was observed in DAPR strains of Enterococcus48,49. However, as shown by our TEM analysis, only one DAPR strain (I-3; Table 1) has thickened CW. Neither the remaining 11 sets of DAPR strains from the group of reduced DAP/VCM susceptibility carrying mprF mutation nor DAPR strain with reduced susceptibility to only DAP carrying lacF mutation showed increased thickness of CW (Table 1). Thus, increased cell wall thickness is not a common phenotype in clinical DAPR strain with cross-reduced susceptibility.
Nonetheless, similar to previous observations7,26,37, changes in the expression of CW-related genes have been identified in both DAPR and VISA strains with cross-reduced susceptibility to DAP and VCM. According to our RNA-Seq differential expression analysis, reduced DAP/VCM susceptibility seems to be associated with altered CW metabolism, although most DAPR strains did not show significant differences in CW sizes compared with their parental strains. This contrasting phenomenon might due in part to the small range of VCM MIC changes observed between DAPS and DAPR strains. In addition, DAP/VCM cross-resistance has been reported in both laboratory-derived and clinical isolates with no phenotypic characteristic of CW thickening31. It is therefore indicated that DAP/VCM cross-resistance is not the result of only one contributing factor; while increased CW thickness is associated with DAP and VCM cross-resistant VISA strains, alteration in membrane surface changes is more likely the causative factor of DAP and VCM cross-resistance in DAPR strains. This hypothesis can be supported by several previous studies that reported that not only CW alteration but also changes in CM properties could be a substantial factor leading to cross-reduced DAP/VCM susceptibility11,12,39,50,51.
Daptomycin is a lipopeptide antibiotic acting on bacterial CM10,11. Membrane depolarization and ion leakage can be observed when the positively charged Ca2+-DAP forms a complex with the negatively charged hydrophilic head group of PG and bactoprenol-bound cell wall precursor in CM11,12,52. Therefore, phenotypic alteration of membrane surface charges via mprF mutation is a commonly reported bacterial evolution to resist positively charged drugs, such as CAMPs and DAP12,39,50,51. Interestingly, VCM molecules contain an ionizable amine and carboxylic group, which also display positive charge when administered53,54. Moreover, disruption of negatively charged wall teichoic acids (WTA) by deletion of the dltABCD operon involved in alanylation of teichoic acids was reported to increase the drug susceptibility of S. aureus Sa113 to both CAMPs, such as α-defensins or nisin, and glycopeptides, such as VCM or teicoplanin55,56. Hence, we postulated that a change in net surface charge as mediated by mprF mutation seems to be able to confer reduced DAP and VCM susceptibility in bacterial strains.
Herein, every DAPR isolate in the group of reduced DAP/VCM susceptibility carrying mprF mutations in different positions, as well as DAPS strain H-3 transformed with mprF mutation, exhibited significant alteration of surface charge as implicated by reduction of cytochrome c binding in these strains compared with the DAPS strains (Table 1). These observations attributed decreased DAP/ VCM susceptibility to reduction of negative cell surface charges. According to our results, this mechanism seems to be regulated by mprF, although rpoB mutations have also been reported to alter surface charges35. The change in bacterial membrane surface charges results from the modification of anionic PG to cationic L-PG by the lysinylation domain of MprF57. DAPR isolates were indeed consistently reported to have increased L-PG production due to mprF mutations23,39,58. In concordance, our results showed that L-PG production in DAPR isolates of the cross-reduced DAP/VCM susceptibility group increased in a mutation site-dependent pattern. The cause-effect relationship between increased L-PG production and mprF mutation is further confirmed in our study by transformation of mutated mprF in the DAPS strain (Table 1). Moreover, RNA-Seq analysis showed changes in gene expression that enhance fatty acid biosynthesis (DAPR strain H-5), which indirectly facilitate the production of L-PG. Among, upregulation of genes involved in generation of F-6P (gatA, tal and pmi) were observed (Supplemental Table 5, Fig. 2). The F-6P is the key substrate for pyruvate biosynthesis, a crucial metabolite of the citric acid cycle required for energy metabolism. The upregulation of this process inevitably increases the intracellular pool of acetyl-CoA59–61, which serves as a precursor for fatty acid biosynthesis catalyzed by acetyl-CoA carboxylase and many acyl-carrier protein62. We postulated that these differential gene expressions will favor alteration of membrane property, since enhanced fatty acid synthesis will facilitate CM biosynthesis and subsequently increase the supply of building block for L-PG production by using acetyl-CoA as a precursor. In support of our hypothesis, mutation in acetyl-CoA synthetase in combination with other mutations has been reported to contribute to DAPR17.
Although different locations of mutations in mprF have been proposed to affect L-PG production and consequently DAP susceptibility23,25, the levels of L-PG production varied even when host cells do not carry mprF mutations or carry the same mprF mutation site, as shown in the study by Mishra et al.63. Many reports also showed that DAPR strains carrying mprF mutation exhibited increased intracellular L-PG production, but the ratio of outer leaflet L-PG is not differ from DAPS strains22,25,64–66, possibly due to reduced intradomain interaction25. Therefore, whether or not increased L-PG production directly affect cross-reduced DAP/VCM susceptibility needs to be further investigated. Nevertheless, our results indicated that changes in surface charge, enhanced levels of L-PG production and alteration of CW metabolism, presumably regulated by mprF, can contribute to reduced susceptibility to both DAP and VCM.
The mprF mutation is not a unique genetic determinant correlated with cross-reduced susceptibility phenotype. Other mutations have been reported to be responsible for reduced DAP and VCM susceptibility in laboratory mutants (RNA polymerase rpoB) and clinical isolates (cardiolipin biosynthesis cls or PG production pgsA)35,46,67–69. In fact, a few DAPR strains with cross-reduced DAP/VCM susceptibility included in our study were found to carry additional mutations besides mprF. Apart from the proposed mechanism of the reduced DAP/VCM susceptibility, the current study deduced another possible pathway conferring reduced susceptibility to only DAP, which is not related to mprF. The single-DAPR strain K-2 carrying only a lacF mutation showed an increased binding of cytochrome c compared to DAPS strain, which was contradictory to DAPR strains of cross-reduced DAP/VCM susceptibility group. This indicated a lack of correlation between increased surface positive charge and lacF mutation-associated DAP resistance, consistent with previous studies which demonstrated not all DAPR isolates showed increment of surface positive charges70,71.
DAPR strain K-2 carrying mutations in lacF, a lactose phosphotransferase system (PTS), had reduced susceptibility to only DAP (Table 1). The association between mutations affecting carbohydrate transportation and resistance to cationic peptide (including DAP) has been demonstrated in S. aureus, E. faecalis, and Listeria monocytogenes72–74. L. monocytogenes carrying mutation in the mannose PTS system showed resistance to bacteriocins, one of the cationic peptides capable of making pore-like structures in the membrane just as DAP, due to lower glucose consumption rate74. A previous study also reported that the bactericidal effect of DAP is enhanced by increased glucose concentration that eventually induces lysis protein activity75. Thus, reduced DAP susceptibility in the K-2 strain carrying a lacF mutation seems to be a result of decreased cell lysis caused by lowered glucose consumption. In addition, DAPR strain K-2 demonstrated increased expression of genes involved in cysteine and methionine metabolism, which generates GSH (Supplemental Table 6, Fig. 3). This compound is commonly known to have antioxidant activity, protecting prokaryotic and eukaryotic cells from oxidative stresses76. However, the exact regulatory pathway(s) of GSH in conferring reduced DAP susceptibility is not experimentally proven, although mutation in GSH has been reported to cause DAP resistance in E. faecalis77. Our results indicated that increased GSH metabolism coupled with reduced cell lysis are the possible cellular adaptations protecting bacteria with lacF mutations from DAP toxicity.
Figure 3.
Gene expression in contribution to DAP resistance in DAPR strain K-2. Acetyl-CoA, precursor for fatty acid metabolism, is produced via carbohydrate (lactose, tagatose, and sucrose) and cysteine methionine metabolism or acetate metabolism. The red arrows refer to gene upregulation. The blue arrows refer to gene downregulation.
This study concluded that cross-reduced susceptibility of MRSA to DAP and VCM is associated with mprF mutations. The reduction of DAP and VCM susceptibility is mainly mediated by alteration of bacterial surface charge and increased L-PG production, while increased CW thickness is marginally involved. We also revealed a novel pathway leading to DAP resistance that is not related to mprF. Moreover, reduced DAP susceptibility without a parallel reduction of VCM susceptibility, as observed in our studied strain, is believed to be caused by alterations in cellular metabolisms ensued from lacF mutations, but the exact mechanisms remain to be elucidated.
Materials and methods
Bacterial strains and drug susceptibility testing
The bacterial isolates used in this study included 12 pairs of DAPS and DAPR strains, each collected from the same patient before and after DAP treatment (Supplemental Table 1), and 32 VISA strains isolated from patients receiving VCM therapy (Supplemental Table 2). All bacteria were kept in a final concentration of 40% glycerol at -80°C. Unless otherwise stated, the bacterial glycerol stocks were revived through cultivation in MH broth (Becton Dickinson, USA) at 37°C with constant agitation.
Two methods of drug susceptibility tests were employed in this study: Etest for determining DAP and VCM MIC for all studied strains, and broth microdilution for determining sensitivity toward DAP and VCM. Etests were performed following the guidelines of the Clinical and Laboratory Standard Institute. Briefly, each bacterial culture with 0.5 McFarland turbidity was streaked onto an MH agar plate, and DAP and VCM Etest strips (bioMérieux, France) were placed on the bacterial lawn. The inhibition zone break points for each isolate were read after 48 h. For the broth microdilution method, the ranges of DAP (0.5, 1, 1.5, 1.75, 2, 2.25, 2.5, and 3 mg/L) and VCM (0.5, 1, 1.5, 2, 2.25, 2.5, 3 and 4 mg/L) were tested against each bacterial culture. According to their susceptibility profile, each strain was classified as single DAP- or VCM-resistance or cross-reduced susceptibility to DAP and VCM.
Growth curve and doubling time
Growth kinetics of bacterial strains in the DAP treatment group were determined. Overnight bacterial culture was adjusted to an optical density (OD) at 600 nm (OD600) of 0.3. Following that, the OD-adjusted culture was diluted 1:1000 with fresh brain–heart infusion broth (Becton Dickinson, USA), yielding a final concentration of 105 colony forming unit (CFU)/mL. Bacterial suspensions were then incubated at 37°C with continuous agitation at 25 rpm in a temperature gradient rocking incubator (TVS126MB; ADVANTEC, Japan). The bacterial density at OD600 was recorded every 5 min over a period of 24 h. Growth curves were then generated by plotting OD measurements against time, and doubling time of bacteria was then determined with the equation described previously7,78.
Population analysis profiling area under the curve (PAP-AUC) analysis
PAP-AUC is adapted from previous studies to confirm the DAP and VCM MICs of clinical MRSA isolates7,79. Briefly, an overnight culture of a MRSA strain was adjusted to an OD600 of 0.3 and serially diluted tenfold over a range of 10−3 to 10−10. Then, 100 µL of each dilution was spread on drug-free MH agar, MH agar with DAP (0.5, 0.75, 1, 1.5, 2, and 3 mg/L), and MH agar with VCM (0.5, 1, 2, 3, and 4 mg/L). After 48 h, the number of bacterial colonies was calculated and plotted semilogarithmically.
DNA extraction and purification
Genomic DNAs of all studied strains were extracted from 20 mL of overnight culture grown in tryptic soy broth (Becton Dickinson, USA) by phenol–chloroform method and purified by DNeasy Blood and Tissue Kit (Qiagen, German) according to the manufacturer’s instructions. The DNA concentration was measured by a NanoDrop Lite spectrophotometer (Thermo Scientific, USA) and PicoGreen dsDNA assay kit (Invitrogen, USA).
Multilocus sequence typing (MLST) and SNP determination by whole-genome sequencing
Each DAPS strain was used as reference sequences for comparison. The integrative analysis of genomic DNA sequence was performed without size selection using Nextera Mate Pair Library Prep Kit following standard protocols and MiSeq instrument (2 × 301 bp) with the MiSeq reagent kit version 3 (Illumina, USA). Quality trimming was performed with FASTQ Toolkit version 2.0.0, and quality-trimmed sequences were assembled by the Velvet de novo assembly version 1.2.10 algorithm.
For DAPR strains, sample libraries were prepared with the Nextera XT DNA Sample Preparation and Index Kits. The prepared DNA libraries were sequenced using MiSeq platform (Illumina, USA) with 300-bp paired end reads. The genetic backgrounds of clinical MRSA isolates were characterized by MLST, which involved determining the sequences of ~ 450-bp internal fragments of housekeeping genes (arcC, aroE, glp, gmk, pta, tpi, and yqiL) and compared with the reference genes on “Center for Genomic Epidemiology” website (https://cge.cbs.dtu.dk/services/MLST/). On the other hand, other gene mutations were identified by mapping the sequenced genomes against corresponding reference sequences by using CLC Genomics Workbench software (Qiagen, German). The sequence mapping satisfied with average coverage reads of over 40 across the whole reference genome. Genome sequences with coverage reads less than 10, or with equal or greater than 60% differences compared to that of reference (for those with coverage greater than 10) were called for analysis as potential variants (SNPs, deletion or insertion mutations). All the potential variants were verified by PCR and Sanger sequencing with an ABI3130 × 1 Genetic Analyzer (Applied Biosystems, USA).
Gene replacement into the chromosome
To investigate the effect of the mprF mutation (L291I), identified in DAPR isolates of each patient, on drug susceptibility, gene replacement was performed using the pKOR1 plasmid30,80. In brief, mprF genes were amplified from each H-1 (DAPS) and H-5 (DAPR) strain with primer sets listed in Supplemental Table 3. The PCR fragments were individually cloned into the pKOR1 plasmid using Gateway BP Clonase II enzyme mix (Thermo Scientific, USA), and recombinant plasmids were selected through CcdB-based positive selection system in Escherichia coli DH5α. The plasmid-carrying wild-type mprF gene was then introduced into DAPR strain H-5, while the mutated mprF gene was transformed into DAPS strain H-3. This was achieved by electroporation using NEPA21 electroporator (NEPAGENE, Japan) following the parameters reported previously81. Chromosomal gene replacement involved single-crossover plasmid integration at 43°C followed by overnight incubation in drug-free medium at 37°C to eliminate the plasmid. Anhydrotetracycline was used to select for non-plasmid-carrying mutants. The presence of gene mutations was confirmed by PCR and targeted gene sequencing with an ABI3130 × 1 Genetic Analyzer (Applied Biosystems, USA).
In vitro induction by stepwise DAP exposure
Overnight bacterial cultures (C-1 and K-1 strains) were streaked onto MH agar supplemented with 50 mg/L calcium and a range of concentrations of DAP (0.5–4 mg/L). After incubation at 37°C for 2 days, the colonies grown on MH agar containing the highest concentration of DAP were picked and then streaked again onto fresh MH agar containing DAP at different concentrations. The colonies that could grow on 4 mg/L DAP were further investigated for their DAP and VCM MICs, along with the presence of mprF or lacF mutation by Sanger sequencing.
Transmission electron microscopy (TEM)
CW thickness of all bacterial isolates from DAP treatment group were determined using TEM as previously described and visualized with TEM (Hitachi H-7600, Japan)82,83. Thirty cells of each bacterial strain were examined for CW thickness measurement at nearly equatorial cut surfaces. The results were presented as means standard deviations.
Evaluation of membrane surface charge
Cytochrome c binding assays were performed as previously84 described to measure membrane surface charges of bacterial isolates from the DAP treatment group and transformed mutants carrying a wild-type or mutated mprF gene. The amount of unbound cytochrome c (Sigma-Aldrich, USA) was determined with spectrophotometry at OD410. Cytochrome c binding values in each DAPR strain were determined from three independent studies with normalization to DAPS strains of the same set.
Determination of L-PG production
PL extraction from clinical MRSA isolates was adapted from the Bligh-Dyer procedure85. Briefly, the pelleted cells of overnight cultures of DAPS / DAPR clinical isolates and transformed mutants were adjusted to an OD620 of 20 and digested with a mixture of chloroform/methanol/water (1.75:3.5:1.4; v/v/v). Chloroform and 0.85% KCl weighing 1.75 mL and 1.6 mL, respectively, were added sequentially to the mixture. The extracted organic layers were concentrated by evaporation before separation with TLC (Silica-Gel 60-W-F254s, Merck, USA) in chloroform/methanol/water (65:25:4; v/v/v). Lysyl-phosphatidylglycerol was visualized by ninhydrin spray (FUJIFILM Wako Pure Chemicals, Japan), while total PLs were detected with molybdenum blue (Merck, USA). The relative amount of L-PG and total PLs in each sample were determined by ImageJ software (Wayne Rasband, USA). The L-PG levels relative to total PLs of DAPR strains were calculated from three independent studies by comparing with DAPS strains of the same set.
RNA extraction and RNA expression analysis
Overnight bacterial cultures (H-3/H-5 and K-1/K-2) diluted 1:100 in 10 mL of MH broth were incubated at 37°C to an OD600 of 0.8. The bacterial pellet was harvested and resuspended with 6 mL of precooled T10E10 buffer (10 mM Tris–HCl, 10 mM EDTA; pH 8.0), followed by the addition of 10 mg/L lysostaphin (Sigma-Aldrich, USA) for complete bacterial lysis. Consequently, 7 mL of acidic-phenol saturated with 20 mM NaOAc (pH 4.8) (FUJIFILM Wako Pure Chemicals, Japan) and 600 µL of 3 M NaOAc (pH 4.8) were added. The mixture was subjected to three cycles of 20 min freezing at -80 °C and 5 min thawing at 65 °C. Bacterial RNA was then extracted by the phenol–chloroform method, followed by ethanol precipitation. The RNA pellet was dissolved with DNase I, recombinant, RNase-free (Roche, Germany) and purified by RNeasy Mini Kit Part 2 (Qiagen, German) before re-extraction with phenol–chloroform and ethanol precipitation. Finally, ribosomal RNAs in total RNA preparations were depleted using the Ribo-Zero rRNA Removal Kit (Illumina, USA). The extracted RNAs were first converted into complementary DNA (cDNA) and subsequently made into double-stranded DNA (dsDNA) by PrimeScript Double Strand cDNA Synthesis Kit (Takara, Japan). The generated dsDNAs were then used as templates for cDNA library preparation using Nextera XT DNA Library Prep Kit (Illumina, USA) as previously described. The fold change of RNA expression between the DAPS and DAPR strains was determined by CLC Genomics Workbench software.
Statistical analysis
Student’s t test was employed for all statistical analyses.
Ethics approval and consent to participate
Ethics approval and consent to participate were not required. All bacteria were isolated from hospitals in Japan as part of the standard patient care and used anonymously.
(To Editors: For consideration on this issue, ethics approval is not required following the ethical guidelines for medical and health research involving human subjects by Ministry of Health, Labour and Welfare, Japan since this study analyzed bacteria which were isolated as a clinical specimen and patients’ personal health information could not be accessed.
(https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/hokabunya/kenkyujigyou/i-kenkyu/index.html).
Supplementary information
Acknowledgments
We greatly appreciate Dr. Yoshitaka Yamamoto from Dokkyo Medical University Koshigaya Hospital, Prof. Intetsu Kobayashi from Toho University, Prof. Harumi Yano from International University of Health and Welfare, Prof. Naohisa Fujita from Kyoto Prefectural University of Medicine, Dr. Go Matsumoto from Shinshu University, Dr. Jun Ogawa and Dr. Susumu Kawanishi from Tsuyama Chuo Hospital, and BEI Resources of the National Institute of Allergy and Infectious Diseases for kindly provided the MRSA isolates included in this study.
Author contributions
K.T. and L.C. designed the study, analyzed the data, and wrote the manuscript. Y.A. and X.T. contributed to acquisition, interpretation of data, and assisted with the preparation of the manuscript. S.W., K.K., Y.S., T.B., F.L., T.S., Y.T., A.H.A., and Y.Z. contributed to data collection and interpretation, and critically revised the manuscript. All authors approved the final version of the manuscript and agreed for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was supported by the Japan Agency for Medical Research and Development (grant No. JP17fm0208028, JP18fm0208028, JP19fm0208028, and JP20fk0108134 to LC), JSPS KAKENHI (Grant No. 18K15149 to KK, 15H05654 and 19K08960 to SW, 17K15691 to YS and 17K19570 to LC), the Takeda Science Foundation (LC), Jichi Medical University Young Investigator Award (YA). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The genome sequence has been deposited at DDBJ/Genbank: PRJDB9008 (BioProject) DRA009427 (Raw data).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-73108-x.
References
- 1.Pantosti A, Sanchini A, Monaco M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007;2:323–334. doi: 10.2217/17460913.2.3.323. [DOI] [PubMed] [Google Scholar]
- 2.Hope R, Livermore DM, Brick G, Lillie M, Reynolds R, et al. Non-susceptibility trends among staphylococci from bacteraemias in the UK and Ireland, 2001–06. J. Antimicrob. Chemother. 2008;62(Suppl 2):ii65–ii74. doi: 10.1093/jac/dkn353. [DOI] [PubMed] [Google Scholar]
- 3.Hiramatsu K. The emergence of Staphylococcus aureus with reduced susceptibility to vancomycin in Japan. Am. J. Med. 1998;104:7S–10S. doi: 10.1016/s0002-9343(98)00149-1. [DOI] [PubMed] [Google Scholar]
- 4.Chesneau O, Morvan A, Solh NE. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J. Antimicrob. Chemother. 2000;45:887–890. doi: 10.1093/jac/45.6.887. [DOI] [PubMed] [Google Scholar]
- 5.Watanakunakorn C. Mode of action and in-vitro activity of vancomycin. J. Antimicrob. Chemother. 1984;14(Suppl D):7–18. doi: 10.1093/jac/14.suppl_d.7. [DOI] [PubMed] [Google Scholar]
- 6.Ferraz V, Dusé AG, Kassei M, Black AD, Ito T, Hiramatsu K. Vancomycin-resistant Staphylococcus aureus occurs in South Africa. S. Afr. Med. J. 2000;90:1113. [PubMed] [Google Scholar]
- 7.Cui L, Ma X, Sato K, et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 2003;41:5–14. doi: 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui L, Iwamoto A, Lian JQ, et al. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents. Chemother. 2006;50:428–438. doi: 10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stryjewski ME, Corey GR. New treatments for methicillin-resistant Staphylococcus aureus. Curr. Opin. Crit Care. 2009;15:403–412. doi: 10.1097/MCC.0b013e32832f0a74. [DOI] [PubMed] [Google Scholar]
- 10.Muraih JK, Pearson A, Silverman J, Palmer M. Oligomerization of daptomycin on membranes. Biochim. Biophys. Acta. 2011;1808:1154–1160. doi: 10.1016/j.bbamem.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Jung D, Rozek A, Okon M, Hancock RE. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem. Biol. 2004;11:949–957. doi: 10.1016/j.chembiol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 12.Schriever CA, Fernández C, Rodvold KA, Danziger LH. Daptomycin: a novel cyclic lipopeptide antimicrobial. Am. J. Health Syst. Pharm. 2005;62:1145–1158. doi: 10.1093/ajhp/62.11.1145. [DOI] [PubMed] [Google Scholar]
- 13.Ernst CM, Staubitz P, Mishra NN, et al. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoSPathog. 2009;5:e102. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui L, Tominaga E, Neoh HM, Hiramatsu K. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50:1079–1082. doi: 10.1128/AAC.50.3.1079-1082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakoulas G, Alder J, Thauvin-Eliopoulos C, Moellering RC, Eliopoulos GM. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 2006;50:1581–1585. doi: 10.1128/AAC.50.4.1581-1585.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishi H, Komatsuzawa H, Fujiwara T, McCallum N, Sugai M. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, Staphylococcus aureus. Antimicrob. Agents Chemother. 2004;48:4800–4807. doi: 10.1128/AAC.48.12.4800-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang SJ, Xiong YQ, Dunman PM, et al. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2009;53:2636–2637. doi: 10.1128/AAC.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slavetinsky CJ, Peschel A, Ernst CM. Alanyl-phosphatidylglycerol and lysyl-phosphatidylglycerol are translocated by the same MprF flippases and have similar capacities to protect against the antibiotic daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012;56:3492–3497. doi: 10.1128/AAC.00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayer AS, Mishra NN, Cheung AL, Rubio A, Yang SJ. Dysregulation of mprF and dltABCD expression among daptomycin-non-susceptible MRSA clinical isolates. J. Antimicrob. Chemother. 2016;71:2100–2104. doi: 10.1093/jac/dkw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst CM, Kuhn S, Slavetinsky CJ, et al. The lipid-modifying multiple peptide resistance factor is an oligomer consisting of distinct interacting synthase and flippase subunits. MBio. 2015;6:e02340. doi: 10.1128/mBio.02340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang SJ, Mishra NN, Rubio A, Bayer AS. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob. Agents Chemother. 2013;57:5658–5664. doi: 10.1128/AAC.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayer AS, Mishra NN, Chen L, Kreiswirth BN, Rubio A, Yang SJ. Frequency and distribution of single-nucleotide polymorphisms within mprF in methicillin-Resistant Staphylococcus aureus clinical isolates and their role in cross-resistance to daptomycin and host defense antimicrobial peptides. Antimicrob. Agents Chemother. 2015;59:4930–4937. doi: 10.1128/AAC.00970-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra NN, Yang SJ, Chen L, et al. Emergence of daptomycin resistance in daptomycin-naïve rabbits with methicillin-resistant Staphylococcus aureus prosthetic joint infection is associated with resistance to host defense cationic peptides and mprF polymorphisms. PLoS ONE. 2013;8:e71151. doi: 10.1371/journal.pone.0071151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst CM, Slavetinsky CJ, Kuhn S, et al. Gain-of-function mutations in the phospholipid flippasemprF confer specific daptomycin resistance. MBio. 2018;9:e00802. doi: 10.1128/mBio.01659-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertsche U, Yang SJ, Kuehner D, et al. Increased cell wall teichoic acid production and D-alanylation are common phenotypes among daptomycin-resistant methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates. PLoS ONE. 2013;8:e67398. doi: 10.1371/journal.pone.0067398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Utaida S, Dunman PM, Macapagal D, et al. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology. 2003;149:2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 28.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol. 2003;49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 29.Taglialegna A, Varela MC, Rosato RR, Rosato AE. VraSR and virulence trait modulation during daptomycin resistance in methicillin-resistant. mSphere. 2019;4:e00557. doi: 10.1128/mSphere.00557-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoji M, Cui L, Iizuka R, et al. walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob. Agents Chemother. 2011;55:3870–3881. doi: 10.1128/AAC.01563-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang SJ, Nast CC, Mishra NN, Yeaman MR, Fey PD, Bayer AS. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob. Agents Chemother. 2010;54:3079–3085. doi: 10.1128/AAC.00122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yarmus M, Mett A, Shapira R. Cloning and expression of the genes involved in the production of and immunity against the bacteriocinlacticin RM. Biochim. Biophys. Acta. 2000;1490:279–290. doi: 10.1016/s0167-4781(00)00012-9. [DOI] [PubMed] [Google Scholar]
- 33.Hafer C, Lin Y, Kornblum J, Lowy FD, Uhlemann AC. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2012;56:5845–5851. doi: 10.1128/AAC.01139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe Y, Cui L, Katayama Y, Kozue K, Hiramatsu K. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol. 2011;49:2680–2684. doi: 10.1128/JCM.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui L, Isii T, Fukuda M, et al. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54:5222–5233. doi: 10.1128/AAC.00437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanesaka I, Fujisaki S, Aiba Y, et al. Characterization of compensatory mutations associated with restoration of daptomycin-susceptibility in daptomycin non-susceptible methicillin-resistant Staphylococcus aureus and the role mprF mutations. J. Infect. Chemother. 2019;25:1–5. doi: 10.1016/j.jiac.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Sieradzki K, Leski T, Dick J, Borio L, Tomasz A. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 2003;41:1687–1693. doi: 10.1128/JCM.41.4.1687-1693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samant S, Hsu FF, Neyfakh AA, Lee H. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J. Bacteriol. 2009;191:1311–1319. doi: 10.1128/JB.01345-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones T, Yeaman MR, Sakoulas G, et al. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 2008;52:269–278. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kilelee E, Pokorny A, Yeaman MR, Bayer AS. Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: implications for daptomycin resistance. Antimicrob. Agents Chemother. 2010;54:4476–4479. doi: 10.1128/AAC.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boneca IG, Huang ZH, Gage DA, Tomasz A. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new beta-N-acetylglucosaminidase activity. J. Biol. Chem. 2000;275:9910–9918. doi: 10.1074/jbc.275.14.9910. [DOI] [PubMed] [Google Scholar]
- 42.Haag AF, Bagnoli F. The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr. Top. Microbiol. Immunol. 2017;409:145–198. doi: 10.1007/82_2015_5019. [DOI] [PubMed] [Google Scholar]
- 43.Libby EA, Reuveni S, Dworkin J. Multisite phosphorylation drives phenotypic variation in (p)ppGpp synthetase-dependent antibiotic tolerance. Nat. Commun. 2019;10:5133. doi: 10.1038/s41467-019-13127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruzin A, Severin A, Moghazeh SL, et al. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. Biochim. Biophys. Acta. 2003;1621:117–121. doi: 10.1016/s0304-4165(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 45.Sabat AJ, Tinelli M, Grundmann H, et al. Daptomycin resistant Staphylococcus aureus clinical strain with novel non-synonymous mutations in the mprF and vraS genes: a new insight into daptomycin resistance. Front Microbiol. 2018;9:2705. doi: 10.3389/fmicb.2018.02705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peleg AY, Miyakis S, Ward DV, et al. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS ONE. 2012;7:e28316. doi: 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roch M, Gagetti P, Davis J, et al. Daptomycin resistance in clinical MRSA strains is associated with a high biological fitness cost. Front Microbiol. 2017;8:2303. doi: 10.3389/fmicb.2017.02303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran TT, Panesso D, Mishra NN, et al. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio. 2013;4:e00281. doi: 10.1128/mBio.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall SA, D., Werth B. J., Nonejuie P.,, et al. Fosfomycin enhances the activity of daptomycin against vancomycin-resistant Enterococci in an in vitro pharmacokinetic-pharmacodynamic model. Antimicrob. Agents Chemother. 2016;60:5716–5723. doi: 10.1128/AAC.00687-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tally FP, Zeckel M, Wasilewski MM, et al. Daptomycin: a novel agent for Gram-positive infections. Expert Opin. Investig. Drugs. 1999;8:1223–1238. doi: 10.1517/13543784.8.8.1223. [DOI] [PubMed] [Google Scholar]
- 51.Rybak MJ, Hershberger E, Moldovan T, Grucz RG. In vitro activities of daptomycin, vancomycin, linezolid, and quinupristin-dalfopristin against Staphylococci and Enterococci, including vancomycin- intermediate and -resistant strains. Antimicrob. Agents Chemother. 2000;44:1062–1066. doi: 10.1128/aac.44.4.1062-1066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grein F, Müller A, Scherer KM, et al. Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 2020;11:1455. doi: 10.1038/s41467-020-15257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson JL, Yalkowsky SH. Reformulation of a new vancomycin analog: an example of the importance of buffer species and strength. AAPS PharmSciTech. 2006;7:E5. doi: 10.1208/pt070105. [DOI] [PubMed] [Google Scholar]
- 54.Gustafson CT, Boakye-Agyeman F, Brinkman CL, et al. Controlled delivery of vancomycin via charged hydrogels. PLoS ONE. 2016;11:e0146401. doi: 10.1371/journal.pone.0146401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 56.Peschel A, Vuong C, Otto M, Götz F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents. Chemother. 2000;44:2845–2847. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staubitz P, Neumann H, Schneider T, Wiedemann I, Peschel A. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 2004;231:67–71. doi: 10.1016/S0378-1097(03)00921-2. [DOI] [PubMed] [Google Scholar]
- 58.Kang KM, Mishra NN, Park KT, et al. Phenotypic and genotypic correlates of daptomycin-resistant methicillin-susceptible Staphylococcus aureus clinical isolates. J. Microbiol. 2017;55:153–159. doi: 10.1007/s12275-017-6509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Everts B, Amiel E, van der Windt GJ, et al. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harper L, Balasubramanian D, Ohneck EA, et al. Responds to the central metabolite pyruvate to regulate virulence. MBio. 2018 doi: 10.1128/mBio.02272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akram M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys. 2014;68:475–478. doi: 10.1007/s12013-013-9750-1. [DOI] [PubMed] [Google Scholar]
- 62.Payne DJ, Warren PV, Holmes DJ, Ji Y, Lonsdale JT. Bacterial fatty-acid biosynthesis: a genomics-driven target for antibacterial drug discovery. Drug Discov. Today. 2001;6:537–544. doi: 10.1016/s1359-6446(01)01774-3. [DOI] [PubMed] [Google Scholar]
- 63.Mishra NN, Bayer AS. Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013;57:1082–1085. doi: 10.1128/AAC.02182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehta S, Cuirolo AX, Plata KB, et al. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2012;56:92–102. doi: 10.1128/AAC.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mishra NN, Bayer AS, Weidenmaier C, et al. Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: relative roles of mprF and dlt operons. PLoS ONE. 2014;9:e107426. doi: 10.1371/journal.pone.0107426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang SJ, Mishra NN, Kang KM, Lee GY, Park JH, Bayer AS. Impact of multiple single-nucleotide polymorphisms within mprF on daptomycin resistance in Staphylococcus aureus. Microb. Drug Resist. 2018;24:1075–1081. doi: 10.1089/mdr.2017.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen FJ, Lauderdale TL, Lee CH, et al. Effect of a point mutation in mprF on susceptibility to daptomycin, vancomycin, and oxacillin in an MRSA clinical strain. Front Microbiol. 2018;9:1086. doi: 10.3389/fmicb.2018.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Camargo IL, Neoh HM, Cui L, Hiramatsu K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob. Agents Chemother. 2008;52:4289–4299. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bæk KT, Thøgersen L, Mogenssen RG, et al. Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob. Agents Chemother. 2015;59:6983–6991. doi: 10.1128/AAC.01303-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mishra NN, Rubio A, Nast CC, Bayer AS. Differential adaptations of methicillin-resistant Staphylococcus aureus to serial in vitro passage in daptomycin: evolution of daptomycin resistance and role of membrane carotenoid content and fluidity. Int. J. Microbiol. 2012;2012:683450. doi: 10.1155/2012/683450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mishra NN, McKinnell J, Yeaman MR, et al. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2011;55:4012–4018. doi: 10.1128/AAC.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tran TT, Panesso D, Gao H, et al. Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 2013;57:261–268. doi: 10.1128/AAC.01454-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Humphries RM, Kelesidis T, Tewhey R, et al. Genotypic and phenotypic evaluation of the evolution of high-level daptomycin nonsusceptibility in vancomycin-resistant Enterococcus faecium. Antimicrob. Agents Chemother. 2012;56:6051–6053. doi: 10.1128/AAC.01318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vadyvaloo V, Snoep JL, Hastings JW, Rautenbach M. Physiological implications of class IIa bacteriocin resistance in Listeria monocytogenes strains. Microbiology. 2004;150:335–340. doi: 10.1099/mic.0.26731-0. [DOI] [PubMed] [Google Scholar]
- 75.Prax M, Mechler L, Weidenmaier C, Bertram R. Glucose augments killing efficiency of daptomycin challenged Staphylococcus aureuspersisters. PLoS ONE. 2016;11:e0150907. doi: 10.1371/journal.pone.0150907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cameron JC, Pakrasi HB. Glutathione facilitates antibiotic resistance and photosystem I stability during exposure to gentamicin in cyanobacteria. Appl. Environ. Microbiol. 2011;77:3547–3550. doi: 10.1128/AEM.02542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller C, Kong J, Tran TT, Arias CA, Saxer G, Shamoo Y. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob. Agents Chemother. 2013;57:5373–5383. doi: 10.1128/AAC.01473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saito M, Katayama Y, Hishinuma T, et al. "Slow VISA," a novel phenotype of vancomycin resistance, found in vitro in heterogeneous vancomycin-intermediate Staphylococcus aureus strain Mu3. Antimicrob. Agents Chemother. 2014;58:5024–5035. doi: 10.1128/AAC.02470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Satola SW, Farley MM, Anderson KF, Patel JB. Comparison of detection methods for heteroresistant vancomycin-intermediate Staphylococcus aureus with the population analysis profile method as the reference method. J. Clin. Microbiol. 2011;49:177–183. doi: 10.1128/JCM.01128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cui L, Neoh HM, Shoji M, Hiramatsu K. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2009;53:1231–1234. doi: 10.1128/AAC.01173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sato'o Y, Aiba Y, Kiga K, et al. Optimized universal protocol for electroporation of both coagulase-positive and -negative Staphylococci. J. Microbiol. Methods. 2018;146:25–32. doi: 10.1016/j.mimet.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 82.Hanaki H, Kuwahara-Arai K, Boyle-Vavra S, Daum RS, Labischinski H, Hiramatsu K. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 83.Cui L, Murakami H, Kuwahara-Arai K, Hanaki H, Hiramatsu K. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 2000;44:2276–2285. doi: 10.1128/aac.44.9.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raafat D, Leib N, Wilmes M, François P, Schrenzel J, Sahl HG. Development of in vitro resistance to chitosan is related to changes in cell envelope structure of Staphylococcus aureus. Carbohyd. Polym. 2017;157:146–155. doi: 10.1016/j.carbpol.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 85.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 86.Cafiso V, Bertuccio T, Purrello S, et al. dltA overexpression: A strain-independent keystone of daptomycin resistance in methicillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents. 2014;43:26–31. doi: 10.1016/j.ijantimicag.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 87.Berti AD, Baines SL, Howden BP, et al. Heterogeneity of genetic pathways toward daptomycin nonsusceptibility in Staphylococcus aureus determined by adjunctive antibiotics. Antimicrob. Agents Chemother. 2015;59:2799–2806. doi: 10.1128/AAC.04990-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence has been deposited at DDBJ/Genbank: PRJDB9008 (BioProject) DRA009427 (Raw data).