Abstract
Early identification of patients at risk of developing preeclampsia (PE) would allow providers to tailor their prenatal management and adopt preventive strategies, such as low-dose aspirin. Nevertheless, no mid-trimester biomarkers have as yet been proven useful for prediction of PE. This study investigates the ability of metabolomic biomarkers in mid-trimester maternal plasma to predict PE. A case–control study was conducted including 33 pregnant women with mid-trimester maternal plasma (gestational age [GA], 16–24 weeks) who subsequently developed PE and 66 GA-matched controls with normal outcomes (mid-trimester cohort). Plasma samples were comprehensively profiled for primary metabolic and lipidomic signatures based on gas chromatography time-of-flight mass spectrometry (GC-TOF MS) and liquid chromatography Orbitrap mass spectrometry (LC-Orbitrap MS). A potential biomarker panel was computed based on binary logistic regression and evaluated using receiver operating characteristic (ROC) analysis. To evaluate whether this panel can be also used in late pregnancy, a retrospective cohort study was conducted using plasma collected from women who delivered in the late preterm period because of PE (n = 13) or other causes (n = 21) (at-delivery cohort). Metabolomic biomarkers were compared according to the indication for delivery. Performance of the metabolomic panel to identify patients with PE was compared also to a commonly used standard, the plasma soluble fms-like tyrosine kinase-1/placental growth factor (sFlt-1/PlGF) ratio. In the mid-trimester cohort, a total of 329 metabolites were identified and semi-quantified in maternal plasma using GC-TOF MS and LC-Orbitrap-MS. Binary logistic regression analysis proposed a mid-trimester biomarker panel for the prediction of PE with five metabolites (SM C28:1, SM C30:1, LysoPC C19:0, LysoPE C20:0, propane-1,3-diol). This metabolomic model predicted PE better than PlGF (AUC [95% CI]: 0.868 [0.844–0.891] vs 0.604 [0.485–0.723]) and sFlt-1/PlGF ratio. Analysis of plasma from the at-delivery cohort confirmed the ability of this biomarker panel to distinguish PE from non-PE, with comparable discrimination power to that of the sFlt-1/PlGF ratio. In conclusion, an integrative metabolomic biomarker panel in mid-trimester maternal plasma can accurately predict the development of PE and showed good discriminatory power in patients with PE at delivery.
Subject terms: Biomarkers, Cardiology, Risk factors
Introduction
Preeclampsia (PE) is a clinical syndrome specific to human pregnancy and the puerperium that can affect virtually every maternal organ system. It is diagnosed by the presence of hypertension with evidence of end-organ involvement, such as proteinuria, thrombocytopenia, renal insufficiency, pulmonary edema, or cerebral symptoms after 20 weeks’ gestation1. Preeclampsia complicates 2–10% of all pregnancies and is a major cause of maternal mortality2.
Because of its clinical significance, the prediction and prevention of PE has been the target of intensive investigation for decades. Numerous biological markers have been evaluated in an effort to predict PE, but results have been inconsistent3–10. Such biomarkers included those related to placental perfusion/vascular resistance, endocrine dysfunction, renal dysfunction, endothelial dysfunction, and oxidative stress11. Overall, these biomarkers have shown poor sensitivity and poor positive-predictive values for the prediction of PE. A test using plasma biomarkers of endothelial dysfunction—specifically the ratio of the anti- and pro-angiogenic factors, soluble fms-like tyrosine kinase-1 (sFlt-1) and placental growth factor (PlGF), respectively—is now commercially available. Although the sFlt-1/PlGF ratio has shown a high degree of accuracy for predicting PE in late pregnancy12, the clinical utility of this test in the mid-trimester remains questionable. There are currently no widely accepted biological markers that have been shown to be reliable, reproducible, and cost-effective in the prediction of PE in early pregnancy, which would be critical if a preventative strategy is to be effective.
The metabolome refers to the complete profile of metabolites within the maternal plasma that reflect ongoing biochemical process at a single point in time, including metabolic substrates, intermediates, and final products. Metabolomic analysis of samples collected from study subjects with/without underlying diseases is thus ideally suited for developing diagnostic tests, prognostic tools, and personalized therapeutic interventions13–15. Metabolomic studies have been conducted for the purpose of biomarker discovery for PE in a range of biological specimens (including serum, urine, and placenta) and different analytical platforms have been used to measure a number of putative biomarker molecule species including amino acid, fatty acid, organic acid, and acylcarnitine16,17, which have been summarized in detail elsewhere16. In brief, several studies with maternal blood in late pregnancy reported metabolic biomarkers for discrimination of preeclampsia18–21, and other previous studies suggested predictive metabolic biomarkers in early pregnancy with the use of metabolomics tools such as nuclear magnetic resonance spectroscopy (NMR), gas-/ liquid chromatography–mass spectrometry (GC– / LC–MS), with variable sensitivities and specificities (30–100%)22–29. Other biologic samples such as urine or placenta in early or late pregnancy has been also explored to examine the underlying pathophysiology of preeclampsia30–34.
However, few studies have explored comprehensive phospholipid profiles in maternal blood. Phospholipids (PLs) are a major constituent of cell membranes and function also as second messengers in intracellular signal transduction cascades. The different composition of fatty acyl chains within PL subtypes determines the biophysical traits of the individual cell membrane and influences a range of biological processes35. Considering that endothelial injury/dysfunction is a key pathogenic feature of PE36, it is biologically plausible that changes in circulating PL profiles might be useful in predicting and/or diagnosing the disease. In addition, abnormal circulating PL profiles have been reported in many diseases, including obesity, hypertension, seizure disorders, and gestational diabetes, all of which are risks factors for PE37–39.
In this study, we analyzed comprehensive plasma PL profiles and primary metabolomic signatures from women who did and did not develop PE. We identified unique metabolomic features in women with PE, which was consistent in both early and late gestational ages, and describe a biomarker panel based on binary logistic regression model that showed robust performance in differentiating women with PE from those without.
Results
Metabolomic biomarkers in the Mid-trimester Cohort
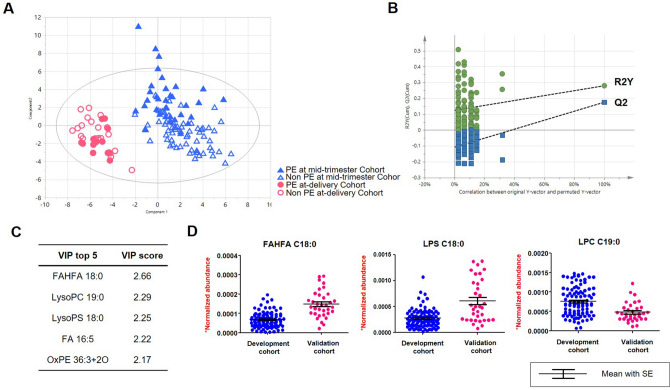
Integrative metabolomic analysis identified and semi-quantified 116 primary metabolites and 213 lipids based on GC-TOF MS and LC-Orbitrap MS, respectively. The profiles were not clearly separated by principal component analysis (PCA) (Figure S1). Supervised multivariate statistics, PLS-DA modeling demonstrated that the primary discriminatory factor was GA at sampling based on the first two latent variables (Fig. 1A,B). Variable importance projection (VIP) analysis determined that the metabolites that contributed most to the model were Fatty Acid Esters of Hydroxy Fatty Acid (FAHFA) C18:0, LysoPC C19:0, and LysoPS C19:0 (Fig. 1C,D). Sixty-one metabolites (18.5%) achieved statistical significance based on one-way ANOVA (adjusted by Benjamini–Hochberg analysis, FDR < 0.05) with post-hoc test (Fisher’s LSD) of which 26 metabolites were significantly different in all 4 comparisons (Table S1). Subsequent PLS-DA showed the PE-unique metabolic profiles in the mid-trimester cohort (R2Y = 0.626 and Q2 = -0.0187) (Figure S2 A,B) and in the at-delivery cohort (R2Y = 0.991 and Q2 = 0.546) (Figure S2 C,D).
Figure 1.
Multivariate statistical modeling of plasma metabolites by partial least squares-discriminant analysis (PLS-DA). (A) The score plot shows that the major discriminatory factor is pregnancy stage. T1 and T2 indicates are the vectors, which explain the two largest degree of variation in the model. R2Y (0.425) is cumulative goodness-of-fit. Q2 (0.179) proposes model predictability. (B) Random permutation plot (100 times). The vertical axis presents R2 (green points) and Q2 (blue points) values of the model. The horizontal axis shows the correlation coefficient between the original and the permuted Y-variable. (C) The top five list of metabolites based on VIP analysis (D) Individual measurements as well as Box-Whisker plots (mean ± SEM) are shown of the metabolites that contributed most to the model.
We first interrogated the clinical characteristics and the metabolic compositional differences in the mid-trimester cohort. Table 1 shows the clinical characteristics of the mid-trimester cohort. Cases and controls were matched for maternal age and GA at blood sampling. As expected, cases (women who developed PE) had an earlier GA at delivery and lower birthweight that those who did not (controls). Neither circulating sFlt-1 concentrations nor sFlt-1/PlGF ratios were different between the two groups, but PlGF levels were significantly lower in women who subsequently developed PE as compared with those that did not (median 43.1 [range, 3.2–387.8] vs 60.3 [11.3–654.4] pg/mL, respectively; p < 0.05).
Table 1.
Clinical characteristics of the study population in the mid-trimester cohort.
| Controls (did not develop preeclampsia) (n = 66) | Cases (developed preeclampsia) (n = 33) | p Value | |
|---|---|---|---|
| Maternal age (years) | 36 (27–43) | 35 (26–44) | NS |
| Nulliparity | 39 (59%) | 20 (61%) | NS |
| BMI before pregnancy | 20.7 (16.4–32.8) (n = 51) | 22.4 (17.6–40.2) (n = 31) | < 0.01 |
| BMI at blood sampling | 22.4 (16.7–36.5) | 23.8 (18.2–38.1) | NS |
| Gestational age at blood sampling (weeks) | 17.4 (16.0–23.4) | 17.6 (16.0–22.6) | NS |
| Gestational age at delivery (weeks) | 39.3 (25.6–42.3) | 37.4 (25.3–41.3) | < 0.001 |
| Birth weight (g) | 3275 (760–4260) | 2670 (450–4700) | < 0.001 |
| Small for gestational age | 4 (6%) | 6 (19%) | NS |
| Sex (male) | 34 (53%) | 16 (49%) | NS |
| Cesarean delivery | 26 (41%) | 16 (49%) | NS |
| Diabetes during pregnancy | 1 (2%) | 0 (0%) | NS |
| Pro- and anti-angiogenic factors | |||
| sFlt-1 (pg/mL) | 887.4 (177.3–3446.2) | 952.2 (273.7–1957.1) | NS |
| PlGF (pg/mL) | 60.3 (11.3–654.4) | 43.1 (3.2–387.8) | < 0.05 |
| sFlt-1/PlGF | 15.1 (0.8–239.2) | 18.0 (1.5–577.8) | NS |
All values are given as median (range) or number (%).
NS, not significant; BMI, body mass index; PlGF, placental growth factor; sFlt-1, soluble fms-like tyrosine kinase-1.
The compositional changes in the metabolic profiles were interrogated based on Student’s t-test in which 6 primary metabolites (1.8%) and 17 lipid molecules (5.2%) were present in plasma at significantly different levels in PE cases compared to healthy controls (p < 0.05) (listed in Table 2). The primary metabolites included glycolysis intermediates (3-phosphoglycerate), nitrogenous compounds (xanthine, glutamate), and palmitoleic acid, all of which were elevated in women who developed PE. Lyxose and propane-1,3-diol were present at significantly lower levels in women who developed PE. Metabolite enrichment analysis suggested that arachidonic acid metabolism, malate-aspartate shuttle, β-alanine metabolism were significantly altered (p value < 0.05) (Figure S3).
Table 2.
Metabolites that were significantly different in the preeclampsia group in the mid-trimester cohort.
| Fold change | p Value | FDR | |
|---|---|---|---|
| Lipid molecules | |||
| LysoPC C19:0 | 0.72 | 0.002 | 0.000 |
| LysoPC C22:1 | 0.53 | 0.010 | 0.000 |
| LysoPE C16:1 | 1.73 | 0.018 | 0.783 |
| LysoPE C17:0 | 1.30 | 0.013 | 0.783 |
| LysoPE C20:0 | 1.46 | 0.044 | 0.783 |
| OxPC C38:4 + 1O | 0.76 | 0.048 | 0.336 |
| OxPI C38:4 + 1O | 0.73 | 0.044 | 0.277 |
| PC C32:1 | 1.37 | 0.007 | 0.783 |
| PC C32:2 | 1.33 | 0.009 | 0.783 |
| PE C23:1e | 0.65 | 0.030 | 0.231 |
| PE C24:1e | 0.46 | 0.031 | 0.231 |
| PE C34:3e | 0.81 | 0.031 | 0.277 |
| PI C36:2 | 0.72 | 0.014 | 0.231 |
| PI C38:3 | 11.23 | 0.041 | 0.783 |
| SM C28:1 | 0.66 | 0.011 | 0.000 |
| SM C30:1 | 1.53 | 0.020 | 0.783 |
| SM C34:1 | 2.22 | 0.044 | 0.783 |
| Primary metabolites | |||
| 3-Phosphoglycerate | 1.37 | 0.037 | 0.783 |
| Glutamate | 1.29 | 0.041 | 0.783 |
| Lyxose | 0.74 | 0.024 | 0.231 |
| Palmitoleic acid | 1.50 | 0.046 | 0.844 |
| Propane-1,3-diol | 0.91 | 0.028 | 0.231 |
| Xanthine | 1.43 | 0.044 | 0.783 |
* Statistical significance is presented by p value and false discovery rate (FDR) against control.
From the lipidomic profiles, phosphatidylinositol (PI) C38:3 was increased the most (11.2 fold-change). Levels of lyso-phosphatidylethanolamine (LysoPEs, C16:1, C17:0, and C20:0) were also measured at higher levels in the PE group, whereas levels of phosphatidylethanolamine (PE) C23:1 and C24:1, and C34:3 were significantly reduced. Similarly, phosphatidylcholine (PC) and LysoPC showed contrasting patterns, with lower contractions of LysoPC C19:0 and C22:1 and higher concentrations of PC C32:1 and C32:2 in the PE group. Other lipid molecules that were differentially regulated included oxidized PLs (OxPC and OxPI) and sphingomyelins (SMs). The identities of the oxidized PLs were validated by predicted spectra based on CFM-ID40.
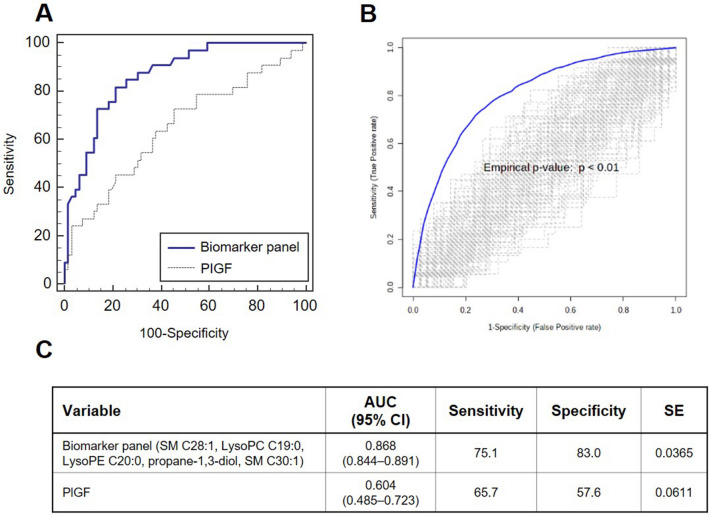
Next, we examined if blood metabolites can discriminate PE from healthy control in mid-trimester cohort. Initially, we tested the metabolite recomposite based on the PLS-DA model; however, the panel did not show good performance (AUC: 0.793) (Figure S4). Alternatively, we explored linearly-recomposited metabolite panel derived based on binary logistic regression analysis. The multivariate logistic model suggested five metabolites (SM C28:1, SM C30:1, LysoPC C19:0, LysoPE C20:0, propane-1,3-diol) and presented a risk of developing PE = -0.626 + (-1.015*SM C28:1) + (1.546*SM C30:1) + (-1.043*LysoPC C19:0) + (0.994*LysoPE C20:0) + (-0.713*propane-1,3-diol) (Figure S5 A). ROC curve analysis confirmed good performance of the metabolic signature (AUC: 0.868, 95% confidence interval [CI]: 0.844–0.891, Sensitivity: 75.1, Specificity: 83.0) (Fig. 2). We also compared the area under the ROC curve between the combined metabolic prediction model and PlGF levels for the ability to predict PE. The combined metabolic model consisting of five metabolites (SM C28:1, SM C30:1, LysoPC C19:0, LysoPE C20:0, propane-1,3-diol) predicted the risk of PE better than PlGF concentrations (AUC [95% CI]: 0.868 [0.844–0.891] vs 0.604 [0.485–0.723]).
Figure 2.
Performance comparison of the metabolic biomarker panel and PlGF for discriminating preeclampsia from healthy controls in the mid-trimester cohort (A) Receiver operating characteristic (ROC) curve analysis. The biomarker panel of 5 discriminatory circulating metabolites were tested against PlGF levels in their ability to predict the development of PE. (B) Validation based on permutation test with 100 time-random sampling (C) Summarized features of ROC curve analysis of two models.
Metabolic biomarkers in the at-delivery cohort
We next performed a comprehensive metabolic profile analysis from maternal plasma collected at delivery. Table 3 shows the clinical characteristics of the at-delivery cohort. Of these, 13 women (38%) were delivered because of PE. With the exception of birth weight, clinical characteristics were not different between women who were delivered for PE or for other indications. Circulating sFlt-1 concentrations and sFlt-1/PlGF ratios were significantly higher and PlGF concentrations significantly lower in the group delivered for PE (Table 3).
Table 3.
Clinical Characteristics of the Study Population in the At-delivery Cohort.
| Group 1 | Group 2 | p Value | |
|---|---|---|---|
| Delivered for reasons other than preeclampsia (n = 21) | Delivered for preeclampsia (n = 13) | ||
| Maternal age (years) | 32 (28–42) | 31 (22–40) | NS |
| Nulliparity | 5 (24%) | 6 (46%) | NS |
| BMI before pregnancy | 20.9 (15.2–29.6) (n = 13) | 22.4 (18.4–25.6) (n = 8) | NS |
| BMI at delivery | 25.9 (20.3–33.3) (n = 17) | 27.7 (24.8–31.0) (n = 10) | NS |
| Gestational age at delivery (weeks) | 35.4 (34.3–36.9) | 35.6 (34.1–36.9) | NS |
| Birth weight (g) | 2660 (1710–3440) | 2160 (1560–3240) | < 0.01 |
| Small for gestational age | 1 (5%) | 2 (15%) | NS |
| Sex (male) | 11 (52%) | 6 (46%) | NS |
| Cesarean delivery | 17 (81%) | 13 (100%) | NS |
| Diabetes during pregnancy | 2 (10%) | 0 (0%) | NS |
| Pro- and anti-angiogenic factors | |||
| sFlt-1 (pg/mL) | 1325.0 (199.5–9884.9) | 2791.6 (723.8–7887.2) | < 0.05 |
| PlGF (pg/mL) | 31.1 (3.0–256.3) | 3.1 (1.6–40.7) | < 0.001 |
| sFlt-1/PlGF | 32.8 (7.1–3278.0) | 856.2 (59.6–3243.8) | < 0.001 |
All values are given as median (range) or number (%).
NS, not significant; BMI, body mass index; PlGF, placental growth factor; sFlt-1, soluble fms-like tyrosine kinase-1.
A comparative analysis of the metabolomic profiles revealed 17 lipids (5.2%) and 7 primary metabolites (2.1%) that were significantly different in women delivered with PE versus no PE (Student’s t-test, listed in Table 4). Of these, the major lipid classes included sphingomyelins (C30:1, C32:1, C33:2, C34:2, and C38:1), that were present at higher levels in the PE versus non-PE group, as well as FAHFA (C18:0), PCs, PE, LysoPC, and LysoPEs, most of which were also significantly higher in the PE group. Similarly, all identified primary metabolites were upregulated in the PE group. Remarkably, the disaccharide, isomaltose, showed a more than two-fold increase in PE. Other primary metabolites of note included 2-deoxytetronic acid, 3,6-anhydro-d-galactose, sugar alcohols (erythritol, xylitol, and myo-inositol), and thymine (Table 4). Isomaltose and erythritol that were originated from dietary resource, have been suggested for the association with disease biomarker, particularly in diabetes41,42.
Table 4.
Metabolites That Were Significantly Different in the Preeclampsia Group in the At-delivery Cohort.
| Metabolites list | Fold change | p Value | FDR |
|---|---|---|---|
| Lipid molecules | |||
| FAHFA C18:0 | 1.41 | 0.028 | 0.331 |
| LysoPC C18:2 | 0.44 | 0.007 | 0.580 |
| LysoPE C15:0 | 0.28 | 0.043 | 0.589 |
| LysoPE C18:2 | 0.31 | 0.037 | 0.589 |
| LysoPE C20:4 | 0.63 | 0.049 | 0.589 |
| OxPC C38:4 + 1O(1Cyc) | 1.72 | 0.003 | 0.000 |
| OxPE C38:3 + 1O | 1.86 | 0.001 | 0.000 |
| OxPI C38:4 + 1O | 1.52 | 0.028 | 0.331 |
| PC C36:5e | 1.65 | 0.042 | 0.331 |
| PC C38:5 | 1.89 | 0.009 | 0.098 |
| PC C40:7 | 1.82 | 0.008 | 0.098 |
| PE C36:4 | 2.73 | 0.010 | 0.098 |
| SM C30:1 | 2.37 | 0.001 | 0.000 |
| SM C32:1 | 1.80 | 0.012 | 0.098 |
| SM C33:2 | 2.63 | 0.033 | 0.331 |
| SM C34:2 | 1.43 | 0.049 | 0.331 |
| SM C38:1 | 2.18 | 0.049 | 0.331 |
| Primary metabolites | |||
| 2-Deoxytetronic acid | 1.55 | 0.035 | 0.193 |
| 3,6-Anhydro-d-galactose | 1.28 | 0.029 | 0.193 |
| Erythritol | 1.31 | 0.043 | 0.315 |
| Isomaltose | 2.37 | 0.028 | 0.193 |
| Myo-inositol | 1.39 | 0.014 | 0.193 |
| Thymine | 1.24 | 0.048 | 0.315 |
| Xylitol | 1.21 | 0.032 | 0.193 |
*Statistical significance is presented by p-value and false discovery rate (FDR) against control.
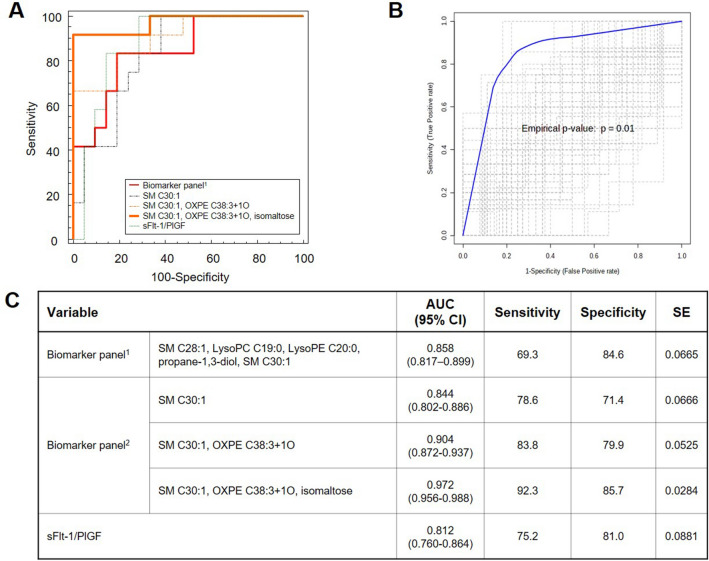
Next, we set out to confirm whether the panel of 5 metabolites identified in the mid-trimester cohort could discriminate between cases with and without PE in the at-delivery cohort. The same panel of 5 compounds (SM C28:1, SM C30:1, LysoPC C19:0, LysoPE C20:0, and propane-1,3-diol) showed good discriminatory performance, with similar AUC to that of the sFlt-1/PlGF ratio (AUC [95% CI]: 0.858 [0.817–0.899] vs AUC [95% CI]: 0.812 [0.760–0.864]) (Fig. 3, Figure S5 B).
Figure 3.
Performance comparison of the metabolic biomarker panels and sFlt-1/PlGF ratio for discriminating preeclampsia from non-preeclampsia in the at-delivery cohort. (A) Receiver operating characteristic (ROC) curve analysis. The same metabolic biomarker derived from the mid-trimester cohort study is re-formulated for at-delivery cohort using binary logistic regression analysis (biomarker panel1). New biomarker panel was generated based metabolite set determined from at-delivery cohort study (biomarker panel2). (B) Validation based on permutation test with 100 time-random sampling (C) Summarized features of ROC curve analysis of the tested models.
Then, we further explored if there is a combination of metabolomic markers that could more accurately distinguish between PE and non-PE in the at-delivery cohort. All metabolites that were significantly different in the PE group were analyzed (Table 4) and prioritized using binary logistic regression. The analysis suggested three consecutive models, which consisted of SM C30:1 alone, SM C30:1 + oxidized PE (OxPE) C38:3, and SM C30:1 + oxidized PE (OxPE) C38:3 + isomaltose. The calculated AUC (95% CI) values were as follows: 0.844 (0.802–0.886), 0.904 (0.872–0.937), and 0.972 (0.956–0.988), respectively. The model with the three metabolites (SM C30:1 + oxidized PE (OxPE) C38:3 + isomaltose) showed the marginal difference (p = 0.08) compared to the sFlt-1/PlGF ratio (0.812 [0.760–0.864]) (Fig. 3). Ten-fold cross validation was performed for each model and provided in Figure S6.
Discussion
In the current study, we comprehensively characterized the metabolome in mid-trimester maternal plasma and identified a panel composed of 5 metabolites that can accurately predict the subsequent development of PE. We then tested this same biomarker signature in a second cohort of women delivered in the late preterm period, and were able to accurately discriminate women who were delivered with PE from those delivered with other indications. In addition, we first reported the significant differences in metabolic phenotype according to gestational age at sampling (mid-trimester and at-delivery), suggesting that these biomarkers may be used for both prediction and diagnosis along with pregnancy period. Particularly, lipid molecules, FAHFA C18:0 and LysoPC C19:0 were highly abundant at the time of delivery.
In mid-trimester population, we obtained predictive model for PE with 5 plasma metabolites based on binary logistic regression following the variable prioritization based on Pattern Searching algorithm43. The regression model presented good predictive performance in the mid-trimester period (16–24 weeks) for the subsequent development of PE. The biomarker signature included SM C28:1, SM C30:1, LysoPC C19:0, LysoPE C20:0, and propane-1,3-diol. Aberrant levels of SMs, which serve as important structural components of cell membranes and as bioactive signal transduction molecules in various cellular response44, have been measured in fetal (umbilical artery) blood45, placentas46, and maternal blood46 collected from pregnancies complicated by PE.
A number of intervention strategies have been proposed to prevent PE including dietary manipulation, exercise, cardiovascular drugs, and antioxidants, but none has been found to be effective. Low-dose aspirin (LDA), which acts in part by inhibiting platelet thromboxane A2 biosynthesis, is the one intervention that has been shown to reduce the development of PE in some high-risk patients47–49. As such, recent guidelines support the use of LDA starting from mid-pregnancy period to prevent PE in high-risk populations, including women with a history of PE in a prior pregnancy, and women with multiple gestations, chronic hypertension, pregestational diabetes, renal disease, or autoimmune diseases1,50. However, the use of LDA in low-risk populations is not currently recommended even though substantial number of the women without these risk factors can develop PE during pregnancy51.
For the reasons outlined above, the early risk assessment of PE development is highly desirable for low-risk population. Indeed, the application of complex algorithms (e.g. maternal historical and demographic risk factors, blood pressure, uterine artery pulsatility index on Doppler velocimetry, and maternal serum biomarkers such as pregnancy-associated plasma protein A [PAPP-A] and PlGF) to identify women at risk for preeclampsia and use of LDA might also lead to a reduction of PE development52. The current study suggests that a new test using maternal plasma biomarkers alone can more accurately identify women at high-risk of PE development and would allow for more timely and appropriate utilization of preventive medications for low-risk pregnant women (those without a history of prior PE and those with no underlying medical conditions). Most guidelines recommend the LDA therapy starting at the late first or early second trimester of pregnancy1,50. Therefore, early and accurate identification of women with high risk for PE is critical to allow for the timely adoption of LDA prophylaxis. In the current study, we developed simple binary logistic regression model composed of 5 plasma metabolites, which accurately predicted the development of PE superior to PIGF. The simpler model using only SMs (SM C28:1 and SM C30:1) also showed good performance for the PE prediction. Moreover, our highly quantitative and comprehensive approach suggested the plausible association between the metabolic features and potential patho-mechanism. For instance, the increases in circulating SMs has been described in a lipidomic analysis of placental microvesicles from abnormal pregnancies and appears to correlate with an increase in inflammation and oxidative stress53. The additional examination of plasma metabolites from the at-delivery cohort implied an underlying aberrant redox regulation given the significant increase in oxidized PLs (OxPC, OxPI, and OxPE) in the PE group. A recent report suggested that this may be due to the enhanced release of the anti-angiogenic factor, endoglin, by circulating exosome in response to exposure to high levels of SMs53. Indeed, in our model, the combination of SM C30:1 and OxPE C38:3 was best able to discriminate between patients with and without PE at delivery. The pathophysiologic mechanism regarding these biomarkers needs to be further investigated for exploring novel therapeutic targets.
The appropriate development of the prediction model would be completed through the validation of the developed model in independent study population, but we could not split the study population into the development and validation set because of small number of cases. Instead we validated the model in the same study population and tested if the developed model can be also used in the identification of preeclampsia patient at delivery. Further studies with large number of cases with separate development and validation population should be performed. For the clinical application, the biomarker also needs to be validated in other cohorts including the group in the first trimester. As guidelines recommend the start of LDA ideally in the late first trimester49,50, the first trimester biomarkers would be clinically more applicable. In addition, the comparative evaluation of prediction accuracy between the metabolic biomarkers and clinical parameters, or the combination of both needs to be evaluated in future studies.
In addition, we evaluated the usefulness of metabolomic panel in at-delivery cohort. The identical biomarker composite developed from the mid-trimester cohort also showed good discrimination between PE and non-PE in the late preterm study population at delivery (at-delivery cohort). Furthermore, the metabolic panel derived from at-delivery cohort showed the outperformance in the discriminant power. For at-delivery cohort, we evaluated the maternal plasma at delivery from patients who delivered in the late preterm period (34–0/7—36–6/7 weeks) as the following reasons: (1) Guidelines recommend that women with severe preeclampsia should be delivered at or after 34 weeks of gestation54. Therefore most patients with severe preeclampsia are intended to deliver at late preterm period; (2) In addition, to compare the pathophysiologic changes in biomaterials of preeclampsia with those of control pregnancy, proper comparison should be performed between pregnant women with preeclampsia and those who delivered at term without any complications, but this comparison can be biased by the difference of gestational age at delivery. To adjust gestational age at delivery, most studies included pregnant women who delivered at the similar gestational age (preterm) after spontaneous preterm labor or rupture of membranes. However, the result from this comparison can be difficult in interpretation, because the changes in biomaterials could be originate from the parturition mechanism in preterm labor/rupture of membranes, as well as from the pathophysiologic mechanism of preeclampsia. To overcome this problem, we included as controls not only women who delivered preterm after spontaneous parturition but also those who delivered preterm because of maternal medical indication in late preterm period.
In conclusion, we demonstrated that a combination of 5 blood metabolites in mid-trimester maternal plasma accurately predicted the development of PE. This unique metabolomic panel was further validated by its accurate predictability that discriminated the patients delivered in the late preterm period for PE from those delivered for other indications.
Methods
Study design
(1) Mid-trimester cohort
The population for this case–control study consisted of 33 healthy pregnant women whose plasma was taken in the mid-trimester period (gestational age [GA] range 16–24 weeks, median 17.4 weeks) and who subsequently developed PE [cases]. Control subjects included 66 pregnant women with mid-trimester plasma samples who did not develop PE and were matched for maternal age and GA at maternal plasma sampling (ratio 1:2) [controls]. Patients with major fetal malformations and multifetal pregnancies were excluded. The maternal plasma metabolite and PL profiles were compared between cases and controls. PE was defined as the development of hypertension with evidences of end-organ involvement such as proteinuria, low platelet count, renal or liver involvement, cerebral symptoms, or pulmonary edema1.
(2) At-delivery cohort
After characterization of the metabolomic biomarker panel, we evaluated the discriminatory power of this panel of biomarkers to identify women with PE at the time of delivery. The latter study population consisted of singleton pregnant women who delivered in the late preterm period (34–0/7 to 36–6/7 weeks), had maternal plasma collected within 3 days of delivery, and had no evidence of chorioamionitis. Study subjects were divided into 2 groups according to the indication for preterm birth (i.e., PE or other causes such as spontaneous preterm labor and maternal and/or fetal indication). The plasma metabolomic biomarker measurements were compared between subjects who did and did not have PE at delivery.
Metabolomic profiling of maternal plasma
Maternal plasma samples were centrifuged at 2,000 rpm for 10 min and the supernatant stored at -70 °C until assayed. Samples were comprehensively analyzed for primary metabolomic and lipidomic signatures using gas chromatography time-of-flight mass spectrometry (GC-TOF MS) and liquid chromatography Orbitrap mass spectrometry (LC-Orbitrap MS) [See Appendix S1 for further details of methodology]55–57. In addition, all analytical procedures, including extraction, reconstitution, and MS analysis were performed in random order to minimize potential systematic errors. The study was approved by the Institutional Review Board of Seoul National University Hospital. All patients provided written informed consent for sample collection and the use of biologic materials for research purposes. All methods were performed in accordance with the relevant guidelines and regulations including Declaration of Helsinki.
Measurement of sFlt-1 and PlGF
For comparison, sFlt-1 and PlGF levels were measured in all samples using a highly sensitive multiplex array in accordance with the instruction of the manufacturer (Meso Scale Discovery (MSD) V-PLEX Angiogenesis Panel 1 Human kit, MSD, USA).
Statistical analysis
For clinical information, continuous data were analyzed using the Mann–Whitney U test and categorical data using the Fisher’s exact test or Chi-square test, as appropriate. Statistical analyses were conducted using the IBM SPSS version 20 (IBM Corp., Armonk, NY, USA).
The integrative metabolomic profiles were pre-processed for data normalization using the MSTUS methodology58 implemented in NOREVA (https://idrb.zju.edu.cn/noreva/)59. MSTUS (MS Total Useful Signal) is a method of summing and adjusting the total intensities of identified metabolites across all samples). Student’s t-test was performed for univariate statistics using EXCEL (Microsoft Office 2010). One-way ANOVA was conducted with post-hoc testing (Fisher’s LSD) as indicated. For adjusting multiple comparisons, the false discovery rate (FDR) was computed (adjusted by Benjamini-Hochberg). Partial least squares-discriminant analysis (PLS-DA) and permutation test were carried out for multivariate statistics using SIMCA 15 (Umetrics AB, Umea, Sweden). Binary logistic regression was performed with forward or enter selection, as appropriate, for predictive model construction using IMB SPSS Statistics for Windows, version 25.0 (IBM Corp.) following auto-scaling procedure. Receiver operating characteristic (ROC) analysis, tenfold cross validation, permutation test, and enrichment analysis were performed using Metaboanalyst 4.0 (https://www.metaboanalyst.ca/).
Supplementary information
Acknowledgment
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science and ICT of Korea (2016M3A9B6902061 and 2018M3A9F3020970) and by Grant 03-2019-0180 from the SNUH research fund.
Author contributions
S.M. L., Y.K., D.Y.L., and J.S.P. had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: S.M.L., J.S.P. and D.Y.L. Acquisition, analysis, or interpretation of data: S.M.L., Y.K., E.M.L., Y.M.J., S.H., S.J.P., C.W.P., E.R.N., D.Y.L., and J.S.P. Drafting of the manuscript: S.M.L., Y.K., J.S.P. and D.Y.L. Critical revision of the manuscript for important intellectual content: S.M.L., Y.K., E.M.L., Y.M.J., S.H., S.J.P., C.W.P., E.R.N., D.Y.L., and J.S.P. Statistical analysis: S.M.L., Y.K., S.J.P., and D.Y.L. Study supervision: J.S.P. and D.Y.L. Guarantor: J.S.P.
Data availability
The datasets analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Seung Mi Lee and Yujin Kang.
Contributor Information
Do Yup Lee, Email: rome73@snu.ac.kr.
Joong Shin Park, Email: jsparkmd@snu.ac.kr.
Supplementary information
is available for this paper at 10.1038/s41598-020-72852-4.
References
- 1.1American College of, O., Gynecologists & Task Force on Hypertension in, P. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists' Task Force on Hypertension in Pregnancy. Obstet Gynecol122, 1122–1131, 10.1097/01.AOG.0000437382.03963.88 (2013). [DOI] [PubMed]
- 2.Khan KS, Wojdyla D, Say L, Gulmezoglu AM, Van Look PF. WHO analysis of causes of maternal death: a systematic review. Lancet. 2006;367:1066–1074. doi: 10.1016/S0140-6736(06)68397-9. [DOI] [PubMed] [Google Scholar]
- 3.3Capriglione, S. et al. Preeclampsia and the challenge of early prediction: reality or utopia? State of art and critical review of literature. J. Mater. Fetal Neonatal. Med., 1–10, 10.1080/14767058.2018.1495191 (2018). [DOI] [PubMed]
- 4.Sunjaya AF, Sunjaya AP. Evaluation of serum biomarkers and other diagnostic modalities for early diagnosis of preeclampsia. J. Family Reprod. Health. 2019;13:56–69. [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy FP, Ryan RM, Chappell LC. Prospective biomarkers in preterm preeclampsia: a review. Pregnancy Hypertens. 2018;14:72–78. doi: 10.1016/j.preghy.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Zhong Y, Zhu F, Ding Y. Serum screening in first trimester to predict pre-eclampsia, small for gestational age and preterm delivery: systematic review and meta-analysis. BMC Pregnancy Childbirth. 2015;15:191. doi: 10.1186/s12884-015-0608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppertz B. An updated view on the origin and use of angiogenic biomarkers for preeclampsia. Expert Rev. Mol. Diagn. 2018;18:1053–1061. doi: 10.1080/14737159.2018.1546579. [DOI] [PubMed] [Google Scholar]
- 8.Wojcik-Baszko D, Charkiewicz K, Laudanski P. Role of dyslipidemia in preeclampsia-a review of lipidomic analysis of blood, placenta, syncytiotrophoblast microvesicles and umbilical cord artery from women with preeclampsia. Prostaglandins Other Lipid Mediat. 2018;139:19–23. doi: 10.1016/j.prostaglandins.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Lv Y, et al. Roles of microRNAs in preeclampsia. J. Cell. Physiol. 2019;234:1052–1061. doi: 10.1002/jcp.27291. [DOI] [PubMed] [Google Scholar]
- 10.Bellos I, Karageorgiou V, Kapnias D, Karamanli KE, Siristatidis C. The role of interleukins in preeclampsia: a comprehensive review. Am. J. Reprod. Immunol. 2018;80:e13055. doi: 10.1111/aji.13055. [DOI] [PubMed] [Google Scholar]
- 11.Duckworth S, et al. Diagnostic biomarkers in women with suspected preeclampsia in a prospective multicenter study. Obstet. Gynecol. 2016;128:245–252. doi: 10.1097/AOG.0000000000001508. [DOI] [PubMed] [Google Scholar]
- 12.Myatt L, et al. Can changes in angiogenic biomarkers between the first and second trimesters of pregnancy predict development of pre-eclampsia in a low-risk nulliparous patient population? BJOG. 2013;120:1183–1191. doi: 10.1111/1471-0528.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gowda GA, et al. Metabolomics-based methods for early disease diagnostics. Expert Rev. Mol. Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Mol. Syst. Biol. 2006;2:52. doi: 10.1038/msb4100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SM, et al. Metabolic biomarkers in midtrimester maternal plasma can accurately predict adverse pregnancy outcome in patients with SLE. Sci. Rep. 2019;9:15169. doi: 10.1038/s41598-019-51285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nobakht MGBF. Application of metabolomics to preeclampsia diagnosis. Syst. Biol. Reprod. Med. 2018;64:324–339. doi: 10.1080/19396368.2018.1482968. [DOI] [PubMed] [Google Scholar]
- 17.Lee SM, et al. Increased biosynthesis and accumulation of cholesterol in maternal plasma, but not amniotic fluid in pre-eclampsia. Sci. Rep. 2019;9:1550. doi: 10.1038/s41598-018-37757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenny LC, et al. Detection and identification of novel metabolomic biomarkers in preeclampsia. Reprod. Sci. 2008;15:591–597. doi: 10.1177/1933719108316908. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee R, Ray CD, Ray S, Dasgupta S, Chaudhury K. Altered metabolic profile in early and late onset preeclampsia: an FTIR spectroscopic study. Pregnancy Hypertens. 2014;4:70–80. doi: 10.1016/j.preghy.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, He P, Tan Y, Xu D. Biomarker identification and pathway analysis of preeclampsia based on serum metabolomics. Biochem. Biophys. Res. Commun. 2017;485:119–125. doi: 10.1016/j.bbrc.2017.02.032. [DOI] [PubMed] [Google Scholar]
- 21.Senyavina NV, Khaustova SA, Grebennik TK, Pavlovich SV. Analysis of purine metabolites in maternal serum for evaluating the risk of gestosis. Bull. Exp. Biol. Med. 2013;155:682–684. doi: 10.1007/s10517-013-2225-y. [DOI] [PubMed] [Google Scholar]
- 22.Bahado-Singh RO, et al. Metabolomics and first-trimester prediction of early-onset preeclampsia. J. Mater. Fetal Neonatal. Med. 2012;25:1840–1847. doi: 10.3109/14767058.2012.680254. [DOI] [PubMed] [Google Scholar]
- 23.Kenny LC, et al. Robust early pregnancy prediction of later preeclampsia using metabolomic biomarkers. Hypertension. 2010;56:741–749. doi: 10.1161/HYPERTENSIONAHA.110.157297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahado-Singh RO, et al. First-trimester metabolomic detection of late-onset preeclampsia. Am. J. Obstet. Gynecol. 2013;208(58):e51–57. doi: 10.1016/j.ajog.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 25.25Bahado-Singh, R. O. et al. Validation of metabolomic models for prediction of early-onset preeclampsia. Am. J. Obstet. Gynecol.213, 530 e531–530 e510, 10.1016/j.ajog.2015.06.044 (2015). [DOI] [PubMed]
- 26.Koster MP, et al. First-trimester serum acylcarnitine levels to predict preeclampsia: a metabolomics approach. Dis. Mark. 2015;2015:857108. doi: 10.1155/2015/857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuc S, et al. Metabolomics profiling for identification of novel potential markers in early prediction of preeclampsia. PLoS ONE. 2014;9:e98540. doi: 10.1371/journal.pone.0098540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odibo AO, et al. First-trimester prediction of preeclampsia using metabolomic biomarkers: a discovery phase study. Prenat. Diagn. 2011;31:990–994. doi: 10.1002/pd.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Austdal M, et al. First trimester urine and serum metabolomics for prediction of preeclampsia and gestational hypertension: a prospective screening study. Int. J. Mol. Sci. 2015;16:21520–21538. doi: 10.3390/ijms160921520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Austdal M, et al. Metabolomic biomarkers in serum and urine in women with preeclampsia. PLoS ONE. 2014;9:e91923. doi: 10.1371/journal.pone.0091923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diaz SO, et al. Second trimester maternal urine for the diagnosis of trisomy 21 and prediction of poor pregnancy outcomes. J. Proteome Res. 2013;12:2946–2957. doi: 10.1021/pr4002355. [DOI] [PubMed] [Google Scholar]
- 32.Austdal M, et al. Metabolic profiles of placenta in preeclampsia using HR-MAS MRS metabolomics. Placenta. 2015;36:1455–1462. doi: 10.1016/j.placenta.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, et al. Impaired mitochondrial fusion, autophagy, biogenesis and dysregulated lipid metabolism is associated with preeclampsia. Exp. Cell Res. 2017;359:195–204. doi: 10.1016/j.yexcr.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 34.Jain S, Jayasimhulu K, Clark JF. Metabolomic analysis of molecular species of phospholipids from normotensive and preeclamptic human placenta electrospray ionization mass spectrometry. Front. Biosci. 2004;9:3167–3175. doi: 10.2741/1470. [DOI] [PubMed] [Google Scholar]
- 35.Wang B, Tontonoz P. Phospholipid remodeling in physiology and disease. Annu. Rev. Physiol. 2019;81:165–188. doi: 10.1146/annurev-physiol-020518-114444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts JM, Hubel CA. The two stage model of preeclampsia: variations on the theme. Placenta. 2009;30(Suppl A):S32–37. doi: 10.1016/j.placenta.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y, et al. A prospective and longitudinal study of plasma phospholipid saturated fatty acid profile in relation to cardiometabolic biomarkers and the risk of gestational diabetes. Am. J. Clin. Nutr. 2018;107:1017–1026. doi: 10.1093/ajcn/nqy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steffen BT, et al. Obesity modifies the association between plasma phospholipid polyunsaturated fatty acids and markers of inflammation: the multi-ethnic study of atherosclerosis. Int. J. Obes. (Lond.) 2012;36:797–804. doi: 10.1038/ijo.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhl O, et al. Effects of obesity and gestational diabetes mellitus on placental phospholipids. Diabetes Res. Clin. Pract. 2015;109:364–371. doi: 10.1016/j.diabres.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 40.Allen F, Pon A, Wilson M, Greiner R, Wishart D. CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014;42:W94–W99. doi: 10.1093/nar/gku436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.41Shao, M. et al. Serum and urine metabolomics reveal potential biomarkers of T2DM patients with nephropathy. Ann. Transl. Med.8 (2020). [DOI] [PMC free article] [PubMed]
- 42.42Ortiz, S. R. & Field, M. S. Mammalian metabolism of erythritol: a predictive biomarker of metabolic dysfunction. Curr. Opin. Clin. Nutrit. Metab. Care (2020). [DOI] [PubMed]
- 43.Lee EM, et al. Highly geographical specificity of metabolomic traits among Korean domestic soybeans (Glycine max) Food Res. Int. 2019;120:12–18. doi: 10.1016/j.foodres.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 44.Melland-Smith M, et al. Disruption of sphingolipid metabolism augments ceramide-induced autophagy in preeclampsia. Autophagy. 2015;11:653–669. doi: 10.1080/15548627.2015.1034414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romanowicz L, Bankowski E. Preeclampsia-associated alterations in sphingolipid composition of the umbilical cord artery. Clin. Biochem. 2009;42:1719–1724. doi: 10.1016/j.clinbiochem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 46.Charkiewicz K, et al. Sphingolipids as a new factor in the pathomechanism of preeclampsia - mass spectrometry analysis. PLoS ONE. 2017;12:e0177601. doi: 10.1371/journal.pone.0177601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Askie LM, Duley L, Henderson-Smart DJ, Stewart LA, Group PC. Antiplatelet agents for prevention of pre-eclampsia: a meta-analysis of individual patient data. Lancet. 2007;369:1791–1798. doi: 10.1016/S0140-6736(07)60712-0. [DOI] [PubMed] [Google Scholar]
- 48.Henderson JT, et al. Low-dose aspirin for prevention of morbidity and mortality from preeclampsia: a systematic evidence review for the US Preventive Services Task Force. Ann. Intern. Med. 2014;160:695–703. doi: 10.7326/M13-2844. [DOI] [PubMed] [Google Scholar]
- 49.ACOG Committee Opinion No. 743 Low-dose aspirin use during pregnancy. Obstet Gynecol. 2018;132:e44–e52. doi: 10.1097/AOG.0000000000002708. [DOI] [PubMed] [Google Scholar]
- 50.50LeFevre, M. L. & Force, U. S. P. S. T. Low-dose aspirin use for the prevention of morbidity and mortality from preeclampsia: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med.161, 819–826, 10.7326/M14-1884 (2014). [DOI] [PubMed]
- 51.Mone F, Mulcahy C, McParland P, McAuliffe FM. Should we recommend universal aspirin for all pregnant women? Am. J. Obstet. Gynecol. 2017;216:141–145. doi: 10.1016/j.ajog.2016.09.086. [DOI] [PubMed] [Google Scholar]
- 52.Rolnik DL, et al. Aspirin versus Placebo in pregnancies at high risk for preterm preeclampsia. N. Engl. J. Med. 2017;377:613–622. doi: 10.1056/NEJMoa1704559. [DOI] [PubMed] [Google Scholar]
- 53.Ermini L, et al. A Single sphingomyelin species promotes exosomal release of endoglin into the maternal circulation in preeclampsia. Sci. Rep. 2017;7:12172. doi: 10.1038/s41598-017-12491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.54Publications Committee, S. f. M.-F. M. & Sibai, B. M. Evaluation and management of severe preeclampsia before 34 weeks' gestation. Am. J. Obstet. Gynecol.205, 191–198, 10.1016/j.ajog.2011.07.017 (2011). [DOI] [PubMed]
- 55.Ji DY, Lee C-W, Park SH, Lee EJ, Lee DY. Simple and robust measurement of blood plasma lysophospholipids using liquid chromatography mass spectrometry. Mass Spectrom. Lett. 2017;8:109–113. [Google Scholar]
- 56.Ji DY, et al. Comparative assessment of Graves’ disease and main extrathyroidal manifestation, Graves’ ophthalmopathy, by non-targeted metabolite profiling of blood and orbital tissue. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park SJ, et al. Integrative metabolomics reveals unique metabolic traits in Guillain-Barré Syndrome and its variants. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dalby AR, et al. Metabolomics data normalization with EigenMS. PLoS ONE. 2014;9:e116221. doi: 10.1371/journal.pone.0116221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B, et al. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017;45:W162–W170. doi: 10.1093/nar/gkx449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.