Abstract
The homochirality of amino acids in living organisms is one of the great mysteries in the phenomena of life. To understand the chiral recognition of amino acids, we have used scanning tunnelling microscopy to investigate the self-assembly of molecules of the amino acid tryptophan (Trp) on Au(111). Earlier experiments showed only homochiral configurations in the self-assembly of amino acids, despite using a mixture of the two opposite enantiomers. In our study, we demonstrate that heterochiral configurations can be favored energetically when l- and d-Trp molecules are mixed to form self-assembly on the Au surface. Using density functional theory calculations, we show that the indole side chain strongly interacts with the Au surface, which reduces the system effectively to two-dimension, with chiral recognition disabled. Our study provides important insight into the recognition of the chirality of amino acid molecules in life.
Subject terms: Scanning probe microscopy, Surface assembly
Introduction
Amino acids containing α-amino (–NH2) and α-carboxyl (–COOH) groups are essential constituents for the synthesis of peptides in a living organism. Except for glycine, the amino acids exist as chiral molecules in two forms, the l- and d-enantiomers. Although the two enantiomers exhibit identical physical and chemical properties, living organisms use only amino acids of l-chirality. This homochirality of the amino acids is one of the great mysteries that need to be explored in life phenomena1. The amino acids are spontaneously self-assembled in space as a result of various non-covalent interactions between the molecules. For understanding the inherent chirality of the self-assembled amino acids, scanning tunnelling microscopy (STM) has been widely used2–19. Kühnle et al. studied the self-assembly of cysteine molecules on a Au(110) surface5. The result showed that homochiral cysteine dimers were stabilized by the formation of three bonds between the same enantiomers. Lingenfelder et al. investigated the interaction between l-phenylalanine (Phe)–l-Phe and d-Phe–d-Phe pairs and found that stereoselectivity plays a crucial role in the formation of the homochiral chains12. These pioneering works highlighted the role of chiral recognition in the self-assembly of amino acids.
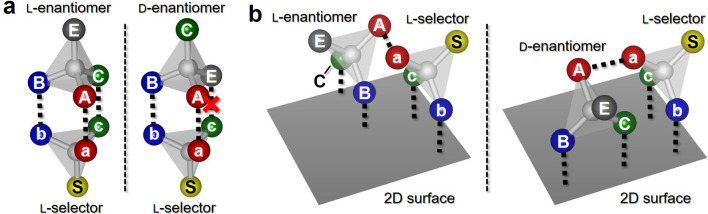
The chiral recognition of molecules is generally discussed in terms of the three-point interaction (TPI) model, derived from the lock-and-key model4,5,12,20–29. Figure 1a shows the illustration of the TPI model. The l-selector uniquely mates with the l-enantiomer when bonds between the molecules are formed at three points (Fig. 1a, left panel). In contrast, the l-selector does not mate firmly with the d-enantiomer because the number of bonds is less than three (right panel). Therefore, the l-selector is energetically favored to connect to the l-enantiomer. Although the TPI model oversimplifies the nature of this process, it provides insight into the origin of the chiral recognition of chiral molecules. Dynamic conformational adjustments between the molecules help to ensure the chiral self-assembly even when the three-point bonds are not precisely defined13,30–38.
Figure 1.
(a) A schematic view of the molecule–molecule interactions based on the three-point interaction model, depending on their molecular chirality. ‘A’, ‘B’, and ‘C’ sites interact with the ‘a’, ‘b’, and ‘c’ sites, respectively. (b) A model of the molecule–molecule interactions on a 2D surface. Only one intermolecular bond is active, between ‘A’ and ‘a’ site, owing to the interactions between the molecules and the surface.
Since chiral recognition is based on stereochemical interaction between the chiral molecules, it is most prominent in the self-assembly in 3-dimensional (3D) space. When the number of dimensions of the system is reduced, the stereochemical interaction is limited. Figure 1b illustrates chiral molecules in 2D space. The molecules are confined to a surface as a result of interactions with the surface. We assume here that only one bond is active in the intermolecular bonding. Unlike the 3D-case in Fig. 1a, there is then no energy difference between an l-selector–l-enantiomer pair (Fig. 1b, left panel) and an l-selector–d-enantiomer pair (right panel), implying that chiral recognition is lost. As a rule, the chirality of self-assembly is significantly affected by dimensions of the system. However, thus far, no experiments have been conducted to determine if chiral recognition can be disabled in lower dimensions. In this study, we have investigated the self-assembly of tryptophan (Trp) molecules with attention to the molecular chirality, using STM. We first deposited a single type (l- or d-) of Trp molecules on a Au(111) substrate, which naturally resulted in homochiral self-assembly. In contrast, when both l- and d-Trp molecules were deposited on the substrate simultaneously, we found that heterochiral structures were predominantly formed, providing evidence of the chiral recognition being broken. By carefully modeling the self-assembled structures using density functional theory (DFT), we demonstrate that the interactions between the Trp molecules and the surface effectively reduces the system to 2D, inducing a breakdown of chiral recognition in the self-assembly of the mixture of l- and d-Trp molecules.
Results
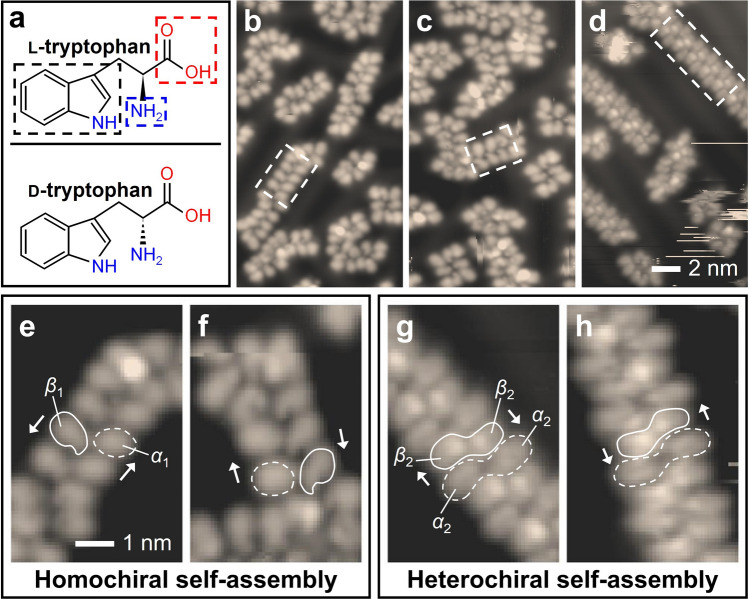
A molecule of Trp, which is one of the essential amino acids used in proteins, contains an α-amino group, an α-carboxyl group, and an indole side chain, as shown in Fig. 2a. According to the bond direction of the α-amino group, Trp molecules are classified into l- or d-enantiomers (Fig. 2a). In the experiment, we deposited l-Trp molecules on the Au(111) surface and observed 1D-like chain structures constructed in the self-assembly (Fig. 2b). Because only l-Trp molecules are used in the self- assembly, the chain structures are naturally homochiral. The deposition of d-Trp molecules on the Au(111) surface led to the formation of similar chain structures but with the opposite chirality (Fig. 2c). This chiral-selective self-assembly motivated us to deposit the l- and d-Trp molecules in equal proportions on a clean Au(111) surface. Remarkably, the homochiral chain structures were not observed in the self-assembly, but heterochiral chain structures were observed, as shown in Fig. 2d, a fact that is confirmed in our DFT calculation.
Figure 2.
(a) Chemical structures of l- and d-Trp molecules. The blue, red, and black-dashed boxes indicate α-amino group, α-carboxyl group, and indole side chain, respectively. Topographic images of homochiral self-assemblies of (b) l- and (c) d-Trp molecules with a 1D chain-like structure (white-dashed box) on Au(111) surface at 77 K, with a bias voltage (Vbias) of − 0.5 V and a tunnelling current (It) of 50 pA. (d) The heterochiral self-assembly obtained by the deposition of a racemic mixture of l- and d-Trp molecules (Vbias = − 1 V, It = 50 pA). Zoom-in images of the self-assembled chain structures constructed by Trp molecules: (e) the l-Trp molecule-constructed and (f) the d-Trp molecule-constructed chain structures. There are two different shapes of adsorbed molecules; ellipse shape (dashed outline, α1) and apostrophe shape (solid outline, β1). (g, h) Heterochiral chain structures in the self-assembly of a racemic mixture of l- and d-Trp molecules. The dashed-outline (α2) and the solid-outline (β2) represent pairs of Trp molecules that appear to be loosely and tightly connected in the image, respectively.
To understand the chirality of self-assembled structures, we studied the conformation of the Trp molecules in the chain structures. Figure 2e depicts an enlarged image of the chain structure of the l-Trp molecules. Two differently-shaped l-Trp molecules are identified in the chain structure; the molecules that resembled the shape of an ellipse (dashed outline) and an apostrophe (solid outline) are denoted as α1 and β1 in Fig. 2e, respectively. The α1 and the β1 molecules form a pair, and such pairs are repeatedly connected along the chain. The structure formed by d-Trp molecules is similar to that formed by l-Trp molecules, as shown in Fig. 2f. However, their chirality is the opposite, reflecting the chirality of the individual l- and d-Trp molecules. Figure 2g,h show the chain structures formed by the racemic mixture of l- and d-Trp molecules. Two different ellipse-shaped Trp molecules are found; the α2 shape is similar to the α1 shape, but the β2 shape is a new type (Fig. 2g). The two β2 molecules are closely bound, as indicated by the solid-outline. In contrast, the two α2 molecules are loosely combined into a pair (the dashed-outline). In the self-assembly of the racemic mixture of l- and d-Trp molecules, we observed two types of chain structures with similar shapes but opposite chirality (Fig. 2g,h), indicating that they are heterochiral. The heterochirality of the racemic mixture is further supported by the cluster structures in Supplementary Information.
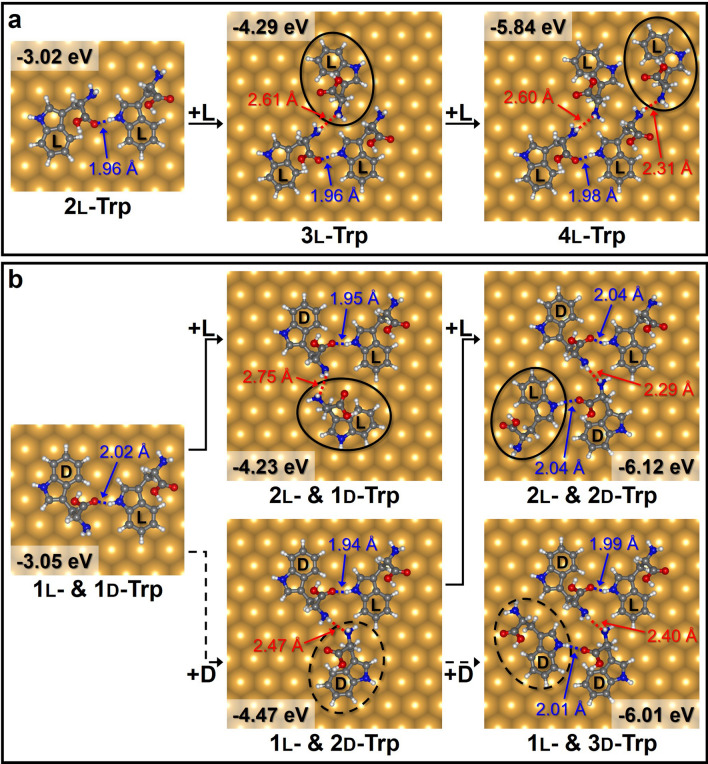
To understand the chain structures at the atomic scale, we investigated the Trp molecular interactions using DFT calculation. Based on the Bader charges of atoms in an l-Trp molecule (Table S1), we designed the various configurations of two Trp molecular interactions (Fig. S1). The adsorption energy (Eads) and the hydrogen bond (HM···XN) length for a given configuration are summarized in Fig. 3a, where XN indicates the X atom (X = H, N, O) in the N group (M, N = A: α-amino, C: α-carboxyl, I: indole). abM-N is the HM···XN interaction between the a- and b-Trp molecules (a, b = l, d chirality). Eads was calculated from the equation: Eads = En × Trp on Au(111) – (n × ETrp + EAu(111)), where En × Trp on Au(111), ETrp, and EAu(111) are the energies of the n × Trp molecules adsorbed on Au(111), the isolated Trp molecule, and the pure Au(111), respectively. In the pair of two l-Trp molecules, the lowest Eads was found to be − 3.02 eV (llI-C, left panel in Fig. 3b) when the OC atom in an l-Trp molecule was close to the HI atom in the other l-Trp molecule (HI···OC interaction). In the pair of l- and d-Trp molecules, a similar HI···OC interaction leads to the lowest Eads of − 3.05 eV (ldI-C, the right panel in Fig. 3b). Although the HI···OC distance of the llI-C pair is shorter than that of the ldI-C pair, the llI-C pair exhibited a slightly higher Eads. To understand this, we investigated the strain energies and the HIb-NI-CIb angles in Trp molecules of the llI-C and ldI-C pairs (Table S2), where HIb and CIb are the HI and CI atoms bonded to the NI atom in a Trp molecule. The right-hand l-Trp molecule (lR) in the llI-C pair produced extra strain energy, owing to the change in the HIb-NI-CIb angle, which is responsible for the higher formation energy of the llI-C pair.
Figure 3.
(a) The adsorption energy and the average hydrogen bond length in a pair of two Trp molecules on Au(111) surface. abM-N indicates the interaction between the M group of an a-Trp molecule and the N group (M, N = A: α-amino, C: α-carboxyl, I: indole) of a b-Trp molecule (a, b = l, d chirality). ab2(M–N) means double hydrogen bonding. The structural detail of each Trp molecular pair is given in Supplementary Information. (b) Perspective and top views of the optimized 2l-Trp and the optimized 1l- & 1d-Trp configurations (the llI-C and ldI-C pairs, respectively).
We constructed a structure of more than two Trp molecules by adding Trp molecules to the configurations found in Fig. 3b. Figure 4a shows the construction of a chain of l-Trp molecules. We attached an l-Trp molecule next to the HI···OC bond of the llI-C pair. When the NA atom in the l-Trp molecule (black solid oval) is close to the HA atom near the HI···OC bond of the llI-C pair, the 3l-Trp configuration is optimized with an Eads of − 4.29 eV. In a subsequent calculation, we added another l-Trp molecule to the 3l-Trp configuration. The HA···NA interaction between the added l-Trp molecule and the 3l-Trp configuration resulted in the optimized 4l-Trp configuration with an Eads of − 5.84 eV.
Figure 4.
Construction process of (a) homochiral and (b) heterochiral structures consisting of four Trp molecules, with the adsorption energies shown. The black solid and dashed ovals indicate l- and d-Trp molecules, respectively. The blue and red dotted lines are the HI···OC and HA···NA bonds, respectively.
The racemic mixed chain structures were also investigated, as shown in Fig. 4b. In separate calculations, l- or d-Trp molecules were added to the stable 1l- & 1d-Trp configuration (the ldI-C pair shown in Fig. 3b). When the NA atom of the added l-Trp molecule was close to the HA atom of the d-Trp molecule in the ldI-C pair, the 2l- & 1d-Trp configuration was optimized with an Eads of − 4.23 eV. The addition of a d-Trp molecule to the ldI-C pair gave the optimized 1l- & 2d-Trp configuration resulting from the HA···NA interaction between the added d-Trp molecule (black dashed oval) and the d-Trp molecule in the ldI-C pair, with a HA···NA distance of 2.47 Å. The optimized configuration required an Eads of − 4.47 eV, which is the lowest Eads among the possible structures consisting of three Trp molecules, because it has the shortest HA···NA distance.
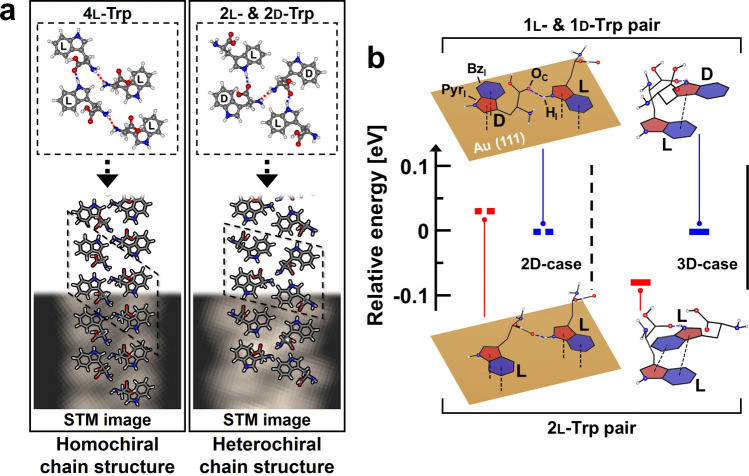
By adding l- or d-Trp molecules to the optimized 1l- & 2d-Trp configuration, we constructed heterochiral structures consisting of four Trp molecules. When the HI atom of the added d-Trp molecule approached the OC atom of the d-Trp molecule that did not participate in the HI···OC interaction, the 1l- & 3d-Trp configuration was optimized with an Eads of − 6.01 eV. However, when an l-Trp molecule was added to the optimized 1l- & 2d-Trp configuration, the additional HI···OC interaction gave rise to the optimized 2l- & 2d-Trp configuration with an Eads of − 6.12 eV. This energy is lower than the Eads of the optimized 1l- & 3d-Trp configuration owing to the shorter total hydrogen bond length. The optimized 2l- & 2d-Trp configuration had two HI···OC and one HA···NA interactions, whereas the optimized 4l-Trp configuration consisted of one HI···OC and two HA···NA interactions. As a result, the optimized 2l- & 2d-Trp configuration showed a lower Eads than the optimized 4l-Trp configuration because the optimized 2l- & 2d-Trp configuration included more HI···OC interactions, which are more stable than HA···NA interactions. The result that the heterochiral configuration was energetically favorable than the homochiral configurations was in agreement with our STM results (Fig. 2d). By using the optimized configuration of 4l-Trp and the 2l- & 2d-Trp molecules as a periodic unit cell, we designed continuous chain structures, as shown in Fig. 5a. Geometrically, the chain structures constructed using the configurations optimized by DFT calculation were in good agreement with the experimentally obtained STM images.
Figure 5.
(a) Modelling of continuous 1D-chain structures using the optimized configurations consisting of four Trp molecules (black-dashed parallelogram as a periodic unit cell), with the STM-measured chain structures. (b) The energy difference between the homochiral (2l-Trp pair) and the heterochiral (1l- & 1d-Trp pair) structures in the 2D-case (on surface) and 3D-case (in vacuum). The energies of 2l-Trp pairs are referenced to those of 1l- & 1d-Trp pairs.
Discussion
Previous studies have shown that homochiral structures are assembled even if a mixture of l- and d-amino acids is deposited on substrates, which indicates that the chiral recognition of amino acids is in play5,12,13. However, this is not necessary in nature, as is demonstrated in our experiments. To understand the breakdown of chiral recognition in our system, we consider the role of the Au(111) surface in the self-assembly. The Trp molecules are adsorbed on this surface by interactions between the indole groups of the molecules and the Au(111) surface. The indole group comprises pyrrole-like (PyrI) and benzene-like (BzI) rings; thus, the energy optimization of the 2l-Trp and 1l- & 1d-Trp pairs is mainly governed by the three interactions: HI···OC, PyrI···Au, and BzI···Au. Among them, the PyrI and BzI rings of the Trp molecule interact strongly with the Au(111) surface. The intermolecular interaction is then guided only by the HI···OC bonding (refer to the Trp molecules in the 2D-case in Fig. 5b), a proof of the concept depicted in Fig. 1b. Hence, it is concluded that the strong interaction between the Trp molecules and the Au(111) surface reduces the system to 2D, with the chiral recognition disabled. To understand how the lock-and-key model in Fig. 1a works for Trp molecules in 3D space, we calculated the interaction between two Trp molecules in the absence of the Au(111) surface. The 2l-Trp molecules are optimized in energy by two PyrI···BzI (π–π stacking) interactions and one HI···OC interaction (refer to the Trp molecules in the 3D-case in Fig. 5b). In contrast, the 1l- and 1d-Trp molecules are restricted to the configuration with one PyrI···BzI interaction and one HI···OC interaction, owing to their steric hindrance. As a result, the energy of the 2l-Trp molecules is lower than that of the 1l- and 1d-Trp molecules, a fact that was postulated as the origin of the chiral recognition20–29,39–42.
In this study, we demonstrated the heterochiral self-assembly of Trp molecules on a Au(111) surface by STM measurements. The deposition of two opposite Trp enantiomers on the Au(111) surface led to a heterochiral chain structure, which differs from the homochiral chain structure obtained from single Trp enantiomers. To understand the construction mechanism of the heterochiral structure, we performed a DFT calculation. Trp molecules were adsorbed on a Au(111) surface, with interaction occurring between the indole group of the Trp molecule and the Au(111) surface. This adsorption condition causes the dimension of system to be effectively reduced to 2D in the self-assembly. As a result, only a limited HI···OC bonding contributes to the molecule–molecule interaction and the chiral recognition of Trp molecules is broken. Our work provides insight into the self-assembly of chiral molecules in low dimensions and deepens our understanding of how and when the chiral recognition can be disabled in our body.
Methods
STM measurements
All experiments were carried out using a home-built variable-temperature STM with a base pressure of 8 × 10−11 Torr. We used a mechanically sharpened Pt-Ir tip for obtaining STM images. The Au(111) substrate was cleaned by repeated cycles of Ar ion sputtering at an Ar pressure of 1.5 × 10−5 Torr (10 min) alternating with annealing at 600 °C (15 min). The Trp molecules (Sigma-Aldrich l-Trp: T0254 and d-Trp: T9753) were deposited on the Au(111) surface using two Knudsen cells; one for l-Trp and the other for d-Trp. Both Knudsen cells were used for the deposition of the racemic mixture of l-Trp and d-Trp. The substrate temperature was maintained at 100 °C during the deposition. The sample was then cooled to 77 K for the STM measurements.
DFT calculations
The initial atomic coordinates of a single Trp molecule for the DFT calculation were obtained from ‘https://pubchem.ncbi.nlm.nih.gov/compound/Tryptophan’. This structure is most stable in its isolated form (Supplementary Information). The DFT calculation was performed using the Vienna Ab-initio Simulation Package (VASP) code43,44, employing the project augmented wave (PAW) method of Blöchl45 implemented in VASP by Kresse and Joubert46. The exchange correlation energy was determined by means of the generalized-gradient approximation (GGA) of Perdew, Burke, and Ernzerhof (PBE) 47. All energies were acquired by considering a non-local van der Waals density functional48 and optPBE49,50. A cutoff energy of 500 eV and 2 × 2 × 1 k-points mesh with zero shift for a slab structure containing a vacuum layer with a thickness of 25 Å were used to generate a plane-wave basis set by the Monkhorst–Pack grid method. Electronic and geometry optimizations were converged when the total energy difference between successive calculation steps was less than 10−4 and 10−3 eV, respectively. All atoms were allowed to relax until the force on each ion was less than 0.02 eV/Å. The Bader analysis method was performed to calculate the electronic charge density of atoms in a Trp molecule51,52. We constructed four-layer Au(111) slabs in 9 × 9 and 10 × 10 super cells using the optimized Au(111) unit cell to investigate the interactions between more than 2 Trp molecules (9 × 9 Au(111) slab: two or three Trp molecules; 10 × 10 Au(111) slab: four Trp molecules). Only the two top layers in the Au(111) slabs were allowed to relax.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary information
Acknowledgements
We are grateful for G. W. Lee and S.-M. Shin for fruitful discussions. This work was supported by the DGIST Start-up Fund Program of the Ministry of Science and ICT (2015010010). The supercomputing resource for this work was supported by Grand Challenging Project of Supercomputing Bigdata Center, DGIST.
Author contributions
J.S. conceived and designed the research. Y.J. performed STM measurements and DFT calculations. Y.J. and H.W.K. analyzed the experimental data. Y.J. and J.K. analyzed the calculated result. All authors discussed the results and contributed to the writing of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongchan Jeong, Email: ycjeong@dgist.ac.kr.
Jungpil Seo, Email: jseo@dgist.ac.kr.
Supplementary information
is available for this paper at 10.1038/s41598-020-73300-z.
References
- 1.Blackmond DG. The origin of biological homochirality. Cold Spring Harbor Persp. Biol. 2010;2:a002147–a002163. doi: 10.1101/cshperspect.a002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao X, Gai Z, Zhao RG, Yang WS, Sakurai T. Adsorption of glycine on cu (001) and related step faceting and bunching. Surf. Sci. 1999;424:L347–L351. [Google Scholar]
- 3.Zhao X, Zhao RG, Yang WS. Scanning tunneling microscopy investigation of l-lysine adsorbed on cu (001) Langmuir. 2000;16:9812–9818. [Google Scholar]
- 4.Chen Q, Richardson NV. Enantiomeric interactions between nucleic acid bases and amino acids on solid surfaces. Nat. Mater. 2003;2:324–328. doi: 10.1038/nmat878. [DOI] [PubMed] [Google Scholar]
- 5.Kühnle A, Linderoth TR, Hammer B, Besenbacher F. Chiral recognition in dimerization of adsorbed cysteine observed by scanning tunnelling microscopy. Nature. 2002;415:891–893. doi: 10.1038/415891a. [DOI] [PubMed] [Google Scholar]
- 6.Kühnle A, Linderoth TR, Besenbacher F. Self-assembly of monodispersed, chiral nanoclusters of cysteine on the au (110)-(1×2) surface. J. Am. Chem. Soc. 2003;125:14680–14681. doi: 10.1021/ja0377403. [DOI] [PubMed] [Google Scholar]
- 7.Kühnle A, Molina LM, Linderoth TR, Hammer B, Besenbacher F. Growth of unidirectional molecular rows of cysteine on a u (110)−(1 × 2) driven by adsorbate-induced surface rearrangements. Phys. Rev. Lett. 2004;93:086101–086104. doi: 10.1103/PhysRevLett.93.086101. [DOI] [PubMed] [Google Scholar]
- 8.Kühnle A, Linderoth TR, Besenbacher F. Enantiospecific adsorption of cysteine at chiral kink sites on au (110)-(1 × 2) J. Am. Chem. Soc. 2006;128:1076–1077. doi: 10.1021/ja0568751. [DOI] [PubMed] [Google Scholar]
- 9.Kühnle A, Linderoth TR, Schunack M, Besenbacher F. l-cysteine adsorption structures on au (111) investigated by scanning tunneling microscopy under ultrahigh vacuum conditions. Langmuir. 2006;22:2156–2160. doi: 10.1021/la052564s. [DOI] [PubMed] [Google Scholar]
- 10.Barlow SM, et al. Supramolecular assembly of strongly chemisorbed size-and shape-defined chiral clusters: S-and R-alanine on cu (110) Langmuir. 2004;20:7171–7176. doi: 10.1021/la049391b. [DOI] [PubMed] [Google Scholar]
- 11.Barlow SM, et al. Polymorphism in supramolecular chiral structures of R-and S-alanine on cu (1 1 0) Surf. Sci. 2005;590:243–263. [Google Scholar]
- 12.Lingenfelder M, et al. Tracking the chiral recognition of adsorbed dipeptides at the single-molecule level. Angew. Chem. Int. Ed. 2007;46:4492–4495. doi: 10.1002/anie.200700194. [DOI] [PubMed] [Google Scholar]
- 13.Eralp T, et al. Global and local expression of chirality in serine on the cu 110 surface. Langmuir. 2010;26:18841–18851. doi: 10.1021/la1036772. [DOI] [PubMed] [Google Scholar]
- 14.Reichert J, et al. l-tyrosine on ag (111): Universality of the amino acid 2d zwitterionic bonding scheme? ACS Nano. 2010;4:1218–1226. doi: 10.1021/nn901669p. [DOI] [PubMed] [Google Scholar]
- 15.Smerieri M, Vattuone L, Kravchuk T, Costa D, Savio L. (s)-glutamic acid on ag (100): Self-assembly in the nonzwitterionic form. Langmuir. 2011;27:2393–2404. doi: 10.1021/la1033993. [DOI] [PubMed] [Google Scholar]
- 16.Kalashnyk N, et al. Scanning tunneling microscopy reveals single-molecule insights into the self-assembly of amyloid fibrils. ACS Nano. 2012;6:6882–6889. doi: 10.1021/nn301708d. [DOI] [PubMed] [Google Scholar]
- 17.Seljamäe-Green RT, et al. Assembly of a chiral amino acid on an unreactive surface:(s)-proline on au (111) Langmuir. 2014;30:3495–3501. doi: 10.1021/la500336c. [DOI] [PubMed] [Google Scholar]
- 18.Abb S, Harnau L, Gutzler R, Rauschenbach S, Kern K. Two-dimensional honeycomb network through sequence-controlled self-assembly of oligopeptides. Nat. Commun. 2016;7:10335. doi: 10.1038/ncomms10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, et al. Single-molecule insights into surface-mediated homochirality in hierarchical peptide assembly. Nat. Commun. 2018;9:2711. doi: 10.1038/s41467-018-05218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Easson LH, Stedman E. Studies on the relationship between chemical constitution and physiological action: Molecular dissymmetry and physiological activity. Biochem. J. 1933;27:1257–1266. doi: 10.1042/bj0271257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogston AG. Interpretation of experiments on metabolic processes, using isotopic tracer elements. Nature. 1948;162:963. doi: 10.1038/162963b0. [DOI] [PubMed] [Google Scholar]
- 22.Booth TD, Wahnon D, Wainer IW. Is chiral recognition a three-point process? Chirality. 1997;9:96–98. [Google Scholar]
- 23.Davankov VA. The nature of chiral recognition: Is it a three-point interaction? Chirality. 1997;9:99–102. [Google Scholar]
- 24.Ahn S, Ramirez J, Grigorean G, Lebrilla CB. Chiral recognition in gas-phase cyclodextrin: Amino acid complexes—is the three point interaction still valid in the gas phase? J. Am. Soc. Mass Spectrom. 2001;12:278–287. doi: 10.1016/S1044-0305(00)00220-8. [DOI] [PubMed] [Google Scholar]
- 25.Bentley R. Diastereoisomerism, contact points, and chiral selectivity: A four-site saga. Arch. Biochem. Biophys. 2003;414:1–12. doi: 10.1016/s0003-9861(03)00169-3. [DOI] [PubMed] [Google Scholar]
- 26.Blankenburg S, Schmidt WG. Long-range chiral recognition due to substrate locking and substrate-adsorbate charge transfer. Phys. Rev. Lett. 2007;99:196107–196110. doi: 10.1103/PhysRevLett.99.196107. [DOI] [PubMed] [Google Scholar]
- 27.Xiao W, et al. Microscopic origin of chiral shape induction in achiral crystals. Nat. Chem. 2016;8:326–330. doi: 10.1038/nchem.2449. [DOI] [PubMed] [Google Scholar]
- 28.Dong L, et al. Chiral pedot-based enantioselective electrode modification material for chiral electrochemical sensing: Mechanism and model of chiral recognition. Anal. Chem. 2017;89:9695–9702. doi: 10.1021/acs.analchem.7b01095. [DOI] [PubMed] [Google Scholar]
- 29.Lu Y, et al. Homochiral mof–polymer mixed matrix membranes for efficient separation of chiral molecules. Angew. Chem. Int. Ed. 2019;58:16928–16935. doi: 10.1002/anie.201910408. [DOI] [PubMed] [Google Scholar]
- 30.Lopinski GP, Moffatt DJ, Wayner DDM, Wolkow RA. Determination of the absolute chirality of individual adsorbed molecules using the scanning tunnelling microscope. Nature. 1998;392:909–911. [Google Scholar]
- 31.Lorenzo MO, Baddeley C, Muryn C, Raval R. Extended surface chirality from supramolecular assemblies of adsorbed chiral molecules. Nature. 2000;404:376–379. doi: 10.1038/35006031. [DOI] [PubMed] [Google Scholar]
- 32.Weckesser J, De Vita A, Barth JV, Cai C, Kern K. Mesoscopic correlation of supramolecular chirality in one-dimensional hydrogen-bonded assemblies. Phys. Rev. Lett. 2001;87:096101–096104. doi: 10.1103/PhysRevLett.87.096101. [DOI] [PubMed] [Google Scholar]
- 33.Barth JV, et al. Stereochemical effects in supramolecular self-assembly at surfaces: 1-d versus 2-d enantiomorphic ordering for pvba and peba on ag (111) J. Am. Chem. Soc. 2002;124:7991–8000. doi: 10.1021/ja0256764. [DOI] [PubMed] [Google Scholar]
- 34.Xu QM, et al. Discriminating chiral molecules of (r)-ppa and (s)-ppa in aqueous solution by ecstm. Angew. Chem. Int. Ed. 2002;114:3558–3561. doi: 10.1002/1521-3773(20020916)41:18<3408::AID-ANIE3408>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Blüm MC, Ćavar E, Pivetta M, Patthey F, Schneider WD. Conservation of chirality in a hierarchical supramolecular self-assembled structure with pentagonal symmetry. Angew. Chem. Int. Ed. 2005;44:5334–5337. doi: 10.1002/anie.200501467. [DOI] [PubMed] [Google Scholar]
- 36.Sundaresan V, Abrol R. Biological chiral recognition: The substrate's perspective. Chirality. 2005;17:S30–S39. doi: 10.1002/chir.20108. [DOI] [PubMed] [Google Scholar]
- 37.Rohde D, Yan CJ, Yan HJ, Wan LJ. From a lamellar to hexagonal self-assembly of bis (4, 4′-(m, m′-di (dodecyloxy) phenyl)-2, 2′-difluoro-1, 3, 2-dioxaborin) molecules: A trans-to-cis-isomerization-induced structural transition studied with stm. Angew. Chem. Int. Ed. 2006;45:3996–4000. doi: 10.1002/anie.200600725. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, et al. Spontaneous chiral resolution in supramolecular assembly of 2, 4, 6-tris (2-pyridyl)-1, 3, 5-triazine on au (111) J. Am. Chem. Soc. 2009;131:5885–5890. doi: 10.1021/ja9001986. [DOI] [PubMed] [Google Scholar]
- 39.Klussmann M, et al. Thermodynamic control of asymmetric amplification in amino acid catalysis. Nature. 2006;441:621–623. doi: 10.1038/nature04780. [DOI] [PubMed] [Google Scholar]
- 40.Shibata T, Morioka H, Hayase T, Choji K, Soai K. Highly Enantioselective Catalytic Asymmetric Automultiplication of Chiral Pyrimidyl Alcohol. J. Am. Chem. Soc. 1996;118:471–472. [Google Scholar]
- 41.Soai K, Shibata T, Morioka H, Choji K. Asymmetric autocatalysis and amplification of enantiomeric excess of a chiral molecule. Nature. 1995;378:767–768. [Google Scholar]
- 42.Shibata T, Choji K, Hayase T, Aizu Y, Soai K. Asymmetric autocatalytic reaction of 3-quinolylalkanol with amplification of enantiomeric excess. Chem. Commun. 1996;10:1235–1236. [Google Scholar]
- 43.Kresse G, Furthmüller J. Efficiency of ab-initio total energy calculations for metals and semiconductors using a plane-wave basis set. Comp. Mater. Sci. 1996;6:15–50. doi: 10.1103/physrevb.54.11169. [DOI] [PubMed] [Google Scholar]
- 44.Kresse G, Furthmüller J. Efficient iterative schemes for ab initio total-energy calculations using a plane-wave basis set. Phys. Rev. B. 1996;54:11169–11186. doi: 10.1103/physrevb.54.11169. [DOI] [PubMed] [Google Scholar]
- 45.Blöchl PE. Projector augmented-wave method. Phys. Rev. B. 1994;50:17953–17978. doi: 10.1103/physrevb.50.17953. [DOI] [PubMed] [Google Scholar]
- 46.Kresse G, Joubert D. From ultrasoft pseudopotentials to the projector augmented-wave method. Phys. Rev. B. 1999;59:1758–1775. [Google Scholar]
- 47.Perdew JP, Burke K, Ernzerhof M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996;77:3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 48.Dion M, Rydberg H, Schröder E, Langreth DC, Van der Lundqvist BI. waals density functional for general geometries. Phys. Rev. Lett. 2004;92:246401–246404. doi: 10.1103/PhysRevLett.92.246401. [DOI] [PubMed] [Google Scholar]
- 49.Klimeš J, Bowler DR, Michaelides A. Chemical accuracy for the van der waals density functional. J. Phys. Condens. Matter. 2009;22:022201–022205. doi: 10.1088/0953-8984/22/2/022201. [DOI] [PubMed] [Google Scholar]
- 50.Klimeš J, Bowler DR, Michaelides AV, der, waals density functionals applied to solids. Phys. Rev. B. 2011;83:195131–195143. [Google Scholar]
- 51.Henkelman G, Arnaldsson A, Jónsson H. A fast and robust algorithm for bader decomposition of charge density. Comput. Mater. Sci. 2006;36:354–360. [Google Scholar]
- 52.Sanville E, Kenny SD, Smith R, Henkelman G. Improved grid-based algorithm for bader charge allocation. J. Comput. Chem. 2007;28:899–908. doi: 10.1002/jcc.20575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.