Highlights
-
•
MRI derived total 23Na concentration differs significantly in glioma subregions.
-
•
Total 23Na concentration could reflect IDH mutation status and tumor grade.
-
•
23Na MRI yields potential non-invasive biomarkers for the treatment of gliomas.
Abbreviations: AUC, area under the curve; CRT, chemoradiotherapy; GBM, glioblastoma WHO IV; Gdce, Gadolinium-contrast-enhancing regions; IDH, isocitrate dehydrogenase; IQR, interquartile range; LGG, lower-grade-glioma WHO I-III; MGMT, O6-methylguanine DNA methyltransferase; 23Na, sodium; Nawm, normal-appearing white matter; ROC, receiver operating characteristic; SD, standard deviation; SNR, signal-to-noise ratio; T, Tesla; 3D-DLCS, 3D Dictionary Learning Compressed Sensing algorithm
Keywords: Glioma, Glioblastoma, IDH mutation, Ultra-high-field MRI, Sodium imaging, Non-invasive biomarker
Abstract
Purpose
This prospective clinical trial investigated sodium (23Na) MRI at 7 Tesla (T) field strength as biomarker for tumor extent, isocitrate dehydrogenase (IDH) mutation and O6-methylguanine DNA methyltransferase (MGMT) promotor methylation in glioma patients.
Methods
28 glioma patients underwent 23Na MRI on a 7T scanner (Siemens Healthcare, Erlangen, Germany) parallel to standard 3T MRI before chemoradiation. Areas of Gadolinium-contrast enhancement (gdce), non-enhancing T2-hyperintensity (regarded as edema), necrosis, and normal-appearing white matter (nawm) were segmented on 3T MRI imaging and were co-registered with the 23Na images. The median total 23Na concentrations of all areas were compared by pairwise t-tests. Furthermore, areas of gdce and edema were merged to yield the whole tumor area without necrosis. Subsequently, the difference in median of the 23Na concentration of this whole tumor area was compared between IDH-mutated and IDH wild-type gliomas as well as MGMT methylated and MGMT not-methylated glioblastomas using Whitney-Mann U-tests. All p-values were corrected after the Bonferroni-Holm procedure.
Results
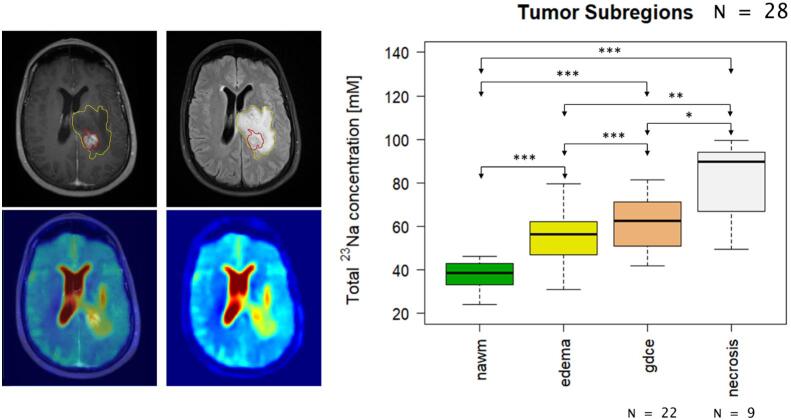
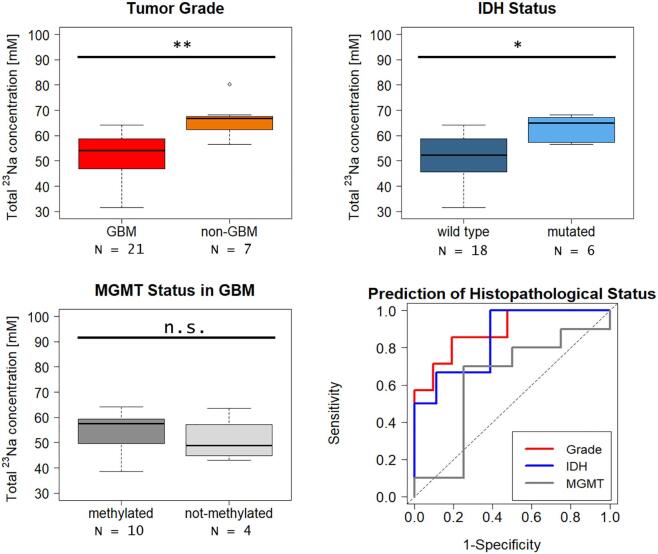
The 23Na concentration increased successively from nawm to necrotic areas (mean ± sd: nawm = 37.84 ± 5.87 mM, edema = 54.69 ± 10.64 mM, gdce = 61.72 ± 12.95 mM, necrosis = 81.88 ± 17.53 mM) and the concentrations differed statistically significantly between all regarded areas (adjusted p-values for all pairwise comparisons < 0.05). Furthermore, IDH-mutated gliomas showed significantly higher 23Na concentrations than IDH wild-type gliomas (median [interquartile range]: IDH wild-type = 52.37 mM [45.98 – 58.56 mM], IDH mutated = 65.02 mM [58.87–67.05 mM], p = 0.039). Among the glioblastomas, there was a trend towards increased 23Na concentration in MGMT methylated tumors that did not reach statistical significance (median [interquartile range]: MGMT methylated = 57.59 mM [50.70 – 59.17 mM], MGMT not methylated = 48.78 mM [45.88 – 53.91 mM], p = 1.0).
Conclusions
23Na MRI correlates with the IDH mutation status and could therefore enhance image guidance towards biopsy sites as wells as image-guided surgery and radiotherapy. Furthermore, the successive decrease of 23Na concentration from central necrosis to normal-appearing white matter suggests a correlation with tumor infiltration.
1. Introduction
Gliomas represent the most frequent primary malignant brain tumors among adults (Ostrom et al., 2017, Louis et al., 2016). Unfortunately, tumor control using multimodal therapeutic strategies is impeded by the diffuse infiltration of gliomas beyond contrast-enhancing regions on clinical MRI already at an early stage of disease (Niyazi et al., 2016, Eidel et al., 2017, Earnest et al., 1988). In general, prognosis of gliomas is unfavorable but differs greatly (Ostrom et al., 2017). The variety of individual patient prognosis strongly depends on tumor grade (Ohgaki and Kleihues, 2005) as well as genetic features such as isocitrate dehydrogenase (IDH) mutations or O6-methylguanine DNA methyltransferase (MGMT) promotor methylation (Olar et al., 2015, Hegi et al., 2005). While IDH mutation is currently regarded as the predominant prognostic factor(Olar et al., 2015), MGMT promotor status predicts the efficacy of chemotherapy and may therefore already be used to select different therapy strategies (Wick et al., 2012, Perry et al., 2017). Hence, the development of imaging methods that yield non-invasive biomarkers of tumor infiltration as well as predictors of histopathological features are highly desirable to support image-guided biopsies and image-guided therapy strategies as well as patient follow-up where serial biopsies are not feasible. During the last decade, advances of ultra-high-field (UHF) MRI techniques have been made that offer increased signal-to-noise ratios (SNR) in X-nuclei MRI (Shah et al., 2016). A promising technique in this field is sodium (23Na) imaging which has shown to correlate with the IDH mutation status in glioma patients (Biller et al., 2016, Shymanskaya et al., 2019) and has been used to monitor changes in brain tumors during radiotherapy (RT) (Huang et al., 2018, Thulborn et al., 2019). The purpose of this study was to investigate 23Na MRI at 7 Tesla (T) as a novel diagnostic tool in a prospective cohort of 28 glioma patients prior to chemoradiotherapy (CRT). We hypothesized that the quantitative 23Na signal correlates with tissue-specific tumor compartments and serves as a non-invasive predictor of tumor grade, IDH mutation and MGMT status.
2. Patients & methods
2.1. Patients
According to the declaration of Helsinki, this study has received approval by the local ethics committee. MRI examinations were performed after written informed consent was obtained from all patients. A total of 30 glioma patients underwent 23Na imaging on a 7 Tesla MRI system before CRT parallel to clinical 3T MRI and CT scans for RT planning. Inclusion criteria were age > 18 years, no ferromagnetic or active implants which are not suitable for 7T MRI, histologically proven diffuse glioma, planned (C)RT, residual tumor burden detectable on clinical MRI after biopsy or resection was performed. Two patients were excluded, one due to the diagnosis of pseudoprogression without necessity for RT after tumor board review of all available clinical data and another patient because of a displacement of the 23Na reference tubes during imaging. 14 patients presented with recurrent tumors diagnosed histopathologically following re-resection (n = 4) or radiographically according to the updated response assessment in neurooncology (RANO) criteria (n = 10). Patient characteristics are summarized in Table 1.
Table 1.
Overview of patient characteristics (N = 28). (IQR = interquartile range, N = total number, wt = wild-type, MPR = maximum possible resection).
| Age | Median [Years] | IQR [Years] |
| Median Age | 53.5 | 20.3 |
| Sex | N | [%] |
| Male | 16 | 57.1 |
| Female | 12 | 42.9 |
| Pathology | N | [%] |
| Oligodendroglioma | 4 | 14.3 |
| Astrocytoma | 3 | 10.7 |
| Glioblastoma | 21 | 75.0 |
| Grade | N | IDH mutated | IDH wt | [%] |
| I | 1| 0 | 0 | 3.6 |
| II | 3 | 3 | 0 | 10.7 |
| III | 3 | 2 | 0 | 10.7 |
| IV | 21 | 1 | 18 | 75% |
| MGMT-Methylation | N | [%] |
| methylated | 14 | 50 |
| not methylated | 4 | 14.3 |
| not available | 10 | 35.7 |
| Relapse | N | [%] |
| Newly diagnosed | 14 | 50.0 |
| Relapse | 14 | 50.0 |
| Surgery | N | [%] |
| Biopsy only | 6 | 21.4 |
| MPR | 22 | 78.6 |
2.2. Sodium MRI
23Na imaging was performed on a 7T research scanner (Siemens Healthcare, Erlangen, Germany) using a double-resonant (1H/23Na) quadrature birdcage coil (RAPID Biomedical, Rimpar, Germany). 23Na data was acquired with a density-adapted 3D radial pulse sequence (Nagel et al., 2009) with a nominal spatial resolution of (3 mm)3 (Nprojections = 4000; TR/TE = 160 ms / 0.35 ms, Tacq = 10:40 min, Treadout = 10 ms). TE was measured as the time difference between the start of the readout and the center of the rectangular 600 µs RF pulse. Image reconstruction was performed with an iterative 3D Dictionary Learning Compressed Sensing algorithm (3D-DLCS) (Behl et al., 2016) (block size B = 3 × 3 × 3; dictionary size D = 80; sample number Nsamp = 500,000; regularization weighting factor µ = 0.5).
Total 23Na concentration was obtained using two reference vials (0.3% and 0.6% NaCl). B1+ and B1− corrections were applied to cope with transmit and receive inhomogeneities. In our case we used the double angle method for the field estimations (Insko and Bolinger, 1993). Because a birdcage coil was used, the principle of reciprocity can be applied (B1+ = B1 −) (Hoult, 2000).
2.3. Clinical 3T MRI and histopathological parameters
All patients received standard of care MRI at a field strength of 3T. This included pre- and post-contrast T1-weighted imaging employing Gadolinium-based contrast agents as well as T2-weighted fluid-attenuated inversion recovery (FLAIR) imaging. Representative MRI protocols are given in supplementary table 1.
Histopathological analysis was performed as part of the clinical routine and encompassed IDH mutation status, as well as MGMT methylation status in the glioblastoma subgroup.
2.4. Post-processing
The clinical 3T MR images were co-registered to the RT planning CT scan employing an automatic multi-modal rigid algorithm in MITK (Nolden et al., 2013). Consecutively, an experienced radiologist (D.P., 7 years of experience in neuroimaging) segmented Gadolinium-contrast-enhancing regions (gdce), T2 FLAIR hyperintense non-enhancing regions and necrosis as visible on clinical 3T MRI. Necrosis encompassed all areas of fluid-isointense signal adjacent to gdce (usually located centrally) on visual inspection of T1-weighted post-contrast images. If resection had been performed previously, the corresponding areas were not treated as necrosis but as resection cavities and were not included in the statistical analysis. The T2 FLAIR hyperintense non-enhancing regions were treated as the clinical peritumoral edema. Furthermore, three adjacent image slices with tumor areas were chosen and one representative ROI was placed in contralateral normal-appearing white matter (nawm) on each slice. These ROIs were merged to yield a more robust representation of nawm. Finally, the segmentations and ROIs defined on the clinical MRI were co-registered to the 7T 23Na MR-images to extract the 23Na concentration values as well as segmentation volumes.
2.5. Qualitative assessment
To assess potential differences of the depiction of tumors between clinical 3T FLAIR MRI and 7T 23Na imaging, we performed a semiquantitative analysis of the 23Na concentration inside the peritumoral edema region. For this purpose, 23Na image window was set 2 standard deviations (SD) around the individual mean 23Na concentration in the edema (edema mean ± 2 SD) for every patient. Hotspot regions were defined inside the edema segmentation according to two criteria: 1) 23Na concentration lies above the upper frame of the window as cut-off value (quantitative) and 2) region is not adjacent to or considerably exceeds the border to the gdce region (qualitative).
2.6. Statistics
The median 23Na concentration was calculated for each segmentation and ROI. Two-sided, paired t-tests were performed pairwise between all segmentations and ROIs to search for significant differences in the median 23Na concentrations between different tissue types. Subsequently, gdce and peritumoral edema were merged resulting in the whole tumor volume segmentation without necrosis or resection cavities. This fusion should include all areas of macroscopically visible tumor infiltration but necrosis to prevent a confounding effect of the high content of cerebrospinal fluid. Whitney-Mann U-tests were used to check a possible difference in the median 23Na concentration of the whole tumor volume without necrotic areas between glioblastomas WHO IV (GBM) and non-GBM WHO I-III, IDH-mutated and IDH wild-type (wt) gliomas as well as MGMT methylated GBM versus not-methylated GBM. Additionally, receiver operating characteristic (ROC) curves were plotted to show the prediction of tumor grade, IDH mutation and MGMT methylation by the median 23Na concentration inside the whole tumor volume. The subsequent ROC analysis encompassed calculation of the area under the curve (AUC) as well as the best thresholds according to Youden’s index with corresponding sensitivity and specificity values. Moreover, 23Na concentration was compared between recurrent and newly diagnosed tumors using a two-sided, unpaired t-test. The global level of significance was set to 0.05, and all p-values were corrected according to the Bonferroni-Holm procedure. In addition, further descriptive analyses of the employed imaging techniques were conducted. Firstly, the signal intensities of the different tumor subcompartments on clinical T1- and T2-weighted imaging were normalized to the nawm signal and compared to each other by pairwise t-tests. Secondly, the correlation between segmentation volumes and median total 23Na concentrations was investigated using scatterplots and Pearson’s r for each subcompartment (intra-group analysis). Finally, segmentation volumes were compared between different tumor subcompartments by pairwise t-tests and the whole tumor volumes excluding necrosis were compared between histopathological subgroups based on Whitney-Mann U-tests (inter-group analysis). All statistical evaluation employed R version 3.6.0 and the pROC as well as the precrec package (Robin et al., 2011, Saito and Rehmsmeier, 2017).
3. Results
The comparison of the 23Na concentration inside the different tumor subregions and normal-appearing white matter (nawm) showed a successive increase from nawm towards central necrosis (23Na concentration mean ± SD: nawm = 37.84 ± 5.87 mM, edema = 54.69 ± 10.64 mM, gdce = 61.72 ± 12.95 mM, necrosis = 81.88 ± 17.53 mM). The concentration differences between all investigated tissue types were statistically significant (see Table 2 and Fig. 1). Moreover, 5 patients presented 23Na concentration hotspots inside the peritumoral edema that did not fully correlate with findings on the corresponding clinical FLAIR MRI on qualitative analysis (supplementary table 2 and supplementary Fig. 1).
Table 2.
Statistical Results: All p-values are given as raw values and after Holm-Bonferroni correction to a global α ≤ 0.05 (in brackets). The best cut-offs in the ROC analysis were based on Youden’s index. (N = total number, SD = standard deviation, nawm = normal-appearing white matter, gdce = Gadolinium-contrast enhancement, IQR = interquartile range, AUC = area under the curve, CI = confidence interval, GBM = glioblastoma, IDH = isocitrate dehydrogenase, wt = wild-type, mut = mutation, MGMT = O6-methylguanine DNA methyltransferase, met = methylated, ND = newly-diagnosed, MPR = maximum possible resection).
|
23Na-concentration [mM] in different tumor areas | |||||
|---|---|---|---|---|---|
|
Region |
Mean ± SD |
p-value for pairwise comparison t-Test |
|||
| Nawm | Edema | Gdce | Necrosis | ||
| Nawm (N = 28) | 37.84 ± 5.87 | – | 5.2 ∙10−10(5.7∙10−9) | 2.7 ∙10−9(2.7∙10−8) | 4.6 ∙10−5(3.7∙10−4) |
| Edema (N = 28) | 54.69 ± 10.64 | – | – | 6.7 ∙10−6(6.0∙10−5) | 4.3 ∙10−4(0.003) |
| Gdce (N = 22) | 61.72 ± 12.95 | – | – | – | 0.0078 (0.039) |
| Necrosis (N = 9) | 81.88 ± 17.53 | – | – | – | – |
| 23Na-concentration [mM] and histopathological parameters | |||||
| Median [IQR] | p-value | AUC (95% CI) | Best cut-off | Sensitivity / Specificity (95% CI) | |
| GBM (N = 21) | 54.18 [46.86–58.77] | 0.0012 (0.0074) | 0.89 (0.75–1) | 60.39 | 0.86 (0.57–1.00)/0.90 (0.52–1.00) |
| Non-GBM (N = 7) | 66.73 [62.39–67.66] | ||||
| IDH mut (N = 6) | 65.02 [58.87–67.05] | 0.0094 (0.0391) | 0.85 (0.67–1) | 55.78 | 1.00 (0.50–1.00)/0.83 (0.50–1.00) |
| IDH wt (N = 18) | 52.37 [45.98 – 58.56] | ||||
| MGMT met (N = 10) | 57.59 [50.70 – 59.17] | 0.5395 (1.0) | 0.63 (0.23–1) | 52.37 | 0.75 (0.25–1.00)/0.80 (0.10 – 1.00) |
| MGMT not-met (N = 4) | 48.78 [45.88 – 53.91] | ||||
| 23Na-concentration [mM] in recurrent disease | |||||
|
Mean ± SD |
p-value |
||||
| Recurrence (N = 14) | 54.56 ± 10.9 | 0.5022 (1.0) | |||
| ND (N = 14) | 57.29 ± 10.4 | ||||
| 23Na-concentration [mM] and different surgical approaches | |||||
|
Median [IQR] |
p-value |
||||
| Biopsy only (N = 6) | 58.35 [57.82 – 59.17] | 0.8916 (1.0) | |||
| MPR (N = 22) | 56.98 [47.55 – 63.18] | ||||
Fig. 1.
Total 23Na concentration inside different tumor areas. Top left: T1-weighted imaging after application of Gadolinium-based contrast agent. Top middle: T2-weighted FLAIR imaging. Bottom left: Fusion of T1-weighted and 23Na MRI. Bottom middle:23Na MRI. The different segmentations are shown on the clinical images: red = Gadolinium-contrast enhancement (gdce), yellow = peritumoral edema. In general, the signal inside the contrast-enhancing region is elevated compared to the non-enhancing peritumoral edema zone. However, there is another hotspot in the peripheral zone of the non-enhancing peritumoral edema, which might reflect an area of increased tumor infiltration. Right: the boxplots show the successive increase of 23Na signal from normal-appearing white matter (nawm) towards central necrosis (*** = p < 0.001, ** = p < 0.01, * = p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Evaluation of the 23Na concentration regarding histopathological features revealed a significantly elevated concentration in non-GBM (WHO I-III) compared to GBM (WHO IV) (23Na concentration median [IQR]: GBM = 54.18 mM [46.86 – 58.77 mM], non-GBM = 66.73 mM [62.39–67.66 mM], p = 0.0074). Correspondingly, IDH mutant gliomas showed significantly increased 23Na concentration as compared to IDH wild-type tumors (23Na concentration median [IQR]: IDH wild type = 52.37 mM [45.98 – 58.56 mM], IDH mutated = 65.02 mM [58.87–67.05 mM], p = 0.0391). Hence, when performing ROC analysis, 23Na MRI was able to predict tumor grade (AUC = 0.89 [95% CI: 0.75 – 1], sensitivity 85.7%, specificity 90.5%) and IDH mutation (AUC = 0.85 [95% CI: 0.67 – 1], sensitivity 100%, specificity 78%).
In the GBM subgroup, MGMT methylated tumors showed a non-significant trend towards increased 23Na concentrations (23Na concentration median [IQR]: MGMT methylated = 57.59 mM [50.70 – 59.17 mM], MGMT not methylated = 48.78 mM [45.88 – 53.91 mM], p = 1.0) and 23Na MRI did not yield a statistically significant predictor of MGMT status in the consecutive ROC analysis (AUC = 0.63 [95% CI: 0.23 – 1]). Fig. 2 yields an overview of the prediction of different histopathological parameters by 23Na concentration. Supplementary Fig. 2 shows corresponding precision recall curves.
Fig. 2.
Total 23Na concentration for different grades and IDH mutation status. Comparison of 23Na concentrations for different histopathological grade (top left), IDH-status (top right) and MGMT-status (bottom left) with the corresponding ROC curves (bottom right). 23Na concentration differs significantly between glioblastoma (GBM, WHO IV) and non-GBM (WHO I-III) with higher values inside non-GBM. Accordingly, significantly higher concentrations can be observed in IDH mutated gliomas. MGMT methylated GBM show a trend towards higher 23Na concentration which does not reach statistical significance. The corresponding ROC curves show both high and statistically significant AUC values for prediction of tumor grade (AUC = 0.89) and IDH mutation (AUC = 0.85), but not for MGMT status (AUC = 0.63). (** = p < 0.01, * = p < 0.05, n.s. = not statistically significant).
Newly diagnosed and recurrent tumors did not differ significantly in their 23Na signal (23Na signal mean ± sd: Recurrent disease = 54.56 ± 10.9 mM, newly diagnosed = 57.29 ± 10.4 mM, p = 1.0). Table 2 summarizes all results of the main statistical analyses.
Supplementary figure 3 and supplementary table 3 give an overview of the normalized signal intensities in different tumor subcompartments on clinical MRI. In contrast to 23Na imaging, T1- and T2-weighted sequences could not show a continuous, statistically significant signal change from the center to the peripheral parts of the tumor.
In the additional volumetric analysis, we could not find significant correlations between the volumes and median total 23Na concentrations inside different tumor subcompartments (Supplementary Figure 4). The volumes of all subcompartments differed significantly from each other, with peritumoral edema > gdce > necrosis. Furthermore, non-GBM and IDH mutated gliomas showed significantly decreased tumor volumes (Supplementary Figure 5).
4. Discussion
In this hypothesis-generating trial, we showed that that the quantitative total 23Na signal correlates with tissue-specific tumor compartments and serves as a non-invasive predictor of tumor grade and IDH mutation. No statistically significant concentration differences were found with respect to MGMT promotor methylation.
4.1. Origins of the elevated 23Na signal
An elevation of the total 23Na signal inside gliomas is generally well known (Turski et al., 1987, Hashimoto et al., 1991, Ouwerkerk et al., 2003) and could be explained by the increased intracellular 23Na content due to malignant growth (Cameron et al., 1980, Zhu et al., 2016, Rotin et al., 1989) as well as the elevated extracellular volumes in gliomas (Zamecnik et al., 2004, Bruehlmeier et al., 2003, Bakay, 1970). Since the intracellular contribution to the total 23Na concentration is relatively small, some authors argue that the elevated 23Na signal predominantly reflects an increase in extracellular volume (Thulborn, 2016), whereas others consider both intra- and extracellular changes equally important (Ouwerkerk et al., 2003). The absolute 23Na concentrations measured in this study are in good agreement with recent studies at 3T and 4T (Shymanskaya et al., 2019; Thulborn et al., 2019), but almost twofold lower than in an earlier study at 1.5T (Ouwerkerk et al., 2003).
4.2. Tumor extent
The observed gradual increase of 23Na concentration from nawm towards the central necrotic subregion of the tumor is supported by previous works which found stronger elevation of 23Na concentration in gdce as compared to edema without reaching statistical significance (Ouwerkerk et al., 2003, Haneder et al., 2015). This might reflect a tumor infiltration reaching from the border of central necrosis towards the nawm, as previously suggested (Thulborn et al., 2019). Consequently, 23Na MRI might yield a quantitative measure of tumor infiltration and thus add more sophisticated information to clinical MRI in treatment planning. However, different biological explanations for elevated 23Na concentration than tumor cell infiltration need to be considered: Firstly, non-infiltrative brain edema also leads to higher total 23Na levels (Hashimoto et al., 1991, Turski et al., 1986), which might interfere with increased 23Na concentration in infiltrated T2 FLAIR hyperintense regions. Similarly, a disruption of the blood brain barrier with consecutively larger extracellular volumes causes elevated 23Na concentrations (Turski et al., 1986), which is an alternative explanation to the high 23Na levels especially inside contrast-enhancing regions. Ultimately, previous histopathological correlations have suggested that contrast-enhancing regions contain significantly more necrotic areas than non-enhancing regions (Eidel et al., 2017), thus higher 23Na concentration inside enhancing regions could also reflect increased amounts of extracellular fluid collections. From a technical perspective, we found that the volumes of all subcompartments differed significantly from each other, which might have affected the measured 23Na concentrations. Especially small segmentation volumes could potentially yield a biased estimate of 23Na concentration due to partial volume effects (Niesporek et al., 2015, Stobbe and Beaulieu, 2018). However, no significant intra-group correlations were found between the segmentation volume and 23Na signal for any subcompartment. Finally, we found several patients with hotspots of 23Na concentration inside the tumor edema that did not fully correlate with findings on clinical FLAIR MRI on qualitative assessment. The significance of those hotspots remains unclear, but they could represent zones of increased tumor infiltration. Since a possible correlation of ultra-high-field 23Na MRI and tumor infiltration could strongly support image-based treatment planning, this hypothesis should be investigated in forthcoming prospective studies.
4.3. Tumor Grade, IDH mutation, and MGMT status
Our findings of significantly increased 23Na concentrations in non-GBM (WHO I-III) and correspondingly IDH-mutated gliomas are supported by earlier studies. Those studies investigated the ratio between total 23Na signal and intracellular 23Na contributions and found a low intracellular to total ratio as predictor of IDH mutation and even progression free survival (Biller et al., 2016, Nagel et al., 2011). The low intracellular to total 23Na signal ratio is in line with the elevated total 23Na signal found in our study. Furthermore, a recent study at 4T in a smaller patient cohort (n = 11) (Shymanskaya et al., 2019) found significantly increased 23Na levels inside IDH-mutated gliomas. The higher 23Na concentration in lower-grade gliomas is somewhat counter-intuitive from a biological standpoint because earlier studies suggested a positive correlation between proliferation rate and intracellular 23Na levels in various tumor cell lines (Cameron et al., 1980, Zhu et al., 2016, Nagy et al., 1983). However, the intracellular 23Na concentration might play a minor role in the constitution of the total 23Na signal (Thulborn, 2016). One possible explanation for the elevated 23Na signal in non-GBM could be that not only the volume, but also the matrix of the extracellular spaces differs between low- and high-grade gliomas (Zamecnik et al., 2004). This could also influence 23Na as the most important counter ion. Moreover, we found that non-GBM and correspondingly IDH mutated gliomas showed significantly decreased tumor volumes. As was already discussed for the tumor subcompartments, a volumetric difference might have affected the measured 23Na concentration.
We could not find a statistically significant difference of 23Na concentration between MGTM-methylated and MGMT-non-methylated GBM, but the MGMT methylated tumors trended towards increased 23Na values. Even though this finding was based on the analysis of a small subgroup (n = 14), it might further support the hypothesis that higher total 23Na concentrations reflect a more favorable tumor biology and, therefore, merits further investigation in larger patient cohorts.
4.4. Limitations
Our study has several limitations that need to be acknowledged.
-
1.
The heterogeneous patient cohort consisting of recurrent and newly diagnosed tumors is a limitation. However, we additionally compared recurrent to newly diagnosed tumors which did not yield a significant difference of 23Na signal among the two groups. This renders an influence of tumor relapse on the 23Na concentrations unlikely.
-
2.
Regarding the relatively small and imbalanced patient cohort, the predictive value of 23Na MRI for tumor grade and IDH mutation found in this study needs validation in future trials with higher patient numbers. Additional precision-recall analysis hinted towards a somewhat decreased precision due to imbalanced groups, but still suggested 23Na concentration as good-fair predictor of tumor grade and IDH mutation.
-
3.
Partial volume effects originating from both, the large voxel sizes and the broad shapes of the point spread function of 23Na MRI, were not corrected in this study. Concentration values, especially for small regions, might be affected by surrounding tissues (Niesporek et al., 2015, Stobbe and Beaulieu, 2018).
-
4.
Finally, 7-Tesla MRI scanners, which are especially advantageous for X-nuclei imaging, are not part of the clinical routine in most hospitals. Yet, their availability is steadily increasing since vendors recently released scanners for clinical use, including 23Na imaging.
5. Conclusion
23Na MRI correlates with the IDH mutation status and could therefore enhance image guidance towards biopsy sites as well as image-guided surgery and radiotherapy. Furthermore, the successive decrease of 23Na concentration from central necrosis to normal-appearing white matter suggests a correlation with tumor infiltration.
Funding
S. R. is funded by the Physician-Scientist Programme of Heidelberg University, Faculty of Medicine.
Disclosure statement
N.G.R.B. is currently employee of Siemens Healthcare in Erlangen, Germany.
CRediT authorship contribution statement
Sebastian Regnery: Conceptualization, Validation, Formal analysis, Investigation, Visualization, Data curation, Writing - original draft, Writing - review & editing. Nicolas G.R. Behl: Methodology, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Tanja Platt: Methodology, Software, Validation, Writing - original draft, Writing - review & editing. Nina Weinfurtner: Investigation, Writing - original draft. Paul Windisch: Investigation, Writing - original draft. Katerina Deike-Hofmann: Investigation, Writing - original draft. Felix Sahm: Data curation, Writing - original draft. Martin Bendszus: Resources, Writing - original draft, Writing - review & editing. Jürgen Debus: Resources, Writing - original draft, Writing - review & editing. Mark E. Ladd: Methodology, Resources, Writing - original draft, Writing - review & editing. Heinz-Peter Schlemmer: Supervision, Resources, Writing - original draft, Writing - review & editing. Stefan Rieken: Writing - original draft, Writing - review & editing. Sebastian Adeberg: Conceptualization, Supervision, Writing - original draft. Daniel Paech: Conceptualization, Validation, Formal analysis, Investigation, Supervision, Project administration, Data curation, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2020.102427.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bakay, L., 1970. The extracellular space in brain tumours. I. Morphological considerations. Brain : J. Neurol. 93(4), 693–698. Epub 1970/01/01. doi: 10.1093/brain/93.4.693. PubMed PMID: 4321422. [DOI] [PubMed]
- Behl, N.G., Gnahm, C., Bachert, P., Ladd, M.E., Nagel, A.M., 2016. Three-dimensional dictionary-learning reconstruction of (23)Na MRI data. Magn Reson Med. 75(4), 1605–1616. Epub 2015/05/21. doi: 10.1002/mrm.25759. PubMed PMID: 25989746. [DOI] [PubMed]
- Biller, A., Badde, S., Nagel, A., Neumann, J.O., Wick, W., Hertenstein, A., et al., 2016. Improved Brain Tumor Classification by Sodium MR Imaging: Prediction of IDH Mutation Status and Tumor Progression. AJNR Am. J. Neuroradiol. 37(1), 66–73. Epub 2015/10/24. doi: 10.3174/ajnr.A4493. PubMed PMID: 26494691. [DOI] [PMC free article] [PubMed]
- Bruehlmeier M., Roelcke U., Blauenstein P., Missimer J., Schubiger P.A., Locher J.T. Measurement of the extracellular space in brain tumors using 76Br-bromide and PET. J. Nuclear Medi.: Official Publication, Soc. Nucl. Med. 2003;44(8):1210–1218. Epub 2003/08/07 PubMed PMID: 12902409. [PubMed] [Google Scholar]
- Cameron I.L., Smith N.K., Pool T.B., Sparks R.L. Intracellular concentration of sodium and other elements as related to mitogenesis and oncogenesis in vivo. Cancer Res. 1980;40(5):1493–1500. Epub 1980/05/01 PubMed PMID: 7370987. [PubMed] [Google Scholar]
- Earnest, F.t., Kelly, P.J., Scheithauer, B.W., Kall, B.A., Cascino, T.L., Ehman, R.L., et al., 1988. Cerebral astrocytomas: histopathologic correlation of MR and CT contrast enhancement with stereotactic biopsy. Radiology. 166(3), 823–827. Epub 1988/03/01. doi: 10.1148/radiology.166.3.2829270. PubMed PMID: 2829270. [DOI] [PubMed]
- Eidel, O., Burth, S., Neumann, J.O., Kieslich, P.J., Sahm, F., Jungk, C., et al., 2017. Tumor Infiltration in Enhancing and Non-Enhancing Parts of Glioblastoma: A Correlation with Histopathology. PLoS One. 12(1), e0169292. Epub 2017/01/20. doi: 10.1371/journal.pone.0169292. PubMed PMID: 28103256; PubMed Central PMCID: PMCPMC5245878. [DOI] [PMC free article] [PubMed]
- Haneder, S., Giordano, F.A., Konstandin, S., Brehmer, S., Buesing, K.A., Schmiedek, P., et al., 2015. (2)(3)Na-MRI of recurrent glioblastoma multiforme after intraoperative radiotherapy: technical note. Neuroradiology. 57(3), 321–326. Epub 2014/11/28. doi: 10.1007/s00234-014-1468-2. PubMed PMID: 25428586. [DOI] [PubMed]
- Hashimoto T., Ikehira H., Fukuda H., Yamaura A., Watanabe O., Tateno Y. In vivo sodium-23 MRI in brain tumors: evaluation of preliminary clinical experience. Am. J. Physiologic Imaging. 1991;6(2):74–80. Epub 1991/01/01 PubMed PMID: 1867865. [PubMed] [Google Scholar]
- Hegi, M.E., Diserens, A.C., Gorlia, T., Hamou, M.F., de Tribolet, N., Weller, M., et al., 2005. MGMT gene silencing and benefit from temozolomide in glioblastoma. The New England journal of medicine. 352(10), 997–1003. Epub 2005/03/11. doi: 10.1056/NEJMoa043331. PubMed PMID: 15758010. [DOI] [PubMed]
- Hoult D.I. The principle of reciprocity in signal strength calculations—a mathematical guide. Concepts Magnetic Resonance. 2000;12(4):173–187. doi: 10.1002/1099-0534(2000)12:4<173::AID-CMR1>3.0.CO;2-Q. [DOI] [Google Scholar]
- Huang, L., Zhang, Z., Qu, B., Cui, Z., Wang, Y., Li, J., et al., 2018. Imaging of Sodium MRI for Therapy Evaluation of Brain Metastase with Cyberknife at 7T: A Case Report. Cureus. 10(4), e2502. Epub 2018/06/22. doi: 10.7759/cureus.2502. PubMed PMID: 29928562; PubMed Central PMCID: PMCPMC6005396. [DOI] [PMC free article] [PubMed]
- Insko E.K., Bolinger L. Mapping of the Radiofrequency Field. J. Magn. Reson., Ser A. 1993;103(1):82–85. doi: 10.1006/jmra.1993.1133. [DOI] [Google Scholar]
- Louis, D.N., Perry, A., Reifenberger, G., von Deimling, A., Figarella-Branger, D., Cavenee, W.K., et al., 2016. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica. 131(6), 803–820. Epub 2016/05/10. doi: 10.1007/s00401-016-1545-1. PubMed PMID: 27157931. [DOI] [PubMed]
- Nagel, A.M., Laun, F.B., Weber, M.A., Matthies, C., Semmler, W., Schad, L.R., 2009. Sodium MRI using a density-adapted 3D radial acquisition technique. Magn. Reson. Med. 62(6), 1565–1573. Epub 2009/10/28. doi: 10.1002/mrm.22157. PubMed PMID: 19859915. [DOI] [PubMed]
- Nagel, A.M., Bock, M., Hartmann, C., Gerigk, L., Neumann, J.O., Weber, M.A., et al., 2011. The potential of relaxation-weighted sodium magnetic resonance imaging as demonstrated on brain tumors. Investigative Radiol. 46(9), 539–547. Epub 2011/05/18. doi: 10.1097/RLI.0b013e31821ae918. PubMed PMID: 21577129. [DOI] [PubMed]
- Nagy I., Lustyik G., Lukacs G., Nagy V., Balazs G. Correlation of malignancy with the intracellular Na+:K+ ratio in human thyroid tumors. Cancer Res. 1983;43(11):5395–5402. Epub 1983/11/01 PubMed PMID: 6616471. [PubMed] [Google Scholar]
- Niesporek S.C., Hoffmann S.H., Berger M.C., Benkhedah N., Kujawa A., Bachert P. Partial volume correction for in vivo (23)Na-MRI data of the human brain. Neuroimage. 2015;112:353–363. doi: 10.1016/j.neuroimage.2015.03.025. Epub 2015/03/24. PubMed PMID: 25797831. [DOI] [PubMed] [Google Scholar]
- Niyazi M., Brada M., Chalmers A.J., Combs S.E., Erridge S.C., Fiorentino A. ESTRO-ACROP guideline “target delineation of glioblastomas”. Radiother. Oncol. 2016;118(1):35–42. doi: 10.1016/j.radonc.2015.12.003. PubMed PMID: 26777122. [DOI] [PubMed] [Google Scholar]
- Nolden, M., Zelzer, S., Seitel, A., Wald, D., Muller, M., Franz, A.M., et al., 2013. The Medical Imaging Interaction Toolkit: challenges and advances : 10 years of open-source development. International journal of computer assisted radiology and surgery. 8(4), 607-Epub 2013/04/17. doi: 10.1007/s11548-013-0840-8. PubMed PMID: 23588509. [DOI] [PubMed]
- Ohgaki H., Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J. Neuropathol. Exp. Neurol. 2005;64(6):479–489. doi: 10.1093/jnen/64.6.479. Epub 2005/06/28 PubMed PMID: 15977639. [DOI] [PubMed] [Google Scholar]
- Olar, A., Wani, K.M., Alfaro-Munoz, K.D., Heathcock, L.E., van Thuijl, H.F., Gilbert, M.R., et al., 2015. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta neuropathologica. 129(4), 585–596. Epub 2015/02/24. doi: 10.1007/s00401-015-1398-z. PubMed PMID: 25701198; PubMed Central PMCID: PMCPMC4369189. [DOI] [PMC free article] [PubMed]
- Ostrom, Q.T., Gittleman, H., Liao, P., Vecchione-Koval, T., Wolinsky, Y., Kruchko, C., et al., 2017. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro Oncol. 19(suppl_5), v1–v88. Epub 2017/11/09. doi: 10.1093/neuonc/nox158. PubMed PMID: 29117289. [DOI] [PMC free article] [PubMed]
- Ouwerkerk, R., Bleich, K.B., Gillen, J.S., Pomper, M.G., Bottomley, P.A., 2003. Tissue sodium concentration in human brain tumors as measured with 23Na MR imaging. Radiology. 227(2), 529–537. Epub 2003/03/29. doi: 10.1148/radiol.2272020483. PubMed PMID: 12663825. [DOI] [PubMed]
- Perry, J.R., Laperriere, N., O'Callaghan, C.J., Brandes, A.A., Menten, J., Phillips, C., et al., 2017. Short-Course Radiation plus Temozolomide in Elderly Patients with Glioblastoma. New England J. Med. 376(11), 1027–1037. Epub 2017/03/16. doi: 10.1056/NEJMoa1611977. PubMed PMID: 28296618. [DOI] [PubMed]
- Robin, X., Turck, N., Hainard, A., Tiberti, N., Lisacek, F., Sanchez, J.C., et al., 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics. 12, 77. Epub 2011/03/19. doi: 10.1186/1471-2105-12-77. PubMed PMID: 21414208; PubMed Central PMCID: PMCPMC3068975. [DOI] [PMC free article] [PubMed]
- Rotin D., Steele-Norwood D., Grinstein S., Tannock I. Requirement of the Na+/H+ exchanger for tumor growth. Cancer Res. 1989;49(1):205–211. Epub 1989/01/01 PubMed PMID: 2535690. [PubMed] [Google Scholar]
- Saito, T., Rehmsmeier, M., 2017. Precrec: fast and accurate precision-recall and ROC curve calculations in R. Bioinformatics (Oxford, England). 33(1), 145–147. Epub 2016/09/04. doi: 10.1093/bioinformatics/btw570. PubMed PMID: 27591081; PubMed Central PMCID: PMCPMC5408773. [DOI] [PMC free article] [PubMed]
- Shah, N.J., Worthoff, W.A., Langen, K.J., 2016. Imaging of sodium in the brain: a brief review. NMR Biomed. 29(2), 162–174. Epub 2015/10/10. doi: 10.1002/nbm.3389. PubMed PMID: 26451752. [DOI] [PubMed]
- Shymanskaya, A., Worthoff, W.A., Stoffels, G., Lindemeyer, J., Neumaier, B., Lohmann, P., et al., 2019. Comparison of [(18)F]Fluoroethyltyrosine PET and Sodium MRI in Cerebral Gliomas: a Pilot Study. Mol. Imaging Biol. Epub 2019/04/17. doi: 10.1007/s11307-019-01349-y. PubMed PMID: 30989437. [DOI] [PubMed]
- Stobbe, R.W., Beaulieu, C., 2018. Calculating potential error in sodium MRI with respect to the analysis of small objects. Magn. Reson. Med. 79(6), 2968–2977. Epub 2017/10/13. doi: 10.1002/mrm.26962. PubMed PMID: 29023969. [DOI] [PubMed]
- Thulborn, K.R., Lu, A., Atkinson, I.C., Pauliah, M., Beal, K., Chan, T.A., et al., 2019. Residual Tumor Volume, Cell Volume Fraction, and Tumor Cell Kill During Fractionated Chemoradiation Therapy of Human Glioblastoma using Quantitative Sodium MR Imaging. Clinical Cancer Res. Off. J. Am. Assoc. Cancer Res. 25(4), 1226–1232. Epub 2018/11/30. doi: 10.1158/1078-0432.ccr-18-2079. PubMed PMID: 30487127. [DOI] [PMC free article] [PubMed]
- Thulborn, K.R., 2016. Quantitative sodium MR imaging: A review of its evolving role in medicine. Neuroimage. Epub 2016/11/29. doi: 10.1016/j.neuroimage.2016.11.056. PubMed PMID: 27890804; PubMed Central PMCID: PMCPMC5443706. [DOI] [PMC free article] [PubMed]
- Turski, P.A., Perman, W.H., Hald, J.K., Houston, L.W., Strother, C.M., Sackett, J.F., 1986. Clinical and experimental vasogenic edema: in vivo sodium MR imaging. Work in progress. Radiology. 160(3), 821–825. Epub 1986/09/01. doi: 10.1148/radiology.160.3.3090615. PubMed PMID: 3090615. [DOI] [PubMed]
- Turski, P.A., Houston, L.W., Perman, W.H., Hald, J.K., Turski, D., Strother, C.M., et al., 1987. Experimental and human brain neoplasms: detection with in vivo sodium MR imaging. Radiology. 163(1), 245–249. Epub 1987/04/01. doi: 10.1148/radiology.163.1.3029803. PubMed PMID: 3029803. [DOI] [PubMed]
- Wick, W., Platten, M., Meisner, C., Felsberg, J., Tabatabai, G., Simon, M., et al., 2012. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 13(7), 707–715. Epub 2012/05/15. doi: 10.1016/s1470-2045(12)70164-x. PubMed PMID: 22578793. [DOI] [PubMed]
- Zamecnik, J., Vargova, L., Homola, A., Kodet, R., Sykova, E., 2004. Extracellular matrix glycoproteins and diffusion barriers in human astrocytic tumours. Neuropathol. Appl. Neurobiol. 30(4), 338–350. Epub 2004/08/13. doi: 10.1046/j.0305-1846.2003.00541.x. PubMed PMID: 15305979. [DOI] [PubMed]
- Zhu, W., Carney, K.E., Pigott, V.M., Falgoust, L.M., Clark, P.A., Kuo, J.S., et al., 2016. Glioma-mediated microglial activation promotes glioma proliferation and migration: roles of Na+/H+ exchanger isoform 1. Carcinogenesis. 37(9), 839–851. Epub 2016/06/12. doi: 10.1093/carcin/bgw068. PubMed PMID: 27287871; PubMed Central PMCID: PMCPMC5008247. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.