Abstract
While therapies targeting deficiencies in the homologous recombination (HR) pathway are emerging as the standard treatment for high grade serous ovarian cancer (HGSOC) patients, this strategy is limited to the ~50% of patients with a deficiency in this pathway. Therefore, patients with HR-proficient tumors are likely to be resistant to these therapies and require alternative strategies. We found that the HR gene Ataxia Telangiectasia Mutated (ATM) is wildtype and its activity is upregulated in HGSOC compared to normal fallopian tube tissue. Interestingly, multiple pathways related to metabolism are inversely correlated with ATM expression in HGSOC specimens, suggesting that combining ATM inhibition with metabolic drugs would be effective. Analysis of FDA-approved drugs from the Dependency Map demonstrated that ATM-low cells are more sensitive to fenofibrate, a PPARα agonist that affects multiple cellular metabolic pathways. Consistently, PPARα signaling is associated with ATM expression. We validated that combined inhibition of ATM and treatment with fenofibrate is synergistic in multiple HGSOC cell lines by inducing senescence. Together, our results suggest that metabolic changes induced by ATM inhibitors are a potential target for the treatment of HGSOC.
Keywords: Cell biology, Bioinformatics, Metabolite, Biochemistry, Molecular biology, Cancer research, PPARa, Cellular senescence, Cellular metabolism, Drug combinations, Homologous recombination
Cell biology; Bioinformatics; Metabolite; Biochemistry; Molecular biology; Cancer research; PPARa; Cellular senescence; Cellular metabolism; Drug combinations; Homologous recombination
1. Introduction
Epithelial ovarian cancer (EOC) remains the most lethal gynecological malignancy [1]. EOCs are divided into multiple subtypes with high grade serous ovarian cancer (HGSOC) as the most common. Most HGSOC patients are diagnosed at advanced stages (III-IV), and the 5-year survival rate for these patients is <30% [1]. Current standard-of-care for HGSOC is debulking surgery followed by platinum-based chemotherapy [2]. While the majority of patients initially respond to therapy, relapse with chemoresistant disease occurs in a significant number of patients [3]. Recurrent disease is treated with poly(ADP)ribose polymerase (PARP) inhibitors if the patient is BRCA mutant or previously responded to platinum-based therapy [4]. Response to platinum and PARP inhibitors occurs due to deficiencies in the DNA damage repair pathway homologous recombination (HR) [5]. Homologous recombination deficiency (HRD) occurs in ~50% of patients and includes loss-of-function mutations in multiple proteins related to HR-mediated DNA repair [4]. Unfortunately, the 50% of HGSOC patients with HR-proficient disease typically do not respond well to current therapies and have worse overall survival [6]. Therefore, identification of therapies to treat this subset of HGSOC patients is urgently needed.
Ataxia Telangiectasia Mutated (ATM) is a serine/threonine kinase that is critical for HR-mediated repair of DNA double strand breaks (DSBs) [7, 8]. Germline mutations in ATM lead to the disorder Ataxia Telangiectasia (A-T), which has a number of pathological consequences, including predisposition to cancer and metabolic dysfunction [7, 8]. Given that A-T patients have a higher risk of cancer and Atm−/− mice develop malignancies [7, 8, 9], ATM has been thought to be a tumor suppressor. However, many tumors rely on elevated DNA repair pathways, and a recent publication demonstrates that ATM is required for tumorigenesis [10]. Additionally, a previous study in HGSOC showed that patients with high nuclear ATM expression have worse survival [11]. Together, these studies suggest that ATM may be an actionable target in the subset of tumors where it is wildtype and elevated. Indeed, ATM inhibitors have been in clinical development for the past two decades [12, 13]. Multiple pre-clinical studies have indicated that ATM inhibitor monotherapy is not likely to be effective [13, 14, 15, 16, 17]. However, combined inhibition of ATM and DNA damaging agents such as PARP inhibitors and irradiation is synergistic [17], and recently a Phase I clinical trial using the ATM inhibitor AZD0156 in combination with a variety of DNA damaging agents has commenced (clinicaltrials.gov). As ATM inhibition also affects metabolic pathways [18, 19, 20], identifying targets beyond DNA damaging agents for combinatorial therapy with ATM inhibitors may therefore open up a new paradigm for treatment.
Peroxisome Proliferator Activated Receptors (PPARs: PPARα, PPARδ, PPARγ) are nuclear receptors and ligand-inducible transcription factors [21, 22]. Downstream targets of the PPARs differ greatly, likely due to dissimilarity in endogenous and exogenous ligands. Activation of PPARα leads to transcription of multiple metabolic genes, such as those related to fatty acid oxidation or inhibition of glycolysis [23]. This has been therapeutically exploited in patients with dyslipidemia by using fenofibrate or clofibrate, exogenous ligands for PPARα [24]. The role of PPARα in cancer is not fully defined, as it is tumor-promoting in rodents but not humans and tumor suppressive in a context- and cancer-type dependent manner [25]. Activation of PPARα by treating cancer cells with fenofibrate or clofibrate alone or in combination with other drugs has been shown to decrease proliferation and survival through a variety of mechanisms [26]. However, the combination of fenofibrate and ATM inhibitors has never been explored.
Here we show that ATM is wildtype and upregulated in HGSOC. Analysis of HGSOC The Cancer Genome Atlas (TCGA) datasets found that multiple metabolic pathways are associated with low ATM. Using data from Dependency Map, we identified the PPARα agonist fenofibrate as an FDA-approved drug that inhibits ATM-low cell survival to a greater extent than ATM-high cancer cells. Indeed, PPARα correlates with ATM expression in HGSOC patient specimens. Consistent with high throughput data, combined inhibition of ATM and treatment with fenofibrate is synergistic in HGSOC cells by inducing senescence. These results provide a proof-of-principle study to used combined inhibition of ATM and treatment with a metabolic drug for HGSOC therapy.
2. Results
2.1. ATM is wildtype and upregulated in HGSOC
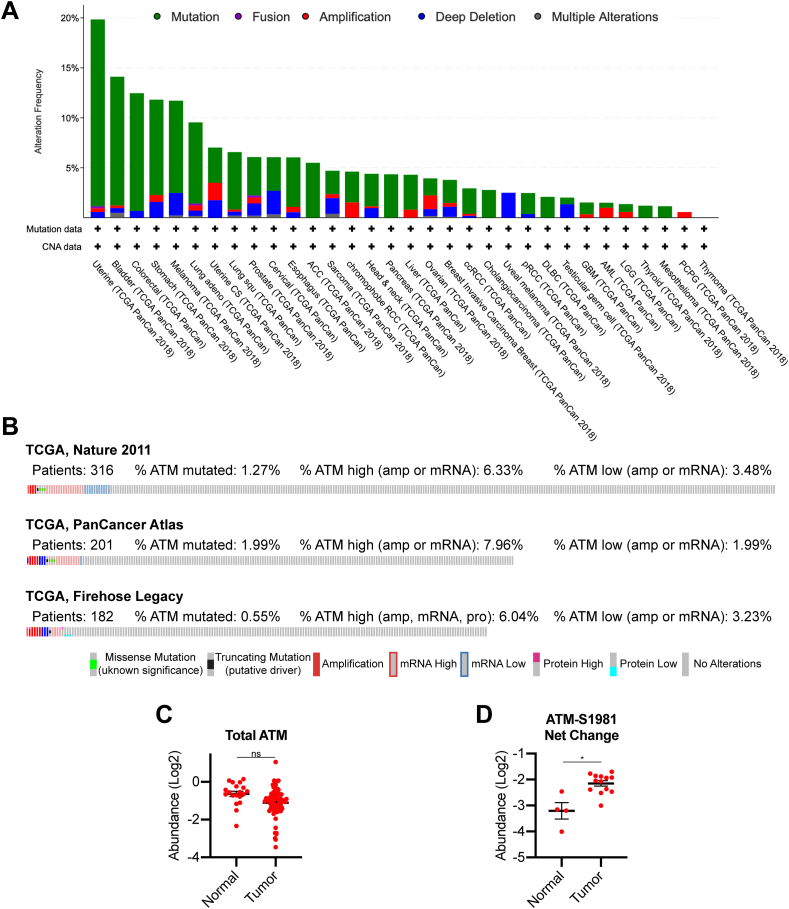
ATM is thought to be a tumor suppressor as mutations in ATM predispose patients to tumorigenesis [7, 8]. However, recent data suggest that ATM expression is required for malignant transformation in response to oncogenic stress [10], suggesting that ATM plays a context-dependent role in tumorigenesis. Analysis of The Cancer Genome Atlas (TCGA; cbioportal.com) data demonstrate that the proportion of patients with ATM mutations varies widely among different tumor types (Figure 1A). Interestingly, although HGSOC is known to have defects in HR [4, 27], ATM is only mutated in ~0.5–2% of these patients (Figure 1B). Compared to normal fallopian tube, the likely cell-of-origin for HGSOC [28, 29], HGSOC specimens do not have markedly increased ATM protein expression (Figure 1C) [30]. However, phosphoproteomics analysis demonstrates that ATM kinase activity is significantly upregulated in HGSOC compared to normal fallopian tube, as indicated by increased S1981 autophosphorylation (Figure 1D). Previous reports have shown that high ATM expression at both the protein and mRNA level is associated with worse overall and progression-free survival [11]. Together, these data suggest that ATM is wildtype and its signaling pathway is upregulated in HGSOC, indicating that it may be an actionable target for these patients.
Figure 1.
ATM is wildtype and upregulated in high grade serous ovarian cancer patients. (A) Analysis of ATM alterations in TCGA PanCancer Atlas Studies. (B) Analysis of ATM alterations in TCGA HGSOC studies. (C) Total ATM protein expression in normal fallopian tube and HGSOC specimens. ns = not significant. (D) Phosphorylated ATM (S1981) protein expression in normal fallopian tube and HGSOC specimens. ∗p < 0.001.
2.2. ATM low HGSOC tumors display a metabolic gene signature that is targetable
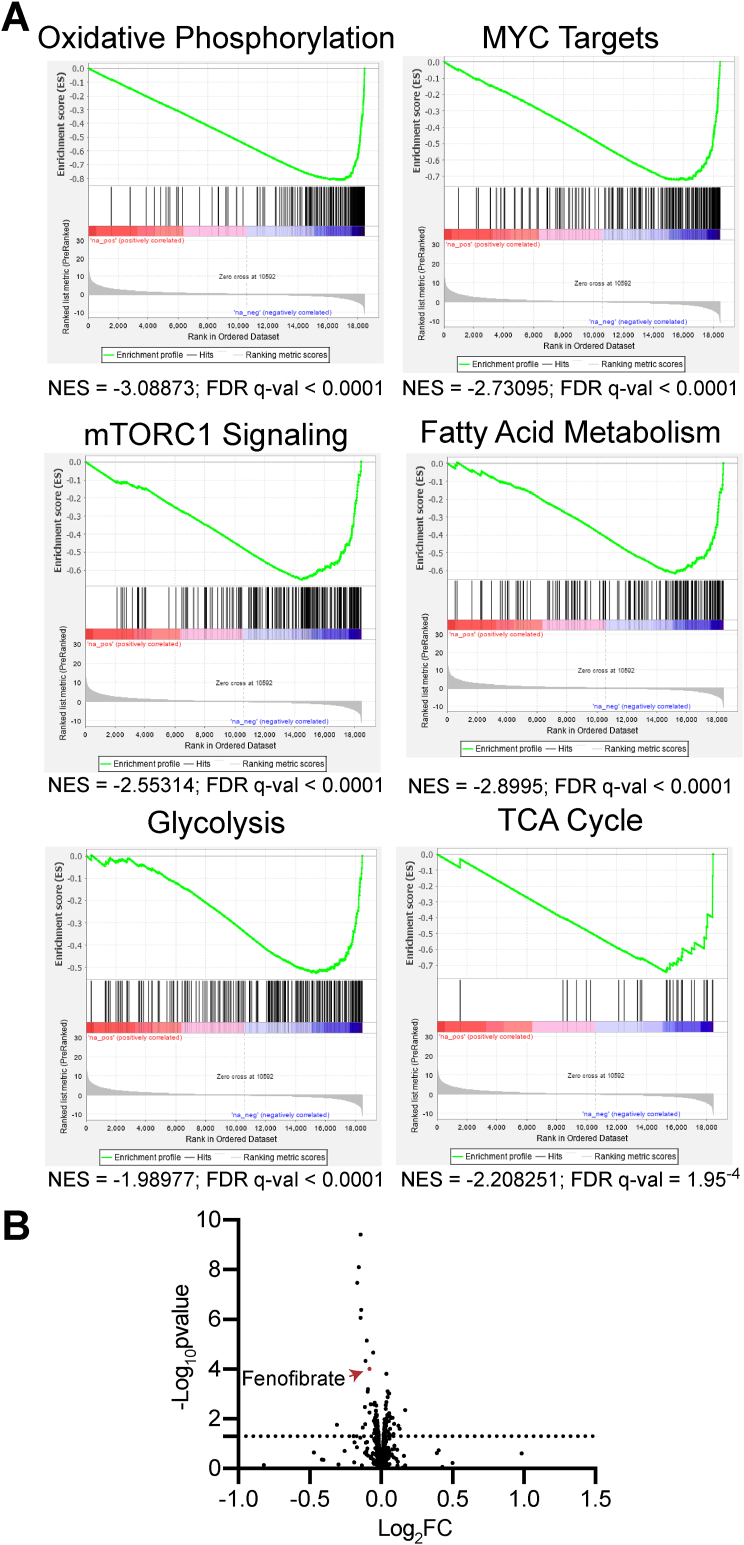
ATM may be an actionable target in HGSOC. However, many preclinical studies have demonstrated that inhibition of ATM as a single agent is not likely to be effective [13, 14, 15, 16, 17]. Combined inhibition of ATM and other therapeutic agents has shown potential to inhibit cancer cell survival both in vitro and in vivo [12, 13, 17, 31]. We reasoned that genes and pathways that are inversely correlated with ATM expression may lead to the identification of novel targets for combinatorial therapeutics. Taking HGSOC data from TCGA (cbioportal.com), we obtained the Spearman's correlation coefficient and q-value of each gene in the RNA-Seq compared to ATM mRNA expression and ran gene set enrichment analysis (GSEA). GSEA suggested that terms related to both cell cycle and DNA damage are negatively correlated with ATM expression as expected (Table S1). Interestingly, we also found that metabolic pathways such as oxidative phosphorylation, MYC signaling, mTORC1 signaling, fatty acid metabolism, glycolysis, and TCA cycle metabolism are negatively correlated with ATM expression, suggesting that targeting these pathways may be synergistic with ATM inhibitors (Figure 2A and Table S1). This is consistent with previous data demonstrating that suppression of ATM, either through mutation, small molecule inhibition, or knockdown, affects multiple metabolic processes [8, 18, 19, 20, 32, 33, 34, 35]. To further investigate potential drug combinations, we took advantage of the Dependency Map database (depmap.org) PRISM drug repurposing screen [36]. We divided the cell lines into ATM high and ATM low based on protein expression (top and bottom 50%), and looked for FDA-approved drugs that kill ATM-low cells to a greater degree (log2FC < 0; FDR<25%). We found 17 “hits” that met these criteria (Figure 2B and Table 1). Of these, the majority were inhibitors of EGFR, which have already been shown to have synergistic effects with ATM inhibitors [37]. Interestingly, fenofibrate was found as one of the hits (Figure 2B and Table 1). Fenofibrate is a PPARα agonist that has previously been shown to inhibit multiple tumor-promoting metabolic pathways including oxidative phosphorylation and glycolysis [26, 38]. These data indicate that inhibition of metabolic pathways using the FDA-approved drug fenofibrate may be a novel therapy to use in combination with ATM inhibitors.
Figure 2.
Low ATM expression correlates with increased metabolic gene signatures and sensitivity to fenofibrate. (A) Spearman's correlation in HGSOC (TCGA, PanCancer Atlas) between ATM mRNA and other gene expression values was obtained from the standard co-expression analysis performed using cBioportal, and GSEA was performed. Negative NES means enrichment of expression in HGSOC specimens with lower ATM expression. (B) Volcano plot of FDA-approved drugs in ATM-low vs. ATM-high cell lines from depmap.org (see Materials and Methods for details on how cell lines were divided by ATM protein expression). Line represents p-value<0.05.
Table 1.
FDA-approved drugs that sensitize ATM-low cells to a greater extent than ATM-high cells.
| Drug Name | Target | Log2FC (ATM-low vs. ATM-high) | FDR |
|---|---|---|---|
| lapatinib | EGFR, ERBB2 | -0.1431668 | 4.54E-07 |
| osimertinib | EGFR | -0.1575673 | 6.22E-06 |
| ibrutinib | BTK | -0.1669926 | 1.95E-05 |
| gefitinib | EGFR | -0.139171 | 0.00012 |
| afatinib | EGFR, ERBB2, ERBB4 | -0.1432169 | 0.000225 |
| icotinib | EGFR | -0.1019151 | 0.001532 |
| olmutinib | EGFR | -0.0559589 | 0.003793 |
| vandetanib | EGFR, VEGFR, RET | -0.1091835 | 0.007295 |
| fenofibrate | PPARA | -0.0821997 | 0.012972 |
| spironolactone | Various | -0.0930345 | 0.067266 |
| erlotinib | EGFR | -0.0937315 | 0.075598 |
| alfacalcidol | VDR | -0.0450776 | 0.177706 |
| bosutinib | Bcr-Abl, Src | -0.0724218 | 0.189293 |
| maxacalcitol | VDR | -0.0500369 | 0.189293 |
| paricalcitol | VDR | -0.0490078 | 0.191315 |
| loperamide | Various | -0.0341769 | 0.19634 |
| dequalinium | Various | -0.1146281 | 0.219346 |
2.3. PPARα signaling is associated with ATM expression in HGSOC
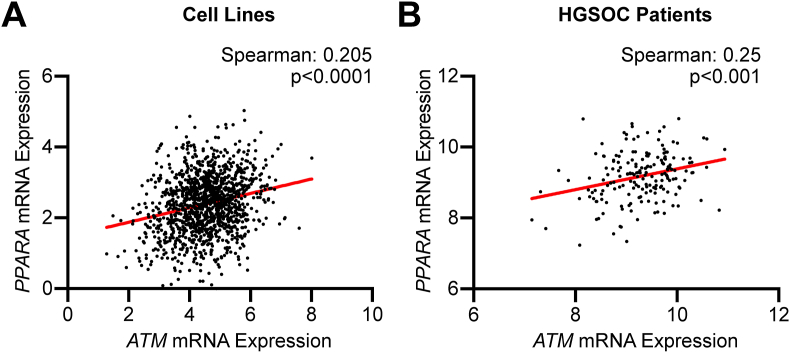
ATM-low cancer cells are slightly, although significantly more sensitive to fenofibrate (Figure 2), suggesting that these cells may have low PPARα signaling. Indeed, there is a positive correlation between ATM and PPARα in both cell lines and HGSOC patient samples (Figure 3A, B), suggesting that ATM-low cancers have low PPARα, which may be therapeutically exploited using fenofibrate.
Figure 3.
PPARα expression is associated with ATM expression in cell lines and HGSOC patient specimens. (A) Correlation between ATM and PPARA (encoding PPARα) expression in cell lines from Dependency Map. (B) Correlation between of ATM and PPARA expression in HGSOC patient specimens from TCGA.
2.4. Combination of ATM inhibitor and fenofibrate is synergistic in HGSOC cells
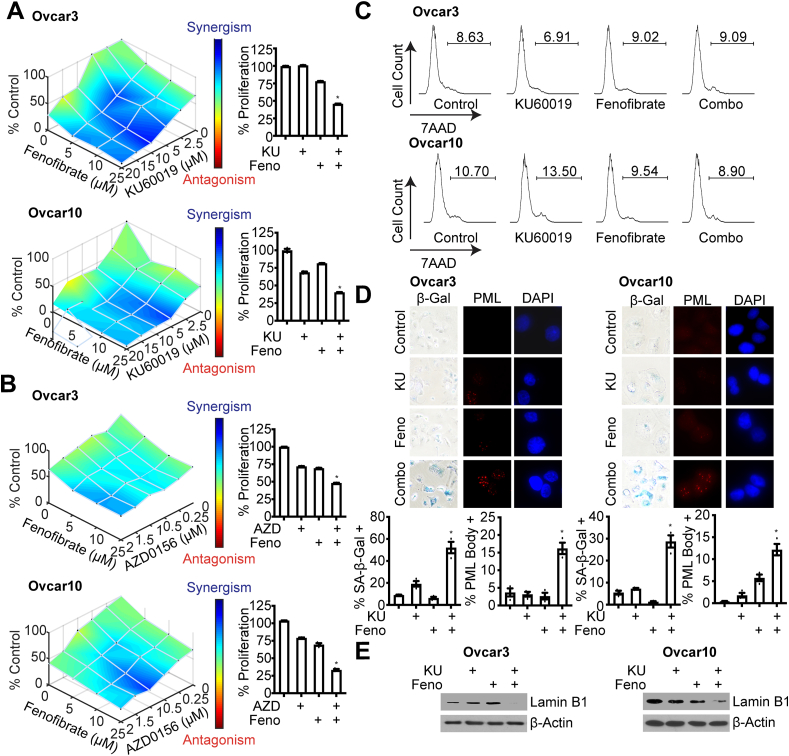
We found that ATM-low cancer cells are more sensitive to fenofibrate (Figure 2) and HGSOC cells with low ATM expression have low PPARα (Figure 3), suggesting that the combination of fenofibrate with ATM inhibitors may be synergistic in HGSOC cells with wildtype ATM. Using two independent HGSOC cells carrying wildtype ATM (cbioportal.com), we found that inhibition of ATM using two small molecule inhibitors (KU60019 and AZD0156) synergized with fenofibrate in multiple HGSOC cell lines to decrease colony formation (Figure 4A, B). While cell death, as determined by 7AAD staining, was not affected (Figure 4C), we observed multiple senescence markers, including increased SA-β-Gal and PML bodies and decreased lamin B1 expression [18, 39, 40, 41, 42, 43, 44, 45] in the combination treated cells (Figure 4D, E). Together, these data demonstrate that the combination of ATM inhibition and fenofibrate treatment is synergistic in HGSOC through induction of senescence.
Figure 4.
Combined inhibition of ATM and treatment with fenofibrate is synergistic in HGSOC cell lines. (A–B) ATM wildtype Ovcar3 and Ovcar10 cells were treated with the ATM inhibitors KU60019 (A) or AZD0156 (B) or the PPARα agonist fenofibrate alone or in combination for 3 days and colonies were stained with crystal violet. n = 3/group, one of 3 experiments is shown. ∗p < 0.001 (C) Cell death was assessed by 7AAD staining. n = 3/group, one of 3 experiments is shown. Data represent mean ± SEM. (D) SA-β-Gal and PML body staining. n = 3/group, one of 3 experiments is shown. Data represent mean ± SEM. ∗p < 0.001. (E) Immunoblot analysis of lamin B1 (uncropped images: Ovcar3: Figure S1; Ovcar10: Figure S2). β-actin was used as a loading control (uncropped images: Ovcar3: Figure S3; Ovcar10: Figure S4). One of 3 experiments is shown.
3. Discussion
Many tumors are addicted to DNA repair signaling [46], which has led to investigation of ATM inhibitors as cancer therapies [12, 13]. These inhibitors are not effective as a monotherapy, demonstrating the need to identify new targets for combinatorial therapeutic strategies. Here, we identified fenofibrate as a potential drug combination for use with ATM inhibitors. This may be due in part to upregulation of multiple metabolic pathways in ATM-low cancers. The combination induced senescence, a stable cell cycle arrest that is considered a positive patient outcome [47, 48, 49]. Together, our results provide rationale for exploration of drugs that modify metabolism as combinatorial therapies with ATM inhibitors.
ATM is an critical mediator of DNA DSB repair through HR [7, 8, 13]. Based on this, and the fact that both A-T patients and Atm knockout mice are predisposed to cancer, it has been well-appreciated that ATM is a tumor suppressor [7, 8, 9]. However, many cancer cells are addicted to DNA damage repair and ATM signaling, and a recent study demonstrated that ATM is required for breast tumorigenesis [10]. This suggests that in some contexts, ATM may act as an oncogene. Indeed, we found that HGSOC patients overwhelmingly harbor wildtype ATM alleles, and ATM kinase activity is upregulated in HGSOC samples compared to fallopian tube (Figure 1). This is consistent with another paper that demonstrated increased ATM nuclear expression in serous ovarian cancer, which was associated with worse survival [11]. Therefore, in the context of the ~50% of HGSOCs with HR-proficient disease, ATM can be considered to act more like an oncogene than tumor suppressor. This suggests that inhibition of ATM may be relevant for a large subset of HGSOC patients. As these patients often have worse survival than HR-deficient patients, identification of novel therapies is a clinical need. Indeed, ATM inhibitors, while not effective on their own, have shown promise in multiple cancer cell types in combination with irradiation or other DNA damaging agents [12, 13, 15, 17].
ATM has multiple functions outside of its role in DNA repair [8, 18, 19, 20, 50]. We found that ATM-low HGSOC specimens showed multiple terms related to metabolism, including oxidative phosphorylation, TCA cycle metabolism, fatty acid metabolism, glycolysis, and signaling related to both MYC and mTORC1 (Figure 2). This is consistent with previous reports from us and others that have shown suppression of ATM alters metabolic functions in multiple ways [8, 18, 19, 20, 32, 33, 34, 35, 51]. For instance, we found that inhibition of ATM increases consumption of multiple metabolites, including glucose, glutamine, and branched chain amino acids [18, 19, 50]. Similarly, A-T patients and A-T patient cells display multiple metabolic phenotypes, including mitochondrial dysfunction, insulin resistance, and an increased susceptibility to both diabetes and cardiovascular disease [7, 8]. Considering ATM inhibitors are not effective as a monotherapy, exploring metabolic vulnerabilities of ATM inhibited cancer cells may lead to additional combinatorial therapeutic strategies.
PPARα has not been well-studied in ovarian cancer. One study found that PPARα is expressed at a higher level in pleural effusions than in primary or metastatic tumors [52]. Interestingly, they also found PPARα to be expressed at a much lower level than either PPARδ or PPARγ, suggesting that PPARα signaling while present may be low in ovarian cancers. Consistently, multiple studies have found that PPARα agonists inhibit ovarian cancer proliferation and growth both in vitro and in vivo through a variety of mechanisms [53, 54, 55]. We also found that at the dose and timing used in this study, the PPARα agonist fenofibrate moderately inhibits HGSOC cell proliferation (Figure 4). Fenofibrate is an FDA-approved with limited toxicity, suggesting that further studies are warranted to determine whether PPARα agonists hold promise for HGSOC therapy.
Here, we found that the combination of ATM inhibition and fenofibrate is synergistic by inducing senescence (Figure 4). Senescence is a tumor suppressive mechanism due to its inhibition of cancer cell proliferation [48, 49, 56]. Recent data from HGSOC patients suggest that senescence occurs in vivo after therapy and is associated with a better outcome [47]. Indeed, other publications have also indicated that senescence is a beneficial therapeutic response [43, 48, 49, 57, 58, 59]. The mechanism of senescence induction by the combination of ATM inhibition and fenofibrate remains to be explored. We found that PPARα expression is associated with ATM expression both in HGSOC patient samples and cell lines (Figure 3), and ATM-low patients have altered metabolic pathways (Figure 2). Thus, is possible that enhanced PPARα signaling using an agonist competes with metabolic pathways that are altered in ATM-low/inhibited cells. For instance, we previously published that ATM suppression increases glucose uptake and utilization [18], whereas fenofibrate decreases this process [26]. Similarly, others have reported that Atm knockout cells display mitochondrial dysfunction [33]. This may be further exacerbated by fenofibrate, which decreases mitochondrial metabolism and oxidative phosphorylation through a variety of mechanisms [26, 38]. Regardless of the mechanism, given that fenofibrate is FDA-approved and has an excellent safety profile, our results provide a proof-of-principle study to combine fenofibrate or other PPARα agonists with ATM inhibitors. Our studies may also suggest that cancers with a high prevalence of ATM mutations (for instance melanoma) may be especially sensitive to PPARα agonists.
4. Conclusions
The present study shows that ATM is wildtype and upregulated in HGSOC, which corresponds to low PPARα expression. ATM-low cells display changes in multiple metabolic pathways that reveal a therapeutic vulnerability to use for combinatorial treatment with ATM inhibitors. The combined inhibition of ATM and treatment with the PPARα agonist fenofibrate was synergistic in HGSOC cell lines. Although further mechanistic and in vivo studies are needed to validate our bioinformatics analyses, our study provides a new potential combination therapy for HGSOCs that are HR-proficient. As multiple metabolic terms were associated with ATM-low HGSOC specimens, we predict that additional metabolic drugs may have synergistic effects with ATM inhibitors.
5. Material and methods
5.1. TCGA database and GSEA analysis
Spearman's correlation in HGSOC (TCGA, PanCancer Atlas) between ATM mRNA and other gene expression values was obtained from the standard co-expression analysis performed using cBioportal [60, 61]. Genes were ranked according to the Spearman correlation coefficient and the p-value of the correlation as follows: -log10(p value)∗sign(log2 correlation coefficient) [62]. Pre-ranked files were used to run pre-ranked GSEA (MSigDB collection KEGG and Reactome) [63] under predefined parameters. Following GSEA documentation: https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/FAQ#Why_does_GSEA_use_a_false_discovery_rate_.28FDR.29_of_0.25_rather_than_the_more_classic_0.05.3F. Terms with a q-value < 0.25 where considered significant.
5.2. Dependency Map data analysis
Raw data from the PRISM drug repurposing screen [36] and reverse phase protein array (RPPA) [36, 64] were downloaded from depmap.org. Cell lines were divided in half based on ATM protein expression from the RPPA (bottom 50%:ATM-low; top 50%: ATM-high). Only “launched” drugs were considered. Drugs were considered “hits” if log2foldchange <0 (ATM-low vs. ATM-high) and FDR <0.25. Other analyses were performed using the depmap.org online tool.
5.3. Cell lines and culture conditions
Ovcar3 and Ovcar10 HGSOC cells were cultured in RPMI-1640 (Corning, Cat# 10-040-CV) supplemented with 5% FBS. All cell lines were cultured in MycoZap and were routinely tested for mycoplasma using a highly sensitive PCR-based method [65]. Tumor cell lines were authenticated using STR Profiling using Genetica DNA Laboratories.
5.4. Colony formation
Cells (5 × 106/well in 12-well plates) were seeded in 1 mL RPMI-1640 supplemented with 5% FBS, allowed to adhere overnight, and washed twice with 1x PBS the next day. Cells were cultured with 0.5 mL serum-free RPMI-1640 for another 24 h and then treated with 2.5–20 μM KU60019 (A8336, ApexBio) or 0.25–2μM AZD0156 (B7822, ApexBio) and a combination of 5–20 μM fenofibrate (F6020-5G, Sigma) in RPMI-1640 supplemented with 0.1% FBS for 3 days. KU60019 and AZD0156 were administered 5 h prior to fenofibrate. Colony formation was visualized by fixing cells in 1% paraformaldehyde for 5 min and staining with 0.05% crystal violet for 20 min. Wells were destained for 5 min in 500 mL 10% acetic acid. Absorbance (590 nm) was measured using a spectrophotometer (Spectra Max 190). Each sample was assessed in triplicate. The synergy studies were further analyzed using Combenefit [66].
5.5. Flow cytometry
Cells (5 × 106/well in 12-well plates) were seeded in 1 mL RPMI-1640 supplemented with 5% FBS, allowed to adhere overnight, and washed twice with 1x PBS the next day. Cells were cultured cell with 0.5 mL serum-free RPMI-1640 for another 24 h. Ovcar3 cells were then treated 1 μM KU60019 and 10μM fenofibrate; Ovcar10 cells were treated 10 μM KU60019 and 25 μM fenofibrate in RPMI-1640 + 0.1% FBS for 3 days. KU60019 was administered 5 h prior to fenofibrate. Cells were harvested by trypsin and washed twice with PBS. Cells were suspended in 0.5 μg/ml 7-AAD (13-6993-T500, Tonbo Biosciences) in 1 mL staining solution [900 μL H2O + 100 μL NaCitrate (380mM)] for 15 min at room temperature. Cells were run on a 10-color FACSCanto flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (Ashland, OR).
5.6. Western blotting
Cell lysates were collected in 1X sample buffer (2% SDS, 10% glycerol, 0.01% bromophenol blue, 62.5 mM Tris, pH 6.8, 0.1 M DTT) and boiled for 10 min at 95 °C. Protein concentration was determined using the Bradford assay. Proteins were resolved using SDS-PAGE gels and transferred to nitrocellulose membranes (Fisher Scientific) (110mA for 2 h at 4 °C). Membranes were blocked with 5% nonfat milk or 4% BSA in TBS containing 0.1% Tween-20 (TBS-T) for 1 h at room temperature. Membranes were incubated overnight at 4 °C in primary antibodies (anti-lamin B1, ab16048, Abcam, 1:5000; anti-β-Actin, A1978, Sigma, 1:10,000) in 4% BSA/TBS + 0.025% sodium azide. Membranes were washed 4 times in TBS-T for 5 min at room temperature after which they were incubated with HRP-conjugated secondary antibodies (Cell Signaling, Danvers, MA) for 1 h at room temperature. After washing 4 times in TBS-T for 5 min at room temperature, proteins were visualized on film after incubation with SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher, Waltham, MA).
5.7. Immunofluorescence
Cells (5 × 106/well in 12-well plates) were seeded in 1 mL RPMI-1640 supplemented with 5% FBS overnight and washed with 1x PBS twice on next day. Cells were cultured with 0.5 mL serum-free RPMI for another 24 h Ovcar3 cells were then treated 1 μM KU60019 and 10 μM fenofibrate; Ovcar10 cells were treated 10 μM KU60019 and 25 μM fenofibrate in RPMI + 0.1% FBS for 3 days. KU60019 was administered 5 h prior to fenofibrate. Cells were fixed in 4% paraformaldehyde for 10min and permeabilized in 0.2% Triton X-100 for 5 min. Cells then were stained with PML (1:200, Santa Cruz, Cat# sc-966) in 3% BSA/PBS at room temperature for 1 h. Cells were further incubated with 0.15 μg/ml DAPI in PBS (1 min), mounted, and sealed. Cells were washed three times and then incubated in Cy3 anti-mouse secondary antibody (1:5000, Jackson ImmunoResearch Labs, Cat# 715-165-150) in 3% BSA/PBS at room temperature for 1 h. Finally, cells were incubated with 0.15 μg/ml DAPI in PBS for 1 min, washed three times with PBS, mounted, and sealed. Images were acquired at room temperature using a Nikon Eclipse 90i microscope with a 20x/0.17 objective (Nikon DIC N2 Plan Apo) equipped with a CoolSNAP Photometrics camera.
5.8. Senescence-associated beta-galactosidase staining
SA-β-Gal staining was performed as previously described [67]. Cells were fixed in 2% formaldehyde/0.2% glutaraldehyde in PBS for 5 min and stained (40 mM Na2HPO4, 150 mM NaCl, 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 1 mg/ml X-gal) overnight at 37 °C in a non-CO2 incubator. Images were acquired at room temperature using an inverted microscope (Nikon Eclipse Ts2) with a 20X/0.40 objective (Nikon LWD) equipped with a camera (Nikon DS-Fi3). Each sample was assessed in triplicate and at least 100 cells per well were counted (>300 cells per experiment).
5.9. Quantification and statistical analysis
GraphPad Prism version 8.0 was used to perform statistical analysis. One-way ANOVA or t-test were used as appropriate to determine p-values of raw data. p-values < 0.05 were considered significant.
Declarations
Author contribution statement
C. Chen: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
K. Aird: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
R. Buj: Performed the experiments; Analyzed and interpreted the data.
E. Dahl and K. Leon: Performed the experiments.
Funding statement
E. Dahl was supported by National Cancer Institute (F31CA236372). K. Leon was supported by National Cancer Institute (F31CA250366). K. Aird was supported by National Cancer Institute (R37CA240625 and R00CA194309), Congressionally Directed Medical Research Programs (W81XWH-18-1-0103), and American Cancer Society (RSG CCG 134157). R. Buj was supported by Penn State Hershey Cancer Institute.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Chi-Wei Chen, Email: chc329@pitt.edu.
Katherine M. Aird, Email: katherine.aird@pitt.edu.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Fig S1.tif.
Fig S2.tif.
Fig S3.tif.
Fig S4.tif.
References
- 1.Torre L.A. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jayson G.C. Ovarian cancer. Lancet. 2014;384(9951):1376–1388. doi: 10.1016/S0140-6736(13)62146-7. [DOI] [PubMed] [Google Scholar]
- 3.Ushijima K. Treatment for recurrent ovarian cancer-at first relapse. J. Oncol. 2010;2010:1687–8469. doi: 10.1155/2010/497429. (Electronic): p. 497429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bitler B.G. PARP inhibitors: clinical utility and possibilities of overcoming resistance. Gynecol. Oncol. 2017;147(3):695–704. doi: 10.1016/j.ygyno.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Andrea A.D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair (Amst) 2018;71:172–176. doi: 10.1016/j.dnarep.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 6.Tumiati M. A functional homologous recombination assay predicts primary chemotherapy response and long-term survival in ovarian cancer patients. Clin. Cancer Res. 2018;24(18):4482–4493. doi: 10.1158/1078-0432.CCR-17-3770. [DOI] [PubMed] [Google Scholar]
- 7.McKinnon P.J. ATM and ataxia telangiectasia. EMBO Rep. 2004;5(8):772–776. doi: 10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKinnon P.J. ATM and the molecular pathogenesis of ataxia telangiectasia. Annu. Rev. Pathol. 2012;7:303–321. doi: 10.1146/annurev-pathol-011811-132509. [DOI] [PubMed] [Google Scholar]
- 9.Barlow C. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86(1):159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 10.Liu X. ATM paradoxically promotes oncogenic transformation via transcriptional reprogramming. Cancer Res. 2020;80(8):1669–1680. doi: 10.1158/0008-5472.CAN-19-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Fatah T.M. ATM, ATR and DNA-PKcs expressions correlate to adverse clinical outcomes in epithelial ovarian cancers. BBA Clin. 2014;2:10–17. doi: 10.1016/j.bbacli.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber A.M., Ryan A.J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Jin M.H., Oh D.Y. ATM in DNA repair in cancer. Pharmacol. Ther. 2019;203:107391. doi: 10.1016/j.pharmthera.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Golding S.E. Dynamic inhibition of ATM kinase provides a strategy for glioblastoma multiforme radiosensitization and growth control. Cell Cycle. 2012;11(6):1167–1173. doi: 10.4161/cc.11.6.19576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batey M.A. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol. Cancer Therapeut. 2013;12(6):959–967. doi: 10.1158/1535-7163.MCT-12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimaki S. Blockade of ataxia telangiectasia mutated sensitizes hepatoma cell lines to sorafenib by interfering with Akt signaling. Cancer Lett. 2012;319(1):98–108. doi: 10.1016/j.canlet.2011.12.043. [DOI] [PubMed] [Google Scholar]
- 17.Riches L.C. Pharmacology of the ATM inhibitor AZD0156: potentiation of irradiation and olaparib responses preclinically. Mol. Cancer Therapeut. 2020;19(1):13–25. doi: 10.1158/1535-7163.MCT-18-1394. [DOI] [PubMed] [Google Scholar]
- 18.Aird K.M. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell Rep. 2015;11(6):893–901. doi: 10.1016/j.celrep.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C.-W. ATM inhibition drives metabolic adaptation via induction of macropinocytosis. bioRxiv. 2020 doi: 10.1083/jcb.202007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahl E.S., Aird K.M. Ataxia-Telangiectasia mutated modulation of carbon metabolism in cancer. Front. Oncol. 2017;7:291. doi: 10.3389/fonc.2017.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michalik L., Wahli W. Peroxisome proliferator-activated receptors: three isotypes for a multitude of functions. Curr. Opin. Biotechnol. 1999;10(6):564–570. doi: 10.1016/s0958-1669(99)00030-0. [DOI] [PubMed] [Google Scholar]
- 22.Berger J., Moller D.E. The mechanisms of action of PPARs. Annu. Rev. Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 23.Rakhshandehroo M. Comparative analysis of gene regulation by the transcription factor PPARalpha between mouse and human. PloS One. 2009;4(8) doi: 10.1371/journal.pone.0006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staels B., Maes M., Zambon A. Fibrates and future PPARalpha agonists in the treatment of cardiovascular disease. Nat. Clin. Pract. Cardiovasc. Med. 2008;5(9):542–553. doi: 10.1038/ncpcardio1278. [DOI] [PubMed] [Google Scholar]
- 25.Tachibana K. The role of PPARs in cancer. PPAR Res. 2008;2008:102737. doi: 10.1155/2008/102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lian X. Anticancer properties of fenofibrate: a repurposing use. J. Cancer. 2018;9(9):1527–1537. doi: 10.7150/jca.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perets R. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell. 2013;24(6):751–765. doi: 10.1016/j.ccr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurman R.J., Shih Ie M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer--shifting the paradigm. Hum. Pathol. 2011;42(7):918–931. doi: 10.1016/j.humpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDermott J.E. Proteogenomic characterization of ovarian HGSC implicates mitotic kinases, replication stress in observed chromosomal instability. Cell Rep. Med. 2020;1(1):100004. doi: 10.1016/j.xcrm.2020.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickson I. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64(24):9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 32.Halaby M.J. ATM protein kinase mediates full activation of Akt and regulates glucose transporter 4 translocation by insulin in muscle cells. Cell. Signal. 2008;20(8):1555–1563. doi: 10.1016/j.cellsig.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Valentin-Vega Y.A. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119(6):1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zakikhani M. Alterations in cellular energy metabolism associated with the antiproliferative effects of the ATM inhibitor KU-55933 and with metformin. PloS One. 2012;7(11) doi: 10.1371/journal.pone.0049513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guleria A., Chandna S. ATM kinase: much more than a DNA damage responsive protein. DNA Repair (Amst) 2016;39:1–20. doi: 10.1016/j.dnarep.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Barretina J. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misumi K. Enhanced gefitinib-induced repression of the epidermal growth factor receptor pathway by ataxia telangiectasia-mutated kinase inhibition in non-small-cell lung cancer cells. Cancer Sci. 2016;107(4):444–451. doi: 10.1111/cas.12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashton T.M. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 2018;24(11):2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 39.Buj R., Leon K.E., Aird K.M. Suppression of p16 alleviates the senescence-associated secretory phenotype. bioRxiv. 2020 doi: 10.18632/aging.202640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dahl E.S. Targeting IDH1 as a pro-senescent therapy in high-grade serous ovarian cancer. Mol. Cancer Res. 2019 doi: 10.1158/1541-7786.MCR-18-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buj R. Suppression of p16 induces mTORC1-mediated nucleotide metabolic reprogramming. Cell Rep. 2019;28(8):1971–1980. doi: 10.1016/j.celrep.2019.07.084. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aird K.M. HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J. Cell Biol. 2016:325–334. doi: 10.1083/jcb.201608026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aird K.M. Identification of ribonucleotide reductase M2 as a potential target for pro-senescence therapy in epithelial ovarian cancer. Cell Cycle. 2014;13(2):199–207. doi: 10.4161/cc.26953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aird K.M. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3(4):1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freund A. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012;23(11):2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nickoloff J.A. Drugging the cancers addicted to DNA repair. J. Natl. Cancer Inst. 2017;109(11) doi: 10.1093/jnci/djx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calvo L. Cellular senescence is a central response to cytotoxic chemotherapy in high-grade serous ovarian cancer. bioRxiv. 2018 [Google Scholar]
- 48.Ewald J.A. Therapy-induced senescence in cancer. J Natl. Cancer Inst. 2010;102(20):1536–1546. doi: 10.1093/jnci/djq364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nardella C. Pro-senescence therapy for cancer treatment. Nat. Rev. Cancer. 2011;11(7):503–511. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- 50.Aird K.M., Zhang R. ATM in senescence. Oncotarget. 2015;6(17):14729–14730. doi: 10.18632/oncotarget.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cosentino C., Grieco D., Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011;30(3):546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson B. Expression of the peroxisome proliferator-activated receptors-alpha, -beta, and -gamma in ovarian carcinoma effusions is associated with poor chemoresponse and shorter survival. Hum. Pathol. 2009;40(5):705–713. doi: 10.1016/j.humpath.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 53.Shigeto T. Peroxisome proliferator-activated receptor alpha and gamma ligands inhibit the growth of human ovarian cancer. Oncol. Rep. 2007;18(4):833–840. [PubMed] [Google Scholar]
- 54.Yokoyama Y. Clofibric acid, a peroxisome proliferator-activated receptor alpha ligand, inhibits growth of human ovarian cancer. Mol. Cancer Therapeut. 2007;6(4):1379–1386. doi: 10.1158/1535-7163.MCT-06-0722. [DOI] [PubMed] [Google Scholar]
- 55.Stebbins K.J. In vitro and in vivo pharmacology of NXT629, a novel and selective PPARalpha antagonist. Eur. J. Pharmacol. 2017;809:130–140. doi: 10.1016/j.ejphar.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 56.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120(4):513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Fatkhutdinov N. Targeting RRM2 and mutant BRAF is a novel combinatorial strategy for melanoma. Mol. Cancer Res. 2016;14(9):767–775. doi: 10.1158/1541-7786.MCR-16-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dahl E.S. Targeting IDH1 as a prosenescent therapy in high-grade serous ovarian cancer. Mol. Cancer Res. 2019;17(8):1710–1720. doi: 10.1158/1541-7786.MCR-18-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bitler B.G. Wnt5a suppresses epithelial ovarian cancer by promoting cellular senescence. Cancer Res. 2011;71(19):6184–6194. doi: 10.1158/0008-5472.CAN-11-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cerami E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao J. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buj R. Suppression of p16 induces mTORC1-mediated nucleotide metabolic reprogramming. Cell Rep. 2019;28(8):1971–1980 e8. doi: 10.1016/j.celrep.2019.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528(7580):84–87. doi: 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uphoff C.C., Drexler H.G. Detection of mycoplasma contaminations. Methods Mol. Biol. 2005;290:13–23. doi: 10.1385/1-59259-838-2:013. [DOI] [PubMed] [Google Scholar]
- 66.Di Veroli G.Y. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32(18):2866–2868. doi: 10.1093/bioinformatics/btw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dimri G.P. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.