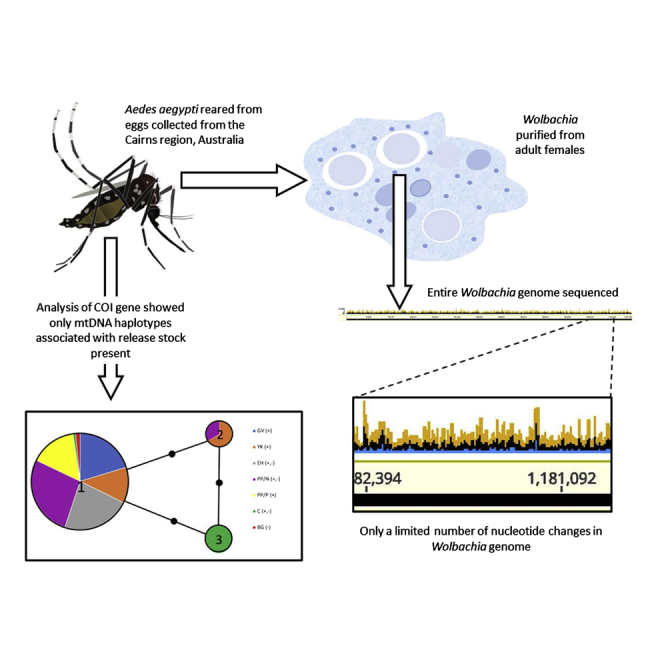
Summary
A dengue suppression strategy based on release of Aedes aegypti mosquitoes infected with the bacterium Wolbachia pipientis is being trialed in many countries. Wolbachia inhibits replication and transmission of dengue viruses. Questions remain regarding the long-term stability of virus-suppressive effects. We sequenced the Wolbachia genome and analyzed Ae. aegypti mitochondrial DNA markers isolated from mosquitoes sampled 2–8 years after releases in the greater Cairns region, Australia. Few changes were detected when Wolbachia genomes of field mosquitoes were compared with Wolbachia genomes of mosquitoes obtained soon after initial releases. Mitochondrial variants associated with the initial Wolbachia release stock are now the only variants found in release sites, highlighting maternal leakage as a possible explanation for rare Wolbachia-negative mosquitoes and not migration from non-release areas. There is no evidence of changes in the Wolbachia genome that indicate selection against its viral-suppressive effects or other phenotypes attributable to infection with the bacterium.
Subject Areas: Biological Sciences, Entomology, Parasitology, Virology, Genomics
Graphical Abstract
Highlights
-
•
Long-term effect of Wolbachia on dengue virus blocking in Aedes aegypti is unknown
-
•
There were minimal changes in Wolbachia genomes up to 8 years post-release
-
•
Mitochondrial DNA indicated rare loss of Wolbachia by maternal leakage
-
•
These results suggest the stability of the Wolbachia genome in field populations
Biological Sciences; Entomology; Parasitology; Virology; Genomics
Introduction
Record levels of dengue virus transmission, coupled with the recent global Zika virus emergency, reflect the difficulties confronting current insecticide-based control programs targeting Aedes aegypti, the primary urban vector of these viruses (Anonymous, 2019; Musso et al., 2019). An alternative strategy involving the release of mosquitoes transinfected with the Wolbachia bacterium is being trialed in a number of countries (Flores and O'Neill, 2018). This approach exploits two phenotypes that the maternally transmitted Wolbachia confers on Ae. aegypti: (1) inhibition of virus transmission and (2) cytoplasmic incompatibility (CI) to drive the bacterium into field populations of mosquitoes. Evidence from field releases with the wMel strain in north Queensland, Australia, and the wAlbB strain in Kuala Lumpur, Malaysia, suggest that Wolbachia can be established in wild populations of A. Aegypti, and this has coincided with a decrease in the incidence of local dengue virus transmission (Nazni et al., 2019; Ryan et al., 2020).
The continued success of Wolbachia-based approaches will depend on the stability of the mosquito-bacteria-virus association, and ongoing monitoring will be essential to identify changes in any of these components or their interaction (Ritchie et al., 2018). Selection pressure may assist to maintain this complex association (Ford et al., 2019). However, evolution of the Ae. aegypti or Wolbachia genome may impact the expression of the virus inhibition or CI phenotypes, effects of Wolbachia on host fitness and thus maintenance of the transinfection, and rates of maternal transfer. Changes to the virus may also potentially lead to escape from blocking effects (Bull and Turelli, 2013). There is evidence for the evolution of such components in natural Wolbachia infections, as in the case of Drosophila simulans flies evolving to overcome negative effects on fecundity due to changes in Wolbachia or other maternally inherited components (Weeks et al., 2007) and the loss of male killing in the butterfly Hypolimnas bolina due to the evolution of suppressors encoded in the host DNA (Hornett et al., 2009).
Any changes in the dynamics of Wolbachia following introduction can also be tracked through changes in mitochondrial (mt) DNA variants associated with the Wolbachia at the time of release compared with variation present in background populations (Yeap et al., 2016). This includes maternal leakage, which results in loss of Wolbachia infection but retention of mtDNA variants, associated with releases being detected in the remaining uninfected individuals in a population, versus ongoing migration, which would introduce new mtDNA variants into this remaining uninfected component. Paternal transmission or horizontal transmission of Wolbachia can also be detected by the presence of new mtDNA variants in the infected component of the population (Yeap et al., 2016).
Here we apply whole-genome sequencing and analysis to determine whether there have been changes in the Wolbachia genome in Ae. aegypti populations in Cairns (Queensland, Australia) and the surrounding suburbs, the locations of the first releases in 2011 until 2017 (Hoffmann et al., 2011). We also examine mtDNA variants associated with the infected and uninfected components of the Ae. aegypti population in locations with different invasion histories, taking advantage of the facts that the original mtDNA variants in the release stock were relatively uncommon (frequency of 8%–13%) in the natural mosquito population where they were released (Yeap et al., 2016) and that uninfected mosquitoes continue to appear in the Ae. aegypti population 10 years after the releases (Hoffmann et al., 2014; Ryan et al., 2020; Schmidt et al., 2018). Taken together, these data indicate stability of the Wolbachia genome after establishment in the local mosquito population and an ongoing low level of maternal leakage in the Wolbachia population rather than migration or paternal/horizontal transmission.
Results and Discussion
Genome Sequencing Reveals Minimal Change in Wolbachia Post-release
As a baseline sample, sequencing of the Wolbachia genome was performed on mosquitoes from a wMel-infected Ae. aegypti colony that originated from mosquitoes collected in 2013 soon after the initial releases. We ascertained that there were eight sites in the genome where there were nucleotide differences between these colony mosquitoes and the wMel reference sequence, which was derived from a Drosophila melanogaster laboratory culture (Table 1) and used in creating the original wMel Ae. aegypti line (Walker et al., 2011). These differences could be a result of changes occurring since the Wolbachia was transinfected from D. melanogaster into the Ae. aegypti line as Wolbachia can evolve in cell lines used before microinjection (Woolfit et al., 2013). Alternatively, as many of these changes disrupted potential or known reading frames and often occurred in homopolymeric runs, they were more likely sequencing errors in the reference.
Table 1.
Nucleotide Differences between Wolbachia from the Colony Mosquitoes and the wMel Reference Sequence (19)
| Base | Genome Site | Change | Type of Change | CDS Position | ORF Effect |
|---|---|---|---|---|---|
| T | 1,006,083 | +T | Insertion | 950 | Frameshift |
| T | 1,020,480 | (T)4 -> (T)5 | Insertion (tandem repeat) | ||
| T | 1,094,461 | (T)2 -> (T)3 | Insertion (tandem repeat) | 29 | Frameshift |
| A | 1,097,797 | T - > A | SNP (transversion) | ||
| - | 1,103,472 | (T)4 -> (T)3 | Deletion (tandem repeat) | 286 | Frameshift |
| T | 1,161,856 | (T)5 -> (T)6 | Insertion (tandem repeat) | 51 | Frameshift |
| – | 1,163,171 | -C | Deletion | 1146 | Frameshift |
| – | 1,177,855 | -C | Deletion | 370 | Frameshift |
Genome sequencing was then performed on field samples. Ae. aegypti were reared from eggs collected in April 2019 from 23 sites in the Cairns region encompassing six suburbs where Wolbachia-infected mosquitoes were released between 2011 and 2017 (Figure 1). Multiple oviposition trap samples were taken at most sites (Table 2). Mosquitoes were reared from collected eggs, and pools of two to five adult female Ae. aegypti were formed from progeny from each trap collection. As a control location, traps were also deployed at Caravonica, a suburb where Wolbachia-infected mosquitoes had not been deployed. However, a pool reared from one of the Caravonica traps was positive for Wolbachia infection, suggesting migration or importation from a release location, and this sample was also included in the analysis. As expected, the eight sites where there were nucleotide differences in the genome of the colony mosquitoes compared with the reference were also present in the genomes of the field-collected mosquitoes (two to five adults per sample).
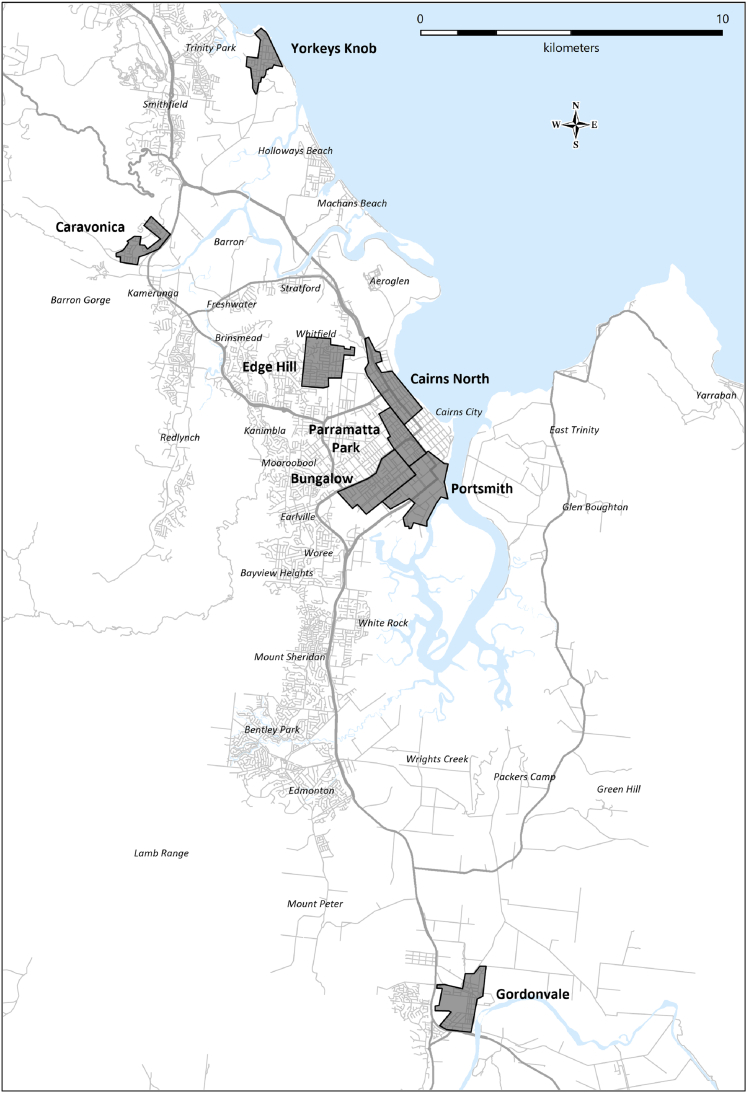
Figure 1.
Sampling Locations in the Greater Cairns Area, North Queensland
Eggs were collected in ovitraps deployed in the suburbs of Gordonvale, Yorkeys Knob, Caravonica, Edge Hill, Cairns North, Paramatta Park, Portsmith, and Bungalow. Note that there were two releases of Wolbachia-infected mosquitoes in two geographically separated zones in Paramatta Park (January 2013 and March 2017) and Cairns North (August 2014 and March 2017). Samples collected from Portsmith were only included in the mitochondrial DNA analysis.
Table 2.
Genome Polymorphisms Identified in Wolbachia Isolated from Aedes aegypti Collected from the Cairns Region, Northern Australia, in 2019
| Majority Nucleotide at 229,585 (%) |
Majority Nucleotide at 1,174,712 (%)a |
||||||
|---|---|---|---|---|---|---|---|
| Release siteb (or Colony) | Release Date | No. Mosquitoes | No. Pools | C | A | T | C |
| Colony (∼400 mosquitoes) | NA | 400 | 1 | 100 | – | 100 | - |
| Caravonica | NA | 2 | 1 | 100 | - | 100 | - |
| Gordonvale | January 2011 | 30 | 6 | 100 | - | 67 | 33 |
| Yorkeys Knob | January 2011 | 5 | 2 | 100 | - | 100 | - |
| Edge Hill | January 2013 | 25 | 5 | 80 | 20 | 80 | 20 |
| Parramatta Park | January 2013 | 8 | 2 | 100 | - | 100 | - |
| Bungalow | July 2014 | 5 | 1 | 100 | 100 | - | |
| Cairns North | August 2014 | 5 | 1 | 100 | - | - | 100 |
| Cairns North | March 2017 | 15 | 3 | 67 | 33 | - | 100 |
| Parramatta Park | March 2017 | 7 | 2 | 100 | - | 100 | - |
NA, not applicable.
Silent mutation to hypothetical ORF.
There were two release dates for different areas of Parramatta Park and North Cairns.
The main observation of importance was the limited number of changes in the Wolbachia genomes of the population sampled up to 8 years post-release. Owing to the nature of samples (i.e., pooled material), a polymorphism was defined as the detection of a nucleotide at a genome site that differed from the colony mosquitoes, and that was present in the majority of reads. There were two such polymorphisms detected. The site showing the most nucleotide change was located at genome position 1,174,712 and was a change from T to C relative to the reference sequence (Table 2). This change was silent (i.e., no amino acid change) for the predicted open reading frame (ORF) for a hypothetical gene (GeneID: 29554797). This polymorphism was present in all samples from Cairns North and was also present in Edge Hill (20% of samples) and Gordonvale (33% of samples). The former two release sites are near each other, so this polymorphism could have arisen at one of these locations and been transferred to the other through dispersal of mosquitoes. Alternatively, it could have arisen in Wolbachia from a geographically disparate release site and been incidentally introduced by human transport (Schmidt et al., 2018), or even arisen independently at both sites. When individual pooled samples were inspected for the presence of the T to C change, this change was detected in most samples, often at a lower frequency (Data S1. Wolbachia genome nucleotide variation, related to Table 2.). For all samples, the overall frequency for this change was 0–100% of reads (average 35%). Interestingly, the T to C change was also detected in the pooled colony mosquito reads at a frequency of 6%, suggesting that this polymorphism may be due to a founder effect resulting from the bottleneck produced by the mosquito release.
A second polymorphism at nucleotide site 229,585 was a C to A change relative to the reference sequence. This change would also result in a leucine to methionine change to a hypothetical 16-amino acid ORF (GeneID: 41335129). However, this is a predicted ORF whose status as a functional gene is currently unclear and biological significance is unknown. This polymorphism was present in 20% of Edge Hill samples and 33% of samples from the Cairns North sites where releases were conducted in 2017. The C to A change was present in a total of three samples with a frequency of 0–99% of reads (average 10%). The colony mosquito pooled sample had a mix of C (94%) and A (6%) at this site, similar to the other polymorphism in that the nucleotide differencing from the reference at that site (i.e., A) occurred at a low frequency. Hence, this polymorphism might also be a result of a founder effect post-release from the original mosquito release.
Nucleotide variation at two other sites on the genome was present but at lower frequencies. At genome position 184,222–184,225 (i.e., 4 nucleotides), there was a change from ACTG to TATA relative to the reference sequence (Data S1; Wolbachia genome nucleotide variation, Related to Table 2.). This change was in the 23S ribosomal RNA sequence of the Wolbachia genome. The TATA sequence was present in most samples in the minority of the reads, and it varied among individual samples with a frequency of 0–24% of reads (average 6%). The significance of this low-frequency change is unclear. A fourth change was a single-nucleotide deletion present in a minority of reads (31%) in only one of the mosquito samples and, hence, was present in at least one of the five mosquitoes in that sample (Data S1; Wolbachia genome nucleotide variation, Related to Table 2.). This would have disrupted an ORF with predicted similarity to the “RDD family” of proteins, which have recently been functionally characterized as an Na+(Li+)/H+ antiporter (Shao et al., 2018). The significance, if any, of the function of this protein to virus suppression is unclear. However, as this change was only observed at a low frequency in one sample, this is potentially a spontaneous mutation unrelated to selection pressure.
Analysis of Mosquito Mitochondrial DNA Reveals Only Rare Maternal Loss of Wolbachia
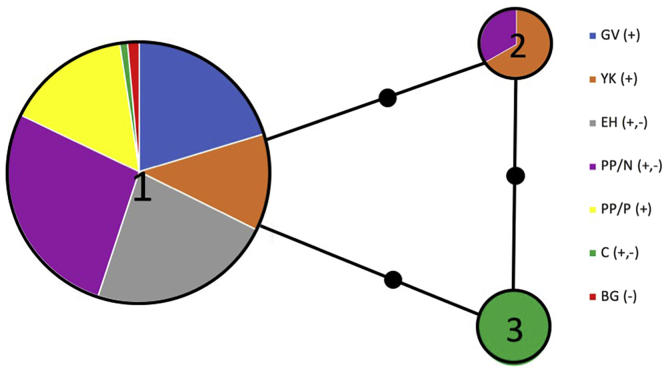
We analyzed the cytochrome oxidase I (COI) region in the mitochondria from both wMel-infected and uninfected Ae. aegypti field samples from Queensland, including the same samples as tested for Wolbachia (Table S1). A 750-bp region (trimmed from 970 bp) on the reverse strand of the COI gene was compared among locations and to the original samples described in Yeap et al. (2016). Three haplotypes were identified (Figure 2, Table S2). Mosquitoes from all locations, except for the pooled negative sample from Caravonica, had haplotype 1. This includes both Wolbachia-positive and Wolbachia-negative samples from all areas at different time periods post-release (Ryan et al., 2020). An infected mosquito from Yorkeys Knob from 2018 and one uninfected mosquito from Parramatta Park/Cairns North also from 2018 had haplotype 2 (Figure 2). This haplotype had previously been collected from one infected mosquito from Yorkeys Knob in 2011 (Yeap et al., 2016) and is likely to have been present in the original release stock. We therefore did not find many other common haplotypes originally present in the population before release (Yeap et al., 2016) in the release areas, reflecting complete replacement of the mtDNA of Ae. aegypti by the variants present in the release stock. This effectively reflects an mtDNA sweep akin to that originally observed in natural populations of Drosophila simulans following a sweep by the wRiv Wolbachia infection (Hale and Hoffmann, 1990; Turelli et al., 1992). Note that we also used the genomic data to examine other variations in the mtDNA across samples, but only found one change in the ND5 gene region in one sample (210323S4) from Cairns North at genome position 6,900, when there was a change from G to A. However, this was not detected in the other Cairns North samples and the other 22 field samples. This lack of variability in the rest of the mtDNA genome is consistent with ddRAD-derived SNP data (9).
Figure 2.
Haplotype Network for COI and Table of Frequencies
Each colored node represents an observed haplotype with circle size indicating the number of individuals with each numbered haplotype. Solid black nodes represent a single base change. Abbreviations: G, Gordonvale; YK, Yorkeys Knob; C, Caravonica; EH, Edge Hill; PP/N, Parramatta Park/Cairns North; PP/P, Parramatta Park/Portsmith; BG, Bungalow. (+), wMel infected; (−), uninfected. Note that Haplotype 1 is equivalent to Haplotype 8 in Yeap et al. (2016), which was found in the wMel release strain and Haplotype 2 is equivalent to Haplotype 10 in Yeap et al. (2016), which was at a low frequency in the wMel release strain.
The pool of five uninfected mosquitoes from Caravonica was haplotype 3, which was also present in the original Cairns population before releases (Yeap et al., 2016). As Caravonica had not been fully invaded by Wolbachia at the time of collection because no releases had been carried out there, with invasion instead relying on natural Wolbachia spread also observed in the rest of Cairns (Schmidt et al., 2017), we expected uninfected individuals in this area to retain the original mtDNA constitution. Haplotype 3 was previously common in the original population (Yeap et al., 2016). We interpret the lack of this haplotype in the release areas as evidence for limited migration of Ae. aegypti into the release locations from other uninfected areas and an ongoing low level of maternal loss of Wolbachia in invaded areas. This leakage is not expected to have much influence on Wolbachia infection levels, which are high because of ongoing cytoplasmic incompatibility, but likely reflects the effects of heat stress in some larval development containers, which can clear Wolbachia infections (Ross et al., 2020). The results also highlight the absence of any detectable paternal transmission or horizontal transmission in the population.
Overall, there appears to be minimal change in the Wolbachia genomes in mosquitoes sampled at the release sites over time. All the polymorphisms identified were rare and resulted in minor changes or occur at a low frequency. There was no clear correlation between the nucleotide frequency and release dates and locations. Thus, there seems to be little evidence for any selection process in the time since release. Importantly, the lack of change in the Wolbachia genome and associated mtDNA variants suggests that the virus-inhibiting and CI-inducing phenotypes are stable and that maternal transmission is imperfect, although leakage rates are relatively low, highlighting the ongoing viability of Wolbachia-based population replacement strategies.
Limitations of the Study
We note three limitations of the study. First, our data indicated minimal changes in the Wolbachia genome. Changes may also occur in the genome of the mosquito host, which could potentially reduce virus blocking. However, previous studies have indicated the persistence of cytoplasmic incompatibility post-releases, with symbiont-induced fitness cost and associated virus inhibition, which suggest a lack of evolutionary changes in host mosquitoes (Frentiu et al., 2014; Hoffmann et al., 2014). Future RNA sequencing studies will attempt to determine any changes in mosquito gene expression. Second, the focus in this study was on SNPs, but we cannot entirely rule out the possibility of large-scale genome rearrangements that may have been missed with our use of pooled mosquito samples. Finally, although the genetic diversity in our mosquito study population is comparable to that of Asia (Rasic et al., 2014), our observations are not necessarily applicable outside Australia. Hence, additional studies are required to determine whether the experience in dengue hyper-endemic regions in Asia or the Americas parallels that in Australia.
Resource Availability
Lead Contact
For additional information please contact David Warrilow (David.Warrilow@health.qld.gov.au).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Raw sequence data are available at NCBI SRA: PRJNA641232.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
Many thanks to Rikki Graham for advice on microbiology. We also thank Jason Axford for Aedes colony material; Michael Townsend, Thomas Schmidt, and Perran Ross for field Aedes aegypti samples; Peter Horne and Joe Davis for providing the map; Heng Lin Yeap for advice on mitochondrial DNA analysis; and Ana Ramirez for providing the image of Aedes aegypti for the graphical abstract (available from: Ramírez, Ana L. (2019): Aedes aegypti mosquito. figshare. Figure. https://doi.org/10.6084/m9.figshare.7155857). Finally, thanks to Cassie Jansen for comments on the manuscript. The Wolbachia genome component was internally funded by Forensic and Scientific Services of Australia, grant number RSS19-001. The mtDNA component of this research was funded by the National Health and Medical Research Council of Australia, grant numbers 1132412 and 1118640.
Author Contributions
B.H., A.A.H, S.A.R., A.F.v.d.H., and D.W. designed research; S.A.R. performed field work; B.H., Q.Y., A.F.v.d.H., and D.W. performed research; A.A.H. contributed reagents/analytic tools; B.H., A.A.H., Q.Y., and D.W. analyzed data; A.F.v.d.H. and D.W. wrote the first draft of the manuscript, and all authors approved the final version.
B.H. and Q.Y. contributed equally to this work.
Declaration of Interests
Although Scott Ritchie is currently employed with the World Mosquito Program, the study reported herein was conducted before commencing in that position. His current employers had no input into the design, outcomes, and interpretation of the work presented in the current study.
Published: October 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101572.
Contributor Information
Ary A. Hoffmann, Email: ary@unimelb.edu.au.
Andrew F. van den Hurk, Email: andrew.vandenhurk@health.qld.gov.au.
David Warrilow, Email: david.warrilow@health.qld.gov.au.
Supplemental Information
References
- Anonymous . Pan American Health Organization/World Health Organization; 2019. Epidemiological Update: Dengue.https://www.paho.org/en/documents/epidemiological-update-dengue-11-november-2019 [Google Scholar]
- Bull J.J., Turelli M. Wolbachia versus dengue: evolutionary forecasts. Evol. Med. Public Health. 2013;2013:197–207. doi: 10.1093/emph/eot018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores H.A., O'Neill S.L. Controlling vector-borne diseases by releasing modified mosquitoes. Nat. Rev. Microbiol. 2018;16:508–518. doi: 10.1038/s41579-018-0025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S.A., Allen S.L., Ohm J.R., Sigle L.T., Sebastian A., Albert I., Chenoweth S.F., McGraw E.A. Selection on Aedes aegypti alters Wolbachia-mediated dengue virus blocking and fitness. Nat. Microbiol. 2019;4:1832–1839. doi: 10.1038/s41564-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu F.D., Zakir T., Walker T., Popovici J., Pyke A.T., van den Hurk A., McGraw E.A., O'Neill S.L. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. Plos Negl. Trop. Dis. 2014;8:e2688. doi: 10.1371/journal.pntd.0002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale L.R., Hoffmann A.A. Mitochondrial DNA polymorphism and cytoplasmic incompatibility in natural populations of Drosophila simulans. Evolution. 1990;44:1383–1386. doi: 10.1111/j.1558-5646.1990.tb05241.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann A.A., Iturbe-Ormaetxe I., Callahan A.G., Phillips B.L., Billington K., Axford J.K., Montgomery B., Turley A.P., O'Neill S.L. Stability of the wMel Wolbachia Infection following invasion into Aedes aegypti populations. Plos Negl. Trop. Dis. 2014;8:e3115. doi: 10.1371/journal.pntd.0003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A.A., Montgomery B.L., Popovici J., Iturbe-Ormaetxe I., Johnson P.H., Muzzi F., Greenfield M., Durkan M., Leong Y.S., Dong Y. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:454–457. doi: 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- Hornett E.A., Charlat S., Wedell N., Jiggins C.D., Hurst G.D. Rapidly shifting sex ratio across a species range. Curr. Biol. 2009;19:1628–1631. doi: 10.1016/j.cub.2009.07.071. [DOI] [PubMed] [Google Scholar]
- Musso D., Ko A.I., Baud D. Zika virus infection - after the pandemic. N. Engl. J. Med. 2019;381:1444–1457. doi: 10.1056/NEJMra1808246. [DOI] [PubMed] [Google Scholar]
- Nazni W.A., Hoffmann A.A., NoorAfizah A., Cheong Y.L., Mancini M.V., Golding N., Kamarul G.M.R., Arif M.A.K., Thohir H., NurSyamimi H. Establishment of Wolbachia strain wAlbB in Malaysian populations of Aedes aegypti for dengue control. Curr. Biol. 2019;29:4241–4248 e4245. doi: 10.1016/j.cub.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasic G., Filipovic I., Weeks A.R., Hoffmann A.A. Genome-wide SNPs lead to strong signals of geographic structure and relatedness patterns in the major arbovirus vector, Aedes aegypti. BMC Genomics. 2014;15:275. doi: 10.1186/1471-2164-15-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie S.A., van den Hurk A.F., Smout M.J., Staunton K.M., Hoffmann A.A. Mission accomplished? We need a guide to the 'post release' World of Wolbachia for aedes-borne disease control. Trends Parasitol. 2018;34:217–226. doi: 10.1016/j.pt.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Ross P.A., Axford J.K., Yang Q., Staunton K.M., Ritchie S.A., Richardson K.M., Hoffmann A.A. Heatwaves cause fluctuations in wMel Wolbachia densities and frequencies in Aedes aegypti. Plos Negl. Trop. Dis. 2020;14:e0007958. doi: 10.1371/journal.pntd.0007958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P.A., Turley A.P., Wilson G., Hurst T.P., Retzki K., Brown-Kenyon J., Hodgson L., Kenny N., Cook H., Montgomery B.L. Establishment of wMel Wolbachia in Aedes aegypti mosquitoes and reduction of local dengue transmission in Cairns and surrounding locations in northern Queensland, Australia. Gates Open Res. 2020;3:1547. doi: 10.12688/gatesopenres.13061.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.L., Barton N.H., Rasic G., Turley A.P., Montgomery B.L., Iturbe-Ormaetxe I., Cook P.E., Ryan P.A., Ritchie S.A., Hoffmann A.A. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol. 2017;15:e2001894. doi: 10.1371/journal.pbio.2001894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.L., Filipovic I., Hoffmann A.A., Rasic G. Fine-scale landscape genomics helps explain the slow spatial spread of Wolbachia through the Aedes aegypti population in Cairns, Australia. Heredity (Edinb) 2018;120:386–395. doi: 10.1038/s41437-017-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Abdel-Motaal H., Chen J., Chen H., Xu T., Meng L., Zhang Z., Meng F., Jiang J. Characterization of a functionally unknown arginine-aspartate-aspartate family protein from Halobacillus andaensis and functional analysis of its conserved arginine/aspartate residues. Front. Microbiol. 2018;9:807. doi: 10.3389/fmicb.2018.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., Hoffmann A.A., McKechnie S.W. Dynamics of cytoplasmic incompatibility and mtDNA variation in natural Drosophila simulans populations. Genetics. 1992;132:713–723. doi: 10.1093/genetics/132.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T., Johnson P.H., Moreira L.A., Iturbe-Ormaetxe I., Frentiu F.D., McMeniman C.J., Leong Y.S., Dong Y., Axford J., Kriesner P. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- Weeks A.R., Turelli M., Harcombe W.R., Reynolds K.T., Hoffmann A.A. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 2007;5:e114. doi: 10.1371/journal.pbio.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfit M., Iturbe-Ormaetxe I., Brownlie J.C., Walker T., Riegler M., Seleznev A., Popovici J., Rances E., Wee B.A., Pavlides J. Genomic evolution of the pathogenic Wolbachia strain, wMelPop. Genome Biol. Evol. 2013;5:2189–2204. doi: 10.1093/gbe/evt169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap H.L., Rasic G., Endersby-Harshman N.M., Lee S.F., Arguni E., Le Nguyen H., Hoffmann A.A. Mitochondrial DNA variants help monitor the dynamics of Wolbachia invasion into host populations. Heredity (Edinb) 2016;116:265–276. doi: 10.1038/hdy.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data are available at NCBI SRA: PRJNA641232.