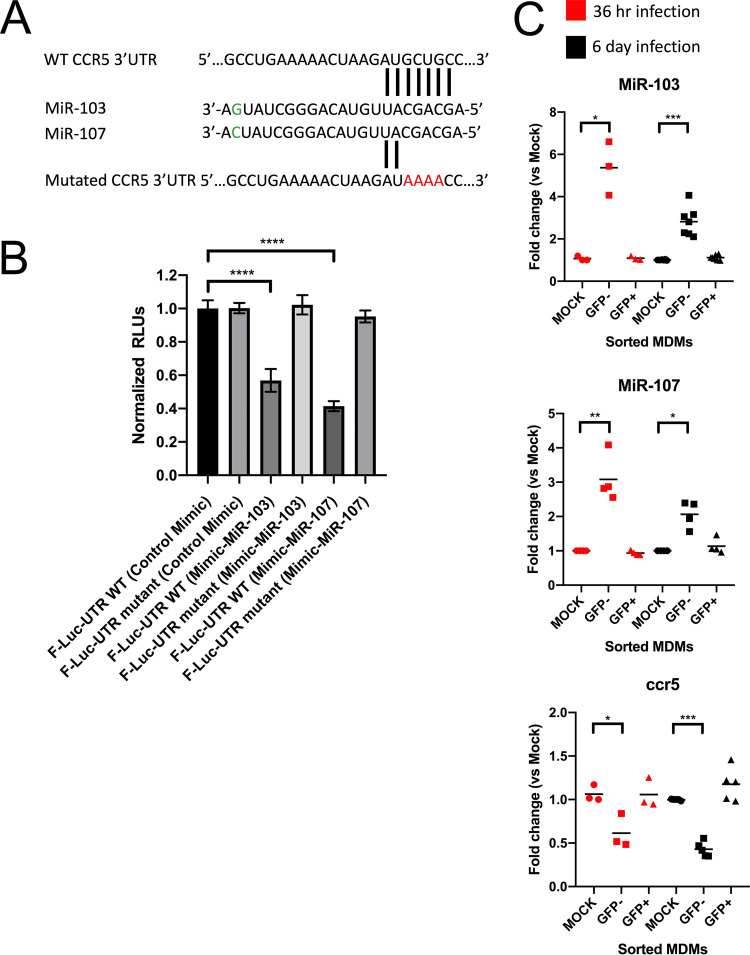
FIG 1.
miR-103 and miR-107 are enhanced and CCR5 is downregulated in bystander MDMs. (A) Nucleotide sequences of miR-103 and miR-107 showing the matching seed sequence nucleotides recognized in the CCR5 mRNA 3′ UTR. The nucleotides changed in the mutated version used in the target validation assay are in red. The single G/C substitution in miR-103 compared to miR-107 is shown in green. The CCR5 nucleotide sequence is from GenBank accession number NM_000579. (B) Target validation assay in wild-type (WT) or mutant firefly luciferase (F-Luc)-CCR5-3′-UTR-transfected HEK293T cells treated with either control, miR-103, or miR-107 mimics. Results are plotted as mean F-Luc activities (relative light units [RLUs]) standardized to the control Renilla Luc activity ± standard deviations (SD). Statistical significance was determined using Student’s t test (n = 4 technical replicates). (C) miR-103, miR-107, and CCR5 expression levels were determined in sorted macrophages derived from 3 to 7 blood donors by real-time qPCR. Bars shown are the mean fold changes compared to mock cells (n = 3 to 7 blood donors; Wilcoxon matched-pairs signed-rank test).