Abstract
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) and represents a global pandemic affecting more than 26 million people and has claimed >870,000 lives worldwide. Diagnostic tests for SARS-COV-2 infection commonly use nasopharyngeal swabs (NPS). As an alternative specimen, we investigated the potential use of the real-time reverse transcriptase PCR (RT-PCR) detection of SARS-COV-2 in saliva samples in large suspected-COVID-19 patients in Kuwait.
NPS and saliva samples pairs were prospectively collected from 891 COVID-19 suspected patients in Kuwait and analyzed using TaqPath™ COVID-19 multiplex RT-PCR.
Of the 891 patients, 38.61 % (344/891) were positive for SARS-CoV-2, 4.83 % (43/891) were equivocal, and 56.56 % (504/891) were negative with NPS by RT-PCR. For saliva, 34.23 % (305/891) were positive for SARS-CoV-2, 3.14 (28/891) were equivocal, and 62.63 % (558/891) were negative. From 344 confirmed cases for SARS-CoV-2 with NPS samples, 287 (83.43 %) (95 % CI, 79.14–86.99) were positive with saliva specimens. Moreover, the diagnostic sensitivity and specificity of RT-PCR for the diagnosis of COVID-19 in saliva were 83.43 % (95 % CI: 79.07–87.20) and 96.71 % (95 % CI: 94.85–98.04 %), respectively. An analysis of the agreement between the NPS and saliva specimens demonstrated 91.25 % observed agreement (κ coefficient = 0.814, 95 % CI, 0.775–0.854).
This study demonstrates that saliva can be a noninvasive specimen for detection of SARS-CoV-2 by RT-PCR.
Keywords: SARS-CoV-2, Nasopharyngeal swab, Saliva, RT-PCR, COVID-19
1. Introduction
In late December 2019, the coronavirus disease (COVID-19) occurred in Wuhan, China, and spread rapidly to become public health emergency of international concern, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection [[1], [2], [3], [4]]. The typical symptoms of COVID-19 are fever, dry cough, myalgias and/or fatigue, loss of smell and taste, sputum production, and shortness of breath [5,6]. At present, there is no specific antiviral treatment recommended for COVID-19, and no vaccine is available [5]. Early diagnosis and isolation of infected individuals will play a vital role in stopping the further escalation of the pandemic [5].
SARS-CoV-2 has been detected in a variety of oral samples, including whole saliva, oral swabs, oropharyngeal swabs, and deep saliva/sputum [[7], [8], [9]]. Nasopharyngeal swabbing, followed by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) technique of the extracted RNA, is the gold standard for detection of SARS-CoV-2 infection [10,11]. However, it has been reported that self-collected of saliva samples in comparison with nasopharyngeal swabs can greatly decrease the chance of exposing healthcare workers to SARS-CoV-2 [8]. The potential use of saliva for the detection of SARS-CoV-2 infection appears clinically useful [[12], [13], [14]], but further validation diagnostic accuracy studies comparing saliva to matched nasopharyngeal swabs (NPS) in positive and negative patients are needed [15]. Moreover, the U.S. Food and Drug Administration (FDA) authorized the first diagnostic test with the option of using home-collected saliva samples for COVID-19 testing [16].
The situation in Kuwait may be under control as the Ministry of Health declared the first case on 24 February 24, 2020. As a result of the 648,051 tests carried out as of September 7, 2020, a total of 90,387 cases were confirmed, and the number of deaths recorded was 546 (https://corona.e.gov.kw/En/). We aimed to investigate the potential use of the RT-PCR detection of SARS-COV-2 in saliva samples as an alternative diagnostic specimen in large suspected-COVID-19 patients in Kuwait.
2. Materials and methods
This was a cross-sectional study conducted among 891 suspected COVID-19 admitted consecutively to the Al-Sabah, Jaber, and Alrazi Hospitals, Kuwait between 19 and 21 July 2020.
The study protocol was approved by the permanent Committee for Coordination of Medical and Health Research, Ministry of health, Kuwait and the study was conducted in accordance with the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the institution's human research committee. NPS and saliva samples pairs were collected from 891-suspected SARS-CoV-2 infection.
To collect NPS, the swab was passed through the nostril until reaching the posterior nasopharynx and removed while rotating. After swabbing, each absorbent swab was placed immediately into a sterile tube with viral transport medium. Whole saliva (≈1.5 mL) was collected after deep cough from the suspected patients into a sterile container. Viscous saliva was added to 300 μL of viral transport media (VTM), mixed vigorously, and then 200 μL of sample was used for RNA isolation. NPS and saliva samples were collected at the same time. All medical staff was equipped with personal protection equipment. Viral RNA was automatically extracted from 200 μL of the NPS and saliva specimens using the MagMax Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA) on KingFisher (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. A negative extraction control (molecular-grade, nuclease-free water) was added to each KingFisher extraction run and carried through to quantitative real-time reverse transcription polymerase chain reaction (RT-PCR). RT-PCR was performed using TaqPath™ COVID-19 multiplex real-time RT-PCR test (the Orf1ab, N and S genes) (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Negative and positive controls were run simultaneously with samples. RT-PCR including cDNA synthesis and PCR amplification of the target sequences was performed on the QuantStudio 5 real-time PCR system. The result was considered positive if cycle threshold (Ct) values were <37 for three SARS-CoV-2 targets (the ORF1ab, the N, and the S genes). Samples positive for one or two targets were considered equivocal.
2.1. Statistical analysis
For categorical variables, the Chi-square test was used. Differences in continuous variables were compared using the Mann-Whitney U test Additionally, sensitivity, specificity, positive and negative predictive values and their 95 % confidence intervals (CI) were calculated to assess diagnostic performance. Agreement between the NPS and saliva specimens for the virus detection ability was assessed using Cohen’s Kappa (κ coefficient). Bland-Altman analysis was used to compare the cycle threshold (Ct) values between NPS and saliva. All P-values were two-sided and P < 0.05 was considered significant. Statistical analyses were performed using GraphPad PRISM version 6.0e (GraphPad Software, San Diego, CA, USA) or MedCalc statistical software.
3. Results
Eight hundred ninety-one sample pairs of NPS and saliva samples were collected (Table 1 ). Of the 891 suspected subjects, 38.61 % (344/891) were positive for SARS-CoV-2, 4.83 % (43/891) were equivocal, and 56.56 % (504/891) were negative with NPS by RT-PCR. For saliva, 34.23 % (305/891) were positive for SARS-CoV-2, 3.14 (28/891) were equivocal, and 62.63 % (558/891) were negative (Table 1). Comparative study between NPS and saliva samples, showed significant difference regarding negative result (P = 0.0091) (Table 1).
Table 1.
Detection rate of SARS-COV-2 by RT-PCR in suspected-COVID-19 patients in saliva specimens.
| Nasopharyngeal swabs (%) | Saliva specimens (%) | p values | |
|---|---|---|---|
| Negative | 504 (56.56) | 558 (62.23) | 0.0091 |
| Equivocal | 43 (4.83) | 28 (3.14) | 0.0693 |
| Positive | 344 (38.61) | 305 (34.23) | 0.0549 |
| Total | 891 (100) | 891 (100) |
In our study, 344 cases were tested positive for NPS nucleic acid detection. Among these 344 patients, there were 287 (83.43 %) (95 % CI, 79.14–86.99 %) were positive with saliva specimens. In 324 COVID-19 patients, based on the result of RT-PCR obtained using NPS, there were 193 (59.57 %) male and 131 (40.4 %) female patients.
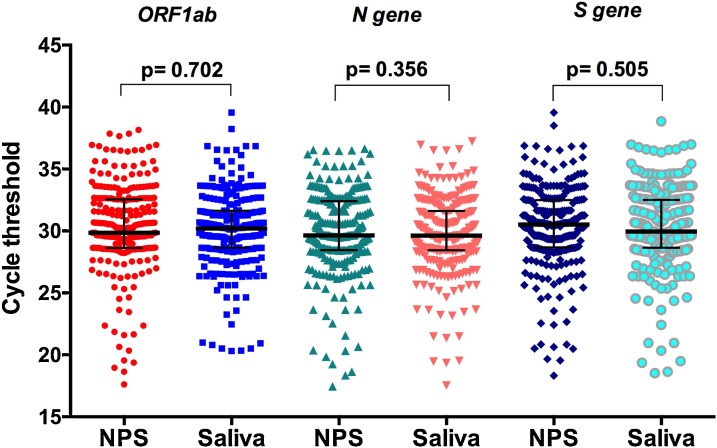
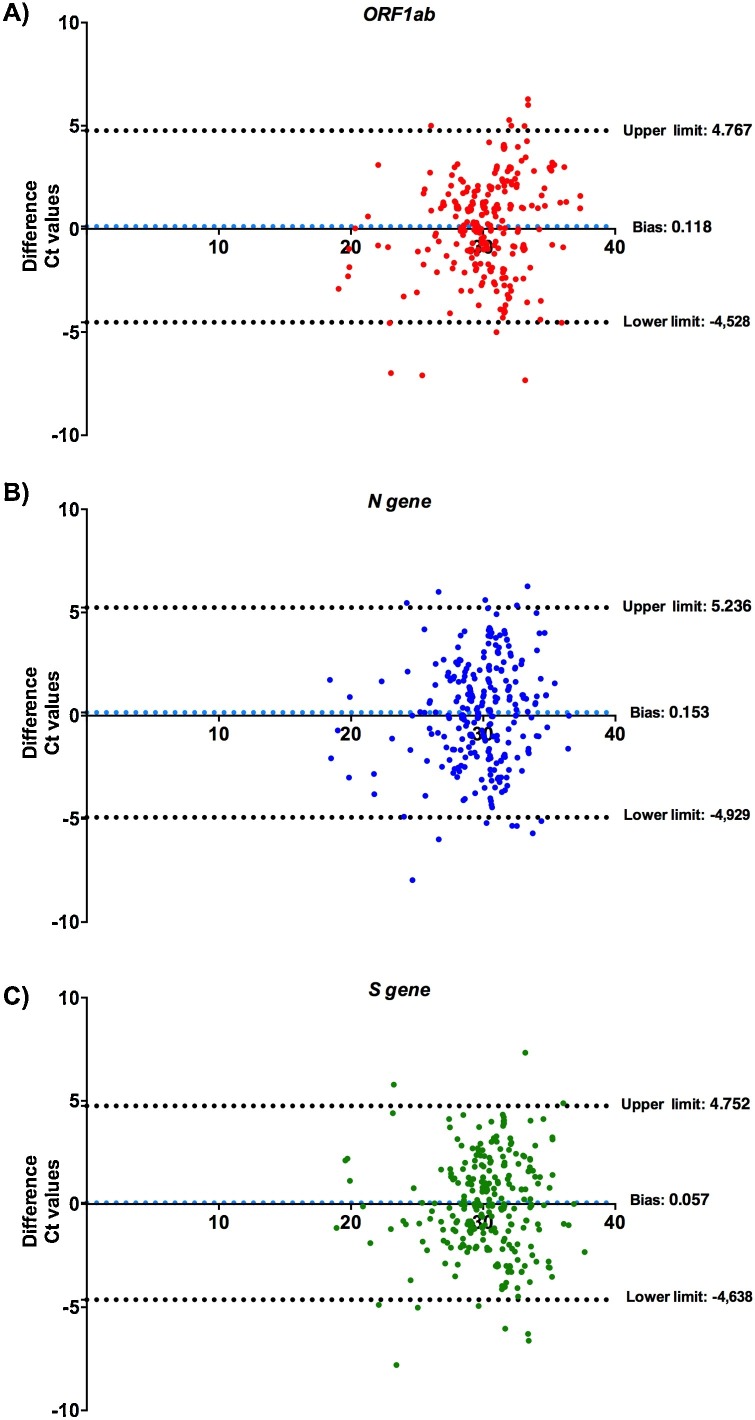
The median Ct values were not significantly different between NPS and saliva (P > 0.05), suggesting same viral loads in both samples (Fig. 1 , Supplementary Fig. 1). Moreover, Bland-Altman analysis performed on the quantitative results obtained by NPS versus saliva and demonstrated similar close agreement (Fig. 2 ).
Fig. 1.
Cycle threshold (Ct) values in nasopharyngeal swabs (NPS) and saliva specimens positive for SARS-CoV-2 for three targets (ORF1ab, N gene, and S gene). Data are presented as median and bars represent the interquartile range. The Mann-Whitney U test was used for comparison.
Fig. 2.
Bland-Altman analysis for the Ct values with NPS and saliva specimens. Consistency analysis of Ct values based on ORF1ab (A), N gene (B), and S gene (C). Dashed lines indicate 95 % limits of agreement.
Additionally, a comprehensive assessment of the diagnostic accuracy of COVID-19 using nucleic acid detection in saliva specimens was performed (Table 2 ). Using NPS RT-PCR as the reference standard, the sensitivity and specificity of RT-PCR for the diagnosis of COVID-19 in saliva were 83.43 % (95 % CI: 79.07–87.20) and 96.71 % (95 % CI: 94.85–98.04 %), respectively. Notably, analysis of the agreement between the NPS and saliva specimens demonstrated 91.25 % observed agreement (κ coefficient = 0.814, 95 % CI, 0.775–0.854).
Table 2.
Assessment of the diagnostic accuracy of COVID-19 in saliva specimens.
| Value (%) | (95 % CI) | |
|---|---|---|
| Sensitivity | 83.43 | (79.07–87.20) |
| Specificity | 96.71 | (94.85–98.04) |
| Positive likelihood ratio | 25.35 | (16.06–40.03) |
| Negative likelihood ratio | 0.17 | (0.14–0.22) |
| Positive predictive value | 94.10 | (90.99–96.18) |
| False discovery rate | 5.90 | (3.63–9.33) |
| Negative predictive value | 90.27 | (87.98–92.17) |
| Accuracy | 91.58 | (89.56–93.32) |
4. Discussion
Throat swabs are the gold standard to diagnose COVID-19. However, NPS is relatively invasive, induce coughing and cause bleeding occasionally, which may increase risks of healthcare workers infection [17]. The use of saliva samples for diagnosis of COVID-19 has many advantages in clinical practice. First, collecting saliva is a non-invasive procedure and rather than nasal or throat swabs avoids patient discomfort. The second advantage of using saliva as specimen is related to possibility of collecting samples outside the hospitals [18]. In addition, saliva can be used to detect SARS-CoV-2 in both symptomatic patients and asymptomatic carriers [19]. There are several limitations to our study. Firstly, we have not evaluated the detection sensitivity regarding COVID-19 severity. Secondly, we don't have enough information regarding clinical data of COVID-19 patients. Moreover, saliva-based testing requires a careful preparation prior to RNA extraction and to get the right volume (minimum 1.5 mL).
In this study, we found that the detection rate of SARS-CoV-2 by saliva RT-PCR was 83.43 %. The result was in accordance with the previous studies and showed that the detection rate of SARS-CoV-2 ranges from 31 % to 100 among COVID-19 patients [8,13,14,[20], [21], [22]]. Moreover, the temporal profile of viral load in saliva reached the peak of viral load during the first week of symptom onset and then declined, but viral RNA could still be detected for 20 days or even longer 20]. Interestingly, Fakheran et al. reported that there is no statistically significant difference between nasopharyngeal and saliva samples regarding viral load [18]. In addition, previous data showed that angiotensin-converting enzyme II (ACE2) —SARS-CoV-2 host-cell receptor — is expressed in the salivary glands and SARS-CoV-2 can be detected in saliva. These results confirm the possibility of the presence of SARS-CoV-2 in saliva [17,23]. Thus, several dental clinics in Japan are currently performing pre-clinical COVID-19 PCR tests using a saliva-based detection kit [24].
Our results showed that the saliva RT- PCR test demonstrated high sensitivity (83.43 %) and specificity (96.71 %) and comparable performance to the current standard of nasopharyngeal swab. This data seem in agreement with previous cross-sectional study (sensitivity and specificity of the saliva sample RT-PCR are 84.2 % and 98.9 %, respectively) [12]. Furthermore, in recent meta-analysis, the sensitivities for SARS-CoV-2 were 91 % (95 % CI, 80–99 %) and 98 % (95 % CI, 89–100 %) for saliva and for NPS samples, respectively [25]. Interestingly, Kim and al. revealed no significant difference in the sensitivity and specificity of saliva- and NPS-based tests for 11 different viral respiratory infections, including coronaviruses [26]. Recent study showed that saliva pools of either five or ten samples did not compromise the detection of SARS-CoV-2 [27]. Moreover, the κ coefficient value showed a strong agreement of the diagnosis between the NPS and saliva specimen similar to previous data [12].
In conclusion, this study brought more evidence that saliva-based testing is a promising alternative sampling for SARS-CoV-2 detection by RT-PCR and could simplify and accelerate COVID-19 diagnosis.
Financial support
This study was supported by Ministry of Health, Kuwait.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
We acknowledge the contributions of other clinical and technical staff of the Al-Sabah, Jaber, and Alrazi Hospitals. We thank Sathishkumar Ramasamy, and Walled Khaled O Thajeil from Yacoub Behbahani center, virology unit.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104652.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. China novel coronavirus I, research t. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;2020(382):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO . 2020. Coronavirus Disease (COVID-19). Weekly Epidemiological Update. [Available from: https://http://www.who.int/docs/default-source/coronaviruse/situation-reports/20200831-weekly-epi-update-3.pdf?sfvrsn=d7032a2a_4 (Accessed 2 September 2020)] [Google Scholar]
- 4.Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezzikouri S., Nourlil J., Benjelloun S., Kohara M., Tsukiyama-Kohara K. Coronavirus disease 2019-Historical context, virology, pathogenesis, immunotherapy, and vaccine development. Hum. Vaccin. Immunother. 2020:1–9. doi: 10.1080/21645515.2020.1787068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan C.H., Faraji F., Prajapati D.P., Boone C.E., DeConde A.S. Association of chemosensory dysfunction and COVID-19 in patients presenting with influenza-like symptoms. Int. Forum Allergy Rhinol. 2020;10:806–813. doi: 10.1002/alr.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;(395):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.To K.K., Tsang O.T., Yip C.C., Chan K.H., Wu T.C., Chan J.M., Leung W.S., Chik T.S., Choi C.Y., Kandamby D.H., Lung D.C., Tam A.R., Poon R.W., Fung A.Y., Hung I.F., Cheng V.C., Chan J.F., Yuen K.Y. Consistent detection of 2019 novel coronavirus in Saliva. Clin. Infect. Dis. 2020;(71):841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J.H., Yip C.C., Poon R.W., Chan K.H., Cheng V.C., Hung I.F., Chan J.F., Yuen K.Y., To K.K. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg. Microbes Infect. 2020;9:1356–1359. doi: 10.1080/22221751.2020.1775133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippi G., Simundic A.M., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin. Chem. Lab. Med. 2020;58:1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 12.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Sungkanuparph S., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin. Microbiol. Infect. 2020;S1198-743X(20) doi: 10.1016/j.cmi.2020.05.001. 30278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., Fasano M., Sessa F., Tettamanti L., Carinci F., Maurino V., Rossi A., Tagliabue A., Baj A. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khurshid Z., Zohaib S., Joshi C., Moin S.F., Zafar M.S., Speicher D.J. Saliva as a non-invasive sample for the detection of SARS-CoV-2: a systematic review. medRxiv preprint. 2020 doi: 10.1101/2020.05.09.20096354. [DOI] [Google Scholar]
- 16.FDA . 2020. Coronavirus (COVID-19) Update: FDA Authorizes First Test for Patient At-Home Sample Collection. Available online: https://http://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-test-patient-home-sample-collection [Accessed 24 August 2020] [Google Scholar]
- 17.Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int. J. Oral Sci. 2020;12:11. doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fakheran O., Dehghannejad M., Khademi A. Saliva as a diagnostic specimen for detection of SARS-CoV-2 in suspected patients: a scoping review. Infect. Dis. Poverty. 2020;9:100. doi: 10.1186/s40249-020-00728-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harikrishnan P. Saliva as a potential diagnostic specimen for COVID-19 testing. J. Craniofac. Surg. 2020 doi: 10.1097/SCS.0000000000006724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W., Du RH Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng S., Yu F., Fan J., Zou Q., Xie G., Yang X., Chen W., Wang Q., Zhang D., Wang R., Feng B., Lin S., Gong R., Ma Z., Lu S., Wang X., Yu L., Xu K., Sheng J., Chen Y. 2020. Saliva As a Diagnostic Specimen for SARS-CoV-2 by a PCR-Based Assay: a Diagnostic Validity Study. Available at SSRN: https://ssrn.com/abstract=3543605 or. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L., Zhao J., Peng J., Li X., Deng X., Geng Z., Shen Z., Guo F., Zhang Q., Jin Y., Wang L., Wang S. 2020. Detection of 2019-nCoV in Saliva and Characterization of Oral Symptoms in COVID-19 Patients. Available at SSRN: https://ssrn.com/abstract=3556665 or. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi Y., Furuchi M., Kamimoto A., Honda K., Matsumura H., Kobayashi R. Saliva-based PCR tests for SARS-CoV-2 detection. J. Oral Sci. 2020;62:350–351. doi: 10.2334/josnusd.20-0267. [DOI] [PubMed] [Google Scholar]
- 25.Czumbel L.M., Kiss S., Farkas N., Mandel I., Hegyi A., Nagy Á, Lohinai Z., Szakács Z., Hegyi P., Steward M.C., Varga G. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front Med. 2020;7:465. doi: 10.3389/fmed.2020.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y.G., Yun S.G., Kim M.Y., Park K., Cho C.H., Yoon S.Y., Nam M.H., Lee C.K., Cho Y.J., Lim C.S. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J. Clin. Microbiol. 2017;55:226–233. doi: 10.1128/JCM.01704-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasomsub E., Watcharananan S.P., Watthanachockchai T., Rakmanee K., Tassaneetrithep B., Kiertiburanakul S., Phuphuakrat A. Saliva sample pooling for the detection of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.26460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.