Abstract
The human retinoblastoma (RB1) protein is a tumor suppressor that negatively regulates cell cycle progression through its interaction with members of the E2F/DP family of transcription factors. However, RB‐related (RBR) proteins are an early acquisition during eukaryote evolution present in plant lineages, including unicellular algae, ancient plants (ferns, lycophytes, liverworts, mosses), gymnosperms, and angiosperms. The main RBR protein domains and interactions with E2Fs are conserved in all eukaryotes and not only regulate the G1/S transition but also the G2/M transition, as part of DREAM complexes. RBR proteins are also important for asymmetric cell division, stem cell maintenance, and the DNA damage response (DDR). RBR proteins play crucial roles at every developmental phase transition, in association with chromatin factors, as well as during the reproductive phase during female and male gametes production and embryo development. Here, we review the processes where plant RBR proteins play a role and discuss possible avenues of research to obtain a full picture of the multifunctional roles of RBR for plant life.
Keywords: Arabidopsis, CDK/cyclin, cell differentiation, chromatin, endoreplication
Subject Categories: Cell Cycle, Plant Biology
A guide to the wealth of plant‐specific functions in asymmetric cell division, stem cell maintenance, regeneration or reproduction fulfilled by homologs of the famous tumor suppressor RB, whose appearance dates back to the earliest eukaryotes.

Introduction
Formation of organs, either during embryogenesis as in animals or post‐embryonically as in most plants, relies on a continuous supply of new cells. Failure to properly coordinate cell division, cell cycle exit into cell differentiation, and cell fate acquisition frequently results in abnormal growth, developmental aberrations, or cell transformation and tumorigenesis. Strict control of cell cycle progression is necessary to achieve the goal of producing two daughter cells. A plethora of studies has demonstrated that progression through G1 phase and into S‐phase, as well as the G2‐to‐M and metaphase‐to‐anaphase transitions, represents key checkpoints during the cell cycle. A crucial role in both transitions is played by cyclin‐dependent kinases (CDK), as first demonstrated in the fission yeast Schizosaccharomyces pombe (Nurse & Bissett, 1981). Later, it was shown that human cells contain homologs of the yeast Cdc2 CDK (Lee & Nurse, 1987). Cdc2 homologs were found also in plant cells, with their phosphorylation state being cell cycle‐dependent (John et al, 1989). These studies paved the way for identifying Cdc2‐like kinases (John et al, 1989; Feiler & Jacobs, 1990; Ferreira et al, 1991), as well as their A‐ and B‐type cyclin partners (Hata et al, 1991; Hemerly et al, 1992; Hirt et al, 1992), in various plant species.
Identification of the CDK/cyclin targets controlling the G1/S transition proved to be a difficult task, which was enlightened by research in cancer. It was found that cells of rare human tumors, such as retinoblastoma or sarcoma, harbor inactivating mutations in the retinoblastoma susceptibility gene, RB1 (Friend et al, 1986; Fung et al, 1987; Lee et al, 1987). RB1 suppresses cell proliferation and can bind oncoviral proteins such as SV40 large T‐antigen (T‐ag; Lee et al, 1987; DeCaprio et al, 1988; Dyson et al, 1989; Ludlow et al, 1989). Moreover, RB1 was found to be phosphorylated in a cell cycle‐dependent manner by CDKs (Lees et al, 1991). Together, these results led to the proposal that RB1 is a regulator of cell cycle progression in G1 (Buchkovich et al, 1989).
The last piece in the initial puzzle was the identification of E2F (for “adenovirus early gene 2 promoter‐binding factor”) as a cellular protein that could form complexes with RB1 (Chellappan et al, 1991). These and other findings served to establish that the RB1 complexes are regulators of the G1/S transition, as well as of the switch from quiescence to proliferation (reviewed in Fischer & Müller, 2017). Other RB1 interactors, primarily D‐type cyclins, bind RB1 through a LxCxE amino acid motif, an interaction that is mimicked by human oncoviral proteins that inactivate RB1's tumor suppressor function (DeCaprio, 2009); however, although the finding of the LxCxE motif in D‐type cyclins proved originally valuable, more recent work deleting this motif in mammalian cells has demonstrated that it is not essential (Landis et al, 2007; Topacio et al, 2019).
Retinoblastoma protein in plants
Back in the mid‐1990s, a couple of apparently unrelated research lines reinforced the hypothesis that plants might share some kind of RB/E2F regulatory module with human cells. D‐type cyclin homologs were identified in several plant species (Dahl et al, 1995; Soni et al, 1995), which had—despite limited amino acid sequence homology with human D‐type cyclins—two key features in common with them: a similar expression pattern during the cell cycle and a highly conserved LxCxE amino acid motif (Soni et al, 1995; Riou‐Khamlichi et al, 1999). These facts strongly suggested that plants might contain proteins that could recognize this motif, as it is the case for human cyclin D and RB1. Independently, geminiviruses, a group of plant DNA viruses that replicate their single‐stranded DNA genome in the nucleus of the infected cell, were found to trigger synthesis of the host cell nuclear DNA upon infection (Nagar et al, 1995). Additionally, wheat dwarf virus (WDV) and other monocotyledonous plant‐infecting geminiviruses were found to encode an LxCxE motif‐containing protein, RepA (Xie et al, 1995). Importantly, this motif required for efficient viral DNA replication was shown to enable interaction of RepA also with human RB1 (Xie et al, 1995), in a manner analogous to human oncoviral proteins. Subsequent studies identified genes encoding retinoblastoma‐related (RBR1) proteins in maize (Grafi et al, 1996; Xie et al, 1996; Ach et al, 1997a), and in an immediate follow‐up to the cloning of multiple plant E2F and DP factors (Ramirez‐Parra et al, 1999; Sekine et al, 1999; Albani et al, 2000; Magyar et al, 2000; Ramirez‐Parra & Gutierrez, 2000; Kosugi & Ohashi, 2002). Remarkably, Arabidopsis was found to encode six E2F family members with two types of domain organization. The first group (E2FA, B and C) possesses the same domains as human E2F1‐5, including DNA‐binding and dimerization domains. The second group (E2FD, E and F, also known as DEL2, 1 and 3, respectively) is unique in that its members contain a duplicated DNA‐binding domain but lack the dimerization domain that allows binding to DNA in the absence of DP factors and fails to activate gene expression (Kosugi & Ohashi, 2002; Mariconti et al, 2002). Proteins homologous to these atypical E2F family members were subsequently also identified in mammalian cells (reviewed in Trimarchi & Lees, 2002; Lammens et al, 2009). Genome‐wide studies identified putative E2F target genes in the Arabidopsis genome (Ramirez‐Parra et al, 2003; Vandepoele et al, 2005; Naouar et al, 2009), a list that—somewhat surprisingly at the time—contained not only bona fide cell cycle control genes, but also genes involved in many other aspects of plant physiology, strongly pointing to a multifunctional role of RBR1. This will be further discussed below.
Evolutionary perspective on plant RBR proteins
The availability of multiple plant genomes has revealed the presence of RBR‐, E2F‐, and DP‐encoding genes in all species analyzed so far (reviewed in detail in Gutzat et al, 2014; Desvoyes et al, 2014). A first conclusion from the genomic data is that the appearance of the RBR‐E2F/DP module preceded the multiple branches of multicellularity that occurred ~800 million years ago, since it is present in unicellular organisms such as Chlamydomonas reinhardtii (Umen & Goodenough, 2001) and Ostreococcus tauri (Robbens et al, 2005), as well as colonial algae such as Volvox carteri (Kianianmomeni et al, 2008; Fig 1). Within the multicellular plant lineages, RBR proteins are present in all of them, and notably, monocotyledonous plants possess several members with different functions (Fig 1; see discussion below); for a more detailed discussion on how the RBR‐E2F/DP module has evolved in plants and animals, please refer to (Desvoyes et al, 2014). It is likely that RBR and other family members may have evolved specific functions in the different plant lineages. Thus, contrary to dicotyledonous plants that encode for a single RBR1 protein, monocotyledonous plants such as maize or rice contain two major RBR subfamilies: one whose members are involved in negative regulation of cell cycle, e.g., maize RBR1 and RBR2, and another involved in endosperm development, e.g., maize RBR3 and RBR4 (Sabelli et al, 2005; Sabelli & Larkins, 2006; Lendvai et al, 2007).
Figure 1. Phylogenetic relationships of RB family members from representative animal and plant lineages.
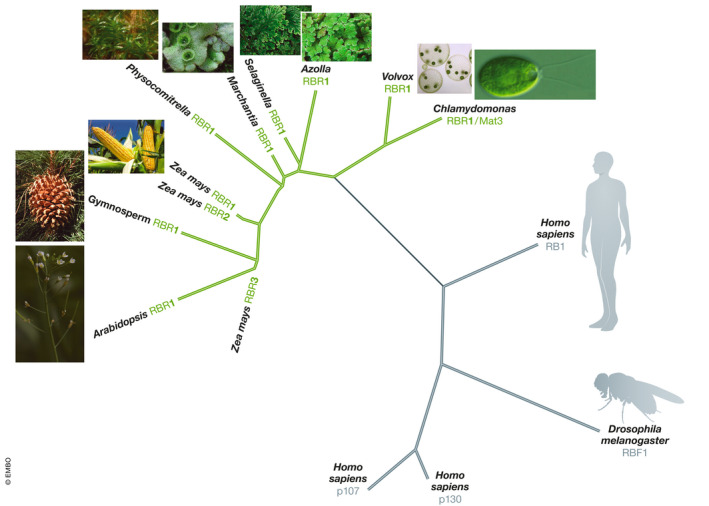
Homo sapiens (Human, mammal); Drosophila melanogaster (Artropoda); Chlamydomonas reinhardtii (Algae, unicellular); Volvox carteri (Algae, colonial); Azolla filliculoides (Fern); Selaginella moellendorffii (Lycophyte); Marchantia polymorpha (Liverwort); Physcomytrella patents (Moss); Pinus silvestris (Gymnosperm); Arabidopsis thaliana (Angiosperm, dicotyledonous); Zea mays (Angiosperm, monocotyledonous).
The current data are consistent with the idea that the RBR‐E2F/DP module is an ancient invention likely present already in the last eukaryotic common ancestor (LECA; Desvoyes et al, 2014). Later, the RBR/E2F‐DP module has been lost in some plant lineages, e.g., Ulvophyceae (De Clerck et al, 2018), and in other eukaryotes, e.g., S. cerevisiae and other yeast (Desvoyes et al, 2014). It is likely that organisms lacking RBR or related proteins use different pathways to regulate G1 progression. In S. cerevisiae, the Whi5 protein (Jorgensen et al, 2002), which is not homologous to animal RB1 or plant RBR1, seems to play analogous functions, whereby increased Cdk activity associated with the G1 cyclin Cln3 mediates activation of the G1 transcription factor heterodimer Swi4/Swi6, also known as SBF. Like RB proteins, Whi5 negatively regulates SBF, and its phosphorylation by Cdk/Cln3 complexes frees SBF to activate its targets (de Bruin et al, 2004; Costanzo et al, 2004). Together, these evolutionary studies reinforce the usefulness of comparative research on key cellular pathways.
Plant RBR1 domains and phosphorylation states
As expected from their ancient evolutionary origin, plant RBR proteins share with animal counterparts not only a relatively high degree of amino acid homology but, more importantly, a similar organization of functional domains. This includes an N‐terminal domain, the central A/B domain that forms the “pocket,” and a C‐terminal domain (Fig 2). A groove in the B domain is necessary for binding of LxCxE‐containing proteins, and together with the C‐terminal domain mediates interaction with E2F (Rubin et al, 2005). In addition, intrinsically disordered regions are present between the A and B domains as well as in the majority of the C‐terminal domain. It is remarkable that these regions contain most of the conserved CDK phosphorylation sites, including T373, involved in driving a more globular structure of this region, and S608, whose phosphorylation prevents interaction with E2F, as described for human RB1 (Burke et al, 2010).
Figure 2. Domain organization of representative proteins of the RB family.
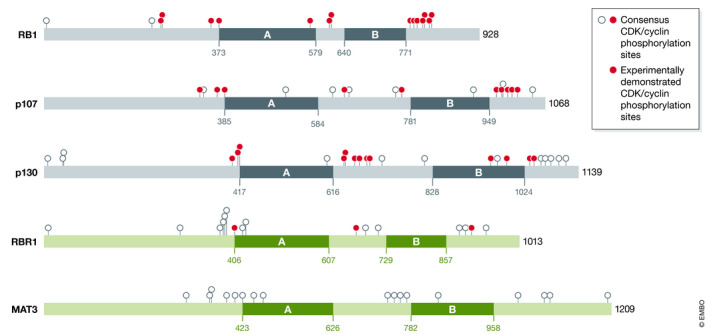
Domain organization of retinoblastoma family proteins comparing the three human proteins (RB1, p107, and p130) with two plant retinoblastoma‐related proteins (Arabidopsis thaliana RBR1 and Chlamydomonas reinhardtii MAT3). The two major domains A and B (green) defining the “pocket” together with the amino acid positions are indicated. The CDK/cyclin consensus phosphorylation sites (empty circles) are also indicated. Those experimentally demonstrated to be phosphorylated are shown (closed red circles). Data are taken from the UNIPROT database (Hansen et al, 2001; Farkas et al, 2002; Rubin, 2013; Willems et al, 2020).
Plant RBR proteins contain putative CDK phosphorylation sites in similar locations, although the role of individual sites on RBR structure and function is not yet known. In spite of this, there are reports on the overall function of RBR1 phosphorylation in cell cycle control in various plant species. The mechanism mediating RBR1 activity through phosphorylation involves the interaction with CDK/cyclin complexes containing many plant D‐type cyclins (Grafi et al, 1996; Nakagami et al, 1999, 2002; Boniotti & Gutierrez, 2001; Gutiérrez et al, 2005; Godínez‐Palma et al, 2017) and either CDKA or CDKB types of kinases (Boniotti & Gutierrez, 2001; Kawamura et al, 2006). The CDK/cyclin activity on RBR1 is cell cycle‐regulated and highest from mid‐G1 phase until the G1/S transition (Boniotti & Gutierrez, 2001; Nakagami et al, 2002; Sanchez et al, 2002; Hirano et al, 2008; Nowack et al, 2012), as well as and in G2 mediated by CDKB1;1 (Kawamura et al, 2006; Nowack et al, 2012). These CDK activities can be suppressed by CDK inhibitors known as Kip‐related proteins (KRPs; Pettkó‐Szandtner et al, 2006). In unicellular algae, RBR1/CDK/cyclin ternary complexes remain bound to chromatin throughout the cell cycle (Olson et al, 2010), but this has not been corroborated in multicellular plants. A functional connection between members of the RBR‐E2F/DP‐CDK/cyclin module has been demonstrated by the finding that activation of PCNA expression by E2F/DP is inhibited by co‐expression of RBR1, an inhibition counteracted by additional expression of cyclin D (Uemukai et al, 2005; Shimizu‐Sato et al, 2008).
Antibodies detecting specific phosphorylated residues in human RB1, such as pS807/pS811, proved useful to identify phosphorylated forms of RBR1 that accumulate after the G1/S transition and, in particular, during the G2 phase (Abrahám et al, 2011; Polit et al, 2012). It is interesting that phosphorylated RBR1 can be detected in the nucleus of interphase cells in the form of granules (Abrahám et al, 2011), although the functional relevance of such foci remains to be determined. Proteomic studies have demonstrated RBR1 phosphorylation in vivo at residues T406, S652, and S911 (Reiland et al, 2009; Willems et al, 2020). The identification of other phosphorylated residues, as well as their functions, remains as a future challenge. RBR1 phosphorylation is important not only for cell cycle progression but also for cell fate determination, as exemplified in the stem cell niche of the root apical meristem (RAM; Cruz‐Ramirez et al, 2012; see discussion below). Plant RBR1 is also a substrate of other kinases, e.g., PIP5K (Dieck et al, 2012) or S6K (Henriques et al, 2010, 2013), but in these cases the mechanistic implications are not fully understood.
RBR in unicellular green organisms
The lack of RB1 orthologs in yeasts and their presence in animals and plants had initially suggested that it may have been an evolutionary acquisition linked to multicellularity. This hypothesis was however ruled out by the identification of RBR1 homologs as well as its E2F/DP partners in the unicellular alga Chlamydomonas reinhardtii (Umen & Goodenough, 2001; Fang et al, 2006). This organism operates a peculiar cell division cycle, since a mother cell grows in G1 to much more than twice its size, and upon reaching a critical size then undergoes multiple cycles of S‐phases and mitoses to produce multiple daughter cells of the size typical for this organism. However, RBR1 does not regulate the length of G1, but instead acts to counter the number of mitotic divisions, a process that further depends on E2F/DP (Umen & Goodenough, 2001; Fang et al, 2006), a SUMO peptidase (Fang & Umen, 2008), and a unique G‐type CDK (Li et al, 2016). Remarkably, periodic expression of cell cycle genes is independent of functional RBR, E2F, and DP in this unicellular alga (Fang et al, 2006), altogether suggesting that an early role in evolution of RBR proteins was likely related to cell size control, rather than cell cycle progression through G1.
Non‐cell cycle functions of RBR proteins are also found in the colonial alga Volvox carteri, where the RBR1 gene has a gender‐specific expression pattern in female cells (Kianianmomeni et al, 2008). In these cells, up to four differentially spliced products of the RBR1 gene have been identified (Kianianmomeni et al, 2008; Hallmann, 2009; Ferris et al, 2010), suggesting a multifunctional role of RBR1 in different processes that still need to be delineated.
Transcriptional waves controlled by RBR1 during the cell cycle
As in animal cells, two major transcriptional waves can be distinguished in plants during G1 and G2 phases, which regulate expression of gene products required for S‐phase and mitotic progression, respectively.
RBR1‐E2F complexes
In the case of G1, several lines of evidence demonstrate that the key role of the RBR1‐E2F module includes the negative RBR1 regulation by CDK/cyclin complexes, the counteracting CDK inhibitors (KRPs for Kip‐related proteins), and the participation of chromatin remodeling enzymes, such as histone acetyl transferases (HATs), and subsequent recruitment of RNA polymerase II at the target promoters (Fischer & Müller, 2017). High levels of E2FA/DPA or E2FC/DPB expression lead to misregulation of E2F target genes and developmental abnormalities in Arabidopsis (De Veylder et al, 2002; del Pozo et al, 2002, 2006). Modest overexpression of E2FA or CYCD3 directly affects expression of their target genes and demonstrated that E2FC does not counteract E2FA‐mediated gene upregulation (see also discussion of E2FC roles in G2, below). Furthermore, cell wall biogenesis depends on E2FA (de Jager et al, 2009) and on E2FF/DEL3 (Ramirez‐Parra & Gutierrez, 2007). Direct RBR1 regulation by CDKA;1 was shown using cdka;1 mutants able to rescue the rbr1 mutant phenotype (Nowack et al, 2012). However, RBR1 is not exclusively regulated by CDKA;1, and S6K was found as another RBR1‐interacting kinase phosphorylating it and repressing cell proliferation by inhibiting E2FB factors (Henriques et al, 2010, 2013). Interestingly, RBR1 can form a stable repressor complex with E2FA but not with E2FB. In the proliferation area, differentiation genes such as CCS52A1 and CCS52A2 (encoding activators of the anaphase promoting complex/cyclosome (APC/C)), are repressed under conditions of high cyclin D/CDK activity (Magyar et al, 2012).
Other upstream regulators included the KRP family of CDK inhibitors, formed by seven members in Arabidopsis, and the F‐box‐like protein FBL17 (Kim et al, 2008; Zhao et al, 2012; Noir et al, 2015). FBL17 is itself regulated by the RBR1/E2F pathway and at the same time (as subunit of an SCF ubiquitin ligase) mediates degradation of several downstream targets of E2F, e.g., CDT1a (Desvoyes et al, 2019). CDK activity is also inhibited by another family of proteins, SIAMESE (SIM) and SIAMESE‐RELATED (SMR), and some SMRs are under control of the TARGET OF RAPAMYCIN (TOR) signaling pathway (Ahmad et al, 2019; Barrada et al, 2019). Therefore, RBR1‐mediated gene expression in G1 is controlled by multiple redundant pathways including kinases, inhibitors, and ubiquitin/proteasome‐dependent degradation, to finely balance the availability of gene products required for G1/S transition and S‐phase progression (Fig 3A and B).
Figure 3. Role of Arabidopsis RBR1 complexes in transcriptional control during the cell cycle.
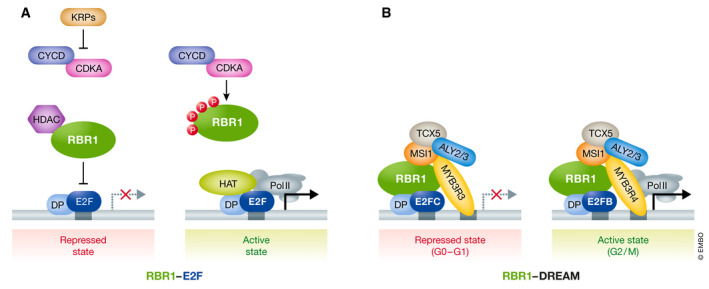
(A) RBR1‐E2F complexes. Genes required for the G1/S transition are bound by E2F‐DP heterodimers located at the E2F binding sites in their promoters (grey box). They are repressed by the retinoblastoma‐related (RBR1) protein in association with histone deacetylases (HDAC). At this stage, CDKA‐cyclin complexes are inactivated by one or more CDK inhibitors (KRPs). In Arabidopsis, E2FA‐C, bound to DP partners, participate in regulation of different gene targets. Later in G1, when CYCD levels are sufficiently high, CDKA phosphorylates RBR1 (small red circles) leaving free the E2F‐DP complexes to transactivate their target genes, in association with histone acetylases (HAT) after recruitment of RNA polymerase II (Pol II). (B) RBR1‐DREAM complexes. RBR1 also participates in other transcriptional regulatory complexes. Briefly, at the repressed state, while CDK are inactive, E2FC is part of the complex together with the repressor MYB3R3 factor and RBR1. E2F and MYB factors bind to different sites in the promoter of target genes (white boxes). The DREAM complex switches to an activator when E2FB and MYB3R4 factors are incorporated (see text for details on composition and function).
DREAM complexes
RB family proteins form parts of multimeric complexes that coordinate transcriptional waves during the cell cycle, originally identified as dREAM in flies (for Drosophila RBF, E2f2 and Multi‐vulval interacting proteins; Lewis et al, 2004), DREAM in mammals (Litovchick et al, 2007), DRM in C. elegans (Harrison et al, 2006), and DREAM‐like in plants (Kobayashi et al, 2015).
In mammalian cells, DREAM complexes are master regulators of the cell cycle that repress gene expression in quiescent cells and in G1 with the participation of the RB‐related pocket proteins p130 and p107 (while RB1 itself is restricted to repressing E2F targets) together with the MuvB core components (LIN‐9, LIN‐37, LIN‐52, LIN‐54 and RBBP4). DREAM regulates gene repression in mammalian G1 phase by two different modes, one dependent on repressor E2F4/5 binding to E2F sites and another on the presence of the DREAM core component LIN‐54 that recognizes a DNA sequence motif called “cell cycle gene homology region” (CHR), also present in the promoters of many G2/M genes. In S‐G2 phase, a different Muv‐Myb complex then forms to regulate genes required for mitosis, a complex assembled by subsequent recruitment of B‐MYB and the forkhead transcription factor FOXM1 (Sadasivam & DeCaprio, 2013; Fischer & Müller, 2017).
In contrast, the Arabidopsis DREAM complex contains both MYB transcription factors and RBR1 alongside orthologues of the core DREAM elements (Fig 3A and B), as shown by mass spectrometry experiments (Kobayashi et al, 2015; Fischer & Müller, 2017; Horvath et al, 2017): ALY2 and ALY3 (ALWAYS EARLY proteins) are orthologues of LIN‐9, TCX5 (Tesmin/TSO1‐like CXC domain protein) is orthologous to LIN54, and MSI1 (MULTI‐COPY SUPRESSOR OF IRA1) is orthologous to RBBP4. While orthologues of LIN‐37 and LIN‐52 have not been found in Arabidopsis, E2FC/B, DPA/B, CDKA (suggestive of phosphorylation as a potential regulator of DREAM activity), and MYB proteins were all found associated with plant DREAM. Arabidopsis contains multiple MYB3R genes (Kobayashi et al, 2015), with MYB3R1 and MYB3R4 regulating the expression of CYCLIN B or KNOLLE (Haga et al, 2007, 2011). The promoters of these G2/M genes contain the MSA (Mitosis‐specific activator) motif, originally identified in tobacco BY‐2 cells (Ito et al, 1998). Analysis of Arabidopsis myb3r3, myb3r5 double mutants (myb3R3/5) showed activation of G2/M genes both in proliferative and mature tissues, supporting a role of Arabidopsis DREAM in gene expression control in G2. A triple myb3R1/3/5 mutant exhibits enlarged organs resulting from increased cell proliferation, revealing that MYB3R3/5 are repressors while MYB3R1 has a dual activator and repressor role (Kobayashi et al, 2015).
Chromatin immunoprecipitation of MYB3R3 has revealed promoters of early cell cycle and mitotic genes that contain E2F and MSA binding sites, respectively, hinting to the existence of distinct DREAM complexes (Kobayashi et al, 2015). In fact, binding sites of RBR1 and MYB3R3 mostly coincide at promoter regions of S‐phase‐ and G2/M‐regulated genes (Bouyer et al, 2018). Furthermore, RBR1, DP, and either repressors E2FC and MYB3R3 or activators E2FB and MYB3R4 are present in the two different complexes acting in G1 and in G2/M, respectively, a situation different from the case of animal cells (Fischer & Müller, 2017). In this regard, it is worth noting that in contrast to animal cells, plant cell G2 phase requires expression of multiple genes regulated by E2F, including CDKB‐type kinases (Boudolf et al, 2004). Additional components of the DREAM complex have been identified by genetic interactions. For example, loss‐of‐function mutations in the TSO1 gene (Andersen et al, 2007), causing overproliferation of meristems and defects in flower development, are suppressed by myb3r1 mutation but not by myb3r4 mutation, although no direct TSO1 interaction with RBR1 was found (Wang et al, 2018).
DREAM target genes appear to extend beyond cell cycle genes. In a recent study, TCX5 was found to repress the expression of MET1, CMT3, DDM1, KYP, and VIMs genes involved in maintenance of DNA methylation (Ning et al, 2020). TCX5 is redundant with its paralogue TCX6, and the double mutant tcx5, tcx6 exhibits increased levels of DNA methylation, primarily at CHG sites. Another recent study identified that SOL1/TCX3 and SOL2/TCX2, two SPEECHLESS targets from the TSO1‐like family, are important regulators of fate transition in the stomatal lineage (see also discussion on RBR1 role in cell fate acquisition in the stomatal lineage). It was hypothesized that they could compete with DREAM for binding sites on DNA (Simmons et al, 2019), but direct evidence for the participation of TSO1/MYB3R1 and SOL1/SOL2 in the DREAM complex is still lacking.
Interplay of RBR1 with chromatin
Early studies in plants, parallel to those in animals, already showed a connection of RBR1 with chromatin components and provided insightful information about its potential functional relevance. Plant RBR1 was found to interact with MSI1, a homolog of human RbAp46/48 (Ach et al, 1997b; Lusser et al, 1999; Rossi et al, 2001), and with the histone deacetylase (HDAC) Rpd3 for repression of target gene expression (Rossi et al, 2003).
Independent studies of flowering control and response to cold stress identified MSI4 (encoded by the FVE gene), a protein that interacts not only with RBR1 but also with HDACs (Ausin et al, 2004; Kim et al, 2004; Pazhouhandeh et al, 2011). Interestingly, fve mutants show increased histone acetylation levels and abnormal silencing of transposable elements (the latter process also affected by RBR1), through effects on cytosine methylation (mC) at CHH and CHG sites (Gu et al, 2011; Xu et al, 2013). Participation in controlling mC levels implies a role in imprinting, which in plants relies on removing silencing marks. Consistent with this, RBR1 interacts with the mC demethylase DEMETER (DME) and represses MET1 (METHYLTRANSFERASE 1), an E2F target gene (Jullien et al, 2008).
Further support for a role of RBR1 in repressing euchromatic genes and in TE silencing comes from more recent genome‐wide mapping of RBR1 binding sites, which colocalize largely with previously identified E2F and MYB3R3 target genes (see also discussion on DREAM; Bouyer et al, 2018). It is worth noting that TEs have amplified E2F binding sites, as revealed by the presence of ~85% of all E2F binding sites in the heterochromatin of Arabidopsis and other Brassicaceae (Henaff et al, 2014).
In addition to the interaction of RBR1 with factors involved in regulating histone acetylation levels, RBR1 also plays a role in maintenance of the repressed state of polycomb (PcG) chromatin. This relies on the physical interaction of RBR1 with PRC2 (polycomb‐repressive complex 2) components such as FIE, CLF, and VNR2 (Mosquna et al, 2004; Guitton & Berger, 2005a; Johnston et al, 2010). Defects in RBR1‐PRC2 interaction result in mutant phenotypes during Arabidopsis gametophyte development and during cell fate acquisition (Johnston et al, 2008, 2010), but elucidation of detailed mechanisms involved in the RBR1‐PRC2 pathway awaits further research (reviewed in Kuwabara & Gruissem, 2014).
RBR1 in the DNA damage response (DDR)
Plant and animal DNA damage responses share several general strategies, but they also exhibit several unique features. In addition to the conserved ATM and ATR pathways, the plant DDR depends on transcriptional activation of plant‐specific target genes such as SOG1, and on epigenetic modifiers (reviewed in Kim, 2019; Nisa et al, 2019). The first hint of the participation of RBR1 in DDR came from the observation that the typical nuclear foci marked by phosphorylated variant histone H2AX (γH2AX) formed after DNA damage contained E2F and depended on an intact RBR1‐binding motif in E2F (Lang et al, 2012). Upon DNA damage, RBR1 and E2FA are recruited to γH2AX foci in an ATM‐ and ATR‐dependent manner (Horvath et al, 2017). This recruitment process stimulated by the plant‐specific CDKB1 not only involves BRCA1, which physically interacts with RBR1, but also the recombinase RAD51 (Biedermann et al, 2017; Horvath et al, 2017). Whether RBR1 serves as a landing pad or as a recruitment factor for DDR proteins that accumulate at sites of genomic DNA damage, or whether it plays additional roles, is not yet fully understood. Many DDR genes are regulated by RBR1 binding to their promoters. Combining transcriptome analysis after DNA damage and genome‐wide analysis of RBR1 binding sites allowed identification of yet uncharacterized DDR genes, such as FIDGETIN‐LIKE‐1 INTERACTING PROTEIN (FLIP) involved in homologous recombination (Bouyer et al, 2018; Fernandes et al, 2018). Moreover, the DREAM complex also participates in the DDR, since repressor MYB3R3 and MYB3R5 suppress expression of a subset of G2/M genes required for cell cycle arrest after DNA damage (Chen et al, 2017). In mutants lacking FBL17, a major regulator of cell cycle progression and endoreplication, DDR genes are constitutively upregulated in the absence of DNA damage (Gentric et al, 2020). This FBL17‐mediated DDR gene regulation is SOG1‐independent, and recruitment of FBL17 to double‐strand breaks (DSB) leads to its colocalization with γH2AX in a RBR1‐dependent manner. Furthermore, since RBR1 colocalizes with either γH2AX or FBL17 in about 5% of cases and the percentage of foci containing all the three proteins is even lower (~1%), it is likely that recruitment of RBR1 and FBL17 to DNA repair foci is a dynamic process (Gentric et al, 2020). The participation of RBR1 in DSB repair may share some mechanistic aspects with its requirement for the production of meiocytes (Zhao et al, 2017b). Furthermore, an rbr1‐2 allele‐bearing mutant with reduced levels of RBR1, despite showing normal vegetative development, displays reduced chiasma formation during meiotic prophase I (Chen et al, 2011).
RBR1 in asymmetric division and stem cell maintenance
Stem cells are crucial elements in the maintenance of cellular homeostasis, as they ensure a continuous supply of pluripotent cells necessary for organogenesis as well as replenishment of damaged cells during regeneration processes. Stem cells divide asymmetrically at a relatively slow rate and give rise to one cell that remains a stem cell and another cell that initiates acquisition of a given fate while maintaining an active division rate. In the case of plants, where cell movement is precluded by the presence of a cell wall, stem cells remain confined within niches, in direct contact with an organizing center that provides the necessary signals to maintain their stemness nature. The best studied stem cell niches in the plant body are located in the shoot and root apical meristems (Sablowski, 2007; Scheres, 2007). Further cells with stem cell potential are dispersed throughout, responsible for the formation of the stomatal lineage, located in the leaf blade (Han & Torii, 2016; Lee & Bergmann, 2019), among others.
The RBR1‐E2F module plays a crucial role in stem cell maintenance. Thus, reduction of RBR1 or increase of E2FA levels in the root apical meristem (RAM) likewise lead to an increase in stem cell number; vice versa, overexpression of RBR1 leads to a severe reduction of stem cell number (Wildwater et al, 2005). RBR1 is also required cell‐autonomously to limit the division of quiescent center cells in the root meristem (Cruz‐Ramirez et al, 2013) Furthermore, a cdka;1 mutant, albeit viable, exhibits pleiotropic developmental abnormalities, demonstrating that CDKA;1 is part of a pathway contributing to stem cell maintenance by controlling the phosphorylation state of RBR1 (Nowack et al, 2012). In fact, the rbr1‐1 mutation can rescue the stem cell defects in the cdka;1 mutant. The precise phospho‐sites required for this RBR1 function remain to be determined.
The asymmetrical nature of stem cell divisions gives rise to two daughter cells that are frequently different in size, but more importantly, one of the daughters acquires a distinct cell fate. In some cases, the following formative divisions are also asymmetrical. In addition to specific transcription factors required for conferring stemness in various plant organs (De Smet & Beeckman, 2011), there is evidence that cell cycle factors act in a coordinated manner. Indeed, there are two examples where RBR1 is involved in the control of asymmetrical cell division (ACD) and terminal cell fate acquisition: formation of endodermis and cortex in the RAM (Wildwater et al, 2005; Bennett & Scheres, 2010) and in the stomatal lineage (Lee & Bergmann, 2019), respectively.
In the distal part of the RAM, the stem cell that gives rise to the endodermis and cortex cell layers, the so‐called cortex/endodermis initial (CEI), first divides anticlinally and asymmetrically to produce one cell that remains a CEI, and another that becomes a cortex/endodermis initial derivative (CEID). This then divides periclinally and again asymmetrically to form the initials of the endodermis and the cortex cell lineages (Fig 4A and B). SCARECROW (SCR) and SHORTROOT (SHR), two members of the GRAS family of transcription factors, are crucial for the maintenance of the stem cell nature of the CEI (Sabatini et al, 2003). An SHR/SCR heterodimer activates expression of various target genes, one of them being CYCLIND6;1 (CYCD6;1), specifically expressed in the CEID (Sozzani et al, 2010; Cruz‐Ramirez et al, 2012). RBR1 interacts with SHR/SCR through the LxCxE motif of SCR, and RBR1 phosphorylation by CYCD6;1‐CDK releases the transcriptional activity of SHR/SCR that triggers ACD (Fig 4A and B). RBR1 inactivation by phosphorylation likely occurs by a sequential activity of CDKB1;1 and CDKA;1, as revealed by the phenotypes of the corresponding mutants (Cruz‐Ramirez et al, 2012; Weimer et al, 2012). CYCA3;4 is another CDK partner controlling formative divisions in connection to RBR1 phosphorylation, as demonstrated by the abnormal root meristem and stomatal lineage pattern in the presence of high levels of this cyclin (Willems et al, 2020). Once ACD has been successfully completed, daughter cells acquire endodermal or cortex fate depending on the presence or absence of SCR, respectively. An extra layer of regulation that resets the regulatory circuit just before mitosis comes with the proteasome‐mediated degradation of at least SCR and RBR1 (Cruz‐Ramirez et al, 2012). In addition, MED31, a subunit of the Mediator complex, also regulates CYCD6;1 in a SCR/SHR‐dependent manner (Zhang et al, 2018), revealing the participation of different mechanisms to ensure proper ACD. However, the identity of certain cell types in the RAM (e.g., columella stem cells) not only depends on RBR1 but also on other regulatory networks, e.g. specific transcription factors such as SOMBRERO (SMB), FEZ, and auxin response factors that function in parallel (Bennett et al, 2014), revealing a large and still incompletely understood complexity, in the RAM stem cell niche. The participation of multiple RBR1‐independent pathways is reinforced by the demonstration that the topoisomerase TOP1a is required for stele stem cell renewal (Zhang et al, 2016), in a manner modulated by ERF115, originally described as a factor required for controlling division of quiescent center cells (Heyman et al, 2013).
Figure 4. Role of Arabidopsis RBR1 in stem cell maintenance and asymmetric cell division.
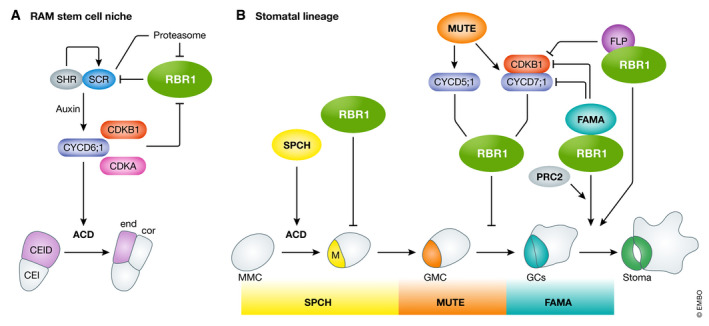
(A) In the stem cell niche of the RAM, the cortex–endodermis initial derivative (CEID) cell undergoes an asymmetric cell division (ACD) that renders the initials of endodermis and cortex already having different fates. This ACD depends on CDK/CYCD6;1, the latter a target of the SHR/SCR heterodimer. The negative regulation of SCR by RBR1 is counteracted by RBR1 phosphorylation. (B) In the stomatal lineage, RBR1 also plays multiple roles including restriction of excessive proliferation of stem cells and the symmetric division of GMC to produce the young guard cells (GCs) as well as the terminal differentiation of GCs into mature guard cells of the stoma. This occurs by association with FAMA and polycomb repressor complex 2 (PRC2). The participation of FLP‐RBR1 in terminal differentiation of GCs is also shown. The three major transcription factors acting in the stomatal lineage, SPCH, MUTE, and FAMA, are shown highlighting their functional windows.
Early studies with RBR1 downregulation showed the need for functional RBR1 levels for proper development of leaf epidermal cells, including the stomatal lineage (Park et al, 2005; Desvoyes et al, 2006; Borghi et al, 2010). Loss of RBR1 function causes overproliferation of cells expressing stomata lineage markers in the leaf epidermis at different developmental stages, consistent with the occurrence of an amplification phase of meristemoids (Fig 4A and B). Moreover, increased expression of E2F targets, e.g., the DNA replication initiation proteins CDT1a or CDC6, leads to increased proliferation of meristemoid cells (Castellano et al, 2001, 2004), underscoring the role of the RBR1‐E2F module in regulating cell division potential in the leaf epidermis.
In addition, formative divisions and cell fate acquisition in the stomatal lineage that give rise to mature stomata require the concerted action of various transcription factors and RBR1 (reviewed in Han & Torii, 2016). Protodermal cells in the epidermis of leaf primordia, the meristemoid mother cells (MMC), accumulate SPEECHLESS (SPCH), a substrate of various MAP kinases and of the protein phosphatase PP2A that together regulate SPCH stability (Bian et al, 2020). SPCH triggers asymmetrical cell division in MMCs, giving rise to a pavement cell and an immature meristemoid (M). At this proliferation stage, RBR1 restricts excess divisions of meristemoids, terminating their stem cell activity by the action of MUTE and leading to differentiation into a guard mother cell (GMC). A single symmetrical division of the guard mother cell, in which RBR1 plays its canonical role as a repressor of the G1/S transition, produces two young guard cells (GC). MUTE promotes CYCD7;1 and CYCD5;1 expression in a narrow window to release RBR1 repression by phosphorylation and, importantly, to assure that GMCs undergo only one cell division (Han et al, 2018; Weimer et al, 2018). It is worth noting that this division is also regulated by SOL1/TCX3 and SOL2/TCX2 (Simmons et al, 2019), members of the CHC family of proteins and putative members of DREAM complexes. The direct involvement of RBR1, SOL1, and SOL2 in a DREAM complex has not yet been experimentally confirmed. Finally, FAMA and FOUR LIPS transcription factors are redundantly required for the final differentiation of stomata guard cells, by inhibiting in an RBR1‐mediated manner the expression of the E2F target CDKB1;1, necessary for the symmetric division of GMC (Boudolf et al, 2004; Lee et al, 2014). The FAMA‐RBR1 complex, mediated by the LxCxE motif of FAMA, represses SPCH expression at late stages of stomatal development. The FAMA‐RBR1 interaction is required for full differentiation of GC and maintenance of their cell fate, a process dependent on the PRC2 complex (Hachez et al, 2011; Matos et al, 2014). Downregulation of RBR1 in stomata and expression of an LxCxE‐mutated FAMA unable to bind RBR1 reactivates SPCH expression in mature stomata and induces ACDs to form a stoma‐in‐stoma phenotype (Lee et al, 2014; Matos et al, 2014). Therefore, RBR1 is important for regulation of both cell cycle and cell fate genes to ensure the correct development of the stomatal lineage.
Cell division and differentiation balance during development
We have discussed various mechanisms and pathways in which RBR1 plays a crucial role at the cellular level. These are mainly related to cell cycle control by virtue of its ability to regulate gene expression at the transcriptional level and via its interaction with chromatin remodeling factors. As a consequence of these activities, RBR1 has also fundamental implications at the developmental and organismal level (Harashima & Sugimoto, 2016). The life cycle of plants, exemplified here by Arabidopsis thaliana (Fig 5), is defined by two main phases. The gametophytic phase consists of production of gametes, double fertilization leading to embryo and endosperm, and development of seeds. The sporophytic phase covers most of plant's life and consists of (i) vegetative growth occurring after seed germination, where most organs are formed post‐embryonically, and (ii) the reproductive phase, when developmental and hormonal signals lead to the formation of flowers where gametes are formed (Fig 5). RBR1 is involved at virtually every stage and is crucial for the developmental phase transitions occurring during the entire life cycle.
Figure 5. Role of RBR1 during the Arabidopsis life cycle.
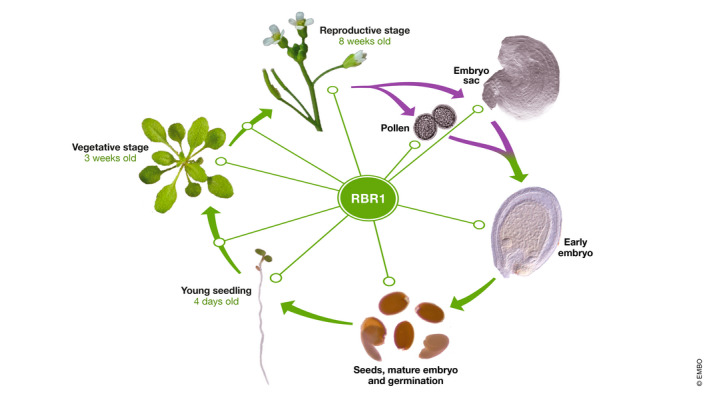
Fully developed embryos present in the seeds grow after germination to produce young seedlings containing cotyledons as well as root and shoot apical meristems responsible for the growth of the root system and the aerial part, respectively. Rosette leaves appear later during vegetative growth. After the transition to the reproductive stage, flowers are produced where some cells specialize into precursors of germinal cells that produce haploid cells after meiosis. These haploid cells divide 2 or 3 times to produce the male and female gametophytes (mature pollen and embryo sac), respectively. After double fertilization, an embryo and the surrounding endosperm are formed inside the seed. Green and violet arrows denote the sporophytic and gametophytic phases, respectively. Most of these developmental stages and transitions are affected by RBR1 (see the text for details).
In the female part, most plants develop gametophytes through three consecutive cell divisions of the haploid products of meiosis, leading to two synergids, one egg cell, one central cell, and three antipodal cells. Here, RBR1 is required to restrict the number of these divisions (Ebel et al, 2004). Embryo formation upon fertilization of the ovule involves rapid cell divisions that are regulated by CYCD7;1 (Sornay et al, 2015), likely through control of RBR1 phosphorylation. At this stage, genes required later during seed maturation, such as LEC1, LEC2, ABI3, or FUSCA, are repressed by E2FA and E2FB, as demonstrated by their upregulation in a e2fa, e2fb double mutant (Leviczky et al, 2019). Surrounding the embryo, endosperm develops after fertilization of the central cell nucleus in a process that is restricted by the interaction of RBR1 with FIE (Mosquna et al, 2004). Furthermore, the possibility of parthenogenetic development of the embryo is restricted by RBR1 through its interaction with MSI1 (Guitton & Berger, 2005a,b). Since RBR1 is expressed across the entire female gametophyte (Ingouff et al, 2006), mechanisms must exist that control RBR1 function in different locations. In monocotyledonous plants, where the endosperm is quantitatively very important, an initial phase of cell proliferation when RBR1 represses the monocot‐specific family member RBR3, precedes the switch to the endoreplication phase (Sabelli et al, 2005; Sabelli & Larkins, 2009). Expression of wheat dwarf virus RepA protein, which blocks RBR1 activity, can up‐regulate RBR3, but not RBR1 (Sabelli et al, 2005). Later, RBR1 is required for repression of the proliferation phase and for promoting the endoreplication phase of endosperm development (Sabelli et al, 2013).
After seed germination, plants initiate the formation of new organs (roots and leaves) and continue doing so during the entire vegetative growth phase (Fig 5). The embryo‐to‐vegetative transition is marked by the switching off and switching on of embryonic and cell cycle genes, respectively. RBR1 participates in both processes but in different ways. Through its interaction with PRC2 complexes, RBR1 facilitates the long‐term maintenance of embryonic genes in a repressed state that depends on establishing a permanent chromatin landscape containing H3K27me3 (Gutzat et al, 2011) and therefore acts as a positive regulator of the embryo‐to‐seedling transition.
Activation of cell cycle gene expression occurs through an increase in CDK/cyclin activity leading to phosphorylation‐dependent inactivation of RBR1's repressor function. This transition has been well studied during maize germination, where changes in the levels of a complex set of D‐type cyclins and KRPs are crucial to control CDK activity (Garza‐Aguilar et al, 2017; Godínez‐Palma et al, 2017). D‐type cyclins directly affect RBR1 phosphorylation levels and the release of E2F activity on target genes required to establish active cell proliferation and to initiate vegetative growth (Sánchez‐Camargo et al, 2020).
The root apex consists of a distal zone in which cells are proliferating, the root apical meristem, and another more proximal area called transition zone, where cells switch to endocycles. E2FA in complex with RBR1 participates in the maintenance of a repressed state of genes such as CCS52A1 and CCS52A2 whose products are needed for the endocycle (Magyar et al, 2012). An analogous situation is found in leaves, where proliferation and endocycle stages are however temporally regulated: In young leaf primordia, most cells are dividing and subsequently enter the endocycle program to increase nuclear ploidy and to expand their size. Nutrient availability is crucial for organ growth in plants. The EbrB‐3 BINDING PROTEIN (EBP1), which is partially regulated by the TOR pathway, promotes root growth in conditions of limiting sucrose supply and counteracts the RBR1‐mediated switch to differentiation (Lokdarshi et al, 2020). Similar to its human counterpart, EBP1 interactors are RNA‐binding proteins that participate in protein biosynthesis, another indication of the role of RBR1 in connecting cell growth and organogenesis in response to nutrient availability. Other pathways connecting RBR1 function to cell growth control, e.g., phosphorylation of RBR1 by casein kinase (Wang et al, 2016), are less well understood at the molecular level.
The role of activator E2F members in the transition from cell proliferation to the endoreplication phase (Fig 5) is balanced by the action of a repressor complex formed by E2FC together with RBR1 (del Pozo et al, 2002). E2FC is degraded by the proteasome, and overexpression of a stable form of E2FC negatively affects cell division by reducing the level of the DNA replication initiation protein CDC6 (Castellano et al, 2001), among other target genes (del Pozo et al, 2002). Conversely, reduction of E2FC leads to the formation of organs with more but smaller cells and a reduction in the nuclear ploidy level (del Pozo et al, 2006). Seedling growth after germination in the absence of light (skotomorphogenesis) is favored by high levels of E2FC and its DPB partner which restricts cell division and promotes endoreplication (del Pozo et al, 2002, 2006; López‐Juez et al, 2007). Upon light availability (photomorphogenesis), the levels of both E2FC and DPB are drastically reduced by proteasome‐dependent degradation mediated by the SCFSKP2a ubiquitin ligase complex, thus allowing to restore active cell proliferation required for organ growth. Trichomes, the specialized epidermal cells, undergo several rounds of endoreplication associated with their genetically defined differentiation program. Among other transcription factors, this depends on the proper expression of GLABRA1 (GL1), a gene than contains E2F binding sites in its promoter and therefore requires RBR1 activity. In this case, a feedback loop is likely active in the protodermal cells that differentiate into trichomes, since genes like RBR1 and SIAMESE (SIM) are direct targets of GL1 and GLABRA3 (GL3; Wenger & Marks, 2008; Morohashi & Grotewold, 2009).
Highly regulated levels of RBR1 are required for proper development of the shoot apical meristem (SAM) and for the growth of leaf primordia. Thus, local and transient overexpression of RBR1 in the SAM causes meristem cells to arrest cell division and initiate differentiation (Wyrzykowska et al, 2006). Later in leaf development, lack of RBR1 function in young leaves primarily engaged in cell proliferation leads to the production of extra cells, while in older leaves where endocycles predominate, an increase in nuclear ploidy is achieved (Park et al, 2005; Desvoyes et al, 2006). In young leaves, RBR1‐E2FB complexes are abundant and of repressive nature, preventing endoreplication and cell differentiation; such complexes disappear concomitantly with leaf development (Henriques et al, 2010, 2013; Őszi et al, 2020). The tissue layer organization of leaves as well as their various cell types are also affected by RBR1. A direct consequence of the RBR1 role in organization of leaf cell layers is its relevance for leaf physiology (Fig 5), as demonstrated by RBR1 control over the amount and distribution of air spaces in‐between mesophyll cells (Dorca‐Fornell et al, 2013; Lehmeier et al, 2017). The regulatory network of the RBR1‐E2F module is also important in other locations, e.g., for differentiation of the vascular system where RBR1 seems to play cell type‐specific roles. Reduction of RBR1 produces a decrease in the number and increase in the size of cortical cells, while the number of xylem parenchyma cells increases (Jordan et al, 2007). This may be due to specific features in the differentiation timing of each cell type. Differentiation of xylem tracheary elements is blocked by XND1, a process that involves XND1 interaction with RBR1 via its LxCxD motif (Zhao et al, 2017a). It is possible that a complex RBR1‐dependent loop operates in tracheary elements, since XND1 is a target of VND7 (Zhong et al, 2010), which in turn is regulated by the repressor E2FC (Taylor‐Teeples et al, 2015). Therefore, it seems that the role of RBR1 in maintaining the balance between cell proliferation and differentiation during xylem development is mediated by XND1, VND7, and possibly other transcription factors, through their direct or indirect interaction with RBR1.
The transition from the vegetative shoot apical meristem to the floral meristem is another crucial phase during plant development (Fig 5). Through its interaction with PRC2 complexes, RBR1 regulates this growth phase transition (Johnston et al, 2008; Jullien et al, 2008; Borghi et al, 2010; Dumbliauskas et al, 2011). In monocotyledonous plants, their multiple RBR members have split roles in flowering. Rice RBR1 is necessary for the establishment and maintenance of the floral meristem and the inner floral organs, by controlling the expression of 10–12 floral homeotic genes, while RBR2 participates in stamen and pollen development (Duan et al, 2019). There are several other processes that are very important for the plant's life. Although less well studied, the transition from dormancy to growth in buds depends on the phosphorylation state of RBR1, with levels of phosphorylated RBR1 being high in dormant buds (Shimizu‐Sato et al, 2008).
RBR1 and pathogen infection
As stated earlier, links between plant viruses (in particular geminiviruses and nanoviruses) and the RBR1 pathways date back to the discovery of RBR1 in plants, when mastreviruses (a geminivirus genus) were found to encode RepA, an RBR1‐interacting protein utilizing a canonical LxCxE motif (Xie et al, 1995; Grafi et al, 1996; Horváth et al, 1998). Later, Rep proteins in begomoviruses (another geminivirus genus) were reported to lack an LxCxE motif but to still interact with RBR1 via a different amino acid motif (Ach et al, 1997a; Gutierrez, 2000; Arguello‐Astorga et al, 2004). A protein binding RBR1 through a canonical motif, Clink, is also found in nanoviruses, a family of plant ssDNA viruses with a multipartite genome (Katul et al, 1998; Aronson et al, 2000). Interactions between geminivirus proteins and the RBR1 pathway are complex and not yet fully understood, since additional viral proteins like AL3/REn (Settlage et al, 2001; Ruhel & Chakraborty, 2019) and C4 (Park et al, 2010; Zeng et al, 2018) also modulate the RBR1 pathway. Moreover, infectivity of geminiviruses requires further interactions with the host cell (McGivern et al, 2005; Arguello‐Astorga et al, 2007).
Irrespective of the whether infections impinge only on the RBR1 pathway or also other pathways, profound changes in the host cells are a primary outcome (Hanley‐Bowdoin et al, 2004; Ascencio‐Ibanez et al, 2008). Modulation of the RBR1 pathway is also seen with various other types of pathogens (Depuydt et al, 2009; Stes et al, 2011; Wen et al, 2012; Villajuana‐Bonequi et al, 2019). An unexpected link between the effector‐triggering immunity (ETI), and its associated programmed cell death, and the RBR1‐ERF module has been reported. The nuclear envelope protein CPR5 negatively regulates ETI by interacting with and suppressing CDK inhibitors, thereby increasing RBR1 hyperphosphorylation and, consequently, the release of E2F activity (Wang et al, 2014). However, further research is needed to better understand the full functional implications of this interaction.
RBR in regeneration and hyperplasia
Regeneration involves a general reprogramming of cells around the wounded area, to first resume cell proliferation and then initiate a process of cell fate acquisition to reform the new organ. It has been shown that the tissue damage response mediated by jasmonic acid is important for stem cell reactivation at least at two complementary levels, one is by impinging on the RBR/SCR pathway and another through activation of CYCD6;1 by the ERF109 transcription factor (Zhou et al, 2019). In fact, members of the ERF/AP2 family of transcription factors appear as key players in the regeneration process as shown for WIND1 (Iwase et al, 2011) and ERF115 (Heyman et al, 2016). Changes in transcriptional activity in the early response to wounding correlate with increased histone acetylation marks (H3K9ac and H3K14ac) deposited by members of the HAG1 family of acetyl transferases in various ERF family genes, including WIND1 (Rymen et al, 2019). Triggering of nuclear reprogramming during regeneration is counterbalanced by mechanisms that prevent unwanted cell dedifferentiation, which in a PRC2‐dependent manner maintain WIND family members like WIND3 and embryonic genes like LEC2 in a repressed state (Ikeuchi et al, 2015). Given the tight link of RBR1 with histone deacetylases and PRC2, it is conceivable that RBR1 also plays a role in regulating regeneration capacity across various plant organs, as demonstrated for the RBR1‐SCR module in the root stem cell niche (Zhou et al, 2019). Thus, the general strategy, although with different molecular players, is similar to the organ regeneration processes described in animals, e.g., in planarians, at the core of whose significant regenerative capacity lies the RB1‐E2F module interacting with chromatin remodeling components (Zhu & Pearson, 2013). This does not mean that the RBR1‐E2F module is the only pathway involved in reprogramming. Others are relevant during dedifferentiation and formation of giant cells in response to root‐knot nematodes through genes controlling lateral root initiation (Olmo et al, 2020) or in plants of different ages through various hormonal pathways (Ye et al, 2020). These pathways frequently serve to regulate the expression of cell cycle genes at the transcriptional level (del Pozo et al, 2005).
A major difference to animals is that high regeneration potential is rather widespread in plants. In addition, their regeneration plasticity is enormous, since they can produce not only particular tissues, but entire organs or entire organisms from a single regeneration event. In all cases, the common path includes dedifferentiation, reprogramming, and cell proliferation and differentiation processes that are extremely well‐coordinated in time and space.
But how do plants deal with extra cells produced during regeneration processes or as a consequence of misregulation of cell proliferation? Plants exhibit a high tolerance to changes in the level and function of key cell cycle genes, including those involved in DNA replication and chromatin dynamics, while such alterations frequently lead to embryonic lethality in animals. Although viruses, bacteria, and fungi can induce tumors in plants (Doonan & Sablowski, 2010), they do not lead to neoplastic transformation as we know it in animals. In fact, loss of the RB1 pathway function is a hallmark of many human cancer cells. In contrast, local loss of RBR1 leads to relatively mild phenotypes in vegetative organs of the adult, e.g., roots and leaves, where visible hyperplasia but no cell transformation occurs (Park et al, 2005; Wildwater et al, 2005; Desvoyes et al, 2006). In these cases, the most frequent behavior of excess cells is to interpret local positional information, acquire the cell fate of surrounding cells, and/or adjust the cell proliferation/endoreplication balance. This eventually allows growth into organs of near‐normal size, containing more but smaller cells. At the same time, this plasticity of plants has to coexist with plant‐specific features, such as the post‐embryonic nature of their organogenesis, and their ability to form organs in a continuous manner throughout a plants’ life. This could be potentially very dangerous when considering risks associated with perturbed cell proliferation control, since it involves numerous dedifferentiation/proliferation/differentiation decisions. In any case, it should be kept in mind that in plants, additional key factors likely contribute to the absence of neoplastic transformation as we understand it in animal cells. These include primarily the specific developmental mode of plants and the absence of metastasis, since cell movements inside the plant body are precluded by the presence of cell walls that maintain plant cells glued together and fixed at the very position where they were originally formed.
Future perspectives
Accumulating evidence gathered over the past 25 years demonstrates that RBR1 participates in multiple pathways, with a variety of roles at different levels of plant physiology. In all cases, its interactions with cellular factors, including transcriptional regulators, chromatin remodeling complexes or other interacting proteins, are at the molecular basis of RBR1 function. However, we still miss detailed insights into the fine regulation of RBR1 interactions, which frequently depend on RBR1 phosphorylation state modulated by various kinases. Therefore, identifying the role of RBR1 phosphorylation at specific sites as well as the CDK/cyclin complexes involved would be necessary to gain more information about how and when during the cell cycle or during differentiation phospho‐RBR1 isoforms exert their action. Such information shall also help to understand the kinetics of complex formation with the different RBR1‐interacting factors. Given the conservation of amino acid sequences around potential phosphorylation sites, a possible way to explore is the use of antibodies available for phosphorylated forms of human RB1 in combination with the generation of site‐specific phosphorylation mutants. These and other protein interactions of RBR1 depend on the presence of intrinsically disordered RBR1 regions, which could be crucial in the formation of protein condensates by liquid‐liquid phase separation.
The understanding of the role of RBR1 complexes with transcription factors of different families is still at its beginning. Following identification of a given interaction, the dynamics of such interaction should provide information about spatial and temporal activity of a given such complex and help to explain the role of RBR1 in different organs and at different developmental stages. Such studies need to be combined with detailed knowledge of RBR1 complex availability, e.g., its protein expression, modification, and degradation. Thus, identification of E2F targets and the various E2F family members in relation to RBR1 will need to be done not only genome‐wide, but also at an individual scale, in order to reveal the basis for their spatial and temporal regulation, which is highly dependent on the target under study. In this context, the role of DREAM complexes and their targets in controlling cell cycle progression and the transition to differentiated states need to be understood at the molecular level. Again, a detailed map of RBR1 interactions and the individual circuitries at the cellular level is still far from being understood. Furthermore, expanding these studies to the response of plants to their environment should be important for the understanding of RBR1 roles at the organismal and population levels.
In summary, there are numerous avenues to be explored at the molecular, cellular, developmental, and environmental levels before we obtain a general picture of the multifunctional roles played by RBR1 in plant physiology. The future should see rich advances in these and other directions.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
The authors are indebted to I. Ruiz‐Trillo, M. Aldea and S. Ramon‐Maiques for discussion on RBR1 evolution, the role of Whi5 protein, and RBR1 structure, respectively, and to E. Martinez‐Salas for comments on the manuscript. Our apologies to colleagues whose publications have not been included here due to space limitations. Research in the C.G. laboratory is funded by grants BIO2017‐92329‐EXP (MICIU), RTI2018‐094793‐B‐I00 (MICIU and FEDER) and ERC‐2018‐AdG_833617 (EU), and by institutional grants from Banco de Santander and Fundación Ramon Areces to the CBMSO.
The EMBO Journal (2020) 39: e105802
References
- Abrahám E, Miskolczi P, Ayaydin F, Yu P, Kotogány E, Bakó L, Otvös K, Horváth GV, Dudits D (2011) Immunodetection of retinoblastoma‐related protein and its phosphorylated form in interphase and mitotic alfalfa cells. J Exp Bot 62: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ach RA, Durfee T, Miller AB, Taranto P, Hanley‐Bowdoin L, Zambryski PC, Gruissem W (1997a) RRB1 and RRB2 encode maize retinoblastoma‐related proteins that interact with a plant D‐type cyclin and geminivirus replication protein. Mol Cell Biol 17: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ach RA, Taranto P, Gruissem W (1997b) A conserved family of WD‐40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell 9: 1595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad Z, Magyar Z, Bögre L, Papdi C (2019) Cell cycle control by the target of rapamycin signalling pathway in plants. J Exp Bot 70: 2275–2284 [DOI] [PubMed] [Google Scholar]
- Albani D, Mariconti L, Ricagno S, Pitto L, Moroni C, Helin K, Cella R (2000) DcE2F, a functional plant E2F‐like transcriptional activator from Daucus carota . J Biol Chem 275: 19258–19267 [DOI] [PubMed] [Google Scholar]
- Andersen SU, Algreen‐Petersen RG, Hoedl M, Jurkiewicz A, Cvitanich C, Braunschweig U, Schauser L, Oh S‐A, Twell D, Jensen EØ (2007) The conserved cysteine‐rich domain of a tesmin/TSO1‐like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana . J Exp Bot 58: 3657–3670 [DOI] [PubMed] [Google Scholar]
- Arguello‐Astorga G, Lopez‐Ochoa L, Kong LJ, Orozco BM, Settlage SB, Hanley‐Bowdoin L (2004) A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma‐related protein. J Virol 78: 4817–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello‐Astorga G, Ascencio‐Ibáñez JT, Dallas MB, Orozco BM, Hanley‐Bowdoin L (2007) High‐frequency reversion of geminivirus replication protein mutants during infection. J Virol 81: 11005–11015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson MN, Meyer AD, Györgyey J, Katul L, Vetten HJ, Gronenborn B, Timchenko T (2000) Clink, a nanovirus‐encoded protein, binds both pRB and SKP1. J Virol 74: 2967–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio‐Ibanez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley‐Bowdoin L (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Alonso‐Blanco C, Jarillo JA, Ruiz‐Garcia L, Martinez‐Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma‐associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Barrada A, Djendli M, Desnos T, Mercier R, Robaglia C, Montané M‐H, Menand B (2019) A TOR‐YAK1 signaling axis controls cell cycle, meristem activity and plant growth in Arabidopsis . Development 146: dev171298 [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B (2010) Root development‐two meristems for the price of one? Curr Top Dev Biol 91: 67–102 [DOI] [PubMed] [Google Scholar]
- Bennett T, van den Toorn A, Willemsen V, Scheres B (2014) Precise control of plant stem cell activity through parallel regulatory inputs. Development 141: 4055–4064 [DOI] [PubMed] [Google Scholar]
- Bian C, Guo X, Zhang Y, Wang L, Xu T, DeLong A, Dong J (2020) Protein phosphatase 2A promotes stomatal development by stabilizing SPEECHLESS in Arabidopsis. Proc Natl Acad Sci USA 117: 13127–13137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann S, Harashima H, Chen P, Heese M, Bouyer D, Sofroni K, Schnittger A (2017) The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J 36: 1279–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti MB, Gutierrez C (2001) A cell‐cycle‐regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J 28: 341–350 [DOI] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Futterer J, Laizet Y, Hennig L, Gruissem W (2010) Arabidopsis RETINOBLASTOMA‐RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L (2004) The plant‐specific cyclin‐dependent kinase CDKB1;1 and transcription factor E2Fa‐DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer D, Heese M, Chen P, Harashima H, Roudier F, Grüttner C, Schnittger A (2018) Genome‐wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLoS Genet 14: e1007797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RAM, McDonald WH, Kalashnikova TI, Yates J, Wittenberg C (2004) Cln3 activates G1‐specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117: 887–898 [DOI] [PubMed] [Google Scholar]
- Buchkovich K, Duffy LA, Harlow E (1989) The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58: 1097–1105 [DOI] [PubMed] [Google Scholar]
- Burke JR, Deshong AJ, Pelton JG, Rubin SM (2010) Phosphorylation‐induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem 285: 16286–16293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, del Pozo JC, Ramirez‐Parra E, Brown S, Gutierrez C (2001) Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13: 2671–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, Boniotti MB, Caro E, Schnittger A, Gutierrez C (2004) DNA replication licensing affects cell proliferation or endoreplication in a cell type‐specific manner. Plant Cell 16: 2380–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR (1991) The E2F transcription factor is a cellular target for the RB protein. Cell 65: 1053–1061 [DOI] [PubMed] [Google Scholar]
- Chen Z, Higgins JD, Hui JTL, Li J, Franklin FCH, Berger F (2011) Retinoblastoma protein is essential for early meiotic events in Arabidopsis. EMBO J 30: 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Takatsuka H, Takahashi N, Kurata R, Fukao Y, Kobayashi K, Ito M, Umeda M (2017) Arabidopsis R1R2R3‐Myb proteins are essential for inhibiting cell division in response to DNA damage. Nat Commun 8: 635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M (2004) CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913 [DOI] [PubMed] [Google Scholar]
- Cruz‐Ramirez A, Diaz‐Trivino S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P et al (2012) A bistable circuit involving SCARECROW‐RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz‐Ramirez A, Diaz‐Trivino S, Wachsman G, Du Y, Arteaga‐Vazquez M, Zhang H, Benjamins R, Blilou I, Neef AB, Chandler V et al (2013) A SCARECROW‐RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol 11: e1001724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl M, Meskiene I, Bogre L, Ha DT, Swoboda I, Hubmann R, Hirt H, Heberle‐Bors E (1995) The D‐type alfalfa cyclin gene cycMs4 complements G1 cyclin‐deficient yeast and is induced in the G1 phase of the cell cycle. Plant Cell 7: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clerck O, Kao SM, Bogaert KA, Blomme J, Foflonker F, Kwantes M, Vancaester E, Vanderstraeten L, Aydogdu E, Boesger J et al (2018) Insights into the evolution of multicvellularity from the sea lettuce genome. Curr Biol 28: 2921–2933 [DOI] [PubMed] [Google Scholar]
- De Smet I, Beeckman T (2011) Asymmetric cell division in land plants and algae: the driving force for differentiation. Nat Rev Mol Cell Biol 12: 177–188 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G et al (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa‐DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM (1988) SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54: 275–283 [DOI] [PubMed] [Google Scholar]
- DeCaprio JA (2009) How the Rb tumor suppressor structure and function was revealed by the study of Adenovirus and SV40. Virology 384: 274–284 [DOI] [PubMed] [Google Scholar]
- Depuydt S, De Veylder L, Holsters M, Vereecke D (2009) Eternal youth, the fate of developing Arabidopsis leaves upon Rhodococcus fascians infection. Plant Physiol 149: 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez‐Parra E, Xie Q, Chua NH, Gutierrez C (2006) Cell type‐specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Mendoza AD, Ruiz‐Trillo I, Gutierrez C (2014) Novel roles of plant RETINOBLASTOMA‐RELATED (RBR) protein in cell proliferation and asymmetric cell division. J Exp Bot 65: 2657–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Noir S, Masoud K, López MI, Genschik P, Gutierrez C (2019) FBL17 targets CDT1a for degradation in early S‐phase to prevent Arabidopsis genome instability. bioRxiv 774109 [Google Scholar]
- Dieck CB, Wood A, Brglez I, Rojas‐Pierce M, Boss WF (2012) Increasing phosphatidylinositol (4,5) bisphosphate biosynthesis affects plant nuclear lipids and nuclear functions. Plant Physiol Biochem 57: 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan JH, Sablowski R (2010) Walls around tumours ‐ why plants do not develop cancer. Nat Rev Cancer 10: 794–802 [DOI] [PubMed] [Google Scholar]
- Dorca‐Fornell C, Pajor R, Lehmeier C, Pérez‐Bueno M, Bauch M, Sloan J, Osborne C, Rolfe S, Sturrock C, Mooney S et al (2013) Increased leaf mesophyll porosity following transient retinoblastoma‐related protein silencing is revealed by microcomputed tomography imaging and leads to a system‐level physiological response to the altered cell division pattern. Plant J 76: 914–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Chen Y, Li W, Pan M, Qu X, Shi X, Cai Z, Liu H, Zhao F, Kong L et al (2019) RETINOBLASTOMA‐RELATED genes specifically control inner floral organ morphogenesis and pollen development in rice. Plant Physiol 181: 1600–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbliauskas E, Lechner E, Jaciubek M, Berr A, Pazhouhandeh M, Alioua M, Cognat V, Brukhin V, Koncz C, Grossniklaus U et al (2011) The Arabidopsis CUL4‐DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J 30: 731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Münger K, Harlow E (1989) The human papilloma virus‐16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243: 934–937 [DOI] [PubMed] [Google Scholar]
- Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Fang S‐C, de los Reyes C, Umen JG (2006) Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet 2: e167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S‐C, Umen JG (2008) A suppressor screen in chlamydomonas identifies novel components of the retinoblastoma tumor suppressor pathway. Genetics 178: 1295–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T, Hansen K, Holm K, Lukas J, Bartek J (2002) Distinct phosphorylation events regulate p130‐ and p107‐mediated repression of E2F‐4. J Biol Chem 277: 26741–26742 [DOI] [PubMed] [Google Scholar]
- Feiler HS, Jacobs TW (1990) Cell division in higher plants: a cdc2 gene, its 34‐kDa product, and histone H1 kinase activity in pea. Proc Natl Acad Sci USA 87: 5397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes JB, Duhamel M, Seguéla‐Arnaud M, Froger N, Girard C, Choinard S, Solier V, De Winne N, De Jaeger G, Gevaert K et al (2018) FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet 14: e1007317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Hemerly AS, Villarroel R, Van Montagu M, Inze D (1991) The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell 3: 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P, Olson BJSC, De Hoff PL, Douglass S, Casero D, Prochnik S, Geng S, Rai R, Grimwood J, Schmutz J et al (2010) Evolution of an expanded sex‐determining locus in Volvox. Science 328: 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Müller GA (2017) Cell cycle transcription control: DREAM/MuvB and RB‐E2F complexes. Crit Rev Biochem Mol Biol 52: 638–662 [DOI] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP (1986) A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646 [DOI] [PubMed] [Google Scholar]
- Fung YK, Murphree AL, T'Ang A, Qian J, Hinrichs SH, Benedict WF (1987) Structural evidence for the authenticity of the human retinoblastoma gene. Science 236: 1657–1661 [DOI] [PubMed] [Google Scholar]
- Garza‐Aguilar SM, Lara‐Núñez A, García‐Ramírez E, Vázquez‐Ramos JM (2017) Modulation of CycD3;1‐CDK complexes by phytohormones and sucrose during maize germination. Physiol Plant 160: 84–97 [DOI] [PubMed] [Google Scholar]
- Gentric N, Masoud K, Journot RP, Cognat V, Chabouté M‐E, Noir S, Genschik P (2020) The F‐box‐like protein FBL17 is a regulator of DNA‐damage response and co‐localizes with RETINOBLASTOMA RELATED 1 at DNA lesion sites. Plant Physiol 183: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godínez‐Palma SK, Rosas‐Bringas FR, Rosas‐Bringas OG, García‐Ramírez E, Zamora‐Zaragoza J, Vázquez‐Ramos JM (2017) Two maize Kip‐related proteins differentially interact with, inhibit and are phosphorylated by cyclin D‐cyclin‐dependent kinase complexes. J Exp Bot 68: 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG Jr (1996) A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc Natl Acad Sci USA 93: 8962–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Jiang D, Yang W, Jacob Y, Michaels SD, He Y (2011) Arabidopsis homologs of retinoblastoma‐associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet 7: e1002366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton AE, Berger F (2005a) Control of reproduction by Polycomb Group complexes in animals and plants. Int J Dev Biol 49: 707–716 [DOI] [PubMed] [Google Scholar]
- Guitton AE, Berger F (2005b) Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr Biol 15: 750–754 [DOI] [PubMed] [Google Scholar]
- Gutierrez C (2000) DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J 19: 792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R, Quiroz‐Figueroa F, Vázquez‐Ramos JM (2005) Maize cyclin D2 expression, associated kinase activity and effect of phytohormones during germination. Plant Cell Physiol 46: 166–173 [DOI] [PubMed] [Google Scholar]
- Gutzat R, Borghi L, Futterer J, Bischof S, Laizet Y, Hennig L, Feil R, Lunn J, Gruissem W (2011) RETINOBLASTOMA‐RELATED PROTEIN controls the transition to autotrophic plant development. Development 138: 2977–2986 [DOI] [PubMed] [Google Scholar]
- Gutzat R, Borghi L, Gruissem W (2014) Emerging roles of RETINOBLASTOMA‐RELATED proteins in evolution and plant development. Trends Plant Sci 17: 139–148 [DOI] [PubMed] [Google Scholar]
- Hachez C, Ohashi‐Ito K, Dong J, Bergmann DC (2011) Differentiation of Arabidopsis guard cells: analysis of the networks incorporating the basic helix‐loop‐helix transcription factor, FAMA. Plant Physiol 155: 1458–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Muller I, Voss U, Jurgens G et al (2007) R1R2R3‐Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Haga N, Kobayashi K, Suzuki T, Maeo K, Kubo M, Ohtani M, Mitsuda N, Demura T, Nakamura K, Jürgens G et al (2011) Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down‐regulation of multiple G2/M‐specific genes in Arabidopsis. Plant Physiol 157: 706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A (2009) Key elements of the retinoblastoma tumor suppressor pathway in Volvox carteri . Commun Integr Biol 2: 396–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S‐K, Torii KU (2016) Lineage‐specific stem cells, signals and asymmetries during stomatal development. Development 143: 1259–1270 [DOI] [PubMed] [Google Scholar]
- Han S‐K, Qi X, Sugihara K, Dang JH, Endo TA, Miller KL, Kim E‐D, Miura T, Torii KU (2018) MUTE directly orchestrates cell‐state switch and the single symmetric division to create stomata. Dev Cell 45: 303–315 [DOI] [PubMed] [Google Scholar]
- Hanley‐Bowdoin L, Settlage SB, Robertson D (2004) Reprogramming plant gene expression: A prerequisite to geminivirus DNA replication. Mol Plant Pathol 5: 149–156 [DOI] [PubMed] [Google Scholar]
- Hansen K, Farkas T, Lukas J, Holm K, Rönnstrand L, Bartek J (2001) Phosphorylation‐dependent and ‐independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J 20: 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H, Sugimoto K (2016) Integration of developmental and environmental signals into cell proliferation and differentiation through RETINOBLASTOMA‐RELATED. Curr Opin Plant Biol 29: 95–103 [DOI] [PubMed] [Google Scholar]
- Harrison MM, Ceol CJ, Lu X, Horvitz HR (2006) Some C. elegans class B synthetic multivulva proteins encode a conserved LIN‐35 Rb‐containing complex distinct from a NuRD‐like complex. Proc Natl Acad Sci USA 103: 16782–16787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata S, Kouchi H, Suzuka I, Ishii T (1991) Isolation and characterization of cDNA clones for plant cyclins. EMBO J 10: 2681–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A, Bergounioux C, Van Montagu M, Inze D, Ferreira P (1992) Genes regulating the plant cell cycle: isolation of a mitotic‐like cyclin from Arabidopsis thaliana. Proc Natl Acad Sci USA 89: 3295–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henaff E, Vives C, Desvoyes B, Chaurasia A, Payet J, Gutierrez C, Casacuberta JM (2014) Extensive amplification of the E2F transcription factor binding sites by transposons during evolution of Brassica species. Plant J 77: 852–862 [DOI] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bogre L (2010) Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1‐E2F pathway activity. EMBO J 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Bögre L (2013) S6K1 and E2FB are in mutually antagonistic regulatory links controlling cell growth and proliferation in Arabidopsis. Plant Signal Behav 8: e24367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, Heyndrickx KS, Van Leene J, Vercauteren I, Vanderauwera S, Vandepoele K, De Jaeger G, Van Der Straeten D et al (2013) ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863 [DOI] [PubMed] [Google Scholar]
- Heyman J, Cools T, Canher B, Shavialenka S, Traas J, Vercauteren I, Van den Daele H, Persiau G, De Jaeger G, Sugimoto K et al (2016) The heterodimeric transcription factor complex ERF115‐PAT1 grants regeneration competence. Nat Plants 2: 16165 [DOI] [PubMed] [Google Scholar]
- Hirano H, Harashima H, Shinmyo A, Sekine M (2008) Arabidopsis retinoblastoma‐related protein 1 is involved in G1 phase cell cycle arrest caused by sucrose starvation. Plant Mol Biol 66: 259–275 [DOI] [PubMed] [Google Scholar]
- Hirt H, Mink M, Pfosser M, Bogre L, Gyorgyey J, Jonak C, Gartner A, Dudits D, Heberle‐Bors E (1992) Alfalfa cyclins: differential expression during the cell cycle and in plant organs. Plant Cell 4: 1531–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath BM, Kourova H, Nagy S, Nemeth E, Magyar Z, Papdi C, Ahmad Z, Sanchez‐Perez GF, Perilli S, Blilou I et al (2017) Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J 36: 1261–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth GV, Pettkó‐Szandtner A, Nikovics K, Bilgin M, Boulton M, Davies JW, Gutiérrez C, Dudits D (1998) Prediction of functional regions of the maize streak virus replication‐associated proteins by protein‐protein interaction analysis. Plant Mol Biol 38: 699–712 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Rymen BH, Harashima H, Shibata M, Ohnuma M, Breuer C, Morao AK, de Lucas M, De Veylder L et al (2015) PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat Plants 1: 15089 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Jullien PE, Berger F (2006) The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell 18: 3491–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A (1998) A novel cis‐acting element in promoters of plant B‐type cyclin genes activates M phase‐specific transcription. Plant Cell 10: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K et al (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- de Jager SM, Scofield S, Huntley RP, Robinson AS, den Boer BGW, Murray JAH (2009) Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol 71: 345–365 [DOI] [PubMed] [Google Scholar]
- John PC, Sek FJ, Lee MG (1989) A homolog of the cell cycle control protein p34cdc2 participates in the division cycle of Chlamydomonas, and a similar protein is detectable in higher plants and remote taxa. Plant Cell 1: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Matveeva E, Kirioukhova O, Grossniklaus U, Gruissem W (2008) A dynamic reciprocal RBR‐PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr Biol 18: 1680–1686 [DOI] [PubMed] [Google Scholar]
- Johnston AJ, Kirioukhova O, Barrell PJ, Rutten T, Moore JM, Baskar R, Grossniklaus U, Gruissem W (2010) Dosage‐sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genet 6: e1000988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CV, Shen W, Hanley‐Bowdoin LK, Robertson DN (2007) Geminivirus‐induced gene silencing of the tobacco retinoblastoma‐related gene results in cell death and altered development. Plant Mol Biol 65: 163–175 [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nishikawa JL, Breitkreutz B‐J, Tyers M (2002) Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F (2008) Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol 6: e194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katul L, Timchenko T, Gronenborn B, Vetten HJ (1998) Ten distinct circular ssDNA components, four of which encode putative replication‐associated proteins, are associated with the faba bean necrotic yellows virus genome. J Gen Virol 79: 3101–3109 [DOI] [PubMed] [Google Scholar]
- Kawamura K, Murray JAH, Shinmyo A, Sekine M (2006) Cell cycle regulated D3‐type cyclins form active complexes with plant‐specific B‐type cyclin‐dependent kinase in vitro. Plant Mol Biol 61: 311–327 [DOI] [PubMed] [Google Scholar]
- Kianianmomeni A, Nematollahi G, Hallmann A (2008) A gender‐specific retinoblastoma‐related protein in volvox carteri implies a role for the retinoblastoma protein family in sexual development. Plant Cell 20: 2399–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Kim HJ, Lee MH, Moon J, Lee I et al (2004) A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana . Nat Genet 36: 167–171 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, Twell D, Nam HG (2008) Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature 455: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Kim J‐H (2019) Chromatin remodeling and epigenetic regulation in plant DNA damage repair. Int J Mol Sci 20: 4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Suzuki T, Iwata E, Nakamichi N, Suzuki T, Chen P, Ohtani M, Ishida T, Hosoya H, Muller S et al (2015) Transcriptional repression by MYB3R proteins regulates plant organ growth. EMBO J 34: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) E2Ls, E2F‐like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J Biol Chem 277: 16553–16558 [DOI] [PubMed] [Google Scholar]
- Kuwabara A, Gruissem W (2014) Arabidopsis Retinoblastoma‐related and Polycomb group proteins: cooperation during plant cell differentiation and development. J Exp Bot 65: 2667–2676 [DOI] [PubMed] [Google Scholar]
- Lammens T, Li J, Leone G, De Veylder L (2009) Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol 19: 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis MW, Brown NE, Baker GL, Shifrin A, Das M, Geng Y, Sicinski P, Hinds PW (2007) The LxCxE pRb interaction domain of cyclin D1 is dispensable for murine development. Cancer Res 67: 7613–7620 [DOI] [PubMed] [Google Scholar]
- Lang J, Smetana O, Sanchez‐Calderon L, Lincker F, Genestier J, Schmit A‐C, Houlné G, Chabouté M‐E (2012) Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol 194: 353–363 [DOI] [PubMed] [Google Scholar]
- Lee MG, Nurse P (1987) Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327: 31–35 [DOI] [PubMed] [Google Scholar]
- Lee WH, Shew JY, Hong FD, Sery TW, Donoso LA, Young LJ, Bookstein R, Lee EY (1987) The retinoblastoma susceptibility gene encodes a nuclear phosphoprotein associated with DNA binding activity. Nature 329: 642–645 [DOI] [PubMed] [Google Scholar]
- Lee E, Lucas JR, Sack FD (2014) Deep functional redundancy between FAMA and FOUR LIPS in stomatal development. Plant J 78: 555–565 [DOI] [PubMed] [Google Scholar]
- Lee LR, Bergmann DC (2019) The plant stomatal lineage at a glance. J Cell Sci 132: jcs228551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JA, Buchkovich KJ, Marshak DR, Anderson CW, Harlow E (1991) The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J 10: 4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmeier C, Pajor R, Lundgren MR, Mathers A, Sloan J, Bauch M, Mitchell A, Bellasio C, Green A, Bouyer D et al (2017) Cell density and airspace patterning in the leaf can be manipulated to increase leaf photosynthetic capacity. Plant J 92: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvai A, Pettkó‐Szandtner A, Csordás‐Tóth E, Miskolczi P, Horváth GV, Györgyey J, Dudits D (2007) Dicot and monocot plants differ in retinoblastoma‐related protein subfamilies. J Exp Bot 58: 1663–1675 [DOI] [PubMed] [Google Scholar]
- Leviczky T, Molnár E, Papdi C, Őszi E, Horváth GV, Vizler C, Nagy V, Pauk J, Bögre L, Magyar Z (2019) E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development 146: dev179333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR (2004) Identification of a Drosophila Myb‐E2F2/RBF transcriptional repressor complex. Genes Dev 18: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu D, López‐Paz C, Olson BJ, Umen JG (2016) A new class of cyclin dependent kinase in Chlamydomonas is required for coupling cell size to cell division. eLife 5: e10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA (2007) Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle‐dependent genes in quiescence. Mol Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- Lokdarshi A, Papdi C, Pettkó‐Szandtner A, Dorokhov S, Scheres B, Magyar Z, von Arnim AG, Bögre L, Horváth BM (2020) ErbB‐3 BINDING PROTEIN 1 regulates translation and counteracts RETINOBLASTOMA RELATED to maintain the root meristem. Plant Physiol 182: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Juez E, Bowyer JR, Sakai T (2007) Distinct leaf developmental and gene expression responses to light quantity depend on blue‐photoreceptor or plastid‐derived signals, and can occur in the absence of phototropins. Planta 227: 113–123 [DOI] [PubMed] [Google Scholar]
- Ludlow JW, DeCaprio JA, Huang CM, Lee WH, Paucha E, Livingston DM (1989) SV40 large T antigen binds preferentially to an underphosphorylated member of the retinoblastoma susceptibility gene product family. Cell 56: 57–65 [DOI] [PubMed] [Google Scholar]
- Lusser A, Eberharter A, Loidl A, Goralik‐Schramel M, Horngacher M, Haas H, Loidl P (1999) Analysis of the histone acetyltransferase B complex of maize embryos. Nucleic Acids Res 27: 4427–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Atanassova A, De Veylder L, Rombauts S, Inze D (2000) Characterization of two distinct DP‐related genes from Arabidopsis thaliana . FEBS Lett 486: 79–87 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Horvath B, Khan S, Mohammed B, Henriques R, De Veylder L, Bako L, Scheres B, Bogre L (2012) Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR‐bound and RBR‐free complexes. EMBO J 31: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D (2002) The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Matos JL, Lau OS, Hachez C, Cruz‐Ramirez A, Scheres B, Bergmann DC (2014) Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA‐RELATED module. Elife 3: e03271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivern DR, Findlay KC, Montague NP, Boulton MI (2005) An intact RBR‐binding motif is not required for infectivity of Maize streak virus in cereals, but is required for invasion of mesophyll cells. J Gen Virol 86: 797–801 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E (2009) A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5: e1000396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquna A, Katz A, Shochat S, Grafi G, Ohad N (2004) Interaction of FIE, a polycomb protein, with pRb: a possible mechanism regulating endosperm development. Mol Genet Genomics 271: 651–657 [DOI] [PubMed] [Google Scholar]
- Nagar S, Pedersen TJ, Carrick KM, Hanley‐Bowdoin L, Robertson D (1995) A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A (1999) Tobacco retinoblastoma‐related protein phosphorylated by a distinct cyclin‐dependent kinase complex with Cdc2/cyclin D in vitro. Plant J 18: 243–252 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma‐related protein by the cyclin D/cyclin‐dependent kinase complex is activated at the G1/S‐phase transition in tobacco. Plant Cell 14: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naouar N, Vandepoele K, Lammens T, Casneuf T, Zeller G, van Hummelen P, Weigel D, Ratsch G, Inze D, Kuiper M et al (2009) Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J 57: 184–194 [DOI] [PubMed] [Google Scholar]
- Ning Y‐Q, Liu N, Su Y‐N, Lan K‐K, Li L, Chen S, He X‐J (2020) DREAM complex suppresses DNA methylation maintenance genes and precludes DNA hypermethylation. Nat Plants 6: 942–956 [DOI] [PubMed] [Google Scholar]
- Nisa M‐U, Huang Y, Benhamed M, Raynaud C (2019) The plant DNA damage response: signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front Plant Sci 10: 653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S, Marrocco K, Masoud K, Thomann A, Gusti A, Bitrian M, Schnittger A, Genschik P (2015) The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F‐Box protein FBL17. Plant Cell 27: 1461–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack MK, Harashima H, Dissmeyer N, Zhao X, Bouyer D, Weimer AK, De Winter F, Yang F, Schnittger A (2012) Genetic framework of cyclin‐dependent kinase function in Arabidopsis. Dev Cell 22: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Nurse P, Bissett Y (1981) Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature 292: 558–560 [DOI] [PubMed] [Google Scholar]
- Olmo R, Cabrera J, Díaz‐Manzano FE, Ruiz‐Ferrer V, Barcala M, Ishida T, García A, Andrés MF, Ruiz‐Lara S, Verdugo I et al (2020) Root‐knot nematodes induce gall formation by recruiting developmental pathways of post‐embryonic organogenesis and regeneration to promote transient pluripotency. New Phytol 227: 200–215 [DOI] [PubMed] [Google Scholar]
- Olson BJ, Oberholzer M, Li Y, Zones JM, Kohli HS, Bisova K, Fang SC, Meisenhelder J, Hunter T, Umen JG (2010) Regulation of the Chlamydomonas cell cycle by a stable, chromatin‐associated retinoblastoma tumor suppressor complex. Plant Cell 22: 3331–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Őszi E, Papdi C, Mohammed B, Petkó‐Szandtner A, Leviczky T, Molnár E, Galvan‐Ampudia C, Khan S, Juez EL, Horváth B et al (2020) E2FB interacts with RETINOBLASTOMA RELATED and regulates cell proliferation during leaf development. Plant Physiol 182: 518–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JA, Ahn JW, Kim YK, Kim SJ, Kim JK, Kim WT, Pai HS (2005) Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J 42: 153–163 [DOI] [PubMed] [Google Scholar]
- Park J, Hwang H‐S, Buckley KJ, Park J‐B, Auh C‐K, Kim D‐G, Lee S, Davis KR (2010) C4 protein of Beet severe curly top virus is a pathomorphogenetic factor in Arabidopsis. Plant Cell Rep 29: 1377–1389 [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh M, Molinier J, Berr A, Genschik P (2011) MSI4/FVE interacts with CUL4‐DDB1 and a PRC2‐like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc Natl Acad Sci USA 108: 3430–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettkó‐Szandtner A, Mészáros T, Horváth GV, Bakó L, Csordás‐Tóth E, Blastyák A, Zhiponova M, Miskolczi P, Dudits D (2006) Activation of an alfalfa cyclin‐dependent kinase inhibitor by calmodulin‐like domain protein kinase. Plant J 46: 111–123 [DOI] [PubMed] [Google Scholar]
- Polit JT, Kaźmierczak A, Walczak‐Drzewiecka A (2012) Cell cycle‐dependent phosphorylation of pRb‐like protein in root meristem cells of Vicia faba . Protoplasma 249: 131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin‐SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Diaz‐Trivino S, Cisneros N, Gutierrez C (2006) The balance between cell division and endoreplication depends on E2FC‐DPB, transcription factors regulated by the ubiquitin‐SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Lopez‐Matas MA, Ramirez‐Parra E, Gutierrez C (2005) Hormonal control of the cell cycle. Physiol Plant 123: 173–183 [Google Scholar]
- Ramirez‐Parra E, Xie Q, Boniotti MB, Gutierrez C (1999) The cloning of plant E2F, a retinoblastoma‐binding protein, reveals unique and conserved features with animal G(1)/S regulators. Nucleic Acids Res 27: 3527–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez‐Parra E, Gutierrez C (2000) Characterization of wheat DP, a heterodimerization partner of the plant E2F transcription factor which stimulates E2F‐DNA binding. FEBS Lett 486: 73–78 [DOI] [PubMed] [Google Scholar]
- Ramirez‐Parra E, Frundt C, Gutierrez C (2003) A genome‐wide identification of E2F‐regulated genes in Arabidopsis. Plant J 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Ramirez‐Parra E, Gutierrez C (2007) E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol 144: 105–120 17351056 [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large‐scale Aranidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou‐Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D‐type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Robbens S, Khadaroo B, Camasses A, Derelle E, Ferraz C, Inzé D, Van de Peer Y, Moreau H (2005) Genome‐wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri. Mol Biol Evol 22: 589–597 [DOI] [PubMed] [Google Scholar]
- Rossi V, Varotto S, Locatelli S, Lanzanova C, Lauria M, Zanotti E, Hartings H, Motto M (2001) The maize WD‐repeat gene ZmRbAp1 encodes a member of the MSI/RbAp sub‐family and is differentially expressed during endosperm development. Mol Genet Genomics 265: 576–584 [DOI] [PubMed] [Google Scholar]
- Rossi V, Locatelli S, Lanzanova C, Boniotti MB, Varotto S, Pipal A, Goralik‐Schramel M, Lusser A, Gatz C, Gutierrez C et al (2003) A maize histone deacetylase and retinoblastoma‐related protein physically interact and cooperate in repressing gene transcription. Plant Mol Biol 51: 401–413 [DOI] [PubMed] [Google Scholar]
- Rubin SM, Gall AL, Zheng N, Pavletich NP (2005) Structure of the Rb C‐terminal domain bound to E2F1‐DP1: a mechanism for phosphorylation‐induced E2F release. Cell 123: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Rubin SM (2013) Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem Sci 38: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhel R, Chakraborty S (2019) Multifunctional roles of geminivirus encoded replication initiator protein. Virus Dis 30: 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Kawamura A, Lambolez A, Inagaki S, Takebayashi A, Iwase A, Sakamoto Y, Sako K, Favero DS, Ikeuchi M et al (2019) Histone acetylation orchestrates wound‐induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun Biol 2: 404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Dante RA, Leiva‐Neto JT, Jung R, Gordon‐Kamm WJ, Larkins BA (2005) RBR3, a member of the retinoblastoma‐related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci USA 102: 13005–13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA (2006) Grasses like mammals? Redundancy and compensatory regulation within the retinoblastoma protein family. Cell Cycle 5: 352–355 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA (2009) The contribution of cell cycle regulation to endosperm development. Sex Plant Reprod 22: 207–219 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Liu Y, Dante RA, Lizarraga LE, Nguyen HN, Brown SW, Klingler JP, Yu J, LaBrant E, Layton TM et al (2013) Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma‐related pathway in maize endosperm. Proc Natl Acad Sci USA 110: 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R (2007) The dynamic plant stem cell niches. Curr Opin Plant Biol 10: 639–644 [DOI] [PubMed] [Google Scholar]
- Sadasivam S, DeCaprio JA (2013) The DREAM complex: master coordinator of cell cycle‐dependent gene expression. Nat Rev Cancer 13: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MP, Torres A, Boniotti MB, Gutierrez C, Vazquez‐Ramo JM (2002) PCNA protein associates to Cdk‐A type protein kinases in germinating maize. Plant Mol Biol 50: 167–175 [DOI] [PubMed] [Google Scholar]
- Sánchez‐Camargo VA, Suárez‐Espinoza C, Romero‐Rodríguez S, Garza‐Aguilar SM, Stam M, García‐Ramírez E, Lara‐Núñez A, Vázquez‐Ramos JM (2020) Maize E2F transcription factors. Expression, association to promoters of S‐phase genes and interaction with the RBR1 protein in chromatin during seed germination. Plant Sci 296: 110491 [DOI] [PubMed] [Google Scholar]
- Scheres B (2007) Stem‐cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol 8: 345–354 [DOI] [PubMed] [Google Scholar]
- Sekine M, Ito M, Uemukai K, Maeda Y, Nakagami H, Shinmyo A (1999) Isolation and characterization of the E2F‐like gene in plants. FEBS Lett 460: 117–122 [DOI] [PubMed] [Google Scholar]
- Settlage SB, Miller AB, Gruissem W, Hanley‐Bowdoin L (2001) Dual interaction of a geminivirus replication accessory factor with a viral replication protein and a plant cell cycle regulator. Virology 279: 570–576 [DOI] [PubMed] [Google Scholar]
- Shimizu‐Sato S, Ike Y, Mori H (2008) PsRBR1 encodes a pea retinoblastoma‐related protein that is phosphorylated in axillary buds during dormancy‐to‐growth transition. Plant Mol Biol 66: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons AR, Davies KA, Wang W, Liu Z, Bergmann DC (2019) SOL1 and SOL2 regulate fate transition and cell divisions in the Arabidopsis stomatal lineage. Development 146: dev171066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Shah ZH, Murray JA (1995) A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7: 85–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sornay E, Forzani C, Forero‐Vargas M, Dewitte W, Murray JAH (2015) Activation of CYCD7;1 in the central cell and early endosperm overcomes cell‐cycle arrest in the Arabidopsis female gametophyte, and promotes early endosperm and embryo development. Plant J 84: 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno‐Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN (2010) Spatiotemporal regulation of cell‐cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stes E, Biondi S, Holsters M, Vereecke D (2011) Bacterial and plant signal integration via D3‐type cyclins enhances symptom development in the Arabidopsis‐Rhodococcus fascians interaction. Plant Physiol 156: 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor‐Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R et al (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517: 571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topacio BR, Zatulovskiy E, Cristea S, Xie S, Tambo CS, Rubin SM, Sage J, Kõivomägi M, Skotheim JM (2019) Cyclin D‐Cdk 4,6 drives cell‐cycle progression via the retinoblastoma proteins's C‐terminal helix. Mol Cell 74: 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA (2002) Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3: 11–20 [DOI] [PubMed] [Google Scholar]
- Uemukai K, Iwakawa H, Kosugi S, de Uemukai S, Kato K, Kondorosi E, Murray JAH, Ito M, Shinmyo A, Sekine M (2005) Transcriptional activation of tobacco E2F is repressed by co‐transfection with the retinoblastoma‐related protein: cyclin D expression overcomes this repressor activity. Plant Mol Biol 57: 83–100 [DOI] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW (2001) Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev 15: 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inze D, De Veylder L (2005) Genome‐wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villajuana‐Bonequi M, Matei A, Ernst C, Hallab A, Usadel B, Doehlemann G (2019) Cell type specific transcriptional reprogramming of maize leaves during Ustilago maydis induced tumor formation. Sci Rep 9: 10227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X (2014) A noncanonical role for the CKI‐RB‐E2F cell‐cycle signaling pathway in plant effector‐triggered immunity. Cell Host Microbe 16: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W‐S, Zhu J, Zhang K‐X, Lü Y‐T, Xu H‐H (2016) A mutation of casein kinase 2 α4 subunit affects multiple developmental processes in Arabidopsis. Plant Cell Rep 35: 1071–1080 [DOI] [PubMed] [Google Scholar]
- Wang W, Sijacic P, Xu P, Lian H, Liu Z (2018) Arabidopsis TSO1 and MYB3R1 form a regulatory module to coordinate cell proliferation with differentiation in shoot and root. Proc Natl Acad Sci USA 115: 3045–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Nowack MK, Bouyer D, Zhao X, Harashima H, Naseer S, De Winter F, Dissmeyer N, Geldner N, Schnittger A (2012) Retinoblastoma related1 regulates asymmetric cell divisions in Arabidopsis. Plant Cell 24: 4083–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Matos JL, Sharma N, Patell F, Murray JAH, Dewitte W, Bergmann DC (2018) Lineage‐ and stage‐specific expressed CYCD7;1 coordinates the single symmetric division that creates stomatal guard cells. Development 145: dev160671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Gao M, Jiao C, Wang Q, Xu H, Walter M, Xu W, Bassett C, Wang X (2012) Characterization and expression analysis of a retinoblastoma‐related gene from Chinese wild Vitis pseudoreticulata . Plant Mol Biol Report 30: 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger JP, Marks MD (2008) E2F and retinoblastoma related proteins may regulate GL1 expression in developing Arabidopsis trichomes. Plant Signal Behav 3: 420–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez‐Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B (2005) The RETINOBLASTOMA‐RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Willems A, Heyman J, Eekhout T, Achon I, Pedroza‐Garcia JA, Zhu T, Li L, Vercauteren I, van de Daele H, van de Cotte B et al (2020) CYCA3;4 is a post‐prophase target of the APC/CCCS52A2 E3‐ligase controlling formative cell divisions in Arabidopsis. Plant Cell 10.1105/tpc.20.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrzykowska J, Schorderet M, Pien S, Gruissem W, Fleming AJ (2006) Induction of differentiation in the shoot apical meristem by transient overexpression of a retinoblastoma‐related protein. Plant Physiol 141: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Suarez‐Lopez P, Gutierrez C (1995) Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: requirement for efficient viral DNA replication. EMBO J 14: 4073–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Sanz‐Burgos AP, Hannon GJ, Gutierrez C (1996) Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J 15: 4900–4908 [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang Y, Stroud H, Gu X, Sun B, Gan E‐S, Ng K‐H, Jacobsen SE, He Y, Ito T (2013) A matrix protein silences transposons and repeats through interaction with retinoblastoma‐associated proteins. Curr Biol CB 23: 345–350 [DOI] [PubMed] [Google Scholar]
- Ye B‐B, Shang G‐D, Pan Y, Xu Z‐G, Zhou C‐M, Mao Y‐B, Bao N, Sun L, Xu T, Wang J‐W (2020) AP2/ERF transcription factors integrate age and wound signals for root regeneration. Plant Cell 32: 226–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Liu X, Yang C, Lai J (2018) Geminivirus C4: interplaying with receptor‐like kinases. Trends Plant Sci 23: 1044–1046 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zheng L, Hong JH, Gong X, Zhou C, Pérez‐Pérez JM, Xu J (2016) TOPOISOMERASE1α acts through two distinct mechanisms to regulate stele and columella stem cell maintenance. Plant Physiol 171: 483–49 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou W, Chen Q, Fang M, Zheng S, Scheres B, Li C (2018) Mediator subunit MED31 is required for radial patterning of Arabidopsis roots. Proc Natl Acad Sci USA 115: 5624–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Harashima H, Dissmeyer N, Pusch S, Weimer AK, Bramsiepe J, Bouyer D, Rademacher S, Nowack MK, Novak B et al (2012) A general G1/S‐phase cell‐cycle control module in the flowering plant Arabidopsis thaliana . PLoS Genet 8: e1002847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Lasses T, Bako L, Kong D, Zhao B, Chanda B, Bombarely A, Cruz‐Ramírez A, Scheres B, Brunner AM et al (2017a) XYLEM NAC DOMAIN1, an angiosperm NAC transcription factor, inhibits xylem differentiation through conserved motifs that interact with RETINOBLASTOMA‐RELATED. New Phytol 216: 76–89 [DOI] [PubMed] [Google Scholar]
- Zhao X, Bramsiepe J, Van Durme M, Komaki S, Prusicki MA, Maruyama D, Forner J, Medzihradszky A, Wijnker E, Harashima H et al (2017b) RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 356: eaaf6532 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye Z‐H (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Zhou W, Lozano‐Torres JL, Blilou I, Zhang X, Zhai Q, Smant G, Li C, Scheres B (2019) A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177: 942–956 [DOI] [PubMed] [Google Scholar]
- Zhu SJ, Pearson BJ (2013) The Retinoblastoma pathway regulates stem cell proliferation in freshwater planarians. Dev Biol 373: 442–452 [DOI] [PubMed] [Google Scholar]


