Abstract
Porcine epidemic diarrhea virus (PEDV) causes an emerging and re-emerging coronavirus disease characterized by vomiting, acute diarrhea, dehydration, and up to 100% mortality in neonatal suckling piglets, leading to huge economic losses in the global swine industry. Vaccination remains the most promising and effective way to prevent and control PEDV. However, effective vaccines for PEDV are still under development. Understanding the genomic structure and function of PEDV and the influence of the viral components on innate immunity is essential for developing effective vaccines. In the current review, we systematically describe the recent developments in vaccine against PEDV and the roles of structural proteins, non-structural proteins and accessory proteins of PEDV in affecting viral virulence and regulating innate immunity, which will provide insight into the rational design of effective and safe vaccines for PEDV or other coronaviruses.
Keywords: Porcine epidemic diarrhea virus, Vaccine, Virulence, Genetics
1. Introduction
Since 2010, highly pathogenic variant porcine epidemic diarrhea virus (PEDV) strains have gradually swept the swine industry worldwide and brought substantial economic losses. Such a huge hazard of the virus is inseparable from its strong transmissibility. Generally, the main transmission route of PEDV is fecal-oral, but airborne transmission via the fecal-nasal route may play a significant role in pig-to-pig and farm-to-farm spread [1]. PEDV mainly infects the porcine intestinal epithelial cells. Then, PEDV colonizing the intestines results in atrophy, necrosis, and shedding of intestinal villi, which affected the absorption of nutrients, leading to vomiting, diarrhea, weight loss, anorexia, and depression [2,3]. When PEDV is excreted into the environment with feces, the contaminated feces may result in a mass epidemic. Obtaining sufficient maternal antibody from the colostrum and milk remains the most promising and effective strategy to protect neonatal suckling piglets against PEDV [4]. Unfortunately, due to the continued mutation of the PEDV genome [5] and the low efficacy to induce mucosal immunity, no effective vaccine has been released yet. Therefore, in-depth research on this disease as well as its pathogens and the rapid development of effective vaccines based on the epidemic strains are urgently needed.
2. PEDV genome structure and functions
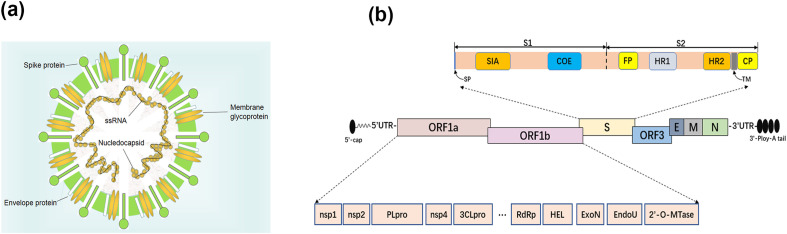
PEDV is an enveloped virus with a single-stranded, positive-sense RNA genome that belongs to the order Nidovirales, family Coronaviridae and genus Alphacoronavirus. The genome of PEDV is approximately 28 kb and arranged in the order of the 5′ untranslated region (5′ UTR), open reading frame 1 a/b (orf1ab), spike (S) protein, accessory proteins (ORF3), envelope (E) protein, membrane (M) protein, nucleocapsid (N) protein, 3′ UTR, and the poly (A) tail (Fig. 1 ). Pp1ab, encoded by the partially overlapping 5′-terminal of PEDV genome, can be cleaved into 16 non-structural proteins (nsp) named nsp1-16 [6]. Among these proteins, the S protein is a type I glycoprotein and a receptor-binding protein that plays an important role in viral entry, virus-host interactions and determining immunogenicity evaluation. The S protein consists of S1 and S2 subunits. The amino terminal of S1 subunit (1–18 aa) is the N-terminal signal peptide, 19–233 aa is the sialic acid binding region, and 499–638 aa is the core neutralizing epitope (COE) region. The S2 subunit contains a fusion peptide (FP, 891–908 aa), two heptad repeat regions HR1 (978–1117 aa) and HR2 (1274–1313 aa), a transmembrane region (1328–1350 aa) and a cytoplasmic domain (CP, 1351–1386 aa) in C-terminal [7,8]. Each structural and non-structural protein play an important role in the viral replication, transcription and translation as well as virus–host cell interactions. Table 1 summarizes the biological roles of PEDV proteins.
Fig. 1.
(a) Schematic structure of the virion of PEDV. (b) Schematic diagram of the genome of PEDV. Structural proteins, including spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins, as well as accessory proteins ORF3 and non-structural proteins derived from ppl1ab, including nsp1-16, papain-like proteinase (PLpro, nsp3), 3C-like proteinase (3CLpro, nsp5), RNA-dependent RNA polymerase (RdRp, nsp12), 5′-to-3′ Helicase (HEL, nsp13), exoribonuclease (ExoN, nsp14), endoribonuclease (EndoU, nsp15), 2′-O-methyltransferases (2′-O-MTase, nsp16). Spike protein, including signal peptide (SP, 1–18 aa), sialic acid-binding region (SIA, 19–233 aa), core neutralizing epitope (COE, 499–638 aa), fusion peptide (FP, 891–908 aa), heptad repeat domain (HR1, 978–1117 aa and HR2, 1274–1313 aa), transmembrane domain (TM, 1328–1350 aa), and cytoplasmic domain (CP, 1351–1386 aa).
Table 1.
The biological roles of different PEDV proteins.
| Classification | Proteins | The biological roles | References |
|---|---|---|---|
| Nonstructural proteins | Nsp1 | A vital virulence factor; suppression of type I interferon and type III interferon production; reduces the early production of proinflammatory cytokine, such as TNFα, TGF-β3, IL-6, IL-15, IL-17, IL-1β | [28,67,129,185] |
| Nsp3 | Deubiquitinates RIG-I and STING; inhibits IFN-β and IFN-λ1 expression. | [129,186] | |
| Nsp4 | Upregulates IL-1α, IL-1β, TNF-α, CCL2, CCL5 and CXCL8 expression. | [187] | |
| Nsp5 | Cleaves NEMO at position 231 (Q231) and inhibits type I interferon production. | [30] | |
| Nsp 6 | Induces autophagy through the PI3K/Akt/mTOR signaling pathway in IPEC-J2 cells. | [188] | |
| Nsp7, nsp8 and nsp9 | Inhibits the IFN-β and IRF3 promoter activities, viral replication | [67,68,129] | |
| Nsp13-16 | Involves in viral replication; inhibition of the type I IFN and the type III IFN; vital virulence factors. | [80,84,85,129,189,190] | |
| Structural proteins | S | Induces neutralizing antibodies; receptor binding and membrane fusion; a vital virulence factor; S1 Spike protein induces cell apoptosis. | [119,191,192] |
| E | Stimulates ER stress and up-regulates the production of IL-6 and IL-8; promotes apoptosis; inhibits the IFN promoter activities. | [95,129] | |
| M | Induces cell cycle arrest at the S-phase; inhibits the IFN promoter activities. | [129,193] | |
| N | Antagonizes IFN-β and IFN-λ Production; prolongs S-phase cell cycle; stimulates ER stress and up-regulates the production of Bcl-2 and IL-8. | [194,195] | |
| Accessory protein | ORF3 | Owns ion channel activity; inhibits apoptosis and promotes autophagy; causes endoplasmic reticulum stress; promotes PEDV replication. | [89,98,196,197] |
2.1. Non-structure proteins
The Nsp1 protein, a unique structure of alphacoronaviruses and betacoronaviruses, is located at the N-terminus of pp1ab [9]. Nsp1 is considered to be a virulence gene of coronavirus, which widely inhibits host genes’ expression to suppress or evade the innate immune response and facilitate viral replication [10]. The nsp1 of severe acute respiratory syndrome coronavirus (SARS-CoV) significantly inhibited host gene expression, including that of type I interferon (IFN), by binding to the ribosome, which led to endonucleolytic cleavage and subsequently promoted host endogenous mRNA degradation in vitro [[11], [12], [13]]. Moreover, abolishing the IFN suppression activity reduced viral virulence in vivo, the deletion of the regions ΔLLRKNGNKG (121–129 aa) and ΔEDYEQNWNTKH (154–164 aa) in nsp1 led to SARS-CoV attenuation in mice. MHV-nsp1 Δ99(829–927 nt) and MHV-nsp1 Δ27 (LLRKxGxKG) were also strongly attenuated but maintained the protection against challenge with the WT MHV virus in vivo [[14], [15], [16], [17], [18]]. IRF1 mediated type III IFN production via interactions with the IFNλ promoter in the nucleus [19,20], and the increase in peroxisome abundance was correlated with an increase in IFNλ mRNA production [21]. NF-κB triggers IFN-β expression by binding to the respective positive regulatory domain (PRD)II element of type I IFNs when NF-κB was released from the NF-κB/IκBα complex and transported to the nucleus after the IκBα was phosphorylated and degraded by the IκB kinase complex [[22], [23], [24]]. IRF3 turned on the expression of type I IFN via forming an IRF3-CBP complex binding to the PRD I/III of the IFN promoter [25,26]. PEDV nsp1 inhibited IFN production by blocking the nuclear translocation of IRF1, reducing the number of peroxisomes, and interfering with the phosphorylation and degradation of IκBα, which could prevent the activation of p65 and suppress the PRD II-mediated NF-κB activity in vitro [27,28]. The highly conserved residues of nsp1 were crucial for type III IFNs and IRF1 suppression activities. The amino acid mutant F44A abolished the suppression activities of nsp1 in vitro, and the PEDV strain containing nsp1 F44A mutation had higher levels of IFN-β, IFN-λ1 and ISG45 in LLC-PK1 cells and reduced the level of fecal virus shedding without significant pathological lesions in the jejunum and ileum in piglets. In addition, the degradation of CBP by nsp1 interrupted the assembly of IRF3 and CREB binding protein (CBP), resulting in the further suppression of IFN and proinflammatory cytokine [29,30]. Crystal structure showed that PEDV nsp1 consists of two α-helices and six β-strands, and six stranded β-barrel folds in the middle of two α-helices. Ribopuromycylation and renilla luciferase reporter assays indicated that 67–71 aa, 78–85 aa, and 103–110 aa of the nsp1 create a stable positive charge functional region that may help PEDV to inhibit host protein synthesis, including that of IFN [31,32]. Nevertheless, the specific mechanism of PEDV nsp1-mediated immunosuppression was entirely unclear and whether PEDV nsp1 inhibited host gene expression in a ribosome-dependent manner similar to that of SARS-CoV and SARS-CoV-2 needs further research [33].
Nsp3 encodes Papain-like protease (PLpro) present in various coronaviruses. The PLpro are responsible for the cleavage of the portion of the ORF1ab polyprotein encoded by the incoming RNA genome, which is essential for viral RNA synthesis [34]. The PLpro usually cleaves nsp1-4 and the cleavage sites can be summarized with LXGG↓ or the similar motif in SARS-CoV and MHV [[35], [36], [37]]. Furthermore, PLpro strongly antagonizes the expression of IFN and interferon-stimulated genes (ISGs). The PLpro of SARS-CoV interacts with IRF3 and inhibits the phosphorylation and nuclear translocation of IRF-3, thereby disrupting the activation of the type I IFN pathway through either Toll-like receptor 3 or retinoic acid-inducible gene I/melanoma differentiation-related gene 5 pathways in vitro [38]. The STING-MAVS-TBK1/IKKε complexes could activate IRF3, which is subsequently transported to the nucleus and induces the expression of type I IFN genes by binding to the PRD(III-I) sequences and the phosphorylation of STING mediated by TBK1 is required for STING-mediated activation of IRF3 [26,28,39,40]. Moreover, dimerization of STING is also critical for self-activation and the ability to induce type I IFN [41]. PLpro encoded by NL63-CoV, SARS-CoV, and PEDV antagonized STING-mediated antiviral innate immunity by disrupting STING dimers and the deubiquitination of RIG-I and STING, and the catalytic mutants (C1729A, H1888A, D1901A) in PEDV nsp3 reduced the inhibition of IFN-β and PRD(III-I)4 promoter activity in vitro, which could be applied to the design of the PEDV attenuated vaccine [42,43]. Moreover, amino acids 590–1215 of TGEV nsp3 have been proven to evade the host immune system by NF-κB pathway [44], whether PEDV uses similar amino acids for immune evasion needs to be further studied.
Contrary to the effect of PEDV nsp3, PEDV nsp4 could upregulate the expression of IL-1α, IL-1β, TNF-α, CCL2, CCL5 and CXCL8 by activating the NF-κB promoter and silencing these cytokine genes inhibited viral replication in vitro [45].
3C-like protease (3CLpro), encoded by the gene for nsp5, processes the viral nsp4–nsp16 part of pp1ab at 11 cleavage sites, which could be summarized with the P2–P1’ consensus motif X-(L/I/V/F/M)-Q↓(S/A/G) and X is any amino acid and ↓ represents the cleavage in SARS-CoV and other coronaviruses [37,[46], [47], [48], [49], [50]]. 3CLpro plays a pivotal role in coronaviruses replication, thus serving as an appealing antiviral drug target. Inhibition of PEDV nsp5 effectively inhibited the replication of PEDV, for instance, the broad-spectrum inhibitor GC376 reduced viral replication via binding the catalytic pocket of PEDV 3CLpro [51,52]. In the process of interaction with the host, NEMO (IKKγ) acting as a signaling adaptor of the RIG-I/MDA5 pathway plays an important role in the phosphorylation of NF-κB and IRF3, which directly activate promoters of type I IFNs [53,54]. PEDV nsp5 cleaved NEMO at position 231 (Q231), which impaired the ability of NEMO to induce IFN production in vitro [55]. The functional coronavirus 3CLpro is dimeric. Each PEDV monomer contains three domains: domain I, domain II, and domain III. Domain I and domain II both exhibit an antiparallel β-barrel structure and domain III consists of five α helices [56]. The substrate catalytic domains of α- and β-coronavirus 3CLpro have two highly conserved residues known as His41 and Cys144 [[57], [58], [59], [60]]. His41 and Cys144 mutating into Ala (H41A, C144A) eliminated the catalytic activity and IFN-β inhibitory capacity of PEDV nsp5 in PK-15 and HEK-293T cells [30]. The antagonistic effect of nsp5 on IFN suggested that it may be related to viral virulence. Besides, the nsp5 T26I/D65G MHV exhibited reduced enzymatic activity and was highly attenuated in IFNAR−/− mice [61]. Whether the PEDV strain with the H41A/C144A in nsp5 exhibits lower 3CLpro activity and reduced viral virulence needs to be further verified by reverse genetics.
Coronavirus nsp7–10 are essential regulatory subunits that are critical for coronavirus replication. The N-terminal region of SARS-CoV nsp8 possessed primase activity and was able to polymerize short RNA oligonucleotides utilized by the primer-dependent nsp12 RdRp during RNA synthesis [62]. The SARS-CoV hexadecameric nsp7–nsp8 supercomplex containing eight copies of nsp8 and eight copies of nsp7 [63] and the feline coronavirus (FCoV) Nsp7-Nsp8 heterotrimer both exhibited noncanonical RdRp activity [64]. The SARS-CoV nsp7-nsp8 supercomplex has a hollow cylindrical structure with different charges distributed on the surface and inside of the cylinder, which ensures that the phosphate backbone of nucleic acids can pass through the channel without electrostatic repulsions. In addition, an average internal diameter of approximately 30 Å could suitably hold the nascent and template strands together to facilitate efficient replication and transcription [63]. Interactions of nsp12 with nsp7 and nsp8 co-factors were crucial to the processivity to the RNA-synthesizing activity of nsp12 and a few mutations of nsp7 and nsp8 residues affected SARS-CoV replication by reducing nsp12 RNA polymerase activity [65,66]. PEDV nsp7 was found to inhibit the IFN-β and IRF3 promoter activities in HeLa cells [67]. Moreover, PEDV nsp8 was shown to suppress type III IFN activities by reduced IRF1 promoter activities in vitro. This fact suggested that nsp7 and nsp8 could be factors, though the specific mechanism of innate immunosuppression remains largely unknown. PEDV nsp9, containing seven antiparallel β-strands and one α-helix, is also critical for viral replication. Nsp9–RNA interactions may stabilize nascent viral RNAs during viral replication or transcription. The dimerization of PEDV nsp9 and its positively charged surface further enhanced the interaction between nsp9 and RNA [37,[68], [69], [70]]. SARS-CoV and MHV nsp10 are involved in viral replication via regulating the nsp14 ExoN and nsp16 2′-O-MTase activities as well as the processing of the nsp4–nsp11 region [[71], [72], [73]]. Nsp13 of 229E-CoV and SARS-CoV possessing both helicases and NTPase activity, which could unwind either DNA or RNA in the 5′-to-3′ direction and utilize all four natural ribonucleotides and nucleotides as substrate, also plays an essential role in viral replication [[74], [75], [76], [77]]. However, there is no direct evidence for the roles of nsp10 and nsp13 in PEDV replication.
CoV nsp14 possesses 3′-to-5′ exoribonuclease (ExoN) activity crucial for CoV RNA genome replication fidelity [78] and N7-methyltransferase (N7-MTase) activity involved in RNA cap formation that is also critical for viral transcribing and translation [79]. Abolishing the N7-MTase activity was lethal to PEDV, but retaining 25% of N7-MTase activity by mutating a single amino acid D350A in nsp14 was viable. In addition, this mutant induced significantly higher expression of both type I and III IFN. These results demonstrated that PEDV nsp14 N7-MTase activity efficiently modulates viral replication, gene expression, and innate immune responses in IPEC-DQ cells [80]. Recombinant rMHV encoding a G332A substitution in the N7-MTase region in nsp14 also displayed delayed replication kinetics and decreased peak titers but higher expression of type I interferon in vitro [81]. Moreover, the severe acute respiratory syndrome (SARS)-CoV, containing inactivation of ExoN by introducing D90A/E92A into the nsp14 resulted in attenuation of pathogenesis, but still protects mice from lethal challenge [82]. These experimental results inspire us to wonder about the immunogenicity and virulence of rPEDV-D350A in piglets.
PEDV nsp15 was identified as an endoribonuclease (EndoU) that plays a pivotal role in evading host response and viral replication [83]. PEDV nsp15 was could subvert the IFN response by the RNA degradation of TBK1 and IRF3 in HEK293T and IPEC-J2 cells. H226, H241 and K282 of PEDV nsp15 were crucial for the EndoU activity [84]. H226A, H241A and K282A in nsp15 probably provide a universal approach for generating live-attenuated vaccine candidates for emerging coronaviruses [85].
Nsp16 has a 2′-O methyltransferase (MTase) activity, which is highly conserved among coronaviruses. This MTase prevents CoV from being recognized by the intracellular sensor via capping of viral RNA [86]. Furthermore, PEDV nsp16 could promote virus replication by downregulating innate immunity mediated by RIG-1 and MDA5 in vitro [87]. Inactivation of PEDV 2′-O-MTase activity attenuated the virus in piglets and induced stronger type I and type III IFN responses in IPEC-DQ cells, which showed that nsp16 potentially provides a universal target for PEDV vaccine development [88].
2.2. Accessory protein
The ORF3 protein is the only PEDV accessory protein located between the S protein and E protein. ORF3, with ion channel activity, was predicted to harbor multiple transmembrane domains, and regulate viral replication and virulence [89,90]. Generally, the full-length ORF3 gene contains 675 nt in wild-type PEDV strains or 624 nt or 626 nt in cell-adapted or attenuated PEDV strains with 49 or 51 nt deletion, such as the classic strain CV777, DR13 and P–5V [91]. Nevertheless, different nucleotides deletion in ORF3 of 85–7 and AVCT12 strains led to early-termination of ORF3 [92,93] and the current study showed that viruses without ORF3 or with truncated ORF3 were completely attenuated in vivo [90,94]. Both full-length and truncated ORF3 could interact with the S protein to regulate viral replication [95]. What's more, PEDV ORF3 also promoted viral replication also by detaining cells at the S-phase, facilitating vesicle formation, the ion channel activity, inhibiting apoptosis and promoting viral autophagy in vitro [89,[96], [97], [98], [99]]. However, the replication of PEDV would be suppressed in cells overexpressing vacuolar protein-sorting-associated protein 36 (VPS36) in a VPS36-ORF3 interaction manner [100].
2.3. Structural proteins
The PEDV S protein, a membrane glycoprotein located at the surface of the virus, exhibits a high degree of genetic diversity and plays a pivotal role in mediating viral entry, inducing neutralizing antibodies and viral virulence in vivo [101]. PEDV was divided into GI and GII types through phylogenetic analysis based on the complete S protein. The GI (e.g. CV777 and DR13) group consists of three subgroups (GIa, GIb and GIc) [102], while the GII group consists two subgroups (GIIa and GIIb). The S gene of GII PEDV contains typical insertion and deletion mutations compared to the GI PEDV, which may significantly affect the immunogenicity of the PEDV strains [103]. This may be the reason why the currently available commercial vaccines based on the attenuated CV777 strain failed to provide effective protection against epidemic GII PEDV strains [104]. Moreover, Lin et al further demonstrated that the neutralizing antibody titers in sera, the colostrum and the milk from the inactivated epidemic YC2014 GII PEDV strain immunized group were significantly higher than the inactivated classical GI CV777 and DR13 groups and sows immunized inactivated YC2014 provided piglets with 100% protection against YC2014 challenge, whereas CV777 or DR13 failed to protect piglets [5]. The molecular weight of PEDV S protein is about 150–220 kDa [105]. Coronaviruses used the homotrimeric spike glycoprotein to bind to their cellular receptors [106]. The PEDV spike monomer comprises a an S1 subunit almost entirely composed of β-sheets and an S2 subunit made up of a series of discontinuous α-helices [107]. The coronavirus S1 subunit binds the cellular receptor, followed by the fusion of viral and cellular membranes mediated the six-helix bundle (6-HB) consisted of the HR1 region and the HR2 region in the S2 subunit [108]. Identifying CoV receptors are essential for the development of drugs and vaccines [[109], [110], [111]]. It has been reported that porcine aminopeptidase (pAPN) was a functional receptor of PEDV [112], but subsequent studies indicated that pAPN only promoted the PEDV infection through its protease activity but not a decisive factor in assisting PEDV to enter into cells [[113], [114], [115]]. Moreover, pAPN knockout pigs were not resistant to PEDV infection, which further demonstrated that pAPN was not a functional receptor for PEDV [116]. The PEDV S gene was considered a major virulence gene, which was verified by reciprocally exchanging the S gene of an attenuated iPEDVPT-P96 and the highly virulent iPEDVPT-P5 [117,118]. Furthermore, the S1 subunit was shown to be an important determinant factor of PEDV virulence [119], and the deletion of the 197-amino-acid in the S1 domain attenuated a highly virulent PEDV in pigs [120]. In addition, the S protein contains two intracellular sorting motifs, YxxΦ and/or KVHVQ at the cytoplasmic tail and deletion of both motifs reduced the virulence of PEDV in piglets [121]. S protein contains multiple B cell epitopes, such as 722SSTFNSTREL731, 748YSNIGVCK755, 764SQYGQVKI771, and 1368GPRLQPY1374 [[122], [123], [124], [125]]. The core neutralization epitope region (COE) of the S1 region has been widely applied to develop PEDV subunit vaccines. Hence, S protein is a major target for the development of coronavirus attenuated live vaccines and subunit vaccines.
The smallest structural protein in PEDV is the E protein with only about 7 kD molecular weight and low homology among different coronaviruses [126]. The E protein plays a vital role in viral assembly and budding [127]. Moreover, the PEDV E protein could induce endoplasmic reticulum (ER) stress and activate the nuclear factor-κB (NF-κB) pathway in vitro [128]. A novel variation with 16–20 aa deletion and an L25P mutation in the transmembrane domain of the PEDV E protein further upregulated the production of the ER stress indicator, improving the expression of IL-6 and IL-8, and promoted apoptosis in vitro [92]. In addition, the PEDV E protein was confirmed to suppress type III IFN expression, but the specific molecular mechanism is still unknown [129]. The virulence of SARS-CoV lacking the E protein was significantly reduced in mice [130]. However, whether the PEDV E protein affects the viral virulence and whether the E protein is absolutely necessary for PEDV remain to be further studied. The E protein is also a diagnostic maker, which will be helpful to the development of novel serological assays as well as the design of vaccines [131].
The M gene, with a total length of 681 bp, encodes an about 27–32 kDa protein. The M protein, a membrane glycoprotein localized throughout the cytoplasm, induced cell cycle arrest at the S-phase via the cyclin A pathway [132]. The PEDV M gene differs significantly between coronaviruses, but is relatively conservative among different PEDV strains [133,134]. Therefore, the M protein is a promising target for establishing various detection methods such as ELISA, RT-qPCR and RT-PCR [135,136]. The B cell epitope 195WAFYVR200 on the M protein screened by hybridoma technology was highly conserved and specific among PEDV viruses [137], nevertheless, antisera from mice immunized TGEV M protein displayed limited TGEV neutralizing activity in vitro, which indicated that PEDV M may also not be a subunit vaccine candidate [138]. Moreover, the M protein is involved in coronavirus envelope assembly [139] and the presence of both the E protein and the M protein is sufficient for the formation and release of virus-like particles (VLP) [140,141]. In addition, the M protein was found to suppress type III IFN promoter activities in vitro with an unclear mechanism [129].
The PEDV N protein is a highly conserved phosphoprotein, with only a few point mutations in different strains. However, the PEDV SH strain had a unique 12 aa (aa 399–410) deletion including an antigenic epitope of the N protein, whereas the deletion had no effect on the immunogenicity or pathogenicity of PEDV in mice and pigs [142]. The N protein has been associated with multiple functions in the viral life cycle, including the regulation of viral RNA synthesis, the packaging of the viral RNA in helical nucleocapsids, and virion assembly [143,144]. In the process of confrontation with host cells, the PEDV N protein not only inhibited cell proliferation by prolonging the S-phase cell cycle [145], but also antagonized the production of IFN-λ by preventing nuclear translocation of NF-κB in IPEC-J2 cells [27,146]. The enhanced replication of PEDV in Vero E6 cells overexpressing the N protein may be related to the inhibition of host response by the N protein [147].
3. PEDV vaccines
3.1. Inactivated and attenuated live vaccines
PEDV traditional vaccines are mainly whole virus vaccines such as inactivated vaccine and live attenuated vaccines. Inactivated vaccines offer several advantages, such as good safety profile and ease of production. However, the immunogenicity of inactivated vaccines could be altered in the inactivation process, which often require administration of multiple doses and booster injections [148]. Live attenuated vaccines are highly immunogenic and single immunization is usually sufficient to induce protective immunity. Nevertheless, the risk of reversion to a virulent wild type limits its applicability [149,150]. The key to developing vaccines is to select low-virulence, high-titer, and highly immunogenic strains. However, the isolation of PEDV strains in vitro is very difficult, especially for the pandemic variable strains, and even if the strains are isolated successfully, they may not necessarily have high titers. As early as 1993, the PEDV tissue-inactivated vaccine could provide piglets with immune protection for up to 6 months [151]. In 1994, the PEDV CV777 strain was attenuated via serial cell culture after 28 passages, providing protection for more than 80% of the piglets after immunization at the Houhai point [152]. Similarly, a PEDV vaccine strain could provide 90% protection after serial 90 passages in vitro [153]. In order to control the disease, China introduced the inactivated TGEV-PEDV bivalent vaccine in 1995 [154]. However, since the inactivated vaccine cannot replicate in the host, multiple immunizations and high immune dose were required. In March 2015, trivalent live attenuated vaccines (PEDV, TGEV and RV) were approved in China. In 2016, researchers produced an inactivated PEDV vaccine using a Korean PEDV epidemic KNU-141112 strain. In six and three weeks prior to farrowing, pregnant sows were immunized intramuscularly with the inactivated adjuvanted monovalent vaccine. The survival rate of piglets vaccinated were able to reach approximately 92% after the virulent strain challenge and diarrhea severity including viral shedding in feces was significantly reduced [155]. The highly virulent epidemic virus strain CT was serially passaged in Vero cells up to 120 passages (P120), and P120 had a higher viral titer and better cell adaptability with more obvious cytopathic effects. In addition, P120 showed significant reductions in clinical symptoms with 100% survival rate in piglets [2].
Although the strains' virulence was attenuated after multiple passages, they strain probably lost partial or total ability to resist the parental strain's challenge. Therefore, some researchers began to modify the PEDV strains with reduced viral virulence but high immunogenicity by reverse genetic operating systems to develop effective live attenuated vaccines. Table 2 summarizes candidates for PEDV live attenuated vaccines based on reverse genetic systems. Mutating the histidine(H) residue at the 226 site in PEDV EndoU into alanine(A) induced early and robust transcriptional activation of type I and type III IFNs, and significantly impaired the viral virulence [156]. Ablating the motif YxxΦEKVHVQ in the C-Terminal and a 197-amino-acid region in the N-terminal domain of S protein likewise impaired the pathogenicity of the highly virulent PEDV icPC22A, but the effectiveness of protection requires further investigation [157,158]. Two recombinant PEDVs were generated by mutating K45-D129-K169-E202 into A (KDKE4A), which inactivated 2′-O-methyltransferase activity of PEDV nsp16, and abolished the endocytosis signal of the S protein (KDKE4A-SYA). The two mutant strains KDKE4A and KDKE4A-SYA replicated less efficiently but induced stronger type I and type III IFNs response in vitro. Moreover, the virulence of KDKE4A and KDKE4A-SYA was significantly reduced and the KDKE4A-SYA strain provided 80% protection rate for piglets [88]. In addition, nsp1, nsp2, nsp3, nsp5, nsp14 and E proteins modified strains are also the candidates for the development of PEDV live attenuated vaccines, since mutating certain sites on these genes significantly reduced the viral virulence, which has been demonstrated in other coronaviruses, such as MHV and SARS-CoV [16,17,[159], [160], [161], [162], [163], [164]]. In a nutshell, introducing mutations into virulence genes and inactivating interferon antagonists by reverse genetic technology are effective strategies for developing effective PEDV live attenuated vaccines.
Table 2.
Candidates for live attenuated vaccines based on reverse genetic systems.
| Proteins | Engineering genes | Animal | virulence | Detection Indicators | References |
|---|---|---|---|---|---|
| Nonstructural proteins | H226A of nsp15 | 7-day-old piglets (n = 8) | Attenuation | (1) Fecal shedding↓ (2) Mortality↓ (3) Villus atrophy ↓ (4) Viral antigen in epithelial cells↓ |
[156] |
| KDKE/AAAA of nsp16 | 4-day-old piglets (n = 7–8) | Attenuation | (1) Mortality rate↓ (2) Fecal consistency scores↓ (3) PEDV shedding in rectal swabs↓ (4) Infectious PEDV titers in the small-intestinal contents↓ (5) Protection against virulent icPC22A challenge |
[198] | |
| F44A of nsp1 H226A and H241A of nsp15 D129A of nsp16 |
3-6-day-old piglets (n = 8–13) | Attenuation | (1)Virus-specific IgG titer and neutralizing antibody titer in sera↓ (2)Clinical signs and fecal viral shedding ↓ |
[185] | |
| Structural proteins | Deletion of YxxΦEKVHVQ of S protein | 5-day-old piglets (n = 3–5) | Attenuation | (1) Villus height to crypt depth (VH/CD) ratios↑ (2) Clinical signs and fecal viral shedding ↓ |
[157] |
| Deletion of 197aa of S protein | 4-day-old piglets (n = 9–11) | Attenuation | (1) Diarrhea rate* (2) Mortality rate ↓ (3) Fecal virus shedding ↓ (4) VH/CD ratios ↑ |
[158] | |
| Accessory proteins | Deletion of ORF3 | 2-3week-old Piglets (n = 3–4) |
Attenuation | (1) Diarrhea rate* (2) Mortality rate ↓ (3) Fecal virus shedding ↓ (4) VH/CD ratios ↑ |
[90] |
3.2. Subunit vaccine
Compared with whole virus vaccines, subunit vaccines have several advantages, including high safety, no contagious viral nucleic acids, and providing a uniform antigen of well-defined nature despite their low immunogenicity, which requires adjuvant or fusion with an immune enhancer to heighten immunogenicity [165,166]. Moreover, establishing a complete subunit vaccine production system is of great significance for the quick response of a sudden epidemic situation. For example, developing an oral vaccine based on the S protein of SARS-CoV-2 in a very short time was attributed to the mature platform of Saccharomyces cerevisiae in the team [167].
The PEDV S protein is the main target for developing subunit vaccines, which is inseparable from its multiple biological roles. In recent years, the researchers have expressed part of the PEDV S protein including COE and S1 or the full length of the S protein using E. coli, Bacillus subtilis, baculovirus, adenovirus, Lactobacillus, and transgenic plants expression systems. Mice or pigs immunized with these vaccines by oral, intramuscular, subcutaneous or intraperitoneal injection produced high levels of IgG and IgA. Table 3 summarizes the subunit vaccines based on the S protein in recent years. In addition, the indeterminate receptor-binding domains (RBDs) and the HR1 and HR2 regions in S2 of PEDV also are potential targets for developing subunit vaccines [168,169]. However, though there are some epitopes in the N and M protein, subunit vaccines based on the PEDV N and M proteins have not yet been reported [137,170]. Furthermore, whether the subunit vaccine based on PEDV non-structural proteins could induce a protective response also needs further verification.
Table 3.
Research progress of PEDV subunit vaccines in recent years.
| Systems | Groups | Animal | Route | Amount of challenge | Detection Indicators | References |
|---|---|---|---|---|---|---|
| Adenovirus | (1) PBS (2) Adenovirus-blank (3) Adenovirus-PEDV-S (4) Inactivated vaccine |
4-week-old pigs (n = 5) | IM | 2 mL 107 TCID50 | (1) IgG and IgA in serum↑ (2) Virus Neutralization (VN) Test↑ (3) Fecal viral shedding↓ (4) Fecal scores↓ |
[199] |
| Silkworm pupae | (1) PBS (2) The S-Bm cell (3) the S-Bm pupae (4) the WT-Bm pupae |
4-week-old pigs (n = 5) | Oral | No | (1) IgG in serum* (2) SIgA in cavity* |
[200] |
| VSV | (1) PBS (2) rVSVMT-S |
pregnant sows (~70 kg, n = 3) | IN IM | Piglets:102 TCID50 | sows: (1) Neutralizing antibodies in the sera↑ (2) Neutralizing antibodies in colostrum or milk↑ Piglets: (3) Neutralizing antibodies in the sera↑ (4) Body weights (10 d after challenge)* (5) Fecal viral shedding (10 d after challenge) ↓ (6) Fecal viral shedding (10 d after challenge) ↓ (7) Mortality (10 d after challenge)↑ |
[201] |
| L. lactis (CH/JLDH/2016) | (1) PBS (2) L. lactis (NZ3900) (3) pNZ8149/NZ3900 (4) pNZ8149-S1/NZ3900 |
6-8-week-old mice(n = 20) | Oral | No | (1) IgG in serum (0, 21, 35, 49 d) ↑ (2) SIgA in stool, small intestine, and cecum (0, 21, 35, 49 d) ↑ (3) Lymphocyte proliferation (35d) ↑ (4) IL-4 and IFN-γ in sera (0, 21, 35, 49 d) ↑ |
[202] |
| Systems | Groups | Animal | Route | Amount of challenge | Detection Indicators | References |
|---|---|---|---|---|---|---|
| B. subtilis | (1) PBS (2)Inactivated PEDV (3)B. subtilis-COE |
5-day-old pigs (n = 4) | Oral | No | (1) The area of PPs↑ (2) The villi length of ileum↑ (3) SIgA in saliva and feces (0, 12, 19, 26, 33 d) ↑ (4) IgG in serum (0, 12, 19, 26, 33 d) ↑ (5) SIgA + T cells and CD3+T cells↑ (6) CD3+CD8+T↑ (7) IL-1β and IL-10 levels↑ (8) Plaque reduction neutralization test↑ |
[203] |
| HEK 293T (PEDV-PT) | (1) Control (2) PEDV S-LTB |
5-week-old pigs (n = 3) | IM | 5 × 105 TCID50 | (1) IgG and IgA in serum (0, 2, 4, 6 w) ↑ (2) SIgA titers in feces (0, 2, 4, 6 w) * (3) Neutralizing antibodies titers (0, 2, 4, 6 w) ↑ (4) Fecal viral load(0, 1, 2, 3, 4, 5 dpi)↓ (5) Clinical scoring of fecal consistency (0, 1, 2, 3, 4, 5 dpi) ↓ |
[204] |
| E. coli (PEDV BM1) | (1) Control (2) NTD 231–501aa (3) NTD 231–501aa/CT |
mice | Oral | (1) IgG1, IgG2a and IgG2b in serum (7 w) ↑ (2) SIgA in feces (7 w) ↑ (3) IgA + cells in the LP (7 w) ↑ |
[205] | |
| HEK 293T (CV777) (LNCT2) |
(1) PBS (2) CV777–S (3) LNCT2-S |
5-week-old mice | SC | No | (1) IgG in serum (0, 14, 21, 28, 42 d) ↑ (2) Neutralizing antibodies titers↑ |
[206] |
| Systems | Groups | Animal | Route | Amount of challenge | Detection Indicators | References |
|---|---|---|---|---|---|---|
| Baculovirus (PEDV-PT) | (1) Medium control (2) S1-Bac (3) S-Bac |
mice (n = 4) 5-week-old pigs (n = 5) |
IM | 5 mL 105 TCID50/mL | Mice: (1) IgG in serum (0, 14, 28 d) ↑ (2) Neutralizing antibodies (0, 14, 28 d) S↑, S1* Pigs: (1) IgG in serum (0, 14, 28 d) ↑ (2) Fecal SIgA (0, 14, 28 d) * (3) Neutralizing antibodies (0, 14, 28 d) S↑, S1* (4) Body weights (7, 21, 35, 49 d) * (5) Stool scores (every day) S↓, S1↑ (6) Fecal viral shedding (every day) S↓, S1↑ |
[207] |
| E. coli (PEDV GDS01) | (1) PBS (2) PBS (used for challenge) (3) rSF (4) COE (5) COE/rSF |
5-week-old pigs (n = 8) |
IM | 7.5 mL 4 × 106 pfu/mL | (1) IgG and IgA in serum (14 d after the last immunization) ↑ (2) SIgA in saliva, snotter and feces (14 d after the last immunization) ↑ (3) Neutralizing antibodies (14 d after the last immunization) ↑ (4) IL-4 and IFN-γ in serum (14 d after the last immunization) ↑ (5) PEDV shedding, fecal scores↓ (6) Pathological lesion↓ |
[208] |
| Systems | Groups | Animal | Route | Amount of challenge | Detection Indicators | References |
|---|---|---|---|---|---|---|
| ORFV (PEDV CO13) | (1) Control (MEM) (2) ORFV-PEDV-S (3) ORFV-PEDV-S (challenge) (4) Live PEDV |
Primiparous gilts (n = 2) |
IM | 2.5 × 102 TCID50 | sows: (1) IgG, IgA and neutralizing antibodies in serum (0, 21, 28, 35, 42, 49, 54, 60, 63, 67, 70, 74 d) ↑ (2) IgG and IgA in milk (1, 3, 7, 10, 14, 17 d) ↑ pigs: (1) IgG and IgA in serum (1 day of farrowing, 3, 7, 10, 14, 17 d) ↑ (2) Clinical scores of fecal consistency (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 d after challenge) ↓ (3) Virus shedding (0, 3、7、10 d after challenge)↓ (4) Mortality of piglets (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 d after challenge) ↓ |
[209] |
| L. lactis | (1) PBS (2) pPG/L393 (3) pPG-COE-DCpep/L393 |
1-day-old piglets (n = 10–15) | Oral | 5 mL 1 × 106 PFU/mL | (1) IgG and IgA in serum (5 d after immunization) ↑ (2) SIgA in feces (5d after immunization) ↑ (3) CD4+IFN-γ+ and CD4+IL-4+T cells (5 d after immunization) ↑ (4) Mortality of piglets (0, 12, 24, 36, 48, 60, 72, 84, 96, 108 h) ↓ (5) Body weights ↓ (6) Virus in jejunum (10–15 min post-mortem) ↓ (7) Virus shedding (0, 12, 24, 36, 48, 60, 72, 84, 96, 108 h) ↓ (8) TLR-4, TLR-9, TGF-β, and TNF-α in splenic lymphocytes and mesenteric lymph nodes↑ |
[210] |
| Systems | Groups | Animal | Route | Amount of challenge | Detection Indicators | References |
|---|---|---|---|---|---|---|
| E. coli (PEDV GDS01) | (1) PBS (2) rSF (3) COE (4) rSF-COE-3D (5) rSF-COE-N (6) rSF-COE-C |
8-week-old mice (n = 15) | IN | No | (1) IgG, IgA, IgG1, IgG2a and IgG2b in serum↑ (2) SIgA in vagina↑ (3) Serum neutralizing antibodies↑ (4) CD3+ CD4+ and CD3 + CD8+ T cells in splenocytes↑ (5) IFN-γ+ and IL-4+T cells in splenocytes↑ |
[211] |
| L. plantarum (S 75-2313 nt) | (1) PBS (2) NC8-pSIP409-pgsA′ (3) NC8/pSIP409/pgsA'/S/Ctrlpep (4) NC8/pSIP409/pgsA′-S/DCpep |
6-week-old mice(n = 6) | Oral | No | (1) IgG and neutralizing antibodies in serum (14, 35, 42 d) ↑ (2) IgG and neutralizing antibodies in feces (14, 35, 42 d) ↑ (3) Secretion levels of S-specific cytokines IL-4, IL-17 and IFN-γ (30 d after the last immunization) ↑ (4) CD 80 and CD 40 of DCs in the LP (30 d after the last immunization) ↑ (5) IgA + B220 + B cells in PPs (30 d after the last immunization) ↑ |
[212] |
| E. coli | (1) HBcAg (149AA) (2) HBcAg + S1(744YSNIGVCK752) (3) HBcAg + S2(756SQYGQVKI771) (4) HBcAg + S3(1371GPRLQPY1377) (5) HBcAg + M(195WAFYVR200) (6) HBcAg + S1/S2/S3/M |
7-week-old mice(n = 8) | IN IP | No | (1) IgG in serum (−1, 13, 27, 42 d) ↑ (2) IgA in serum (−1, 13, 27, 42 d) * (3) IgG of anti-S1, S2, S3 and M in serum (42 d) ↑ (4) Neutralizing antibodies (42 d) S1↑, S2, S3 and M* |
[213] |
| Systems | Groups | Animal | Route | Amount of challenge | Detection Indicators | References |
|---|---|---|---|---|---|---|
| L. lactis | (1) PBS (2) L. lactis (3) L. lactis/6D/COE (4) L. lactis/GFP/6D/COE |
6-week-old mice (n = 15) | Oral | No | (1) IgG in serum (0, 7, 14, 21, 28, 35 d) ↑ (2) SIgA in feces and vaginas (0, 7, 14, 21, 28, 35 d) ↑ (3) Neutralizing antibodies (35 d) ↑ (4) Lymphocyte proliferation (35 d) ↑ |
[214] |
| SPV (SQ2014) (S 386-815aa) |
(1) PBS (2) wtSPV (3) rSPV-St (4) inactivated SQ2014 (5) inactivated CV777 |
1-month-old pigs (n = 3) | Oral SC IM |
1 mL 1 × 106 PFU | (1) IgG and IgA in serum (0, 7, 14, 21, 28, 35, 42, 49 d) ↑ (2) Neutralizing antibodies (0, 7, 14, 21, 28, 35, 42, 49 d) ↑ (3) The concentrations of IL-4 and IFN-γ (30 d) ↑ (4) Mortality↓ |
[215] |
| Baculovirus | (1) PBS (2) VLP (3) Inactivated PEDV |
6-week-old mice (n = 10) | IM | No | (1) Neutralizing antibodies (2, 4, 6 w) ↑ (2) IFN-γ+ and IL-4+CD4+T cells (4 w) ↑ |
[216] |
| 293T PEDV MN (S 21-737aa) |
(1) Adjuvant Control (2) Non-targeted PEDVsAg/adjuvant (3) Langerin-targeted PEDV S/adjuvant/TD (4) Langerin-targeted PEDV S/adjuvant/IM |
4-week-old pigs (n = 7–8) | TD IM |
No | (1) IFN-γ+CD4pos and CD8pos T (0, 7, 28, 35 d)↑ (2) IgG and IgA in serum (21, 28, 35, 42 d) * (3) SIgA in feces (35 d) * (4) Neutralizing antibodies * |
[217] |
| Systems | Groups | Animal | Route | Amount of challenge | Detection Indicators | References |
|---|---|---|---|---|---|---|
| Yeast (CV777) | (1) PBS (2) Yeast/EGFP (3) Yeast/S1 |
3-week-old mice (n = 10) Piglets (n = 10) |
Oral | No | (1) Mice: IgG in serum and SIgA in feces (0, 14, 28 d) ↑ (2) Piglets: SIgA in feces (3, 5, 7, 14 d) ↑ |
[218] |
| ORFV | (1) ORF-GFP/TC + IM (2) ORFV-PEDV-S/TC (3) ORFV-PEDV-S/IM |
3-week-old pigs (n = 4) | TC IM |
2 × 105 TCID50 | (1) IgG and IgA in serum (0, 7, 14, 21, 35, 42, 49, 53, 56, 60 d)TC*,IM↑ (2) Clinical scores of fecal consistency TC*, IM↑ (3) Fecal viral load(0, 3, 5, 7, 9, 11, 14 dpc)TC↑,IM↓ (4) Neutralizing antibodies (0, 3, 7, 10, 14 dpc)TC*,IM↑ |
[219] |
| HEK 293T (USA/Colorado/2013) | (1) TriAdj (2) PEDV S1/TriAdj |
sows (n = 1) | IM | 4-day-old pigs 3 × 102 TCID50 |
(1) IgG in serum of sows↑ (2) Neutralizing antibodies in serum of sows (0, 11, 28, 35 d) ↑ (3) IgG and IgA in colostrum↑ (4) IgG in serum of piglets↑ (5) Neutralizing antibodies in serum of piglets↑ (6) Fecal viral load (0–10 d) ↓ (7) Fecal scores (0–10 d) ↓ (8) The weight of each piglet (0–10 d) * (9) Mortality (0–10 d) ↓ |
[220] |
| Abbreviations in tables. IM: intramuscularly; TC: transcutaneous; TD: Transdermal; IN: intranasally; IP: intraperitoneal; S-Bm: the recombinant baculovirus; VSV: Vesicular stomatitis virus; rVSVMT-S: the rVSV plasmid with PEDV S gene; L. lactis: Lactococcus lactis; B. subtilis: Bacillus subtilis; LTB: heat-labile enterotoxin; SC: subcutaneously; NTD: N-terminal Domain of the Spike Protein of Porcine Epidemic Diarrhea; CT: cholera toxin; LP: lamina propria; ORFV: The parapoxvirus Orf virus; VLP: viruses like particle; SPV: Swinepox virus; rSF: recombined Salmonella flagellin; E. coli: Escherichia coli; ORFV: orf virus; L. plantarum: Lactobacillus plantarum; *: unchanged; ↑: up; ↓: down. | ||||||
3.3. Nucleic acid vaccines
Although the cycle of developing inactivated vaccines is short., inactivated vaccines immunogenicity is poor. Live attenuated vaccines have good immunogenicity, but with poor security and long development cycle. Continual mutation of the PEDV genome, mixed infection with other viruses and huge economic losses all require effective vaccines with a very short development cycle. Hence, nucleic acid vaccines emerged, including DNA vaccines and mRNA vaccines. The advantages of these vaccines are low cost, high safety, short lead time and simple design [171,172]. The most important is that the established platform is universal. Once the platform has been established, all that is required is the synthesizing and inserting of the sequence of the core neutralizing antigen gene into an appropriate expression vector, which might be useful for the development of vaccines against emerging pandemic infectious diseases in a considerably shortened time [173]. However, the bivalent DNA vaccine generated by combining PEDV with and an RV or TGEV have been developed but not yet been applied to clinical trials, possibly because of the poor immunogenicity [174,175]. Although the effectiveness of DNA vaccines has been improved by optimizing codons, selecting appropriate promoters, optimizing plasmid vector backbones, and adding molecular adjuvants, some breakthroughs are still required to be made to allow DNA vaccines to play a more significant role in preventing disease [172]. RNA vaccines have great application prospects. Five advanced candidates have recently been moved into clinical development, including mRNA-1273 from Moderna, Ad5-nCoV from CanSino Biologicals, INO-4800 from Inovio, LV-SMENP-DC and pathogen-specific aAPC from Shenzhen Geno-Immune Medical Institute [176]. As early as 2014, the company of Harrisvaccines in the United States used its SirraVaxSM technology to produce PEDV mRNA vaccines, and this platform technology is conditionally licensed in the USA. Compared to DNA vaccines, mRNA vaccines possess higher immune potency but more instability in vivo [171]. Self-amplifying RNA (saRNA) encoding the alphaviral replicase and a gene of the viruses enables replication of the RNA in the cytoplasm [177,178]. The sa-RNA vaccines based on influenza hemagglutinin was more effective than the mRNA vaccine in mice [179] and the sa-RNA vaccines encoding the SARS-CoV-2 spike protein encapsulated within a lipid nanoparticle also induced high neutralizing antibody titers in mice [180]. Therefore, the application of sa-RNA vaccines and corresponding delivery vectors will probably break the bottlenecks in the development of PEDV or other coronaviruses mRNA vaccines.
4. Conclusion
The current PEDV vaccines may have failed to prevent the PEDV epidemic situation because the virus is constantly variable and the vaccines cannot induce sufficient mucosal immunity. The SIgA secreted by intestinal mucosal epithelial cells was able to neutralize viruses at the invasion site. On the other hand, SlgA in the colostrum and milk of sows play a decisive role in protecting neonatal suckling piglets against PEDV. At present, two strategies have been used to induce mucosal immunity. The first one involves applying suitable carriers for delivering vaccines by mucosal routes; a whole PEDV attenuated strain entrapped in the enteric material hydroxypropyl methyl cellulose phthalate (HPMCP) successfully induced both mucosal immunity and systemic immunity after oral immunization in weaned piglets [181,182]. PEDV killed vaccine antigens (KAg) entrapped in PLGA particles was delivered intranasally to sows at 28 and 14 days prior to farrowing. Pregnant sows immunized with PLGA-KAg had the highest serum lgG and lgA levels, colostrum IgA antibody titers, and the strongest cellular immune response [183]. The second was that intramuscularly administrating vaccines with appropriate adjuvants. Recently, the researchers developed an inactivated vaccine that could induce SIgA and IgG in 5-week-old pigs only by intramuscular injection of inactivated PEDV antigen combined with CCL25, CCL27 and CCL28 proteins [184].
In summary, the application of reverse genetic operating systems, mucosal immune enhancement adjuvants and mucosal delivery vehicles may have a great significance for the development of both traditional vaccines and new vaccines of PED.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
This work was supported by the National Key R&D Project of China (2017YFD0501102, 2017YFD0500605), Key R&D Project in Shaanxi Province of China (2019NY-076) and the Youth Innovation Team of Shaanxi Universities.
References
- 1.Li Y., Wu Q., Huang L., Yuan C., Wang J., Yang Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018;9:3811. doi: 10.1038/s41467-018-06056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Y., Li W., Zhou Q., Li Q., Xu Z., Shen H., Chen F. Characterization and pathogenicity of Vero cell-attenuated porcine epidemic diarrhea virus CT strain. Virol. J. 2019;16:121. doi: 10.1186/s12985-019-1232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas J.T., Chen Q., Gauger P.C., Gimenez-Lirola L.G., Sinha A., Harmon K.M., Madson D.M., Burrough E.R., Magstadt D.R., Salzbrenner H.M., Welch M.W., Yoon K.J., Zimmerman J.J., Zhang J. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in naive conventional neonatal and weaned pigs. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leidenberger S., Schroder C., Zani L., Auste A., Pinette M., Ambagala A., Nikolin V., de Smit H., Beer M., Blome S. Virulence of current German PEDV strains in suckling pigs and investigation of protective effects of maternally derived antibodies. Sci. Rep. 2017;7:10825. doi: 10.1038/s41598-017-11160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin H., Chen L., Gao L., Yuan X., Ma Z., Fan H. Epidemic strain YC2014 of porcine epidemic diarrhea virus could provide piglets against homologous challenge. Virol. J. 2016;13:68. doi: 10.1186/s12985-016-0529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wicht O., Li W., Willems L., Meuleman T.J., Wubbolts R.W., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014;88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B.J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016;226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo P.C., Huang Y., Lau S.K., Yuen K.-Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lokugamage K.G., Narayanan K., Huang C., Makino S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012;86:13598–13608. doi: 10.1128/JVI.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez-Guardeno J.M., Regla-Nava J.A., Nieto-Torres J.L., DeDiego M.L., Castano-Rodriguez C., Fernandez-Delgado R., Perlman S., Enjuanes L. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei L., Ying S., Baojun L., Yi Y., Xiang H., Wenli S., Zounan S., Deyin G., Qingyu Z., Jingmei L., Guohui C. Attenuation of mouse hepatitis virus by deletion of the LLRKxGxKG region of Nsp1. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zust R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel R., Eskdale J., Gallagher G. Regulation of IFN-lambda1 promoter activity (IFN-lambda1/IL-29) in human airway epithelial cells. J. Immunol. 2011;187:5636–5644. doi: 10.4049/jimmunol.1003988. [DOI] [PubMed] [Google Scholar]
- 20.Ueki I.F., Min-Oo G., Kalinowski A., Ballon-Landa E., Lanier L.L., Nadel J.A., Koff J.L. Respiratory virus-induced EGFR activation suppresses IRF1-dependent interferon lambda and antiviral defense in airway epithelium. J. Exp. Med. 2013;210:1929–1936. doi: 10.1084/jem.20121401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odendall C., Dixit E., Stavru F., Bierne H., Franz K.M., Durbin A.F., Boulant S., Gehrke L., Cossart P., Kagan J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden M.S., Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napetschnig J., Wu H. Molecular basis of NF-kappaB signaling. Annu. Rev. Biophys. 2013;42:443–468. doi: 10.1146/annurev-biophys-083012-130338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verstrepen L., Bekaert T., Chau T.L., Tavernier J., Chariot A., Beyaert R. TLR-4, IL-1R and TNF-R signaling to NF-kappaB: variations on a common theme. Cell. Mol. Life Sci. 2008;65:2964–2978. doi: 10.1007/s00018-008-8064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honda K., Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 26.Hermant P., Michiels T. Interferon-lambda in the context of viral infections: production, response and therapeutic implications. J Innate Immun. 2014;6:563–574. doi: 10.1159/000360084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2018;92 doi: 10.1128/JVI.01677-17. e01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q., Ma J., Yoo D. Inhibition of NF-kappaB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology. 2017;510:111–126. doi: 10.1016/j.virol.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q., Ma J., Yoo D. Inhibition of NF-κB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology. 2017;510:111–126. doi: 10.1016/j.virol.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Z., Ye G., Deng F., Wang G., Cui M., Fang L., Xiao S., Fu Z.F., Peng G. Structural basis for the inhibition of host gene expression by porcine epidemic diarrhea virus nsp1. J. Virol. 2018;92 doi: 10.1128/JVI.01896-17. e01896-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Z., Wang G., Yang Y., Shi J., Fang L., Li F., Xiao S., Fu Z.F., Peng G. A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J. Biol. Chem. 2019;294:13606–13618. doi: 10.1074/jbc.RA119.009713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Sturzel C.M., Frohlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanjanahaluethai A., Baker S.C. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 2000;74:7911–7921. doi: 10.1128/jvi.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.-T.K., Baker S.C. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B., Clementz M.A., Banach B.S., Li K., Baker S.C., Chen Z. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong B., Yang Y., Li S., Wang Y.Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H.B. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H., Baker S.C., Feng L., Chen Z. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94:1554. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B., Clementz M.A., Banach B.S., Li K., Baker S.C. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Sun A., Sun Y., Zhang S., Xia T., Guo T., Hao Z., Sun L., Jiang Y., Qiao X., Cui W., Tang L., Xu Y., Li Y., Wang L. Porcine transmissible gastroenteritis virus inhibits NF-kappaB activity via nonstructural protein 3 to evade host immune system. Virol. J. 2019;16:97. doi: 10.1186/s12985-019-1206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu L., Dong J., Wang Y., Zhang P., Liu Y., Zhang L., Liang P., Wang L., Song C. Porcine epidemic diarrhea virus nsp4 induces pro-inflammatory cytokine and chemokine expression inhibiting viral replication in vitro. Arch. Virol. 2019;164:1147–1157. doi: 10.1007/s00705-019-04176-2. [DOI] [PubMed] [Google Scholar]
- 46.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 47.Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 49.Gao F., Ou H.Y., Chen L.L., Zheng W.X., Zhang C.T. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553:451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sturman L.S., Holmes K.V. The molecular biology of coronaviruses. Adv. Virus Res. 1983;28:35–112. doi: 10.1016/S0065-3527(08)60721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Y., Lei Y., Ye G., Sun L., Fang L., Xiao S., Fu Z.F., Yin P., Song Y., Peng G. Identification of two antiviral inhibitors targeting 3C-like serine/3C-like protease of porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus. Vet. Microbiol. 2018;213:114–122. doi: 10.1016/j.vetmic.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye G., Wang X., Tong X., Shi Y., Fu Z.F., Peng G. Structural basis for inhibiting porcine epidemic diarrhea virus replication with the 3C-like protease inhibitor GC376. Viruses. 2020;12 doi: 10.3390/v12020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 54.Yoneyama M., Fujita T. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 2007;18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 55.Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye G., Deng F., Shen Z., Luo R., Zhao L., Xiao S., Fu Z.F., Peng G. Structural basis for the dimerization and substrate recognition specificity of porcine epidemic diarrhea virus 3C-like protease. Virology. 2016;494:225–235. doi: 10.1016/j.virol.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 58.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng X., StJohn S.E., Osswald H.L., O'Brien A., Banach B.S., Sleeman K., Ghosh A.K., Mesecar A.D., Baker S.C. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 2014;88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J. Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng Z., Deng F., Shi K., Ye G., Wang G., Fang L., Xiao S., Fu Z., Peng G. Dimerization of coronavirus nsp9 with diverse modes enhances its nucleic acid binding affinity. J. Virol. 2018;92 doi: 10.1128/JVI.00692-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Donaldson E.F., Sims A.C., Graham R.L., Denison M.R., Baric R.S. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J. Virol. 2007;81:6356–6368. doi: 10.1128/JVI.02805-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3'-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5'-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5'-to-3' polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu Y., Cai H., Lu M., Ma Y., Li A., Gao Y., Zhou J., Gu H., Li J., Gu J. Porcine epidemic diarrhea virus deficient in RNA cap guanine-N-7 methylation is attenuated and induces higher type I and III interferon responses. J. Virol. 2020;94 doi: 10.1128/JVI.00447-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Case J.B., Ashbrook A.W., Dermody T.S., Denison M.R. Mutagenesis of S-Adenosyl-l-Methionine-Binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J. Virol. 2016;90:7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Graham R.L., Becker M.M., Eckerle L.D., Bolles M., Denison M.R., Baric R.S. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012;18:1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., Zust R., Hwang M., V'Kovski P., Stalder H., Marti S., Habjan M., Cervantes-Barragan L., Elliot R., Karl N., Gaughan C., van Kuppeveld F.J., Silverman R.H., Keller M., Ludewig B., Bergmann C.C., Ziebuhr J., Weiss S.R., Kalinke U., Thiel V. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Y., Zhang H., Shi Z., Chen J., Li M., Shi H., Shi D., Guo L., Feng L. Porcine epidemic diarrhea virus nsp15 antagonizes interferon signaling by RNA degradation of TBK1 and IRF3. Viruses. 2020;12 doi: 10.3390/v12060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deng X., van Geelen A., Buckley A.C., O'Brien A., Pillatzki A., Lager K.M., Faaberg K.S., Baker S.C. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 2019;93 doi: 10.1128/JVI.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Menachery V.D., Debbink K., Baric R.S. Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments. Virus Res. 2014;194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi P., Su Y., Li R., Liang Z., Dong S., Huang J. PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 2019;265:57–66. doi: 10.1016/j.virusres.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hou Y., Ke H., Kim J., Yoo D., Su Y., Boley P., Chepngeno J., Vlasova A.N., Saif L.J., Wang Q. Engineering a live attenuated porcine epidemic diarrhea virus vaccine candidate via inactivation of the viral 2'-O-methyltransferase and the endocytosis signal of the spike protein. J. Virol. 2019;93 doi: 10.1128/JVI.00406-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H., Yu W., Xu K., Bian C., Fischer W.B., Schwarz W., Feng L., Sun B. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. mBio. 2016;7:e01451–15. doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155:1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun M., Ma J., Yu Z., Pan Z., Lu C., Yao H. Identification of two mutation sites in spike and envelope proteins mediating optimal cellular infection of porcine epidemic diarrhea virus from different pathways. Vet. Res. 2017;48:44. doi: 10.1186/s13567-017-0449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jengarn J., Wongthida P., Wanasen N., Frantz P.N., Wanitchang A., Jongkaewwattana A. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J. Gen. Virol. 2015;96:2206–2218. doi: 10.1099/vir.0.000184. [DOI] [PubMed] [Google Scholar]
- 94.Lee S., Son K.Y., Noh Y.H., Lee S.C., Choi H.W., Yoon I.J., Lee C. Genetic characteristics, pathogenicity, and immunogenicity associated with cell adaptation of a virulent genotype 2b porcine epidemic diarrhea virus. Vet. Microbiol. 2017;207:248–258. doi: 10.1016/j.vetmic.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun M., Ma J., Yu Z., Pan Z., Lu C., Yao H. Identification of two mutation sites in spike and envelope proteins mediating optimal cellular infection of porcine epidemic diarrhea virus from different pathways. Vet. Res. 2017;48:44. doi: 10.1186/s13567-017-0449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zou D., Xu J., Duan X., Xu X., Li P., Cheng L., Zheng L., Li X., Zhang Y., Wang X. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet. Microbiol. 2019;235:209–219. doi: 10.1016/j.vetmic.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Si F., Hu X., Wang C., Chen B., Wang R., Dong S., Yu R., Li Z. Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses. 2020;12:214. doi: 10.3390/v12020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaewborisuth C., He Q., Jongkaewwattana A. The accessory protein ORF3 contributes to porcine epidemic diarrhea virus replication by direct binding to the spike protein. Viruses. 2018;10 doi: 10.3390/v10080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ye S., Li Z., Chen F., Li W., Guo X., Hu H., He Q. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Gene. 2015;51:385–392. doi: 10.1007/s11262-015-1257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaewborisuth C., Yingchutrakul Y., Roytrakul S., Jongkaewwattana A. Porcine epidemic diarrhea virus (PEDV) ORF3 interactome reveals inhibition of virus replication by cellular VPS36 protein. Viruses. 2019;11 doi: 10.3390/v11040382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen Y., Zhang Z., Li J., Gao Y., Zhou L., Ge X., Han J., Guo X., Yang H. Porcine epidemic diarrhea virus S1 protein is the critical inducer of apoptosis. Virol. J. 2018;15:170. doi: 10.1186/s12985-018-1078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hsueh F.C., Lin C.N., Chiou H.Y., Chia M.Y., Chiou M.T., Haga T., Kao C.F., Chang Y.C., Chang C.Y., Jeng C.R. Updated phylogenetic analysis of the spike gene and identification of a novel recombinant porcine epidemic diarrhoea virus strain in Taiwan. Transboundary Emerg. Dis. 2019;67:417–430. doi: 10.1111/tbed.13365. [DOI] [PubMed] [Google Scholar]
- 103.Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu J., Chai X., Cheng Y., Xing G., Liao A., Du L., Wang Y., Lei J., Gu J., Zhou J. Molecular characteristics of the spike gene of porcine epidemic diarrhoea virus strains in Eastern China in 2016. Virus Res. 2018;247:47–54. doi: 10.1016/j.virusres.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 105.Wanitchang A., Saenboonrueng J., Kaewborisuth C., Srisutthisamphan K., Jongkaewwattana A. A single V672F substitution in the spike protein of field-isolated PEDV promotes cell–cell fusion and replication in VeroE6 cells. Viruses. 2019;11:282. doi: 10.3390/v11030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 107.Wrapp D., McLellan J.S. The 3.1-angstrom cryo-electron microscopy structure of the porcine epidemic diarrhea virus spike protein in the prefusion conformation. J. Virol. 2019;93 doi: 10.1128/JVI.00923-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.K., Wang Q., Du L., Tan W., Wilson I.A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han Y., Kral P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904 e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X.F., Li J., Zhang N.N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D.Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.F., Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li B., Ge J., Li Y. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang J., Guo L., Yang L., Xu J., Zhang L., Feng L., Chen H., Wang Y. Metalloprotease ADAM17 regulates porcine epidemic diarrhea virus infection by modifying aminopeptidase N. Virology. 2018;517:24–29. doi: 10.1016/j.virol.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ji C.-M., Wang B., Zhou J., Huang Y.-W. Aminopeptidase-N-independent entry of porcine epidemic diarrhea virus into Vero or porcine small intestine epithelial cells. Virology. 2018;517:16–23. doi: 10.1016/j.virol.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li W., Luo R., He Q., van Kuppeveld F.J., Rottier P.J., Bosch B.-J. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res. 2017;235:6–13. doi: 10.1016/j.virusres.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Whitworth K.M., Rowland R.R., Petrovan V., Sheahan M., Cino-Ozuna A.G., Fang Y., Hesse R., Mileham A., Samuel M.S., Wells K.D. Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgenic Res. 2019;28:21–32. doi: 10.1007/s11248-018-0100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang D., Ge X., Chen D., Li J., Cai Y., Deng J., Zhou L., Guo X., Han J., Yang H. The S gene is necessary but not sufficient for the virulence of porcine epidemic diarrhea virus novel variant strain BJ2011C. J. Virol. 2018;92:e00603–e00618. doi: 10.1128/JVI.00603-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kao C.-F., Chang H.-W. Investigation of the role of the spike protein in reversing the virulence of the highly virulent taiwan porcine epidemic diarrhea virus pintung 52 strains and its attenuated counterpart. Viruses. 2020;12:41. doi: 10.3390/v12010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Suzuki T., Terada Y., Enjuanes L., Ohashi S., Kamitani W. S1 subunit of spike protein from a current highly virulent porcine epidemic diarrhea virus is an important determinant of virulence in piglets. Viruses. 2018;10 doi: 10.3390/v10090467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suzuki T., Terada Y., Enjuanes L., Ohashi S., Kamitani W. S1 subunit of spike protein from a current highly virulent porcine epidemic diarrhea virus is an important determinant of virulence in piglets. Viruses. 2018;10:467. doi: 10.3390/v10090467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shirato K., Maejima M., Islam M.T., Miyazaki A., Kawase M., Matsuyama S., Taguchi F. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhea virus, but promotes its infectivity via aminopeptidase activity. J. Gen. Virol. 2016;97:2528–2539. doi: 10.1099/jgv.0.000563. [DOI] [PubMed] [Google Scholar]
- 122.Cruz D.J., Kim C.J., Shin H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sun D., Feng L., Shi H., Chen J., Cui X., Chen H., Liu S., Tong Y., Wang Y., Tong G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Okda F.A., Lawson S., Singrey A., Nelson J., Hain K.S., Joshi L.R., Christopher-Hennings J., Nelson E.A., Diel D.G. The S2 glycoprotein subunit of porcine epidemic diarrhea virus contains immunodominant neutralizing epitopes. Virology. 2017;509:185–194. doi: 10.1016/j.virol.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kong N., Meng Q., Jiao Y., Wu Y., Zuo Y., Wang H., Sun D., Dong S., Zhai H., Tong W., Zheng H., Yu H., Tong G., Xu Y., Shan T. Identification of a novel B-cell epitope in the spike protein of porcine epidemic diarrhea virus. Virol. J. 2020;17:46. doi: 10.1186/s12985-020-01305-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kuo L., Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003;77:4597–4608. doi: 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu X., Zhang H., Zhang Q., Dong J., Liang Y., Huang Y., Liu H.-J., Tong D. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2018;92 doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jimenez-Guardeno J.M., Nieto-Torres J.L., DeDiego M.L., Regla-Nava J.A., Fernandez-Delgado R., Castano-Rodriguez C., Enjuanes L. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lei X.-M., Yang Y.-L., He Y.-Q., Peng L., Zhao P., Xu S.-Y., Cao H., Fang P., Qiu W., Qin P. Specific recombinant proteins of porcine epidemic diarrhea virus are immunogenic, revealing their potential use as diagnostic markers. Vet. Microbiol. 2019;236:108387. doi: 10.1016/j.vetmic.2019.108387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu X., Zhang H., Zhang Q., Dong J., Huang Y., Tong D. Porcine epidemic diarrhea virus M protein blocks cell cycle progression at S-phase and its subcellular localization in the porcine intestinal epithelial cells. Acta Virol. 2015;59:265–275. doi: 10.4149/av_2015_03_265. [DOI] [PubMed] [Google Scholar]
- 133.Kim O., Chae C. In situ hybridization for the detection and localization of porcine epidemic diarrhea virus in the intestinal tissues from naturally infected piglets. Vet. Pathol. 2000;37:62–67. doi: 10.1354/vp.37-1-62. [DOI] [PubMed] [Google Scholar]
- 134.Arndt A.L., Larson B.J., Hogue B.G. A conserved domain in the coronavirus membrane protein tail is important for virus assembly. J. Virol. 2010;84:11418–11428. doi: 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fan J.-H., Zuo Y.-Z., Shen X.-Q., Gu W.-Y., Di J.-M. Development of an enzyme-linked immunosorbent assay for the monitoring and surveillance of antibodies to porcine epidemic diarrhea virus based on a recombinant membrane protein. J. Virol Methods. 2015;225:90–94. doi: 10.1016/j.jviromet.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou X., Zhang T., Song D., Huang T., Peng Q., Chen Y., Li A., Zhang F., Wu Q., Ye Y. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol. Cell. Probes. 2017;33:36–41. doi: 10.1016/j.mcp.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 137.Zhang Z., Chen J., Shi H., Chen X., Shi D., Feng L., Yang B. Identification of a conserved linear B-cell epitope in the M protein of porcine epidemic diarrhea virus. Virol. J. 2012;9:225. doi: 10.1186/1743-422X-9-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pulford D.J., Britton P. Expression and cellular localisation of porcine transmissible gastroenteritis virus N and M proteins by recombinant vaccinia viruses. Virus Res. 1991;18:203–217. doi: 10.1016/0168-1702(91)90019-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bos E.C., Luytjes W., van der Meulen H.V., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang X.-W., Wang M., Zhan J., Liu Q.-Y., Fang L.-l., Zhao C.-y., Jiang P., Li Y.-F., Bai J. Pathogenicity and immunogenicity of a new strain of porcine epidemic diarrhea virus containing a novel deletion in the N gene. Vet. Microbiol. 2020;240:108511. doi: 10.1016/j.vetmic.2019.108511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y., Liu H.-J., Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shan Y., Liu Z.-q., Li G.-w., Chen C., Luo H., Liu Y.-j., Zhuo X.-h., Shi X.-f., Fang W.-h., Li X.-l. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-λ production by blocking the nuclear factor-κB nuclear translocation. J. Zhejiang Univ. Sci. B. 2018;19:570–580. doi: 10.1631/jzus.B1700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liwnaree B., Narkpuk J., Sungsuwan S., Jongkaewwattana A., Jaru-Ampornpan P. Growth enhancement of porcine epidemic diarrhea virus (PEDV) in Vero E6 cells expressing PEDV nucleocapsid protein. PLoS One. 2019;14 doi: 10.1371/journal.pone.0212632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.DeZure A.D., Berkowitz N.M., Graham B.S., Ledgerwood J.E. Whole-inactivated and virus-like particle vaccine strategies for chikungunya virus. J. Infect. Dis. 2016;214:S497–S499. doi: 10.1093/infdis/jiw352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lauring A.S., Jones J.O., Andino R. Rationalizing the development of live attenuated virus vaccines. Nat. Biotechnol. 2010;28:573–579. doi: 10.1038/nbt.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent Advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang M., Ma S., Zhou J., Feng L., Tong Y., Yu W., Wei F., Cui X., Huang S., Liu C. Development of the inactivated vaccine of porcine epidemic diarrhea virus. Chin. Anim. Infect. Dis. 1993;5:17–19. [Google Scholar]
- 152.Ma S., Wang M., Zhou J., Feng L. Adaptation of porcine epidemic diarrhea virus to Vero cells and evaluation of the inactivated vaccine against porcine epidemic diarrhea virus. Chin. Anim. Infect. Dis. 1994;2:15–18. [Google Scholar]
- 153.Tong Y., Feng L., Li W., Wang M., Ma S. Development of attenuated vaccine strain of porcine epidemic diarrhea virus. Chin. Anim. Infect. Dis. 1998;20:329–332. [Google Scholar]
- 154.Ma S., Wang M., Feng L., Li W. Development of bi-combined inactivated vaccine against transmissible gastroenteritis virus and porcine epidemic diarrhea virus. Chin. Anim. Infect. Dis. 1995;2:15–18. [Google Scholar]
- 155.Baek P.-S., Choi H.-W., Lee S., Yoon I.-J., Lee Y.J., Lee S., Lee C. Efficacy of an inactivated genotype 2b porcine epidemic diarrhea virus vaccine in neonatal piglets. Vet. Immunol. Immunopathol. 2016;174:45–49. doi: 10.1016/j.vetimm.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Deng X., van Geelen A., Buckley A.C., O'Brien A., Pillatzki A., Lager K.M., Faaberg K.S., Baker S.C. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 2019;93:e02000–e02018. doi: 10.1128/JVI.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hou Y., Meulia T., Gao X., Saif L.J., Wang Q. Deletion of both the tyrosine-based endocytosis signal and the endoplasmic reticulum retrieval signal in the cytoplasmic tail of spike protein attenuates porcine epidemic diarrhea virus in pigs. J. Virol. 2019;93 doi: 10.1128/JVI.01758-18. e01758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hou Y., Lin C.-M., Yokoyama M., Yount B.L., Marthaler D., Douglas A.L., Ghimire S., Qin Y., Baric R.S., Saif L.J. Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 2017;91 doi: 10.1128/JVI.00227-17. e00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Graham R.L., Sims A.C., Brockway S.M., Baric R.S., Denison M.R. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 2005;79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mielech A.M., Deng X., Chen Y., Kindler E., Wheeler D.L., Mesecar A.D., Thiel V., Perlman S., Baker S.C. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J. Virol. 2015;89:4907–4917. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Deng X., StJohn S.E., Osswald H.L., O'Brien A., Banach B.S., Sleeman K., Ghosh A.K., Mesecar A.D., Baker S.C. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 2014;88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7 doi: 10.1128/mBio.01721-16. e01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jimenez-Guardeno J.M., Nieto-Torres J.L., DeDiego M.L., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., Enjuanes L. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Du L., Tai W., Zhou Y., Jiang S. Vaccines for the prevention against the threat of MERS-CoV. Expert Rev. Vaccines. 2016;15:1123–1134. doi: 10.1586/14760584.2016.1167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yap Y.K., Smith D.R. Strategies for the plant-based expression of dengue subunit vaccines. Biotechnol. Appl. Biochem. 2010;57:47–53. doi: 10.1042/BA20100248. [DOI] [PubMed] [Google Scholar]
- 167.Koch L. A platform for RNA virus cloning. Nat. Rev. Genet. 2020;21:388. doi: 10.1038/s41576-020-0246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhao P., Wang B., Ji C.-M., Cong X., Wang M., Huang Y.-W. Identification of a peptide derived from the heptad repeat 2 region of the porcine epidemic diarrhea virus (PEDV) spike glycoprotein that is capable of suppressing PEDV entry and inducing neutralizing antibodies. Antivir. Res. 2018;150:1–8. doi: 10.1016/j.antiviral.2017.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhu X., Liu Q., Du L., Lu L., Jiang S. Receptor-binding domain as a target for developing SARS vaccines. J. Thorac. Dis. 2013;5:S142. doi: 10.3978/j.issn.2072-1439.2013.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang K., Xie C., Zhang J., Zhang W., Yang D., Yu L., Jiang Y., Yang S., Gao F., Yang Z. The identification and characterization of two novel epitopes on the nucleocapsid protein of the porcine epidemic diarrhea virus. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep39010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccines. 2016;15:313–329. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Naik R., Peden K. Regulatory considerations on the development of mRNA vaccines. Curr. Top. Microbiol. Immunol. 2020 doi: 10.1007/82_2020_220. [DOI] [PubMed] [Google Scholar]
- 174.Yin Y., Zhu L., Liu P., Zhao J., Fan Y., Sun X., Xu Z. Evaluation on the efficacy and immunogenicity of recombinant DNA plasmids expressing S gene from porcine epidemic diarrhea virus and VP7 gene from porcine rotavirus. Braz. J. Microbiol. 2019;50:279–286. doi: 10.1007/s42770-018-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang Y., Zhang X., Liao X., Huang X., Cao S., Wen X., Wen Y., Wu R., Liu W. Construction of a bivalent DNA vaccine co-expressing S genes of transmissible gastroenteritis virus and porcine epidemic diarrhea virus delivered by attenuated Salmonella typhimurium. Virus Gene. 2016;52:354–364. doi: 10.1007/s11262-016-1316-z. [DOI] [PubMed] [Google Scholar]
- 176.Thanh Le T., Andreadakis Z., Kumar A., Gomez Roman R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 177.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26:363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T., O'Hagan D.T., Singh M., Mason P.W., Valiante N.M., Dormitzer P.R., Barnett S.W., Rappuoli R., Ulmer J.B., Mandl C.W. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H., Reece S.T., Sahin U., Tregoning J.S. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wen Z., Xu Z., Zhou Q., Li W., Wu Y., Du Y., Chen L., Zhang Y., Xue C., Cao Y. Oral administration of coated PEDV-loaded microspheres elicited PEDV-specific immunity in weaned piglets. Vaccine. 2018;36:6803–6809. doi: 10.1016/j.vaccine.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 182.Choe S., Song S., Piao D., Park G.N., Shin J., Choi Y.J., Kang S.K., Cha R.M., Hyun B.H., Park B.K., An D.J. Efficacy of orally administered porcine epidemic diarrhea vaccine-loaded hydroxypropyl methylcellulose phthalate microspheres and RANKL-secreting L. lactis. Vet. Microbiol. 2020;242:108604. doi: 10.1016/j.vetmic.2020.108604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li B., Du L., Yu Z., Sun B., Xu X., Fan B., Guo R., Yuan W., He K. Poly (d,l-lactide-co-glycolide) nanoparticle-entrapped vaccine induces a protective immune response against porcine epidemic diarrhea virus infection in piglets. Vaccine. 2017;35:7010–7017. doi: 10.1016/j.vaccine.2017.10.054. [DOI] [PubMed] [Google Scholar]
- 184.Hsueh F.C., Chang Y.C., Kao C.F., Hsu C.W., Chang H.W. Intramuscular immunization with chemokine-adjuvanted inactive porcine epidemic diarrhea virus induces substantial protection in pigs. Vaccines. 2020;vol. 8 doi: 10.3390/vaccines8010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Deng X., Buckley A.C., Pillatzki A., Lager K.M., Faaberg K.S., Baker S.C. Inactivating three interferon antagonists attenuates pathogenesis of an enteric coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00565-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H., Baker S.C., Feng L., Chen Z. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yu L., Dong J., Wang Y., Zhang P., Liu Y., Zhang L., Liang P., Wang L., Song C. Porcine epidemic diarrhea virus nsp4 induces pro-inflammatory cytokine and chemokine expression inhibiting viral replication in vitro. Arch. Virol. 2019;164:1147–1157. doi: 10.1007/s00705-019-04176-2. [DOI] [PubMed] [Google Scholar]
- 188.Lin H., Li B., Liu M., Zhou H., He K., Fan H. Nonstructural protein 6 of porcine epidemic diarrhea virus induces autophagy to promote viral replication via the PI3K/Akt/mTOR axis. Vet. Microbiol. 2020;244:108684. doi: 10.1016/j.vetmic.2020.108684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fang S., Chen B., Tay F.P., Ng B.S., Liu D.X. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology. 2007;358:136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shi P., Su Y., Li R., Liang Z., Dong S., Huang J. PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Virus Res. 2019;265:57–66. doi: 10.1016/j.virusres.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chen Y., Zhang Z., Li J., Gao Y., Zhou L., Ge X., Han J., Guo X., Yang H. Porcine epidemic diarrhea virus S1 protein is the critical inducer of apoptosis. Virol. J. 2018;15:170. doi: 10.1186/s12985-018-1078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xu X.G., Zhang H.L., Zhang Q., Dong J., Huang Y., Tong D.W. Porcine epidemic diarrhea virus M protein blocks cell cycle progression at S-phase and its subcellular localization in the porcine intestinal epithelial cells. Acta Virol. 2015;59:265–275. doi: 10.4149/av_2015_03_265. [DOI] [PubMed] [Google Scholar]
- 194.Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shan Y., Liu Z.Q., Li G.W., Chen C., Luo H., Liu Y.J., Zhuo X.H., Shi X.F., Fang W.H., Li X.L. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-lambda production by blocking the nuclear factor-kappaB nuclear translocation. J. Zhejiang Univ. - Sci. B. 2018;19:570–580. doi: 10.1631/jzus.B1700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zou D., Xu J., Duan X., Xu X., Li P., Cheng L., Zheng L., Li X., Zhang Y., Wang X., Wu X., Shen Y., Yao X., Wei J., Yao L., Li L., Song B., Ma J., Liu X., Wu Z., Zhang H., Cao H. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet. Microbiol. 2019;235:209–219. doi: 10.1016/j.vetmic.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Si F., Hu X., Wang C., Chen B., Wang R., Dong S., Yu R., Li Z. Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses. 2020;12 doi: 10.3390/v12020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hou Y., Ke H., Kim J., Yoo D., Su Y., Boley P., Chepngeno J., Vlasova A.N., Saif L.J., Wang Q. Engineering a live attenuated porcine epidemic diarrhea virus vaccine candidate via inactivation of the viral 2'-O-methyltransferase and the endocytosis signal of the spike protein. J. Virol. 2019;93:e00406–e00419. doi: 10.1128/JVI.00406-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Liu X., Zhao D., Zhou P., Zhang Y., Wang Y. Evaluation of the efficacy of a recombinant adenovirus expressing the spike protein of porcine epidemic diarrhea virus in pigs. BioMed Res. Int. 2019:2019. doi: 10.1155/2019/8530273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chang C.-Y., Hsu W.-T., Tsai P.-S., Chen C.-M., Cheng I.-C., Chao Y.-C., Chang H.-W. Oral administration of porcine epidemic diarrhea virus spike protein expressing in silkworm pupae failed to elicit immune responses in pigs. Amb. Express. 2020;10:1–10. doi: 10.1186/s13568-020-0952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ke Y., Yu D., Zhang F., Gao J., Wang X., Fang X., Wang H., Sun T. Recombinant vesicular stomatitis virus expressing the spike protein of genotype 2b porcine epidemic diarrhea virus: a platform for vaccine development against emerging epidemic isolates. Virology. 2019;533:77–85. doi: 10.1016/j.virol.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Guo M., Yi S., Guo Y., Zhang S., Niu J., Wang K., Hu G. Construction of a recombinant lactococcus lactis strain expressing a variant porcine epidemic diarrhea virus S1 gene and its immunogenicity analysis in mice. Viral Immunol. 2019;32:144–150. doi: 10.1089/vim.2018.0108. [DOI] [PubMed] [Google Scholar]
- 203.Wang J., Huang L., Mou C., Zhang E., Wang Y., Cao Y., Yang Q. Mucosal immune responses induced by oral administration recombinant Bacillus subtilis expressing the COE antigen of PEDV in newborn piglets. Biosci. Rep. 2019;39 doi: 10.1042/BSR20182028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chang Y.-C., Chang C.-Y., Tsai P.-S., Chiou H.-Y., Jeng C.-R., Pang V.F., Chang H.-W. Efficacy of heat-labile enterotoxin B subunit-adjuvanted parenteral porcine epidemic diarrhea virus trimeric spike subunit vaccine in piglets. Appl. Microbiol. Biotechnol. 2018;102:7499–7507. doi: 10.1007/s00253-018-9110-6. [DOI] [PubMed] [Google Scholar]
- 205.Kim S.-H., Cho B.-H., Lee K.-Y., Jang Y.-S. N-terminal domain of the spike protein of porcine epidemic diarrhea virus as a new candidate molecule for a mucosal vaccine. Immune Netw. 2018;vol. 18 doi: 10.4110/in.2018.18.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wang X., Chen J., Shi D., Shi H., Zhang X., Yuan J., Jiang S., Feng L. Immunogenicity and antigenic relationships among spike proteins of porcine epidemic diarrhea virus subtypes G1 and G2. Arch. Virol. 2016;161:537–547. doi: 10.1007/s00705-015-2694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chang C.-Y., Hsu W.-T., Chao Y.-C., Chang H.-W. Display of porcine epidemic diarrhea virus spike protein on baculovirus to improve immunogenicity and protective efficacy. Viruses. 2018;10:346. doi: 10.3390/v10070346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li Q., Xu Z., Wu T., Peng O., Huang L., Zhang Y., Xue C., Wen Z., Zhou Q., Cao Y. A flagellin-adjuvanted PED subunit vaccine improved protective efficiency against PEDV variant challenge in pigs. Vaccine. 2018;36:4228–4235. doi: 10.1016/j.vaccine.2018.05.124. [DOI] [PubMed] [Google Scholar]
- 209.Joshi L.R., Okda F.A., Singrey A., Maggioli M.F., Faccin T.C., Fernandes M.H., Hain K.S., Dee S., Bauermann F.V., Nelson E.A. Passive immunity to porcine epidemic diarrhea virus following immunization of pregnant gilts with a recombinant orf virus vector expressing the spike protein. Arch. Virol. 2018;163:2327–2335. doi: 10.1007/s00705-018-3855-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hou X., Jiang X., Jiang Y., Tang L., Xu Y., Qiao X., Liu M., Cui W., Ma G., Li Y. Oral immunization against PEDV with recombinant Lactobacillus casei expressing dendritic cell-targeting peptide fusing COE protein of PEDV in piglets. Viruses. 2018;10:106. doi: 10.3390/v10030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Li Q., Peng O., Wu T., Xu Z., Huang L., Zhang Y., Xue C., Wen Z., Zhou Q., Cao Y. PED subunit vaccine based on COE domain replacement of flagellin domain D3 improved specific humoral and mucosal immunity in mice. Vaccine. 2018;36:1381–1388. doi: 10.1016/j.vaccine.2018.01.086. [DOI] [PubMed] [Google Scholar]
- 212.Huang K.-Y., Yang G.-L., Jin Y.-B., Liu J., Chen H.-L., Wang P.-B., Jiang Y.-L., Shi C.-W., Huang H.-B., Wang J.-Z. Construction and immunogenicity analysis of Lactobacillus plantarum expressing a porcine epidemic diarrhea virus S gene fused to a DC-targeting peptide. Virus Res. 2018;247:84–93. doi: 10.1016/j.virusres.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gillam F., Zhang J., Zhang C. Hepatitis B core antigen based novel vaccine against porcine epidemic diarrhea virus. J. Virol Methods. 2018;253:61–69. doi: 10.1016/j.jviromet.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 214.Yu M., Wang L., Ma S., Wang X., Wang Y., Xiao Y., Jiang Y., Qiao X., Tang L., Xu Y. Immunogenicity of eGFP-marked recombinant Lactobacillus casei against transmissible gastroenteritis virus and porcine epidemic diarrhea virus. Viruses. 2017;9:274. doi: 10.3390/v9100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yuan X., Lin H., Li B., He K., Fan H. Efficacy and immunogenicity of recombinant swinepox virus expressing the truncated S protein of a novel isolate of porcine epidemic diarrhea virus. Arch. Virol. 2017;162:3779–3789. doi: 10.1007/s00705-017-3548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wang C., Yan F., Zheng X., Wang H., Jin H., Wang C., Zhao Y., Feng N., Wang T., Gao Y. Porcine epidemic diarrhea virus virus-like particles produced in insect cells induce specific immune responses in mice. Virus Gene. 2017;53:548–554. doi: 10.1007/s11262-017-1450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Subramaniam S., Cao D., Tian D., Cao Q.M., Overend C., Yugo D.M., Matzinger S.R., Rogers A.J., Heffron C.L., Catanzaro N. Efficient priming of CD4 T cells by Langerin-expressing dendritic cells targeted with porcine epidemic diarrhea virus spike protein domains in pigs. Virus Res. 2017;227:212–219. doi: 10.1016/j.virusres.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang X., Wang Z., Xu H., Xiang B., Dang R., Yang Z. Orally administrated whole yeast vaccine against porcine epidemic diarrhea virus induced high levels of IgA response in mice and piglets. Viral Immunol. 2016;29:526–531. doi: 10.1089/vim.2016.0067. [DOI] [PubMed] [Google Scholar]
- 219.Hain K.S., Joshi L.R., Okda F., Nelson J., Singrey A., Lawson S., Martins M., Pillatzki A., Kutish G.F., Nelson E.A. Immunogenicity of a recombinant parapoxvirus expressing the spike protein of Porcine epidemic diarrhea virus. J. Gen. Virol. 2016;97:2719–2731. doi: 10.1099/jgv.0.000586. [DOI] [PubMed] [Google Scholar]
- 220.Makadiya N., Brownlie R., van den Hurk J., Berube N., Allan B., Gerdts V., Zakhartchouk A. S1 domain of the porcine epidemic diarrhea virus spike protein as a vaccine antigen. Virol. J. 2016;13:57. doi: 10.1186/s12985-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]