Highlights
-
•
Severe COVID-19 is mediated by both a cytokine storm and a thrombotic storm.
-
•
HUFA balance in part determines inflammatory and thrombotic potential.
-
•
Genetic polymorphisms in the HUFA synthetic pathway define arachidonic acid levels.
-
•
HUFA diet/supplements may be particularly important for fast desaturators.
Keywords: Sars-cov-2, Covid-19, Highly unsaturated fatty acids, Fads gene cluster, Rs66698963, Arachidonic acid
Abstract
COVID-19 symptoms vary from silence to rapid death, the latter mediated by both a cytokine storm and a thrombotic storm. SARS-CoV (2003) induces Cox-2, catalyzing the synthesis, from highly unsaturated fatty acids (HUFA), of eicosanoids and docosanoids that mediate both inflammation and thrombosis. HUFA balance between arachidonic acid (AA) and other HUFA is a likely determinant of net signaling to induce a healthy or runaway physiological response. AA levels are determined by a non-protein coding regulatory polymorphisms that mostly affect the expression of FADS1, located in the FADS gene cluster on chromosome 11. Major and minor haplotypes in Europeans, and a specific functional insertion-deletion (Indel), rs66698963, consistently show major differences in circulating AA (>50%) and in the balance between AA and other HUFA (47–84%) in free living humans; the indel is evolutionarily selective, probably based on diet. The pattern of fatty acid responses is fully consistent with specific genetic modulation of desaturation at the FADS1-mediated 20:3→20:4 step. Well established principles of net tissue HUFA levels indicate that the high linoleic acid and low alpha-linoleic acid in populations drive the net balance of HUFA for any individual. We predict that fast desaturators (insertion allele at rs66698963; major haplotype in Europeans) are predisposed to higher risk and pathological responses to SARS-CoV-2 could be reduced with high dose omega-3 HUFA.
Graphical abstract
Introduction
The global COVID-19 pandemic caused by the SARS-CoV-2 virus is an enigma in part because of its extraordinary range of symptoms, from complete silence that may include person-to-person through-air transmissibility to respiratory failure and death within days of diagnosis. The term “cytokine storm”, previously limited to technical journals, has made its way into the popular press because of the severity of runaway inflammation [1]. An additional defining event is widespread thrombosis, with pathological manifestations in the lung similar to SARS and MERS [2]. Platelet–fibrin thrombi in small arterial vessels are consistent with a coagulopathy. In the end stage, multiple organ failure with severe liver damage is found, consistent with thrombotic microangiopathy [3, 4]. Both inflammation and thrombosis are mediated by signaling molecules derived from highly unsaturated fatty acids (HUFA) or more precisely the relative mix of them present at any one time in membranes.
Beyond the general inflammatory/prothrombotic potential of the HUFA milieu, the spike protein of the (2003) SARS-CoV virus induces cylooxygenase-2 (COX-2), one of the key synthetic enzymes for eicosanoid synthesis [5]. Moreover, induction of COX-2 may be required for efficient early stage replication of mouse hepatitis virus, also a coronavirus [6]. Insofar as this is true for SARS-CoV-2, individuals with a robust COX-2 response to viral COX-2 induction and thus supportive of the rapid replication of the virus, would be particularly susceptible to a prothrombotic/proinflammatory HUFA milieu. Inhibition of COX-2 via known inhibitors (e.g. celecoxib (Celebrex®)) at the early infection stage would be expected to reduce viral replication. Selective COX-2 inhibitors are effective against arthritis as are high dose omega-3 HUFA [7], and randomized controlled trial (RCT) evidence indicates regular consumption of omega-3 rich salmon in the context of an “anti-inflammatory diet portfolio” reduces rheumatoid arthritis symptoms [8]. Most selective COX-2 inhibitors were removed from the market because of enhanced thrombotic events, ascribed to rebalancing of the eicosanoid milieu toward thromboxanes [9] and thromboxane A2 production via COX-1 [10]. Because severe COVID-19 appears to be an inherently prothrombotic event, we would not expect selective COX-2 inhibitors to be effective against it and possibly exacerbate symptoms. However, a balanced HUFA milieu may be particularly important for avoidance of a COX-2-enhanced cytokine storm or the hypercoagulopathy with features characteristic of a thrombotic storm [7]. Inherited genetic risk factors can enhance or have an additive effect in increasing the risk of thrombotic events during hypercoagulable periods such as severe COVID-19. For instance, increased risk of thrombotic events and recurrence occur in patients with homozygous and compound heterozygous for Factor V Leiden (FVL) and prothrombin G20210A mutations [11, 12]. Patients with FVL mutation displayed higher levels of d-dimer and fibrinogen-fibrin degradation products in plasma [13].
We review here some of the latest genetic, nutritional, and related factors that drive the balance of membrane HUFA toward or away from proinflammatory, prothrombotic potential, that may contribute to the severity of disease.
HUFA function as components to impart physiochemical properties in membranes and lipid droplets, and as precursors for signaling molecules. Physicochemical properties are most important for support of normal membrane function. Phospholipid molecular profiles adapt based on the nutritional supply of the energetically precious fatty acids according to principles yet to be fully determined; changes in dietary fatty acids certainly influence the fatty acid profile of the membrane. Signaling is mediated by HUFA release from membranes via phospholipases. In some cases this is the rate limiting step for synthesis, while in others the free fatty acid (FFA) milieu precedes activation of the synthetic enzymes [14]. In either case, the mixture of precursor HUFA for eicosanoid and other oxylipin signaling molecules is critical as they are competitive substrates for biosynthesis.
Oxylipins, more specifically eicosanoids and docosanoids, modulate many aspects of overall physiological function. The relative mixture of circulating HUFA has long been known to mediate clotting and inflammation via eicosanoids. In the last two decades, the role of docosanoids in the resolution phase of inflammation has been at least partially elucidated. Based on numerous well established principles, the relative mix of membrane HUFA shifts the balance on the continuum of pro- vs anti-inflammatory state. HUFA-derived signaling also influence thrombotic tendency.
Among the HUFA, a guiding principle is that arachidonic acid (AA) is precursor to mostly prothrombotic/pro-inflammatory signaling molecules, while the omega-3 HUFA eicosapentaenoic acid (EPA), docosapentaenoic acid (DPA), and docosahexaenoic acid (DHA) are generally antithrombotic/anti-inflammatory. The omega-3 s generally inhibit AA conversion to signaling molecules, and the omega-3 s are themselves converted to mediators that promote the resolution phase of inflammation. Also generally antagonistic to the actions of AA-derived products is the omega-6 HUFA 20:3n-6, dihomo-gamma-linolenic acid (DGLA), the immediate precursor of AA via FADS1-catalyzed 5-desaturation; it is also precursor to the anti-inflammatory [15, 16], potently vasodilatory, PGE1 [17]. We recognize that the concept that excess AA predisposes to disease while omega-3 s counteract this effect is a simplification of complex biochemistry for which studies of intermediate markers are ambiguous, especially for inflammation with respect to cardiovascular disease [18, 19], though recent high dose studies such as with EPA show important effects [20]. However, strong and consistent evidence supports the contention that diet and genetics influence the HUFA tissue profile as well as the biosynthesis of HUFA.
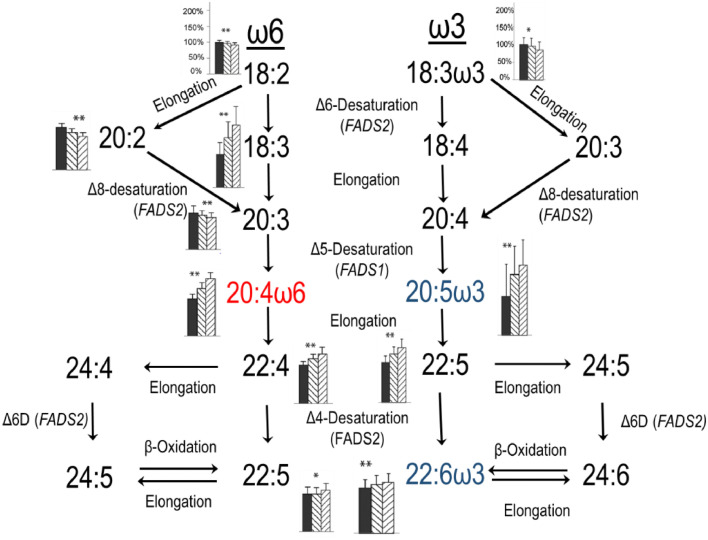
The biochemical pathways from precursor 18 carbon polyunsaturated fatty acids (PUFA) linoleic acid (LA, 18:2n-6) and alpha-linolenic acid (ALA, 18:3n-3) are presented in Fig. 1 . Representations of the PUFA families transformed by the pathway in parallel are common and aptly illustrate that the two HUFA families are subject to the same sets of genes/enzymes and are thus in competition. A misleading aspect of this representation leads to the incorrect assumption that an increase in any upstream precursor causes an increase in products. From a net tissue content point of view, this is not the case. Many studies continue to be designed only to rediscover what has been known for decades.
-
•
Omega-3. In apparently healthy, well-nourished adult humans, no amount of ALA or any precursor of DHA increases circulating [21] or breastmilk [22] DHA when LA is constant. Most of the precursors 18:3 (ALA), 18:4, and 20:5 have been tested multiple times [21], DPA at least once [23]. In other words, when ALA is increased against constant LA, the ratio of LA to ALA changes but circulating DHA does not. In rodents, increases in tissue DHA are small, tissue dependent, and is greater for HUFA [24], [25], [26] than ALA [27]. Clearly then, it is not true that the amount of HUFA depends on the omega-6 to omega-3 ratio, whether or not HUFA are present.
-
•
Omega-6. Similarly, changes in dietary LA cause, at most, minor changes in tissue AA [27].
Fig. 1.
Biochemical pathways for biosynthesis of highly unsaturated fatty acids from the 18 carbon precursors. The bar graphs alongside fatty acids are the relative amounts in plasma PL in the three genotypes (DD, ID, II, left to right) at the ra66698963 Indel locus in a population of 1504 free living participants in Beijing. **p<0.05 for all bars different from one another; *p<0.05 for ID different than II.
However, neither the amount of DHA nor AA is “tightly regulated”, in the same sense as for instance calcium concentrations are tightly regulated. Both DHA and AA concentrations are easily altered over wide ranges by dietary intervention without acute effects, and even by genotype.
-
•
DHA, DPA, and EPA circulating concentrations and LA respond reciprocally. Reduction in dietary LA increases all omega-3 HUFA including DHA [28, 29].
-
•
AA concentrations decrease in response to decrease in LA below about 4% of calories, depending on the ratio of LA to ALA [29].
Put another way, only 2 ways are known to increase circulating DHA status in humans: 1) consume (preformed) DHA, or 2) lower dietary LA.
The levels of specific tissue fatty acids are maintained by numerous mechanisms within broad ranges. Mammalian PUFA requirements arise because the capacity to introduce double bonds between the n-6 and n-7 positions, and the n-3 and n-4 positions in any normal (straight chain) fatty acid present in plants and lower animals has been lost in mammals (n is the number of carbons in the linear chain), presumably because mammalian diets always contain sufficient quantities of these precursors. The inaccurately named “essential fatty acids” linoleic acid (18:2n-6; LA) and alpha-linolenic acid (18:3n-3; ALA) supply most of this part of the molecule because of their prevalence in the food supply. They are inaccurately named for many reasons [30], [31], [32], among which unlike other nutrients labeled “essential” they can be completely replaced in a healthy diet [33, 34]. Functional characterization molecular studies in the last 20 years have shown that the fatty acid desaturase (FADS1 and FADS2, located at 11q12–13.1) and elongase genes (ELOVL2 at 6p24.2 and ELOVL5 at 6p12.1) mediate the endogenous biosynthesis of HUFA. A third similar gene, FADS3 (also at 11q13.1), introduces a cis double bond into a 4(E) (trans) ceramide at the 14–15 position to yield sphingadienine [35], and may have some role in altering fatty acid levels in development [36]. Our purpose is to integrate the known biochemical and genetic control of HUFA levels to describe in part how changes in diet in the context of genotype alter the mixture of HUFA available for signaling, thereby defining the range of inflammatory and thrombotic potential of individuals, with an eye toward possible relevance to insults such as COVID-19. We first review modern genetics relevant to HUFA biosynthesis.
Human FADS and ELOVL genomic localization and structure
The paralogous FADS gene cluster evolved by gene duplication events, and localizes to the long arm of human chromosome 11 at the cytogenetic location 11q12–13.1 [37]. The three FADS (FADS1, FADS2 and FADS3), cluster within 92.6 kb genomic region flanked by FEN1 and RAB3IL1 (NCBI release# GRCh38.p13). FADS1 spans 17.3 kb, FADS2 spans 51.1 kb and FADS3 spans 18.7 kb of genomic DNA (NCBI release# GRCh38.p13). The classical, functional, FADS1 and FADS2 both consist of 12 exons and encode 444-amino acid (aa) peptides with estimated molecular mass of 52.0 kDa and 52.3 kDa, respectively [37], [38], [39]. A high degree of polypeptide sequence identity exists between FADS1 and FADS2 (61%). In the current NCBI database, human FADS1 (GenBank accession NM_013402) is represented by 501-aa protein (NP_037534.5), which is 57-aa more than the functional and classical FADS1 (444 aa). The function of 501-aa protein is not yet characterized. FADS3, is the third member of the FADS gene family, also with 12 exons and encoding a putative protein of 445-aa with estimated molecular mass of 51.1 kDa [37]. All three FADS contain the N-terminal highly conserved cytochrome b5 domain (HPGG) as electron donor and three histidine motifs HDFGH, HFQHH, QIEHH (FADS1); HDYGH, HFQHH, QIEHH (FADS2); and HDLGH, HFQHH, QIEHH (FADS3). All three FADS are extensively spliced and the splice variants are evolutionarily conserved [40], [41], [42], [43].
The elongation of very long chain fatty acids-like (ELOVL) family consists of seven enzymes (ELOVL 1–7). ELOVL2 and ELOVL5 are involved in the biosynthesis of HUFA less than 24 carbons in length [44]. ELOVL2 localizes to the short arm of chromosome 6 at the cytogenetic location 6p24.2, spans 64 kb of genomic DNA, encodes 296-aa protein (NM_017770.4; NP_060240.3). In silico analysis of the protein sequence of ELOVL2 using a simple modular architecture research tool (SMART) software [45, 46] shows seven transmembrane regions (spanning residues 28–50, 62–84, 116–135, 142–164, 179–201, 208–225, and 235–254). ELOVL2 includes four amino acid motifs characteristic of elongases (KSVEFLDT, HVYHH, HILMYSYY and TQAQLVQ). ELOVL5 localizes to the short arm of chromosome 6 at the cytogenetic location 6p12.1, spans 82 kb of genomic DNA, encodes 299-aa protein (NM_021814.5; NP_068586.1). In silico analysis of peptide sequence of ELOVL5 using SMART software shows seven transmembrane regions (spanning residues 26–48, 65–87, 110–132, 139–158, 168–187, 207–224, and 229–251). ELOVL5 includes four amino acid motifs characteristic of elongases (KLIEFMDT, HVYHH, HVLMYSYY and TQGQLLQ).
FADS and ELOVL biochemical functions
Fatty acid desaturases (FADS2, FADS1) perform dehydrogenation reactions with strict stereo and regioselectivity resulting in the introduction of cis (Z) double bonds (DB) into fatty acid hydrocarbon chains [47], [48], [49]. The classical FADS2 is a multifunctional enzyme (Table 1 ) which catalyzes even numbered Δ4-, Δ6- and Δ8-desaturation towards normal even chain fatty acids (n-ECFA), normal odd chain fatty acid (n-OCFA) and monomethyl branched chain fatty acids (BCFA) substrates including eight polyunsaturates, two monounsaturates, one saturate, one n-OCFA, and four BCFA [44, [49], [50], [51], [52], [53]]. Human FADS2 mediates the conversion of 16:0 to 16:1n-10 but has no enzymatic activity towards 18:0 [44]. FADS1 has shown Δ5-desaturation activity towards five polyunsaturates and Δ7-desaturation activity towards one monounsaturate [44, 52, 53] . ELOVL2 has substrate specificity for four PUFA with chain lengths 20 and 22 carbons [54], while ELOVL5 has substrate specificity for four PUFA with chain lengths 18 and 20 carbons [54].
Table 1.
FADS2 catalyzes reactions of at least 16 substrates at three positions.
| Desaturation Position | Substrate |
|---|---|
| Δ4-desaturation |
22:5n-3→22:6n-3 22:4n-6→22:5n-6 |
| Δ6-desaturation |
18:3n-3→18:4n-3 18:2n-6→18:3n-6 18:1n-9→18:2n-9 24:5n-3→24:6n-3 24:4n-6→24:5n-6 16:0→16:1n-10 17:0→17:1n-11 iso16:0→iso6Z-16:1 iso18:0→iso6Z-18:1 iso17:0→iso6Z-17:1 anteiso17:0→anteiso6Z-17:1 |
| Δ8-desaturation |
20:1n-9→20:2n-9 20:3n-3→20:4n-3 20:2n-6→20:3n-6 |
11q13 Genomic Region. Human chromosome 11q13 is known to contain hotspots for viral integration, harbor fragile sites, copy number variations (CNV), and several disease phenotypes, including several types of cancers [55], [56], [57], [58]. The 11q13 locus is well known to contain fragile sites that are prone to breakage. This region has a high frequency of repeats, low GC content, two folate-sensitive fragile sites, potential secondary structures, and a high incidence of cytogenetic and molecular alterations in several cancers [55, [58], [59], [60], [61]]. Structural variations (SV), which alter chromosomal structure and the DNA copy number, are increasingly recognized as major contributors to genome variability [62]. Copy number changes at 11q13 region are associated with several cancer phenotypes such as primary melanoma [63], breast cancer [64], [65], [66], [67], [68], [69], [70], head and neck cancers [71], [72], [73], bladder, lung, oral squamous cell, liver and esophageal cancers [57, 74], cervical cancer [61], endocrine tumors (ETs) of pancreas and duodenum [75], ovarian cancer [74] and prostate cancer [76]. In certain disease conditions, the 11q13 copy number changes span several mega bases (Mb) of DNA [77, 78]. Recently, our prediction of an alternative FADS2 mediated MUFA pathway [149] was confirmed in liver and lung cancer cells [79]. FADS2 mediated the production of sapienic acid (16:1n-10) [79, 80]. Anomalous loss of PUFA desaturase activity has been reported in several cancer cells, including MCF7, HeLa and K562 [53, 81, 82]. The loss of FADS2 activity causes cancer cells to shut down HUFA biosynthesis thereby affecting eicosanoid and docosanoid precursor synthesis and normal cell signaling [53, 81]. Earlier we have shown that this metabolic defect can be corrected by transfection of functional FADS2 [53]. Despite the key importance of FADS2 and the loss of its function in several cancer cells, the consequences of FADS2 loss have not been investigated despite the emergence of 11q13 amplicon.
FADS and ELOVL Genetics. Candidate gene studies and genome-wide association studies (GWAS) both yield strong evidence that FADS and ELOVL genes are related to PUFA levels [83], [84], [85], [86], [87], [88], [89], [90] and human disease phenotypes, e.g. cardiovascular disease (CVD) [91, 92], major depressive disorder [93, 94], metabolic syndrome [88, 95], obesity [96], atopic dermatitis [83], aging [97, 98], spinocerebellar ataxia 38 (SCA38) [99]. FADS genetic studies also showed population differences in the capacity to biosynthesize HUFA [95, 100]. Traditional human populations have been described that are analogous to herbivores (vegans) and carnivores (Natives of the Canadian Arctic) [101]. Due to the loss of FADS2 activity, carnivores, such as cats, have lost the metabolic capacity to synthesize HUFA, presumably due to the ubiquitous presence of AA and DHA in a meat-based diet. In contrast, herbivores intake very little AA and DHA and must have a robust metabolic capacity to synthesize all they need, especially at life stages of high demand such as the brain growth spurt.
Evidence from FADS genetic studies indicates that the regulatory non-protein coding SNPs are strongly associated with interindividual differences in converting precursors to physiologically active HUFA, especially AA, EPA and DHA [85]. We cite a handful of examples. A FADS association study showed a 11 SNP haplotype found in 26% participants is associated with 29% of the variation in AA levels [83, 85], subsequently replicated by many other groups [85, 95, [102], [103], [104], [105], [106]]. A distinct FADS haplotype defined by 28 SNPs is associated with an enhanced ability to biosynthesize HUFA [107]. FADS SNPs are associated with maternal, infant, and child health phenotypes, for instance, plasma lipids in infants [108], gestational duration [109], perinatal depression [110], attention/hyperactivity and learning [111, 112], cognition and intelligence [113, 114] and blood pressure in young children [115]. A recent study showed mothers who were heterozygous or homozygous for the minor alleles of FADS1 and FADS2 SNPs had infants with a greater head circumference and birth weight [116]. A GWAS showed a striking adaptation of the Greenland Inuits attributed to high n-3 FA intake [117], recently expanded to the first Americas [118]. The InCHIANTI GWAS identified a minor allele of rs174537 accounting for 18.6% genetic variance in AA levels, confirmed in an independent sample from the GOLDN study [119]. A FADS2-promoter SNP (rs968567) enhancing FADS2 expression was discovered [120]. A whole genome analysis of the Chinese population showed rs28456 to be associated with AA [121]. ELOVL2 and ELOVL5 maternal genetic variants were associated with PUFA levels in breast milk [89]. Taken together these disparate studies all point to HUFA pathway polymorphisms as key modulators of health.
Insertion-Deletion (Indel) Polymorphisms and rs66698963. In humans, highly polymorphic insertion/deletions (Indel) are the second most frequent polymorphisms after SNPs and are increasingly recognized as functional contributors of genetic variation influencing human phenotypes [122]. Largely because of the technical difficulties in genotyping and calling Indels from short-read sequencing, their functional effects are understudied [122]. One example in the FADS gene cluster appears to be a very strong modulator of circulating PUFA levels and is under selective pressure, presumably by diet. The rs66698963 is a 22 base pair (bp) regulatory polymorphism in FADS2 intron 1 nearby a sterol response element (SRE), discovered by our group [123]. Experimentally determined rs66698963 genotype frequencies from our two clinical trials [100, 124] show among South Asian Indians that II alleles are found in 67.5%, in Chinese 45.8%, whereas in a US population it was only 18% (Table 2 ). Commonly reported non-protein coding SNPs within FADS2 (rs174575, rs174570 and rs1535), and thus the nearby Indel rs66698963 are associated with increased IQ scores, blood FA levels and complex diseases. SNPs rs174575 and rs174570 are within 600 and 6000 bp upstream from the FADS2 Indel, respectively [84, 117, 125]. In humans, common SNP variants are often found to follow Indels [126], suggesting that rs174575, rs174570 and/or rs1535 are tags for the functional genomic Indel that directly modulates binding at the nearby SRE. Our early data established that under basal conditions, human lymphoblasts with the DD genotype had the lowest expression of FADS1, with II the highest and ID in the middle [123]. Subsequent studies of circulating PUFA in free living humans, that is, apparently healthy individuals sampled but not enrolled in any study, confirmed the prediction that the reaction catalyzed by FADS1, 20:3→20;4 (DGLA→AA), DD would favor precursors and II favor products [100, 124].
Table 2.
Experimentally determined rs66698963 genotype frequencies from three countries.
| I/I (%) | I/D (%) | D/D (%) | |
|---|---|---|---|
| Indians (South Asia) |
67.5 | 29.5 | 3 |
| Americans | 18 | 39 | 43 |
| Chinese | 45.8 | 42.4 | 11.8 |
Importantly, genotype at this locus exerts positive selectivity likely based on the PUFA in their diets, whether primarily plant-based 18 carbon precursors that must be converted to HUFA, or ample HUFA consumed from animal and seafood [100]. Positive selectivity directly indicates that those with a genotype mismatched to their conditions of existence leave fewer offspring. From a contemporary health perspective, we interpret this to mean that genotypes mismatched to diet are dangerous, whether due to chronic disease or pandemic events.
We return to the human endogenous biosynthesis pathway from 18:2n-6 and 18:3n-3 PUFA to HUFA (Fig. 1). Using data from a study in 1504 participants in Beijing [124], we post next to each PUFA the experimentally determined plasma PL FA levels for each rs66698963 genotype normalized to the DD genotype (Fig. 1). The genes mediating the synthesis of HUFA contribute wide variability to the efficiency of HUFA synthesis. The Δ6-desaturase [D6D coded by FADS2] metabolizes both 18:2n-6 and 18:3n-3, resulting in the synthesis of 6,9,12–18:3 and 6,9,12,15–18:4, respectively. This initial D6D step is widely regarded as the rate-limiting for HUFA biosynthesis, however, recent data indicate that other steps in the pathway can limit HUFA levels [44]. Both AA and EPA can be further elongated and desaturated to yield 22:5n-6 and DHA, respectively. We established that the last step is via Δ4-desaturation [51], also mediated by FADS2 similar to many other organisms [127, 128]; the widely accepted peroxisomal pathway worked out in rodents is also shown and is likely to operate as well [129].
Clear patterns are evident from rs66698963 genotypes (Fig. 1). The n-6 PUFA 18:2, 20:2, and 20:3, and n-3 PUFA 18:3 all decrease from DD to ID to II, which we refer to as a precursor pattern. In contrast, all other measured PUFA increase from DD to ID to II, a product pattern. Organized another way, the precursor-product pattern pairs are observed in the reactions mediated by FADS2 (18:2n-6→18:3n-6) and FADS1 (20:3n-6→20:4n-6), but not the desaturations of the C22 HUFA. We previously showed that the I allele is associated with an increase in FADS1 gene expression but activation of the nearby SRE (sterol response element) caused both FADS1 and FADS2 expression to increase [123], which predicts the precursor-product pattern if expression is taken as a proxy for activity. Taken together, these data are consistent with the I allele increasing desaturase activity in a dose dependent manner for FADS1.,
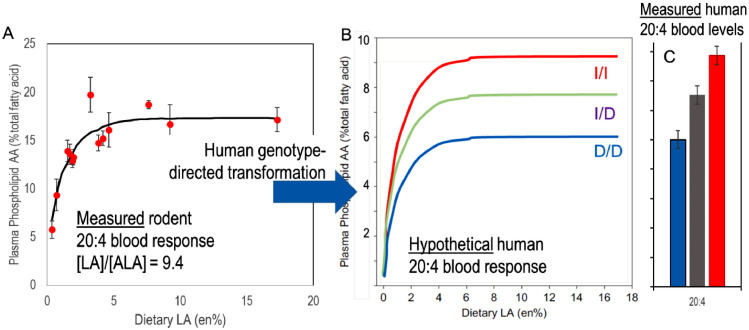
Fig. 2 A and 2B are a hypothetical merging of experiment with theory to explain the role of human genetics on circulating AA (20:4) levels in individuals. Studied in rats [29], panel A shows the response of plasma phospholipid AA with increasing dietary LA, at a dietary ratio of LA to ALA (omega-6 to omega-3) of about 9.4. This latter value is similar to the LA/ALA ratio in the edible oil supply of industrialized nations consuming primarily seed oils rich in LA, including all industrialized nations at the close of the 20th century, and most developing nations at the present time. PL AA rises rapidly from 5% to over 15% where it plateaus at 3–4% of diet LA. The current intake of LA in the US is about 17 g/d for adult men [US DGA 2015] or 7.6 en%, down from a peak of 20 g/d or 9 en% in 2011–2012 [130]. Ascribing this decrease to the introduction of high oleic oils, as is likely, ALA is also lower in most high oleic oils and thus the ratio of about 9.4 likely holds and the curves apply to the changed levels. In rats, these data show that circulating AA levels would not have changed with this reduction, illustrating the central point that not all changes in precursor fatty acids result in a change in tissue fatty acids. Though no data of similar density of LA intake are available for humans, the similarity of the enzymes between rats and humans, and the similar response of human plasma AA to reductions in LA, is strong support for the hypothesis that the human curve is similar [131].
Fig. 2.
A) Measured response of rat plasma PL AA (20:4n-6) as a function of dietary PUFA, LA and ALA in a fixed proportion of 9.4:1. The plasma AA rises to reach a plateau at 3–4%en (energy). B) Hypothetical response of the three rs66698963 Indel genotypes to increasing dietary PUFA. Three different plateau levels are achieved corresponding to C) the measured blood levels found in the Chinese cohort. Panel A redraw from Gibson, PLEFA 2013.
The rs66698963 indel polymorphism appears to be unique to modern humans. Both Neanderthal and Denisovan DNA has the I/I (insertion) genotype [100]. Models of its response must either study the polymorphism directly or rely on hypothetical inferences about responses from well controlled animal studies.
Noting the strong relationship between circulating AA and the indel genotype, we cast a hypothesis about the relative response of the three human genotypes (panel B). Data on AA levels for free-living humans (n = 1504) in the Beijing China region showed a strong relationship to genotype (panel C). The ratio of 20:4/(20:5 + 22:6) is 32% greater for I/I compared to D/D. We note that the measured level of plasma phospholipid LA averaged 36% which is higher than most Western populations (e.g. [132]) and thus suggests that their intake of LA is well into the AA plateau region, above about 4% LA. Bridging the two datasets are hypothetical curves intended to describe the plasma PL AA response as a function of dietary LA intake directed by genotype. Our hypothetical curves predict that at a LA/ALA ratio of 9.4 or greater, AA levels measured for the three genotypes reflect plateau AA levels. This hypothesis also explains why the relationship of AA to genotype is remarkably strong.
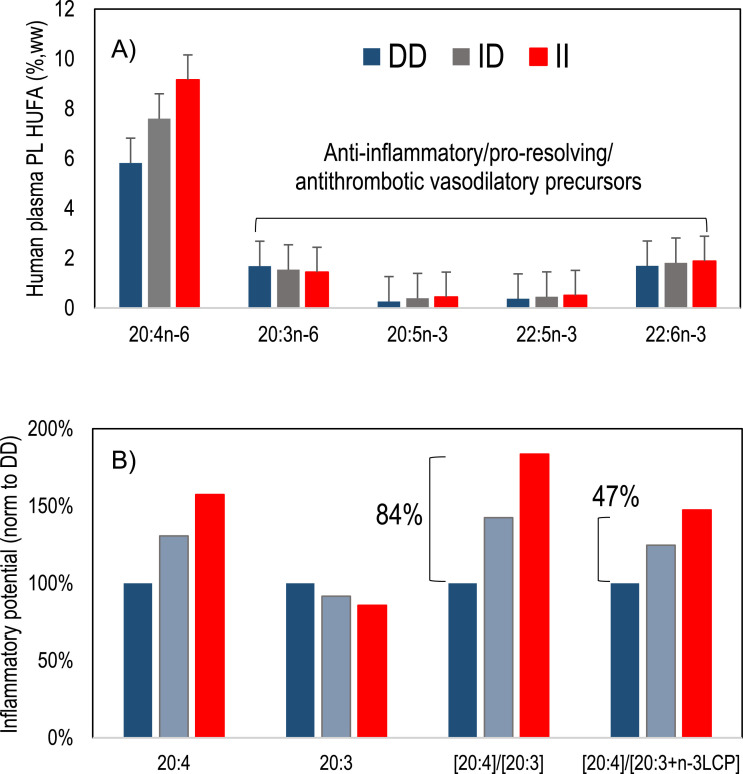
As noted in the introduction, the relative amounts of AA and other HUFA is regarded as a key determinant of net signaling for inflammation and thrombosis. We calculate the relative ratios of AA (20:4) vs its FADS1 mediated precursor and vs all downstream HUFA (Fig. 3 A and 3B). Our results show that the ratio of 20:4 to 20:3 nearly doubles, increasing 84% from DD to II genotype. Similarly, but less intuitively obvious, the ratio of 20:4 to the sum of other HUFA increases by 47% from DD to II. The extent to which these differences predispose free living humans toward or away from disease is not known, but presents the compelling hypothesis to test whether one homozygous genotype predisposes to chronic disease.
Fig. 3.
Human plasma HUFA relative to rs66698963 Indel genotype and inflammatory potential. A) AA (20:4n-6) increases dramatically by genotype (similar to Fig. P1C), as well as all n-3 HUFA), while 20:3n-6 decreases. B) Normalized to their respective DD labels, the ratio of AA to precursor 20:3 is different by 84% while the ratio of 20:4 to the sum of all anti-inflammatory/proresolving precursors increases by 47%. High dietary 18:2n-6 drives the excess 20:4 increase for the I allele.
Why FADS genotypes are important: translation
It is widely assumed that human endogenous biosynthesis of DHA from precursors is inherently near zero. This is not correct: the factor driving very low DHA synthesis is high intakes of LA in the background diets in all studies – abnormal compared to any known human diet prior to industrialization and the rise of high LA seed oils. LA is antagonistic with the last steps of DHA biosynthesis competing for FADS2 for the last desaturation prior to DHA synthesis by either accepted pathway. Reduction of LA removes an effective block and circulating DHA rises. Key data from our RCT in Malawi [133] demonstrate that a conventional ready-to-use-therapeutic food (RUTF) high in LA causes a 25% drop in DHA over 4 weeks, while a high oleic (HO)-RUTF with balanced LA and ALA and with oleic acid replacing LA stabilizes DHA and causes a 60% increase in EPA, by reducing metabolism competition of LA with ω3.
We envision that individuals with II genotype will have a greater drop in DHA when put on diets high in LA, similar to conventional RUTF. Twentieth century commercially produced conventional seed oils, sunflower, safflower, peanut, grapeseed, cottonseed and corn, contain high levels of LA; individuals with a genetic make-up that rapidly converts precursors to products are likely to be vulnerable to AA-related ill-health when adopting a diet rich in LA which severely reduces the synthesis of anti-inflammatory ω3 HUFA. As importantly, n-6 PUFA compete with, and antagonize against, incorporation of ω3 PUFA into tissues [134], inhibiting incorporation of structural n-3 PUFA that are especially concentrated in neural tissue [135]. A re-evaluation of the traditional diet-heart hypothesis shows that with LA rich oils cholesterol decreases but the risk of death increases (-30 mg/dl cholesterol = +22% CVD death) [136]. Population differences at the 11q13 genomic locus exist and can contribute to significant genetic variability. RCT studies should take into consideration this important factor. Finally, the 21st century has seen widespread and increasing use of conventionally bred HO oilseeds with oleic acid replacing LA; the sole exception is HO canola in which oleic replaces ALA. Nearly 100% of US sunflower oil is HO, many peanuts (used in M&Ms®[137], for instance), and the major US oilseed crop, soybean, is rapidly increasing. These changes are driven mostly by the increased shelf-life and frying life of high oleic oils, though improved ω3 status through lower LA is likely an advantage.
Though ELOVL5 and ELOVL2 both elongate polyunsaturates, ELOVL2 is specific to elongation of C20 and C22[138]. The expression of the ELOVL2 gene is different between European and Chinese populations in at least one GWAS [139], and ELOVL2 polymorphisms improve the correlation with circulating fatty acid levels [119, 140]. Importantly, considerable work has implicated methylation as a silencer of ELOVL2 expression [97, 141]. Methylation is tissue specific [142] and has recently been revealed as a molecular regulator of aging in the HUFA-rich retina [143]. Methylation at the ELOVL2 locus has been proposed as a measure of again in forensics [98] and the principle appears to be similar for chimpanzees[144]. The plasma kinetics of 13C-DHA is strongly related to age[145], while EPA is only slightly related to age and AA is not [146, 147]. While these fatty acids are not immediate products of ELOVL2, it is tempting to speculate that methylation of this gene with aging plays a role. To our knowledge, no other gene in the biochemical pathway has been repeatedly related to aging.
Conclusions
Regulatory non-coding SNPs and rs66698963 Indel within FADS and ELOVL genes mediating the endogenous synthesis of HUFA are strongly associated with inter-individual differences in converting essential fatty acid precursors to physiologically active HUFA. Methylation clearly has a role in ELOVL2 function that would influence synthesis of EPA and possibly DHA.
Covid clearly is more dangerous as individuals age, and as well as those with preexisting illnesses. Although omega-3 status is higher in older Americans, rising each decade to plateau in after age 60 [148], effects on metabolism with aging may compromise HUFA metabolism during illness. The weight of evidence suggests that HUFA balance has some role to play in most illnesses and especially those that are known to be related to HUFA-derived signaling molecules such as inflammation and thrombosis. The shift of modern diets to higher n-6 over n-3 is recognized as a major risk factor for lifestyle-related diseases, such as cardiovascular disease and diabetes via inflammation and specific issues. More importantly, n-6 LA in many seed oils never present in human diets prior to industrialization overwhelm and suppress synthesis, tissue incorporation, and downstream signaling of n-3 EPA and DHA. Individuals with high-converting genotypes (e.g. II genotype = fast desaturators) are likely to show a marked increase in proinflammatory, prothrombotic ω6 AA, possibly based on other genetic factors. They are particularly at risk.
Author statement
All authors contributed equally to this work.
Acknowledgement
The authors are grateful to anonymous referee 2 for insightful suggestions.
Footnotes
★Sources of support. None.
★★✰★✰Declaration of Interest. All authors declare no conflict of interest.
References
- 1.Wehmeyer P. A Covid Drug to Help Fight the Storm. Wall Street Journal, New York. 2020 [Google Scholar]
- 2.Carsana L., Sonzogni A., Nasr A., Rossi R.S., Pellegrinelli A., Zerbi P., Rech R., Colombo R., Antinori S., Corbellino M., Galli M., Catena E., Tosoni A., Gianatti A., Nebuloni M. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosmuller H., Traxler S., Bitzer M., Haberle H., Raiser W., Nann D., Frauenfeld L., Vogelsberg A., Klingel K., Fend F. The evolution of pulmonary pathology in fatal COVID-19 disease: an autopsy study with clinical correlation. Virchows Arch. 2020 doi: 10.1007/s00428-020-02881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rapkiewicza A.V., Mai X.C., Carsons S.E., Pittalugac s., D.K.D.E. J.S.Berger, Thomas S., Adler N.M., Charytan D.M., Gasmi B., Hochman J.S., Reynolds H.R. Megakaryocytes and platelet-fibrin thrombi characterize multi-organ thrombosis at autopsy in COVID-19: a case series. E Clin Med. 2020 doi: 10.1016/j.eclinm.2020.100434. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu M., Gu C., Wu J., Zhu Y. Amino acids 1 to 422 of the spike protein of SARS associated coronavirus are required for induction of cyclooxygenase-2. Virus Genes. 2006;33:309–317. doi: 10.1007/s11262-005-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raaben M., Einerhand A.W., Taminiau L.J., van Houdt M., Bouma J., Raatgeep R.H., Buller H.A., de Haan C.A., Rossen J.W. Cyclooxygenase activity is important for efficient replication of mouse hepatitis virus at an early stage of infection. Virol J. 2007;4:55. doi: 10.1186/1743-422X-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbar U., Yang M., Kurian D., Mohan C. Omega-3 Fatty Acids in Rheumatic Diseases: a Critical Review. J Clin Rheumatol. 2017;23:330–339. doi: 10.1097/RHU.0000000000000563. [DOI] [PubMed] [Google Scholar]
- 8.Vadell A.K.E., Barebring L., Hulander E., Gjertsson I., Lindqvist H.M., Winkvist A. Anti-inflammatory Diet In Rheumatoid Arthritis (ADIRA)-a randomized, controlled crossover trial indicating effects on disease activity. Am J Clin Nutr. 2020;111:1203–1213. doi: 10.1093/ajcn/nqaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bing R.J., Lomnicka M. Why do cyclo-oxygenase-2 inhibitors cause cardiovascular events? J Am Coll Cardiol. 2002;39:521–522. doi: 10.1016/s0735-1097(01)01749-1. [DOI] [PubMed] [Google Scholar]
- 10.Crescente M., Armstrong P.C., Kirkby N.S., Edin M.L., Chan M.V., Lih F.B., Jiao J., Maffucci T., Allan H.E., Mein C.A., Gaston-Massuet C., Cottrell G.S., Mitchell J.A., Zeldin D.C., Herschman H.R., Warner T.D. Profiling the eicosanoid networks that underlie the anti- and pro-thrombotic effects of aspirin. FASEB J. 2020 doi: 10.1096/fj.202000312R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kujovich J.L. Factor V Leiden thrombophilia. Genet Med. 2011;13:1–16. doi: 10.1097/GIM.0b013e3181faa0f2. [DOI] [PubMed] [Google Scholar]
- 12.Federici E.H., Al-Mondhiry H. High risk of thrombosis recurrence in patients with homozygous and compound heterozygous factor V R506Q (Factor V Leiden) and prothrombin G20210A. Thromb Res. 2019;182:75–78. doi: 10.1016/j.thromres.2019.07.030. [DOI] [PubMed] [Google Scholar]
- 13.Elmas E., Suvajac N., Jilma B., Weiler H., Borggrefe M., Dempfle C.E. Factor V Leiden mutation enhances fibrin formation and dissolution in vivo in a human endotoxemia model. Blood. 2010;116:801–805. doi: 10.1182/blood-2009-03-213215. [DOI] [PubMed] [Google Scholar]
- 14.Fitzpatrick F.A., Soberman R. Regulated formation of eicosanoids. J Clin Invest. 2001;107:1347–1351. doi: 10.1172/JCI13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fantone J.C., Marasco W.A., Elgas L.J., Ward P.A. Anti-inflammatory effects of prostaglandin E1: in vivo modulation of the formyl peptide chemotactic receptor on the rat neutrophil. J Immunol. 1983;130:1495–1497. [PubMed] [Google Scholar]
- 16.Kirtland S.J. Prostaglandin E1: a review. Prostaglandins Leukot Essent Fatty Acids. 1988;32:165–174. doi: 10.1016/0952-3278(88)90168-8. [DOI] [PubMed] [Google Scholar]
- 17.Jain A., Iqbal O.A. StatPearls; 2020. Alprostadil. Treasure Island (FL) [PubMed] [Google Scholar]
- 18.Johnson G.H., Fritsche K. Effect of dietary linoleic acid on markers of inflammation in healthy persons: a systematic review of randomized controlled trials. J Acad Nutr Diet. 2012;112 doi: 10.1016/j.jand.2012.03.029. 1029-1041, 1041 e1021-1015. [DOI] [PubMed] [Google Scholar]
- 19.Maki K.C., Eren F., Cassens M.E., Dicklin M.R., Davidson M.H. omega-6 Polyunsaturated Fatty Acids and Cardiometabolic Health: current Evidence, Controversies, and Research Gaps. Adv Nutr. 2018;9:688–700. doi: 10.1093/advances/nmy038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatt D.L., Steg P.G., Miller M., Brinton E.A., Jacobson T.A., Ketchum S.B., Doyle R.T., Jr., Juliano R.A., Jiao L., Granowitz C., Tardif J.C., Ballantyne C.M., Investigators R.-.I. Cardiovascular Risk Reduction with Icosapent Ethyl for Hypertriglyceridemia. N Engl J Med. 2019;380:11–22. doi: 10.1056/NEJMoa1812792. [DOI] [PubMed] [Google Scholar]
- 21.Brenna J.T., Salem N., Jr., Sinclair A.J., Cunnane S.C. A. International Society for the Study of Fatty, I. Lipids, alpha-Linolenic acid supplementation and conversion to n-3 long-chain polyunsaturated fatty acids in humans. Prostaglandins Leukot Essent Fatty Acids. 2009;80:85–91. doi: 10.1016/j.plefa.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Francois C.A., Connor S.L., Bolewicz L.C., Connor W.E. Supplementing lactating women with flaxseed oil does not increase docosahexaenoic acid in their milk. Am J Clin Nutr. 2003;77:226–233. doi: 10.1093/ajcn/77.1.226. [DOI] [PubMed] [Google Scholar]
- 23.Liu G., Gibson R.A., Callahan D., Guo X.F., Li D., Sinclair A.J. Pure omega 3 polyunsaturated fatty acids (EPA, DPA or DHA) are associated with increased plasma levels of 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (CMPF) in a short-term study in women. Food Funct. 2020;11:2058–2066. doi: 10.1039/c9fo02440a. [DOI] [PubMed] [Google Scholar]
- 24.Kaur G., Begg D.P., Barr D., Garg M., Cameron-Smith D., Sinclair A.J. Short-term docosapentaenoic acid (22:5 n-3) supplementation increases tissue docosapentaenoic acid, DHA and EPA concentrations in rats. Br J Nutr. 2010;103:32–37. doi: 10.1017/S0007114509991334. [DOI] [PubMed] [Google Scholar]
- 25.Drouin G., Catheline D., Guillocheau E., Gueret P., Baudry C., Le Ruyet P., Rioux V., Legrand P. Comparative effects of dietary n-3 docosapentaenoic acid (DPA), DHA and EPA on plasma lipid parameters, oxidative status and fatty acid tissue composition. J Nutr Biochem. 2019;63:186–196. doi: 10.1016/j.jnutbio.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 26.Drouin G., Rioux V., Legrand P. The n-3 docosapentaenoic acid (DPA): a new player in the n-3 long chain polyunsaturated fatty acid family. Biochimie. 2019;159:36–48. doi: 10.1016/j.biochi.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Tu W.C., Muhlhausler B.S., Yelland L.N., Gibson R.A. Correlations between blood and tissue omega-3 LCPUFA status following dietary ALA intervention in rats. Prostaglandins Leukot Essent Fatty Acids. 2013;88:53–60. doi: 10.1016/j.plefa.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Mohrhauer H., Holman R.T. Effect of Linolenic Acid Upon the Metabolism of Linoleic Acid. J Nutr. 1963;81:67–74. doi: 10.1093/jn/81.1.67. [DOI] [PubMed] [Google Scholar]
- 29.Gibson R.A., Neumann M.A., Lien E.L., Boyd K.A., Tu W.C. Docosahexaenoic acid synthesis from alpha-linolenic acid is inhibited by diets high in polyunsaturated fatty acids. Prostaglandins Leukot Essent Fatty Acids. 2013;88:139–146. doi: 10.1016/j.plefa.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Cunnane S.C. The Canadian Society for Nutritional Sciences 1995 Young Scientist Award Lecture. Recent studies on the synthesis, beta-oxidation, and deficiency of linoleate and alpha-linolenate: are essential fatty acids more aptly named indispensable or conditionally dispensable fatty acids? Can J Physiol Pharmacol. 1996;74:629–639. [PubMed] [Google Scholar]
- 31.Cunnane S.C., Guesnet P. Linoleic acid recommendations–A house of cards. Prostaglandins Leukot Essent Fatty Acids. 2011;85:399–402. doi: 10.1016/j.plefa.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Brenna J.T. Long-chain polyunsaturated fatty acids and the preterm infant: a case study in developmentally sensitive nutrient needs in the United States. Am J Clin Nutr. 2016;103:606S–615S. doi: 10.3945/ajcn.114.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlson S.J., O'Loughlin A.A., Anez-Bustillos L., Baker M.A., Andrews N.A., Gunner G., Dao D.T., Pan A., Nandivada P., Chang M., Cowan E., Mitchell P.D., Gura K.M., Fagiolini M., Puder M. A Diet With Docosahexaenoic and Arachidonic Acids as the Sole Source of Polyunsaturated Fatty Acids Is Sufficient to Support Visual, Cognitive, Motor, and Social Development in Mice. Front Neurosci. 2019;13:72. doi: 10.3389/fnins.2019.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nehra D., Le H.D., Fallon E.M., Carlson S.J., Woods D., White Y.A., Pan A.H., Guo L., Rodig S.J., Tilly J.L., Rueda B.R., Puder M. Prolonging the female reproductive lifespan and improving egg quality with dietary omega-3 fatty acids. Aging Cell. 2012;11:1046–1054. doi: 10.1111/acel.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karsai G., Lone M., Kutalik Z., Brenna J.T., Li H., Pan D., von Eckardstein A., Hornemann T. FADS3 is a Delta14Z sphingoid base desaturase that contributes to gender differences in the human plasma sphingolipidome. J Biol Chem. 2020;295:1889–1897. doi: 10.1074/jbc.AC119.011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J.Y., Qin X., Liang A., Kim E., Lawrence P., Park W.J., Kothapalli K.S.D., Brenna J.T. Fads3 modulates docosahexaenoic acid in liver and brain. Prostaglandins Leukot Essent Fatty Acids. 2017;123:25–32. doi: 10.1016/j.plefa.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marquardt A., Stohr H., White K., Weber B.H. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 38.Cho H.P., Nakamura M.T., Clarke S.D. Cloning, expression, and nutritional regulation of the mammalian Delta-6 desaturase. J Biol Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 39.Cho H.P., Nakamura M., Clarke S.D. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J Biol Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- 40.Park W.J., Kothapalli K.S., Reardon H.T., Kim L.Y., Brenna J.T. Novel fatty acid desaturase 3 (FADS3) transcripts generated by alternative splicing. Gene. 2009;446:28–34. doi: 10.1016/j.gene.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park W.J., Reardon H.T., Tyburczy C., Kothapalli K.S., Brenna J.T. Alternative splicing generates a novel FADS2 alternative transcript in baboons. Mol Biol Rep. 2010;37:2403–2406. doi: 10.1007/s11033-009-9750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park W.J., Kothapalli K.S., Reardon H.T., Lawrence P., Qian S.B., Brenna J.T. A novel FADS1 isoform potentiates FADS2-mediated production of eicosanoid precursor fatty acids. J Lipid Res. 2012;53:1502–1512. doi: 10.1194/jlr.M025312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brenna J.T., Kothapalli K.S., Park W.J. Alternative transcripts of fatty acid desaturase (FADS) genes. Prostaglandins Leukot Essent Fatty Acids. 2010;82:281–285. doi: 10.1016/j.plefa.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J.Y., Kothapalli K.S., Brenna J.T. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr Opin Clin Nutr Metab Care. 2016;19:103–110. doi: 10.1097/MCO.0000000000000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I., Doerks T., Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I., Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hashimoto K., Yoshizawa A.C., Okuda S., Kuma K., Goto S., Kanehisa M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J Lipid Res. 2008;49:183–191. doi: 10.1194/jlr.M700377-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Shanklin J., Guy J.E., Mishra G., Lindqvist Y. Desaturases: emerging models for understanding functional diversification of diiron-containing enzymes. J Biol Chem. 2009;284:18559–18563. doi: 10.1074/jbc.R900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z., Park H.G., Wang D.H., Kitano R., Kothapalli K.S.D., Brenna J.T. Fatty acid desaturase 2 (FADS2) but not FADS1 desaturates branched chain and odd chain saturated fatty acids. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865 doi: 10.1016/j.bbalip.2019.158572. [DOI] [PubMed] [Google Scholar]
- 50.Park W.J., Kothapalli K.S., Lawrence P., Tyburczy C., Brenna J.T. An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J Lipid Res. 2009;50:1195–1202. doi: 10.1194/jlr.M800630-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park H.G., Park W.J., Kothapalli K.S., Brenna J.T. The fatty acid desaturase 2 (FADS2) gene product catalyzes Delta4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells. FASEB J. 2015;29:3911–3919. doi: 10.1096/fj.15-271783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park H.G., Engel M.G., Vogt-Lowell K., Lawrence P., Kothapalli K.S., Brenna J.T. The role of fatty acid desaturase (FADS) genes in oleic acid metabolism: FADS1 Delta7 desaturates 11-20:1 to 7,11-20:2. Prostaglandins Leukot Essent Fatty Acids. 2018;128:21–25. doi: 10.1016/j.plefa.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park W.J., Kothapalli K.S., Lawrence P., Brenna J.T. FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS ONE. 2011;6:e28186. doi: 10.1371/journal.pone.0028186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guillou H., Zadravec D., Martin P.G., Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Koreth J., Bakkenist C.J., McGee J.O. Chromosomes, 11Q and cancer: a review. J Pathol. 1999;187:28–38. doi: 10.1002/(SICI)1096-9896(199901)187:1<28::AID-PATH166>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 56.Schroder A.R., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 57.Huang X., Gollin S.M., Raja S., Godfrey T.E. High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells. Proc Natl Acad Sci U S A. 2002;99:11369–11374. doi: 10.1073/pnas.172285799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Debacker K., Winnepenninckx B., Longman C., Colgan J., Tolmie J., Murray R., van Luijk R., Scheers S., Fitzpatrick D., Kooy F. The molecular basis of the folate-sensitive fragile site FRA11A at 11q13. Cytogenet Genome Res. 2007;119:9–14. doi: 10.1159/000109612. [DOI] [PubMed] [Google Scholar]
- 59.Zainabadi K., Jain A.V., Donovan F.X., Elashoff D., Rao N.P., Murty V.V., Chandrasekharappa S.C., Srivatsan E.S. One in four individuals of African-American ancestry harbors a 5.5kb deletion at chromosome 11q13.1. Genomics. 2014;103:276–287. doi: 10.1016/j.ygeno.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puspurs A.H., Baker E., Callen D.F., Fratini A., Sutherland G.R. Translocation breakpoint in t(11;14) in B-cell leukemia is not at the rare fragile site at 11q13.3. Cancer Genet. Cytogenet. 1988;31:25–30. doi: 10.1016/0165-4608(88)90006-4. [DOI] [PubMed] [Google Scholar]
- 61.Zainabadi K., Benyamini P., Chakrabarti R., Veena M.S., Chandrasekharappa S.C., Gatti R.A., Srivatsan E.S. A 700-kb physical and transcription map of the cervical cancer tumor suppressor gene locus on chromosome 11q13. Genomics. 2005;85:704–714. doi: 10.1016/j.ygeno.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 62.Hurles M.E., Dermitzakis E.T., Tyler-Smith C. The functional impact of structural variation in humans. Trends Genet. 2008;24:238–245. doi: 10.1016/j.tig.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerami P., Jewell S.S., Pouryazdanparast P., Wayne J.D., Haghighat Z., Busam K.J., Rademaker A., Morrison L. Copy number gains in 11q13 and 8q24 [corrected] are highly linked to prognosis in cutaneous malignant melanoma. The Journal of molecular diagnostics: JMD. 2011;13:352–358. doi: 10.1016/j.jmoldx.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karlsson E., Waltersson M.A., Bostner J., Perez-Tenorio G., Olsson B., Hallbeck A.L., Stal O. High-resolution genomic analysis of the 11q13 amplicon in breast cancers identifies synergy with 8p12 amplification, involving the mTOR targets S6K2 and 4EBP1. Genes Chromosomes Cancer. 2011;50:775–787. doi: 10.1002/gcc.20900. [DOI] [PubMed] [Google Scholar]
- 65.Lambrechts D., Truong T., Justenhoven C., Humphreys M.K., Wang J., Hopper J.L., Dite G.S., Apicella C., Southey M.C., Schmidt M.K., Broeks A., Cornelissen S., van Hien R., Sawyer E., Tomlinson I., Kerin M., Miller N., Milne R.L., Zamora M.P., Perez J.I., Benitez J., Hamann U., Ko Y.D., Bruning T., Network G., Chang-Claude J., Eilber U., Hein R., Nickels S., Flesch-Janys D., Wang-Gohrke S., John E.M., Miron A., Winqvist R., Pylkas K., Jukkola-Vuorinen A., Grip M., Chenevix-Trench G., Beesley J., Chen X., k. Investigators, Australian Ovarian Cancer Study G., Menegaux F., Cordina-Duverger E., Shen C.Y., Yu J.C., Wu P.E., Hou M.F., Andrulis I.L., Selander T., Glendon G., Mulligan A.M., Anton-Culver H., Ziogas A., Muir K.R., Lophatananon A., Rattanamongkongul S., Puttawibul P., Jones M., Orr N., Ashworth A., Swerdlow A., Severi G., Baglietto L., Giles G., Southey M., Marme F., Schneeweiss A., Sohn C., Burwinkel B., Yesilyurt B.T., Neven P., Paridaens R., Wildiers H., Brenner H., Muller H., Arndt V., Stegmaier C., Meindl A., Schott S., Bartram C.R., Schmutzler R.K., Cox A., Brock I.W., Elliott G., Cross S.S., Fasching P.A., Schulz-Wendtland R., Ekici A.B., Beckmann M.W., Fletcher O., Johnson N., Silva Idos S., Peto J., Nevanlinna H., Muranen T.A., Aittomaki K., Blomqvist C., Dork T., Schurmann P., Bremer M., Hillemanns P., Bogdanova N.V., Antonenkova N.N., Rogov Y.I., Karstens J.H., Khusnutdinova E., Bermisheva M., Prokofieva D., Gancev S., Jakubowska A., Lubinski J., Jaworska K., Durda K., Nordestgaard B.G., Bojesen S.E., Lanng C., Mannermaa A., Kataja V., Kosma V.M., Hartikainen J.M., Radice P., Peterlongo P., Manoukian S., Bernard L., Couch F.J., Olson J.E., Wang X., Fredericksen Z., Alnaes G.G., Kristensen V., Borresen-Dale A.L., Devilee P., Tollenaar R.A., Seynaeve C.M., Hooning M.J., Garcia-Closas M., Chanock S.J., Lissowska J., Sherman M.E., Hall P., Liu J., Czene K., Kang D., Yoo K.Y., Noh D.Y., Lindblom A., Margolin S., Dunning A.M., Pharoah P.D., Easton D.F., Guenel P., Brauch H. 11q13 is a susceptibility locus for hormone receptor positive breast cancer. Hum Mutat. 2012;33:1123–1132. doi: 10.1002/humu.22089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamdi Y., Ben Rekaya M., Jingxuan S., Nagara M., Messaoud O., Benammar Elgaaied A., Mrad R., Chouchane L., Boubaker M.S., Abdelhak S., Boussen H., Romdhane L. A genome wide SNP genotyping study in the Tunisian population: specific reporting on a subset of common breast cancer risk loci. BMC Cancer. 2018;18:1295. doi: 10.1186/s12885-018-5133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luen S.J., Asher R., Lee C.K., Savas P., Kammler R., Dell’Orto P., Biasi O.M., Demanse D., JeBailey L., Dolan S., Hackl W., Thuerlimann B., Viale G., Colleoni M., Regan M.M., Loi S. Association of Somatic Driver Alterations With Prognosis in Postmenopausal, Hormone Receptor-Positive, HER2-Negative Early Breast Cancer: a Secondary Analysis of the BIG 1-98 Randomized Clinical Trial. JAMA Oncol. 2018;4:1335–1343. doi: 10.1001/jamaoncol.2018.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ooi A., Inokuchi M., Horike S.I., Kawashima H., Ishikawa S., Ikeda H., Nakamura R., Oyama T., Dobash Y. Amplicons in breast cancers analyzed by multiplex ligation-dependent probe amplification and fluorescence In Situ hybridization. Hum Pathol. 2018 doi: 10.1016/j.humpath.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 69.Kutasovic J.R., McCart Reed A.E., Males R., Sim S., Saunus J.M., Dalley A., McEvoy C.R., Dedina L., Miller G., Peyton S., Reid L., Lal S., Niland C., Ferguson K., Fellowes A.P., Al-Ejeh F., Lakhani S.R., Cummings M.C., Simpson P.T. Breast cancer metastasis to gynaecological organs: a clinico-pathological and molecular profiling study. J Pathol Clin Res. 2019;5:25–39. doi: 10.1002/cjp2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park H.G., Zhang J.Y., Foster C., Sudilovsky D., Schwed D.A., Mecenas J., Devapatla S., Lawrence P., Kothapalli K.S.D., Brenna J.T. A Rare Eicosanoid Precursor Analogue, Sciadonic Acid (5Z,11Z,14Z-20:3), Detected In Vivo in Hormone Positive Breast Cancer Tissue. PLEFA. 2018 doi: 10.1016/j.plefa.2018.1005.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muller D., Millon R., Velten M., Bronner G., Jung G., Engelmann A., Flesch H., Eber M., Methlin G., Abecassis J. Amplification of 11q13 DNA markers in head and neck squamous cell carcinomas: correlation with clinical outcome. Eur J Cancer. 1997;33:2203–2210. doi: 10.1016/s0959-8049(97)00198-6. [DOI] [PubMed] [Google Scholar]
- 72.Williams M.E., Gaffey M.J., Weiss L.M., Wilczynski S.P., Schuuring E., Levine P.A. Chromosome 11Q13 amplification in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 1993;119:1238–1243. doi: 10.1001/archotol.1993.01880230084013. [DOI] [PubMed] [Google Scholar]
- 73.Jin C., Jin Y., Gisselsson D., Wennerberg J., Wah T.S., Stromback B., Kwong Y.L., Mertens F. Molecular cytogenetic characterization of the 11q13 amplicon in head and neck squamous cell carcinoma. Cytogenet Genome Res. 2006;115:99–106. doi: 10.1159/000095228. [DOI] [PubMed] [Google Scholar]
- 74.Brown L.A., Kalloger S.E., Miller M.A., Shih Ie M., McKinney S.E., Santos J.L., Swenerton K., Spellman P.T., Gray J., Gilks C.B., Huntsman D.G. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47:481–489. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 75.Debelenko L.V., Zhuang Z., Emmert-Buck M.R., Chandrasekharappa S.C., Manickam P., Guru S.C., Marx S.J., Skarulis M.C., Spiegel A.M., Collins F.S., Jensen R.T., Liotta L.A., Lubensky I.A. Allelic deletions on chromosome 11q13 in multiple endocrine neoplasia type 1-associated and sporadic gastrinomas and pancreatic endocrine tumors. Cancer Res. 1997;57:2238–2243. [PubMed] [Google Scholar]
- 76.Zheng S.L., Stevens V.L., Wiklund F., Isaacs S.D., Sun J., Smith S., Pruett K., Wiley K.E., Kim S.T., Zhu Y., Zhang Z., Hsu F.C., Turner A.R., Johansson J.E., Liu W., Kim J.W., Chang B.L., Duggan D., Carpten J., Rodriguez C., Isaacs W., Gronberg H., Xu J. Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol Biomarkers Prev. 2009;18:1815–1820. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodriguez C., Hughes-Davies L., Valles H., Orsetti B., Cuny M., Ursule L., Kouzarides T., Theillet C. Amplification of the BRCA2 pathway gene EMSY in sporadic breast cancer is related to negative outcome. Clin Cancer Res. 2004;10:5785–5791. doi: 10.1158/1078-0432.CCR-03-0410. [DOI] [PubMed] [Google Scholar]
- 78.Karlseder J., Zeillinger R., Schneeberger C., Czerwenka K., Speiser P., Kubista E., Birnbaum D., Gaudray P., Theillet C. Patterns of DNA amplification at band q13 of chromosome 11 in human breast cancer. Genes Chromosomes Cancer. 1994;9:42–48. doi: 10.1002/gcc.2870090108. [DOI] [PubMed] [Google Scholar]
- 79.Vriens K., Christen S., Parik S., Broekaert D., Yoshinaga K., Talebi A., Dehairs J., Escalona-Noguero C., Schmieder R., Cornfield T., Charlton C., Romero-Perez L., Rossi M., Rinaldi G., Orth M.F., Boon R., Kerstens A., Kwan S.Y., Faubert B., Mendez-Lucas A., Kopitz C.C., Chen T., Fernandez-Garcia J., Duarte J.A.G., Schmitz A.A., Steigemann P., Najimi M., Hagebarth A., Van Ginderachter J.A., Sokal E., Gotoh N., Wong K.K., Verfaillie C., Derua R., Munck S., Yuneva M., Beretta L., DeBerardinis R.J., Swinnen J.V., Hodson L., Cassiman D., Verslype C., Christian S., Grunewald S., Grunewald T.G.P., Fendt S.M. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566:403–406. doi: 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Snaebjornsson M.T., Schulze A. Tumours use a metabolic twist to make lipids. Nature. 2019;566:333–334. doi: 10.1038/d41586-019-00352-1. [DOI] [PubMed] [Google Scholar]
- 81.Jaudszus A., Degen C., Barth S.W., Klempt M., Schlormann W., Roth A., Rohrer C., Sauerwein H., Sachse K., Jahreis G. Loss of FADS2 function severely impairs the use of HeLa cells as an in vitro model for host response studies involving fatty acid effects. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0115610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grammatikos S.I., Subbaiah P.V., Victor T.A., Miller W.M. Diversity in the ability of cultured cells to elongate and desaturate essential (n-6 and n-3) fatty acids. Ann N Y Acad Sci. 1994;745:92–105. doi: 10.1111/j.1749-6632.1994.tb44366.x. [DOI] [PubMed] [Google Scholar]
- 83.Schaeffer L., Gohlke H., Muller M., Heid I.M., Palmer L.J., Kompauer I., Demmelmair H., Illig T., Koletzko B., Heinrich J. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 84.Lattka E., Illig T., Koletzko B., Heinrich J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr. Opin. Lipidol. 2010;21:64–69. doi: 10.1097/MOL.0b013e3283327ca8. [DOI] [PubMed] [Google Scholar]
- 85.Koletzko B., Reischl E., Tanjung C., Gonzalez-Casanova I., Ramakrishnan U., Meldrum S., Simmer K., Heinrich J., Demmelmair H. FADS1 and FADS2 Polymorphisms Modulate Fatty Acid Metabolism and Dietary Impact on Health. Annu Rev Nutr. 2019;39:21–44. doi: 10.1146/annurev-nutr-082018-124250. [DOI] [PubMed] [Google Scholar]
- 86.Morales E., Bustamante M., Gonzalez J.R., Guxens M., Torrent M., Mendez M., Garcia-Esteban R., Julvez J., Forns J., Vrijheid M., Molto-Puigmarti C., Lopez-Sabater C., Estivill X., Sunyer J. Genetic variants of the FADS gene cluster and ELOVL gene family, colostrums LC-PUFA levels, breastfeeding, and child cognition. PLoS ONE. 2011;6:e17181. doi: 10.1371/journal.pone.0017181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fujii T.M.M., Norde M.M., Fisberg R.M., Marchioni D.M.L., Ordovas J.M., Rogero M.M. FADS1 and ELOVL2 polymorphisms reveal associations for differences in lipid metabolism in a cross-sectional population-based survey of Brazilian men and women. Nutr Res. 2020;78:42–49. doi: 10.1016/j.nutres.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 88.Coltell O., Sorli J.V., Asensio E.M., Barragan R., Gonzalez J.I., Gimenez-Alba I.M., Zanon-Moreno V., Estruch R., Ramirez-Sabio J.B., Pascual E.C., Ortega-Azorin C., Ordovas J.M., Corella D. Genome-Wide Association Study for Serum Omega-3 and Omega-6 Polyunsaturated Fatty Acids: exploratory Analysis of the Sex-Specific Effects and Dietary Modulation in Mediterranean Subjects with Metabolic Syndrome. Nutrients. 2020;12 doi: 10.3390/nu12020310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu Y., Wang Y., Tian H., Lu T., Yu M., Xu W., Liu G., Xie L. DHA intake interacts with ELOVL2 and ELOVL5 genetic variants to influence polyunsaturated fatty acids in human milk. J Lipid Res. 2019;60:1043–1049. doi: 10.1194/jlr.M090951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alsaleh A., Maniou Z., Lewis F.J., Hall W.L., Sanders T.A., O'Dell S.D. ELOVL2 gene polymorphisms are associated with increases in plasma eicosapentaenoic and docosahexaenoic acid proportions after fish oil supplement. Genes Nutr. 2014;9:362. doi: 10.1007/s12263-013-0362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Malerba G., Schaeffer L., Xumerle L., Klopp N., Trabetti E., Biscuola M., Cavallari U., Galavotti R., Martinelli N., Guarini P., Girelli D., Olivieri O., Corrocher R., Heinrich J., Pignatti P.F., Illig T. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43:289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 92.Martinelli N., Girelli D., Malerba G., Guarini P., Illig T., Trabetti E., Sandri M., Friso S., Pizzolo F., Schaeffer L., Heinrich J., Pignatti P.F., Corrocher R., Olivieri O. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am J Clin Nutr. 2008;88:941–949. doi: 10.1093/ajcn/88.4.941. [DOI] [PubMed] [Google Scholar]
- 93.Cribb L., Murphy J., Froud A., Oliver G., Bousman C.A., Ng C.H., Sarris J. Erythrocyte polyunsaturated fatty acid composition is associated with depression and FADS genotype in Caucasians. Nutr Neurosci. 2018;21:589–601. doi: 10.1080/1028415X.2017.1327685. [DOI] [PubMed] [Google Scholar]
- 94.Haghighi F., Galfalvy H., Chen S., Huang Y.Y., Cooper T.B., Burke A.K., Oquendo M.A., Mann J.J., Sublette M.E. DNA methylation perturbations in genes involved in polyunsaturated Fatty Acid biosynthesis associated with depression and suicide risk. Front Neurol. 2015;6:92. doi: 10.3389/fneur.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sergeant S., Hugenschmidt C.E., Rudock M.E., Ziegler J.T., Ivester P., Ainsworth H.C., Vaidya D., Case L.D., Langefeld C.D., Freedman B.I., Bowden D.W., Mathias R.A., Chilton F.H. Differences in arachidonic acid levels and fatty acid desaturase (FADS) gene variants in African Americans and European Americans with diabetes or the metabolic syndrome. Br J Nutr. 2012;107:547–555. doi: 10.1017/S0007114511003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khamlaoui W., Mehri S., Hammami S., Hammouda S., Chraeif I., Elosua R., Hammami M. Association Between Genetic Variants in FADS1-FADS2 and ELOVL2 and Obesity, Lipid Traits, and Fatty Acids in Tunisian Population. Clin Appl Thromb Hemost. 2020;26 doi: 10.1177/1076029620915286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Garagnani P., Bacalini M.G., Pirazzini C., Gori D., Giuliani C., Mari D., Di Blasio A.M., Gentilini D., Vitale G., Collino S., Rezzi S., Castellani G., Capri M., Salvioli S., Franceschi C. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11:1132–1134. doi: 10.1111/acel.12005. [DOI] [PubMed] [Google Scholar]
- 98.Spolnicka M., Pospiech E., Peplonska B., Zbiec-Piekarska R., Makowska Z., Pieta A., Karlowska-Pik J., Ziemkiewicz B., Wezyk M., Gasperowicz P., Bednarczuk T., Barcikowska M., Zekanowski C., Ploski R., Branicki W. DNA methylation in ELOVL2 and C1orf132 correctly predicted chronological age of individuals from three disease groups. Int J Legal Med. 2018;132:1–11. doi: 10.1007/s00414-017-1636-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Di Gregorio E., Borroni B., Giorgio E., Lacerenza D., Ferrero M., Lo Buono N., Ragusa N., Mancini C., Gaussen M., Calcia A., Mitro N., Hoxha E., Mura I., Coviello D.A., Moon Y.A., Tesson C., Vaula G., Couarch P., Orsi L., Duregon E., Papotti M.G., Deleuze J.F., Imbert J., Costanzi C., Padovani A., Giunti P., Maillet-Vioud M., Durr A., Brice A., Tempia F., Funaro A., Boccone L., Caruso D., Stevanin G., Brusco A. ELOVL5 Mutations Cause Spinocerebellar Ataxia 38. Am J Hum Genet. 2014;95:209–217. doi: 10.1016/j.ajhg.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kothapalli K.S., Ye K., Gadgil M.S., Carlson S.E., O'Brien K.O., Zhang J.Y., Park H.G., Ojukwu K., Zou J., Hyon S.S., Joshi K.S., Gu Z., Keinan A., Brenna J.T. Positive Selection on a Regulatory Insertion-Deletion Polymorphism in FADS2 Influences Apparent Endogenous Synthesis of Arachidonic Acid. Mol Biol Evol. 2016;33:1726–1739. doi: 10.1093/molbev/msw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gibson R.A., Sinclair A.J. Are Eskimos obligate carnivores? Lancet. 1981;1:1100. doi: 10.1016/s0140-6736(81)92263-7. [DOI] [PubMed] [Google Scholar]
- 102.Bokor S., Dumont J., Spinneker A., Gonzalez-Gross M., Nova E., Widhalm K., Moschonis G., Stehle P., Amouyel P., De Henauw S., Molnar D., Moreno L.A., Meirhaeghe A., Dallongeville J., Group H.S. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51:2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xie L., Innis S.M. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J Nutr. 2008;138:2222–2228. doi: 10.3945/jn.108.096156. [DOI] [PubMed] [Google Scholar]
- 104.Zietemann V., Kroger J., Enzenbach C., Jansen E., Fritsche A., Weikert C., Boeing H., Schulze M.B. Genetic variation of the FADS1 FADS2 gene cluster and n-6 PUFA composition in erythrocyte membranes in the European Prospective Investigation into Cancer and Nutrition-Potsdam study. Br J Nutr. 2010;104:1748–1759. doi: 10.1017/S0007114510002916. [DOI] [PubMed] [Google Scholar]
- 105.Rzehak P., Thijs C., Standl M., Mommers M., Glaser C., Jansen E., Klopp N., Koppelman G.H., Singmann P., Postma D.S., Sausenthaler S., Dagnelie P.C., van den Brandt P.A., Koletzko B., Heinrich J., K.s. group, L.s. group Variants of the FADS1 FADS2 gene cluster, blood levels of polyunsaturated fatty acids and eczema in children within the first 2 years of life. PLoS ONE. 2010;5:e13261. doi: 10.1371/journal.pone.0013261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mathias R.A., Vergara C., Gao L., Rafaels N., Hand T., Campbell M., Bickel C., Ivester P., Sergeant S., Barnes K.C., Chilton F.H. FADS genetic variants and omega-6 polyunsaturated fatty acid metabolism in a homogeneous island population. J Lipid Res. 2010;51:2766–2774. doi: 10.1194/jlr.M008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ameur A., Enroth S., Johansson A., Zaboli G., Igl W., Johansson A.C., Rivas M.A., Daly M.J., Schmitz G., Hicks A.A., Meitinger T., Feuk L., van Duijn C., Oostra B., Pramstaller P.P., Rudan I., Wright A.F., Wilson J.F., Campbell H., Gyllensten U. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet. 2012;90:809–820. doi: 10.1016/j.ajhg.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lauritzen L., Amundsen I.D., Damsgaard C.T., Lind M.V., Schnurr T.M., Hansen T., Michaelsen K.F., Vogel U. FADS and PPARG2 Single Nucleotide Polymorphisms are Associated with Plasma Lipids in 9-Mo-Old Infants. J Nutr. 2019;149:708–715. doi: 10.1093/jn/nxy323. [DOI] [PubMed] [Google Scholar]
- 109.Bernard J.Y., Pan H., Aris I.M., Moreno-Betancur M., Soh S.E., Yap F., Tan K.H., Shek L.P., Chong Y.S., Gluckman P.D., Calder P.C., Godfrey K.M., Chong M.F., Kramer M.S., Karnani N., Lee Y.S. Long-chain polyunsaturated fatty acids, gestation duration, and birth size: a Mendelian randomization study using fatty acid desaturase variants. Am J Clin Nutr. 2018;108:92–100. doi: 10.1093/ajcn/nqy079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xie L., Innis S.M. Association of fatty acid desaturase gene polymorphisms with blood lipid essential fatty acids and perinatal depression among Canadian women: a pilot study. J Nutrigenet Nutrigenomics. 2009;2:243–250. doi: 10.1159/000255636. [DOI] [PubMed] [Google Scholar]
- 111.Brookes K.J., Chen W., Xu X., Taylor E., Asherson P. Association of fatty acid desaturase genes with attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2006;60:1053–1061. doi: 10.1016/j.biopsych.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 112.Caspi A., Williams B., Kim-Cohen J., Craig I.W., Milne B.J., Poulton R., Schalkwyk L.C., Taylor A., Werts H., Moffitt T.E. Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proc Natl Acad Sci U S A. 2007;104:18860–18865. doi: 10.1073/pnas.0704292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Steer C.D., Lattka E., Koletzko B., Golding J., Hibbeln J.R. Maternal fatty acids in pregnancy, FADS polymorphisms, and child intelligence quotient at 8 y of age. Am J Clin Nutr. 2013;98:1575–1582. doi: 10.3945/ajcn.112.051524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lauritzen L., Sorensen L.B., Harslof L.B., Ritz C., Stark K.D., Astrup A., Dyssegaard C.B., Egelund N., Michaelsen K.F., Damsgaard C.T. Mendelian randomization shows sex-specific associations between long-chain PUFA-related genotypes and cognitive performance in Danish schoolchildren. Am J Clin Nutr. 2017;106:88–95. doi: 10.3945/ajcn.117.152595. [DOI] [PubMed] [Google Scholar]
- 115.Wolters M., Dering C., Siani A., Russo P., Kaprio J., Rise P., Moreno L.A., De Henauw S., Mehlig K., Veidebaum T., Molnar D., Tornaritis M., Iacoviello L., Pitsiladis Y., Galli C., Foraita R., Bornhorst C., IDEFICS and I. Family consortia The role of a FADS1 polymorphism in the association of fatty acid blood levels, BMI and blood pressure in young children-Analyses based on path models. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0181485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yeates A.J., Zavez A., Thurston S.W., McSorley E.M., Mulhern M.S., Alhamdow A., Engstrom K., Wahlberg K., Strain J.J., Watson G.E., Myers G.J., Davidson P.W., Shamlaye C.F., Broberg K., van Wijngaarden E. Maternal Long-Chain Polyunsaturated Fatty Acid Status, Methylmercury Exposure, and Birth Outcomes in a High-Fish-Eating Mother-Child Cohort. J Nutr. 2020 doi: 10.1093/jn/nxaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fumagalli M., Moltke I., Grarup N., Racimo F., Bjerregaard P., Jorgensen M.E., Korneliussen T.S., Gerbault P., Skotte L., Linneberg A., Christensen C., Brandslund I., Jorgensen T., Huerta-Sanchez E., Schmidt E.B., Pedersen O., Hansen T., Albrechtsen A., Nielsen R. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 118.Amorim C.E., Nunes K., Meyer D., Comas D., Bortolini M.C., Salzano F.M., Hunemeier T. Genetic signature of natural selection in first Americans. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1620541114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tanaka T., Shen J., Abecasis G.R., Kisialiou A., Ordovas J.M., Guralnik J.M., Singleton A., Bandinelli S., Cherubini A., Arnett D., Tsai M.Y., Ferrucci L. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lattka E., Eggers S., Moeller G., Heim K., Weber M., Mehta D., Prokisch H., Illig T., Adamski J. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res. 2010;51:182–191. doi: 10.1194/jlr.M900289-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Du Z., Ma L., Qu H., Chen W., Zhang B., Lu X., Zhai W., Sheng X., Sun Y., Li W., Lei M., Qi Q., Yuan N., Shi S., Zeng J., Wang J., Yang Y., Liu Q., Hong Y., Dong L., Zhang Z., Zou D., Wang Y., Song S., Liu F., Fang X., Chen H., Liu X., Xiao J., Zeng C. Whole Genome Analyses of Chinese Population and De Novo Assembly of A Northern Han Genome. Genomics Proteomics Bioinformatics. 2019 doi: 10.1016/j.gpb.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Montgomery S.B., Goode D.L., Kvikstad E., Albers C.A., Zhang Z.D., Mu X.J., Ananda G., Howie B., Karczewski K.J., Smith K.S., Anaya V., Richardson R., Davis J., Genomes Project C., MacArthur D.G., Sidow A., Duret L., Gerstein M., Makova K.D., Marchini J., McVean G., Lunter G. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Reardon H.T., Zhang J., Kothapalli K.S., Kim A.J., Park W.J., Brenna J.T. Insertion-deletions in a FADS2 intron 1 conserved regulatory locus control expression of fatty acid desaturases 1 and 2 and modulate response to simvastatin. Prostaglandins Leukot Essent Fatty Acids. 2012;87:25–33. doi: 10.1016/j.plefa.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li P., Zhao J., Kothapalli K.S.D., Li X., Li H., Han Y., Mi S., Zhao W., Li Q., Zhang H., Song Y., Brenna J.T., Gao Y. A regulatory insertion-deletion polymorphism in the FADS gene cluster influences PUFA and lipid profiles among Chinese adults: a population-based study. Am J Clin Nutr. 2018;107:867–875. doi: 10.1093/ajcn/nqy063. [DOI] [PubMed] [Google Scholar]
- 125.Lattka E., Illig T., Heinrich J., Koletzko B. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes? Clinical nutrition. 2010;29:277–287. doi: 10.1016/j.clnu.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 126.Lu J.T., Wang Y., Gibbs R.A., Yu F. Characterizing linkage disequilibrium and evaluating imputation power of human genomic insertion-deletion polymorphisms. Genome Biol. 2012;13:R15. doi: 10.1186/gb-2012-13-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li Y., Monroig O., Zhang L., Wang S., Zheng X., Dick J.R., You C., Tocher D.R. Vertebrate fatty acyl desaturase with Delta4 activity. Proc Natl Acad Sci U S A. 2010;107:16840–16845. doi: 10.1073/pnas.1008429107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oboh A., Kabeya N., Carmona-Antonanzas G., Castro L.F.C., Dick J.R., Tocher D.R., Monroig O. Two alternative pathways for docosahexaenoic acid (DHA, 22:6n-3) biosynthesis are widespread among teleost fish. Sci Rep. 2017;7:3889. doi: 10.1038/s41598-017-04288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Voss A., Reinhart M., Sankarappa S., Sprecher H. The metabolism of 7,10,13,16,19-docosapentaenoic acid to 4,7,10,13,16,19-docosahexaenoic acid in rat liver is independent of a 4-desaturase. J Biol Chem. 1991;266:19995–20000. [PubMed] [Google Scholar]
- 130.Raatz S.K., Conrad Z., Jahns L. Trends in linoleic acid intake in the United States adult population: NHANES 1999-2014. Prostaglandins Leukot Essent Fatty Acids. 2018;133:23–28. doi: 10.1016/j.plefa.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 131.Strandjord S.E., Lands B., Hibbeln J.R. Validation of an equation predicting highly unsaturated fatty acid (HUFA) compositions of human blood fractions from dietary intakes of both HUFAs and their precursors. Prostaglandins Leukot Essent Fatty Acids. 2018;136:171–176. doi: 10.1016/j.plefa.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chandra A., Rosjo H., Svensson M., Vigen T., Ihle-Hansen H., Orstad E.B., Ronning O.M., Lyngbakken M.N., Nygard S., Berge T., Schmidt E.B., Omland T., Tveit A., Eide I.A. Plasma linoleic acid levels and cardiovascular risk factors: results from the Norwegian ACE 1950 Study. Eur J Clin Nutr. 2020 doi: 10.1038/s41430-020-0641-4. [DOI] [PubMed] [Google Scholar]
- 133.Hsieh J.C., Liu L., Zeilani M., Ickes S., Trehan I., Maleta K., Craig C., Thakwalakwa C., Singh L., Brenna J.T., Manary M.J. High-Oleic Ready-to-Use Therapeutic Food Maintains Docosahexaenoic Acid Status in Severe Malnutrition. J. Pediatr. Gastroenterol. Nutr. 2015;61:138–143. doi: 10.1097/MPG.0000000000000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alvheim A.R., Malde M.K., Osei-Hyiaman D., Lin Y.H., Pawlosky R.J., Madsen L., Kristiansen K., Froyland L., Hibbeln J.R. Dietary linoleic acid elevates endogenous 2-AG and anandamide and induces obesity. Obesity. 2012;20:1984–1994. doi: 10.1038/oby.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Diau G.Y., Hsieh A.T., Sarkadi-Nagy E.A., Wijendran V., Nathanielsz P.W., Brenna J.T. The influence of long chain polyunsaturate supplementation on docosahexaenoic acid and arachidonic acid in baboon neonate central nervous system. BMC Med. 2005;3:11. doi: 10.1186/1741-7015-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ramsden C.E., Zamora D., Majchrzak-Hong S., Faurot K.R., Broste S.K., Frantz R.P., Davis J.M., Ringel A., Suchindran C.M., Hibbeln J.R. Re-evaluation of the traditional diet-heart hypothesis: analysis of recovered data from Minnesota Coronary Experiment (1968-73) Bmj. 2016;353:i1246. doi: 10.1136/bmj.i1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Menayang A. Mars pledges to use 100% high-oleic peanuts in M&Ms, Munch Bar, by end of 2017. FOOD navigator-usa.com. 2017 [Google Scholar]
- 138.Gregory M.K., Cleland L.G., James M.J. Molecular basis for differential elongation of omega-3 docosapentaenoic acid by the rat Elovl5 and Elovl2. J Lipid Res. 2013;54:2851–2857. doi: 10.1194/jlr.M041368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hu Y., Li H., Lu L., Manichaikul A., Zhu J., Chen Y.D., Sun L., Liang S., Siscovick D.S., Steffen L.M., Tsai M.Y., Rich S.S., Lemaitre R.N., Lin X. Genome-wide meta-analyses identify novel loci associated with n-3 and n-6 polyunsaturated fatty acid levels in Chinese and European-ancestry populations. Hum Mol Genet. 2016;25:1215–1224. doi: 10.1093/hmg/ddw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Illig T., Gieger C., Zhai G., Romisch-Margl W., Wang-Sattler R., Prehn C., Altmaier E., Kastenmuller G., Kato B.S., Mewes H.W., Meitinger T., de Angelis M.H., Kronenberg F., Soranzo N., Wichmann H.E., Spector T.D., Adamski J., Suhre K. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kananen L., Marttila S., Nevalainen T., Jylhava J., Mononen N., Kahonen M., Raitakari O.T., Lehtimaki T., Hurme M. Aging-associated DNA methylation changes in middle-aged individuals: the Young Finns study. BMC Genomics. 2016;17:103. doi: 10.1186/s12864-016-2421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Slieker R.C., Relton C.L., Gaunt T.R., Slagboom P.E., Heijmans B.T. Age-related DNA methylation changes are tissue-specific with ELOVL2 promoter methylation as exception. Epigenetics Chromatin. 2018;11:25. doi: 10.1186/s13072-018-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen D., Chao D.L., Rocha L., Kolar M., Nguyen Huu V.A., Krawczyk M., Dasyani M., Wang T., Jafari M., Jabari M., Ross K.D., Saghatelian A., Hamilton B.A., Zhang K., Skowronska-Krawczyk D. The lipid elongation enzyme ELOVL2 is a molecular regulator of aging in the retina. Aging Cell. 2020;19:e13100. doi: 10.1111/acel.13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ito H., Udono T., Hirata S., Inoue-Murayama M. Estimation of chimpanzee age based on DNA methylation. Sci Rep. 2018;8:9998. doi: 10.1038/s41598-018-28318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Plourde M., Chouinard-Watkins R., Vandal M., Zhang Y., Lawrence P., Brenna J.T., Cunnane S.C. Plasma incorporation, apparent retroconversion and beta-oxidation of 13C-docosahexaenoic acid in the elderly. Nutr Metab (Lond) 2011;8:5. doi: 10.1186/1743-7075-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leveille P., Chouinard-Watkins R., Windust A., Lawrence P., Cunnane S.C., Brenna J.T., Plourde M. Metabolism of uniformly labeled (13)C-eicosapentaenoic acid and (13)C-arachidonic acid in young and old men. Am J Clin Nutr. 2017;106:467–474. doi: 10.3945/ajcn.117.154708. [DOI] [PubMed] [Google Scholar]
- 147.Chappus-McCendie H., Chevalier L., Roberge C., Plourde M. Omega-3 PUFA metabolism and brain modifications during aging. Prog Neuropsychopharmacol Biol Psychiatry. 2019;94 doi: 10.1016/j.pnpbp.2019.109662. [DOI] [PubMed] [Google Scholar]
- 148.Harris W.S., Pottala J.V., Varvel S.A., Borowski J.J., Ward J.N., McConnell J.P. Erythrocyte omega-3 fatty acids increase and linoleic acid decreases with age: observations from 160,000 patients. Prostaglandins Leukot Essent Fatty Acids. 2013;88:257–263. doi: 10.1016/j.plefa.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 149.Park H.G., Kothapalli K.S.D., Park W.J., DeAllie C., Liu L., Liang A., Lawrence P., Brenna J.T. Palmitic acid (16:0) competes with omega-6 linoleic and omega-3-linolenic acids for FADS2 mediated 6-desaturation. Biochim Biophys Acta. 2016;1861(2):91–97. doi: 10.1016/j.bbalip.2015.11.007. PMID: 26597785. [DOI] [PMC free article] [PubMed] [Google Scholar]