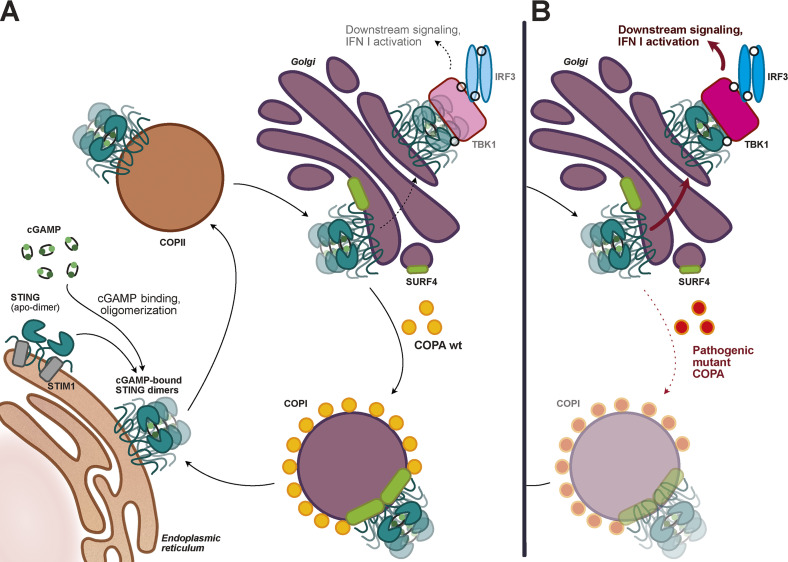
COPA mutations prevent STING’s Golgi–ER trafficking and promote STING activation and interferonopathy.
Abstract
Two studies published in this issue of JEM, by Lepelley et al. (https://doi.org/10.1084/jem.20200600) and Deng et al. (https://doi.org/10.1084/jem.20201045), and two additional manuscripts by Mukai et al. (https://doi.org/10.1101/2020.05.20.107664 Preprint v1) and Steiner et al. (https://doi.org/10.1101/2020.07.09.194399 Preprint v1) demonstrate that COPA syndrome–associated high interferon titers are linked to mutations in COPA preventing STING’s retrieval from the Golgi back to the ER and thereby causing chronic immune activation.
COPA syndrome is an inflammatory Mendelian disease caused by missense mutations in the coatomer subunit α (COPA) protein, which is part of coat protein complex I (COPI) and as such involved in the retrograde Golgi-to-ER trafficking (Brandizzi and Barlowe, 2013). COPA patients exhibit a broad range of clinical symptoms, including, for example, interstitial lung disease and joint inflammation (Watkin et al., 2015). Recently, the pathogenesis of the disease has been associated with high titers of type I IFNs, and parallels have been drawn with the phenotype of another previously characterized autoinflammatory syndrome, STING (stimulator of IFN genes)-associated vasculopathy with onset in infancy (SAVI; Volpi et al., 2018). STING is a key signaling molecule involved in the induction of type I IFNs downstream of the DNA sensor cyclic GMP-AMP synthase (cGAS; Ablasser and Hur, 2020). Upon activation by the cGAS-catalyzed product cyclic GMP-AMP (cGAMP) or other cyclic dinucleotides, STING traffics from the ER to the Golgi, a step which is both necessary and sufficient for the recruitment and activation of TBK1 and IRF3 and, finally, de novo gene expression. In SAVI, mutations in STING allow for its constitutive trafficking in the absence of an external stimulus (Dobbs et al., 2015).

Insights from Sophie Rivara and Andrea Ablasser.
Given the similarities in the clinical presentations of SAVI and COPA syndrome on the one hand and the overlapping aspect of ER–Golgi/Golgi–ER trafficking on the other hand, the four groups (Deng et al., 2020; Lepelley et al., 2020; Mukai et al., 2020 Preprint; Steiner et al., 2020 Preprint) investigated whether the missing link between COPA mutations and high type I IFN titers might be STING.
The group of Yanick J. Crow and colleagues (Lepelley et al., 2020) extensively characterized nine carriers of pathogenic COPA variants. In doing so, they confirmed the similarities between COPA syndrome and SAVI: interstitial lung disease and chronic up-regulation of type I IFN, as evidenced by activated STAT1 signaling as well as high expression levels of several IFN-induced genes (ISGs). Their observations also confirm the remarkably high nonpenetrance of COPA syndrome, with three out of nine carriers being asymptomatic despite elevated type I IFN levels compared with healthy controls. To work out the molecular mechanism underlying the aberrant type I IFN response, the authors studied cell lines depleted for COPA. This led them to discover that knocking down COPA mimicked the phenotype of COPA mutations in their heightened type I IFN response and, more important, that absence of STING (STING KO cells), but not mitochondrial antiviral signaling protein (MAVS, an innate immune adaptor essential for cytosolic RNA sensing also signaling through IRF3 and downstream type I IFN production), abolished this response. Intriguingly, cells depleted of cGAS also showed a diminished type I IFN response when knocked down for COPA, thus pointing toward some sort of baseline activation of cGAS that sets into motion STING cycling between the ER and the Golgi.
As part of COPI, COPA participates in the cargo sorting for retrograde Golgi-to-ER trafficking (Brandizzi and Barlowe, 2013). It hence has to associate with proteins it sends back toward the ER. To test for a potential interaction with STING, Lepelley et al. (2020) performed immunoprecipitation experiments and confirmed an interaction between STING and COPA, which was reduced when COPA mutants were used. The authors mapped the transmembrane region of STING as critical for this interaction and, furthermore, demonstrated by confocal fluorescence microscopy that expression of COPA mutants caused STING to accumulate in the Golgi, whereas COPA overexpression did not affect the localization of STING in the ER.
In their complementary work, the group of Anthony K. Shum and colleagues (Deng et al., 2020) highlight additional insights into the mechanism underlying STING’s involvement in the disease and, in addition, provide in vivo proof-of-concept studies on potential pharmacological treatment options. After confirming STING’s activation (phosphorylation and localization to the Golgi) in lung fibroblasts from a COPA patient, they show, similar to the results of Lepelley et al. (2020), that STING associates with COPA, which is reduced when tested with mutant COPA. Given that STING lacks the COPA-recognized dilysine motif, their results pointed toward another (transmembrane) protein acting as an adaptor to mediate the STING–COPA interaction. In search for a putative adaptor, the authors mined the literature of published STING interaction partners containing a C-terminal dilysine motif to then focus on SURF4, a candidate previously proven to shuttle between the Golgi and the ER via the COPI machinery (Adolf et al., 2019). In support of their candidate, they found that SURF4 indeed coprecipitated with STING and COPA, that its association with COPA was drastically reduced when disrupting the C-terminal dilysine motif, and that less STING was pulled down by COPA in the absence of SURF4.
The authors then used a CopaE241K/+ mouse model to study systemic effects of COPA-mediated STING activation. They noticed perturbed T cell development, which is reminiscent of the SAVI phenotype in mice and may be due to the proapoptotic function of STING in lymphocytes (Gulen et al., 2017; Siedel et al., 2020). Moreover, peripheral lymphoid organs also displayed elevated levels of ISGs and immune dysregulation. Of note, the embryonic lethality of homozygous CopaE241K/E241K mice could be rescued by a deletion of STING (CopaE241K/+×Stinggt/gt parents). Based on these findings, Deng et al. (2020) reasoned that STING inhibition could have therapeutic benefits. Treating splenocytes from CopaE241K/+ mice and peripheral blood mononuclear cells from a COPA syndrome patient with small molecular inhibitors of STING (Haag et al., 2018) or with a JAK-STAT inhibitor (targeting a signaling step downstream of type I IFN production) indeed decreased ISG levels, but only the STING inhibitors reduced type I IFN expression.
Putting together the results of these concordant publications, an intriguing addition to the traditional model of STING trafficking and regulation appears to be emerging: low basal cGAS-mediated STING activation triggers constitutive STING trafficking toward the Golgi, but robust downstream type I IFN pathway activation is prevented by the back-shuttling of STING toward the ER. This retrograde trafficking, mediated by the COPI machinery, involves the sorting protein COPA, which through the help of the adaptor SURF4 can encapsulate STING into COPI vesicles. Upon disruption of this Golgi-to-ER trafficking, which can be caused in a dominant-negative fashion by pathogenic COPA variants, STING now being trapped in the Golgi initiates an immune response similar to the one observed in SAVI (see figure).
The STING cycle. (A) In resting cells, a basal level of cGAMP promotes the COPII (coatomer complex II)-mediated trafficking of STING from the ER to the Golgi, where STING is recognized by COPA via the adaptor SURF4 to be sent back to the ER through the COPI machinery. (B) In patients carrying a pathogenic COPA mutant, the COPA-driven cargo sorting of STING into COPI vesicles is impaired, leading to STING accumulation in the Golgi, increased downstream signaling, and type I IFN production.
Recent advances in the understanding of STING trafficking regulation included the identification of several STING interactors, notably STIM1, involved in its retention in the ER (Srikanth et al., 2019), and STEEP (STING ER exit protein), reportedly essential to load STING into COPII vesicles (Zhang et al., 2020). However, whereas these and other previous publications center around the ER enrichment and the anterograde ER-to-Golgi trafficking of STING, the work published in Lepelley et al., 2020 and Deng et al., 2020 in this issue of JEM as well as the work in Mukai et al., 2020 (Preprint) and Steiner et al., 2020 (Preprint) highlight that the regulation of STING “going back”—overlooked so far—is as important as the one set in place of “going forward.” For future work, it would be interesting to investigate whether a similar adaptor–effector tandem (SURF–COPA equivalent) exists for COPII and anterograde trafficking.
One might also wonder whether there is a link between this new “basal STING turnover” model and the observed low COPA syndrome penetrance. Indeed, inter-individual discrepancies in basal cGAS-STING activation levels might explain the different outcomes for pathogenic COPA variants carriers.
The four publications also bear important clinical consequences. At present, the therapeutic options for the treatment of COPA patients are mostly based on steroid-based traditional immunosuppression, but this is insufficient to prevent irreversible lung damage. The emerging link between COPA syndrome and high type I IFN burden has motivated studies on JAK-STAT inhibitors for the treatment of COPA patients (Tsui et al., 2018). The present studies further rationalize that anti-IFN drugs can be an efficacious approach to alleviate COPA syndrome symptoms while also pointing toward the potential of inhibiting the STING pathway as a more direct means to dampen the immunopathology. More generally, the papers highlight the importance of the ER–Golgi trafficking axis as a key inflammatory regulator, whose modulation or fine-tuning might also be considered in the development of new drugs.
All in all, the insight provided in Deng et al., 2020, Lepelley et al., 2020, Mukai et al., 2020 (Preprint), and Steiner et al., 2020 (Preprint) not only opens the door to novel treatment approaches for COPA syndrome, but it also advances understanding on the intricate mechanisms regulating STING activation and trafficking.
Acknowledgments
A. Ablasser is a member of the scientific advisory board of IFM Therapeutics and scientific co-founder of IFM Due.
References
- Ablasser, A., and Hur S.. 2020. Nat. Immunol. 10.1038/s41590-019-0556-1 [DOI] [PubMed] [Google Scholar]
- Adolf, F., et al. 2019. Cell Rep. 10.1016/j.celrep.2018.12.041 [DOI] [PubMed] [Google Scholar]
- Brandizzi, F., and Barlowe C.. 2013. Nat. Rev. Mol. Cell Biol. 10.1038/nrm3588 [DOI] [PubMed] [Google Scholar]
- Deng, Z., et al. 2020. J. Exp. Med. 10.1084/jem.20201045 [DOI] [Google Scholar]
- Dobbs, N., et al. 2015. Cell Host Microbe. 10.1016/j.chom.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulen, M.F., et al. 2017. Nat. Commun. 10.1038/s41467-017-00573-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, S.M., et al. 2018. Nature. 10.1038/s41586-018-0287-8 [DOI] [Google Scholar]
- Lepelley, A., et al. 2020. J. Exp. Med. 10.1084/jem.20200600 [DOI] [Google Scholar]
- Mukai, K., et al. 2020. bioRxiv. 10.1101/2020.05.20.107664 (Preprint posted May 21, 2020) [DOI]
- Siedel, H., et al. 2020. Clin. Immunol. 10.1016/j.clim.2020.108466 [DOI] [PubMed] [Google Scholar]
- Srikanth, S., et al. 2019. Nat. Immunol. 10.1038/s41590-018-0287-8 [DOI] [Google Scholar]
- Steiner, A., et al. 2020. bioRxiv. 10.1101/2020.07.09.194399 (Preprint posted July 9, 2020) [DOI]
- Tsui, J.L., et al. 2018. ERJ Open Res. 10.1183/23120541.00017-2018 [DOI] [Google Scholar]
- Volpi, S., et al. 2018. Clin. Immunol. 10.1016/j.clim.2017.10.001 [DOI] [Google Scholar]
- Watkin, L.B., et al. 2015. Nat. Genet. 10.1038/ng.3279 [DOI] [Google Scholar]
- Zhang, B., et al. 2020. Nat. Immunol. 10.1038/s41590-020-0730-5 [DOI] [Google Scholar]