Abstract
Resistant cultivars have played important roles in controlling Fusarium wilt disease, but the roles of rhizosphere interactions among different levels of resistant cultivars are still unknown. Here, two phenotypes of cucumber, one resistant and one with increased susceptibility to Fusarium oxysporum f.sp. cucumerinum (Foc), were grown in the soil and hydroponically, and then 16S rRNA gene sequencing and nontargeted metabolomics techniques were used to investigate rhizosphere microflora and root exudate profiles. Relatively high microbial community evenness for the Foc-susceptible cultivar was detected, and the relative abundances of Comamonadaceae and Xanthomonadaceae were higher for the Foc-susceptible cultivar than for the other cultivar. FishTaco analysis revealed that specific functional traits, such as protein synthesis and secretion, bacterial chemotaxis, and small organic acid metabolism pathways, were significantly upregulated in the rhizobacterial community of the Foc-susceptible cultivar. A machine-learning approach in conjunction with FishTaco plus metabolic pathway analysis revealed that four organic acids (citric acid, pyruvate acid, succinic acid, and fumarate) were released at higher abundance by the Foc-susceptible cultivar compared with the resistant cultivar, which may be responsible for the recruitment of Comamonadaceae, a potential beneficial microbial group. Further validation demonstrated that Comamonadaceae can be “cultured” by these organic acids. Together, compared with the resistant cultivar, the susceptible cucumber tends to assemble beneficial microbes by secreting more organic acids.
Subject terms: Secondary metabolism, Soil microbiology
Introduction
Fusarium wilt disease is a persistent and widespread soil-borne disease worldwide. For a long time, breeders have been developing many resistant varieties that generally express high levels of disease resistance genes and/or produce active proteins to defend against the Fusarium oxysporum fungal pathogen1. In recent years, increasing numbers of studies have shown that certain beneficial microorganisms can be recruited by plants in their rhizosphere to resist the invasion of pathogens2. For example, Berendsen et al.3 indicated that Arabidopsis thaliana could recruit three bacterial species in the rhizosphere upon foliar pathogen infection; Kwak et al.4 found that the tomato variety Hawaii 7996 could recruit members of the flavobacterium strain TRM1 to suppress disease. Natural disease-suppressive soils even formed via increases in the abundance of beneficial microorganisms (e.g., Pseudomonas and Bacillus) by monoculture during a long period5,6. These beneficial microorganisms can produce secondary metabolites such as 2,4-diacetylphloroglucinol to antagonize pathogens, thus improving the disease-suppressive ability of the soil. Other studies have shown that root exudates such as citric acid, malic acid and fumaric acid could recruit beneficial rhizosphere microorganisms7–10. Moreover, it was recently shown that specialized triterpenes from A. thaliana could recruit and maintain an A. thaliana-specific microbiota11.
Plant breeders have begun to consider the contribution of rhizosphere microbes in the development of resistant varieties9, including the regulation of the output of root exudates by controlling ABC transporters12 and attempt to transfer microorganisms to the next generation by implanting microbes into flowers13. Since the interactions occurring in the rhizosphere between plants and microorganisms are relatively complex and variable, breeding work that encompasses the rhizosphere microbiome is progressing slowly.
Due to the different root secretion patterns even within the same crop, the composition of the rhizosphere microbial community is variable. Interestingly, crops sensitive to pathogens tend to form disease-suppressive soils more easily than do resistant varieties5. Further studies indicated that traditional susceptible cultivars tend to maintain stronger interactions between plant and beneficial soil microorganisms compared to those of modern resistant varieties14–17. Karasov et al.17 found that the diversity of Pseudomonas in wild A. thaliana leaves was abundant. Recently, a study of domesticated plants seemed to reveal a relatively limited microbiota assembly compared to that of their wild counterparts. Furthermore, the genetic diversity of crop microbiota is likely reduced compared to that of wild plants18. Plant domestication probably alters root exudate profiles and thus impacts rhizosphere microbial community structure and function19. Whether rhizosphere microbial resistance can compensate for the weakness of plant resistance is relatively unknown. Moreover, which kinds of rhizospheric interactions occur during rhizosphere microbial resistance are unclear.
Here, we grew two types of cucumber with contrasting phenotypes (resistant and highly susceptible to Fusarium wilt disease) in the soil and hydroponically to provide controlled and in situ experimental data on these cultivars’ exudates and their rhizobacterial communities. To identify the specific roles of exudate compounds in the recruitment of beneficial bacteria, we employed statistical analyses to evaluate amplicon sequences of the 16S rRNA genes and used nontargeted metabolomics. We aimed to address (i) whether disease-susceptible cultivars have advantages over resistant cultivars in terms of beneficial bacterial enrichment; (ii) whether these interactions are caused by changes in root exudates; and (iii) if so, which types of exudate compounds are responsible for recruitment.
Results
Recruitment of rhizosphere bacterial communities of two cucumber cultivars
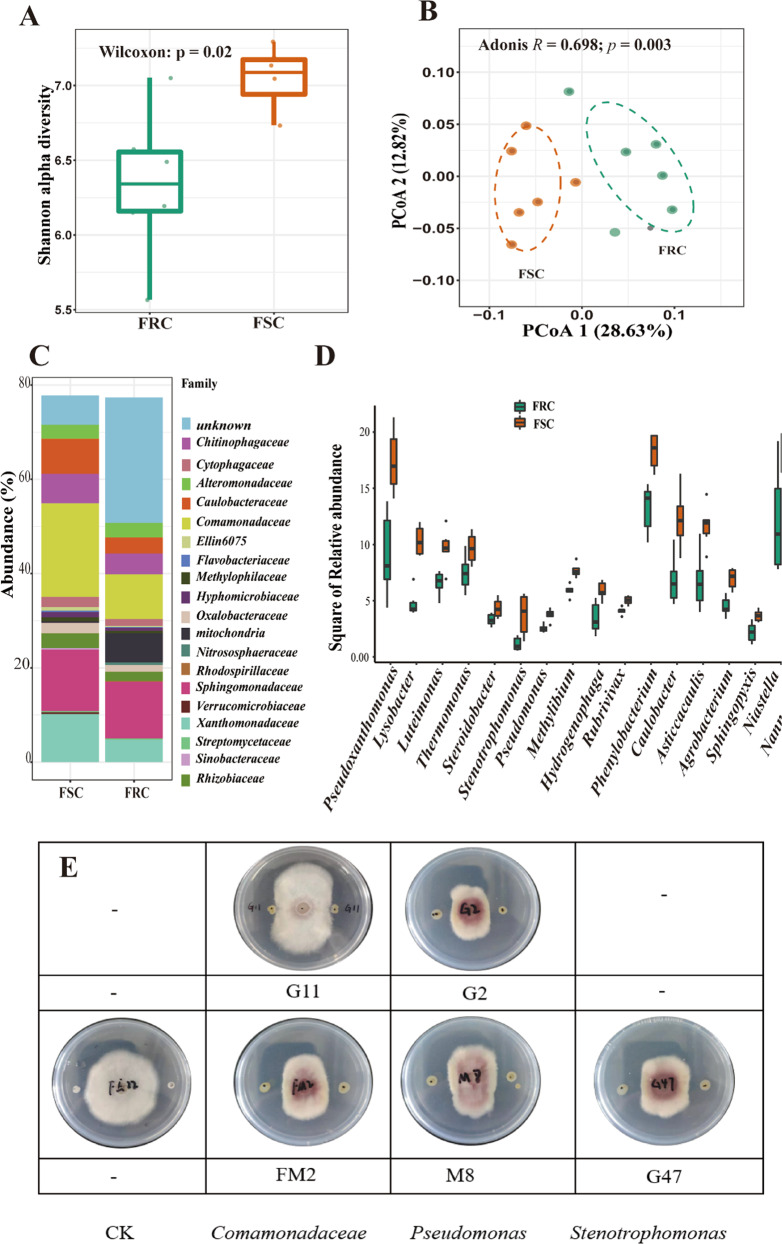
Bacterial communities in the rhizosphere of two cucumber cultivars were characterized with Illumina HiSeq sequencing. In total, 1,010,289 high-quality sequences were obtained, and each sample contained between 66,236 and 107,307 (84,191 ± 12,531) reads. All the sequences clustered into 8023 operational taxonomic units (OTUs) with 97% similarity. The OTU numbers for each cultivar were 6224 in the Foc-susceptible cucumber (FSC) and 6830 in the Foc-resistant cucumber (FRC), and the most abundant phyla were Proteobacteria (64.4%), Cyanobacteria (12.2%), Bacteroidetes (9.2%), Acidobacteria (4.2%), Actinobacteria (3.5%), and Verrucomicrobia (1.9%). We rarefied (without replacement) 89,948 sequences for each sample to calculate the Shannon index, which is often used to assess the evenness and abundance of microbial communities. The rhizosphere soil of Foc-susceptible cucumber exhibited higher bacterial community evenness than did the Foc-resistant cucumber (Wilcoxon test, p < 0.05; Fig. 1a). Principal coordinate analysis (PCoA) with the Bray-Curtis distance showed a significant (p = 0.003, R = 0.89 in Adonis) difference in the rhizosphere communities between the Foc-resistant cucumber and the Foc-susceptible cucumber (Fig. 1b).
Fig. 1. Analysis of rhizosphere bacterial communities between two cultivars.
a The Shannon-Wiener index of Foc-resistant cultivar (green) and Foc-susceptible cultivar (orange) rhizosphere bacterial communities calculated with all clustered OTUs. The horizontal bars within the boxes represent the medians. The tops and bottoms of the boxes represent the 75th and 25th quartiles, respectively. All outliers were plotted as individual points. b Principal coordinate analysis (PCoA) with Bray-Curtis dissimilarity performed on the taxonomic profile (OTU level for a 16S rRNA dataset) of the rhizosphere of the two cucumber cultivars. The R-value and P-value were evaluated via the Adonis test. c Relative abundance (%) of members of the major bacterial phyla, excluding low-abundance OTUs (mean abundance <0.02%), present in the rhizosphere microbial communities of Foc-resistant cultivar (FRC) or Foc-susceptible cultivar (FSC) samples. d The relative abundance (%) of members of specific genera enriched in the rhizosphere soil of the FSC and of the entire genus present here was significantly different (t-test: p<0.05) between the two cultivars. e Inhibitory effects of six isolates on F. oxysporum belonging to Comamonadaceae (G11, FM2), Pseudomonas (M8, G2), and Stenotrophomonas (G47).
Comparative analysis of the rhizosphere microbiome between the Foc-resistant and Foc-susceptible cucumber cultivars revealed a distinctly different abundance of specific groups at the family level (Fig. 1c), with a higher abundance of the families Comamonadaceae and Xanthomonadaceae in the rhizosphere of the Foc-susceptible cultivar compared with the Foc-resistant cultivar (Fig. 1c and Supplementary Table 1). At the lower taxonomical level, we found that the abundance of genera belonging to Comamonadaceae, likely Methylibium, Hydrogenophaga, and Rubrivivax, and some genera belonging to Xanthomonadaceae, such as Pseudoxanthomonas, Lysobacter, Stenotrophomonas, and Pseudomonas, were significantly increased in the rhizosphere soil of the Foc-susceptible cucumber compared to the Foc-resistant cucumber (Supplementary Table 2).
We obtained 16 different isolates from the treatments of the FSC and FRC; these isolates belong to the Bacillaceae, Pseudomonadaceae, Comamonadaceae, Xanthomonadaceae, Enterobacteriaceae, Oxalobacteraceae, and Weeksellaceae. Through an inhibition assay of F. oxysporum, we found that five strains of Comamonadaceae (strains G11 and FM2), Pseudomonas (strains M8 and G2), and Stenotrophomonas (strain G47) could reduce the growth of F. oxysporum in vitro (Fig. 1e). It was also found that these five strains obtained more single colonies in the FSC (Supplementary Table 3). Correspondingly, the abundance of genera belonging to Comamonadaceae and of the Stenotrophomonas and Pseudomonas genera significantly increased in the rhizosphere soil of Foc-susceptible cucumber. This suggested that Foc-susceptible cucumber cultivars could recruit more beneficial microbes to resist F. oxysporum.
Root exudate profiles of the two cultivars
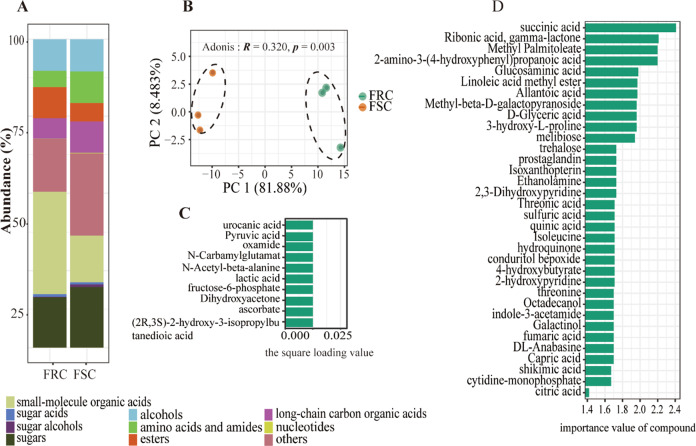
Root exudates of the Foc-resistant and Foc-susceptible cucumber cultivars were analyzed by gas chromatography-time of flight mass spectrometry (GC-TOF-MS) after collection from a sterile hydroponic system. A total of 708 chromatographic peaks were detected, and 236 compounds were identified across all the samples. Then, we divided compounds into several categories based on their chemical nature, namely, sugars (28 compounds), sugar alcohols (3), sugar acids (7), small-molecule organic acids (22), nucleotides (4), long-chain organic acids (14), esters (18), alcohols (16), amino acids and amides (28), and others (97; Fig. 2a). The overall exudation patterns of the Foc-susceptible cucumber plants were found to be distinct (p = 0.043, R = 0.320 in Adonis) from those of the Foc-resistant cultivar (Fig. 2b). The relative abundance of 157 compounds of the 236 total identified compounds was significantly (p < 0.05) different between the two cultivars. Specifically, 79 compounds showed higher relative abundance in the FSC, and 78 compounds showed higher relative abundance in the FRC.
Fig. 2. Analysis of exudation profiles between two cultivars.
a Relative abundance (%) of 10 compounds present in the Foc-resistant cultivar (FRC) or Foc-susceptible cultivar (FSC) root exudates. The data were obtained with peak areas from the GC-MS analysis. b PCA plot of the root exudate profiles of two cucumber cultivars evaluated with GC-MS: FRC=rhizosphere of the resistant cultivar; FSC=rhizosphere of the susceptible cultivar. The P-values were evaluated via the Adonis test (Adonis: p=0.003). c The top 10 (top 4% of 236 total compounds) compounds sorted by loading variable importance from the PCA of the two cucumber cultivars. The 10 compounds are ranked in descending order of importance to the loading matrix. d The top 35 marker compounds were identified by applying random forest classification of their relative abundances in root exudates of two cucumber cultivars. The marker compounds are ranked in descending order of importance with respect to the accuracy of the model.
To further explore the major compounds that caused the difference in root exudate profiles between the two cultivars, a Principal Component Analysis (PCA) loading matrix extracted and random forests classification approach were applied. Loading matrix of the PC1 axis which explained 81.9% of variance, from principal components analysis (PCA) were extracted to identify the top 10 compounds (top 4% of total 236 compounds) that were important to differentiate the exudate profiles of the two cultivars (Fig. 2c). These compounds included pyruvic acid and lactic acid (short-chain carbon organic acids), two amino acids (N-carbamylglutamate and N-acetyl-beta-alanine), and six other compounds, including (2 R,3 S)-2-hydroxy-3-isopropylbutanedioic acid, fructose-6-phosphate, urocanic acid, oxamide, ascorbate, and dihydroxyacetone (Supplementary Table 4). For the random forest classifier (R package RandomForest, ntree = 1000), the first 35 of the 236 compounds (top 15%) sorted by loading variable importance were selected (Fig. 2d and Supplementary Table 5). Then, the top 10 important compounds selected from the PCA and the top 35 important compounds from the random forest classification were further evaluated for significant differences between the cultivars using a t-test with p < 0.05 and a log2(fold change) <1.5. Finally, 34 compounds were recognized as playing the most important role in the separation of the two root exudate profiles (Supplementary Table 6).
Functional features of root exudates and functional shifts mediated by microbial alterations in the rhizosphere of FSC
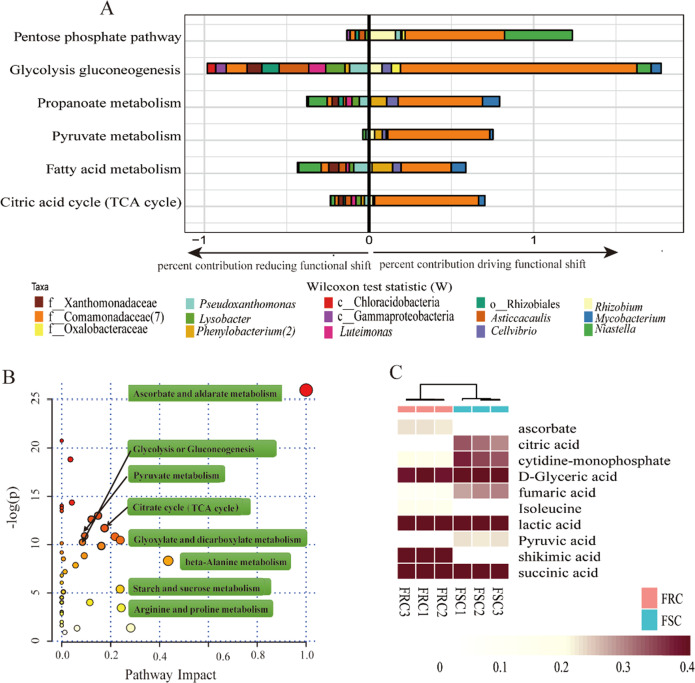
It was shown that root exudates were significantly associated with rhizosphere microorganisms according to Mantel’s test (R = 0.5966, p = 0.003, Supplementary Table 7). Pathway enrichment analysis was performed to further exploration whether root exudates of the Foc-susceptible cultivar are involved in the recruitment of beneficial bacteria. The results revealed that small organic acid metabolism pathways (such as pyruvate acid metabolism and the citric acid cycle) and amino acid metabolic pathways (valine, leucine and isoleucine biosynthesis and glycine, serine and threonine metabolism) were significantly (Wilcoxon test: p < 0.05) different (Fig. 3b and Supplementary Table 8). Then, 24 compounds involved in these different pathways were selected to check if they were covered in the 34 represented compounds causing major differences in the root exudate profiles. Ten compounds were ultimately selected to represent the core difference in root exudate profiles and metabolic pathways (Supplementary Table 9). Among these 10 compounds, six were small organic acids, including four that mainly participate in pyruvate acid metabolism and the citric acid cycle (Fig. 3c).
Fig. 3. Prediction of the major pathways mediated by microbial communities and KEGG pathway enrichment analysis of root exudates between two cultivars.
a To identify the shifts in rhizosphere communities caused by these potential beneficial bacteria enriched in the Foc-susceptible cultivar rhizosphere, deconvolution of significant community-wide functional shifts into individual taxonomic contributions was performed. The right bar plot represents relative contributions driving functional shifts by the taxa of Foc-susceptible samples, and the left bar plot represents relative contributions reducing functional shifts by the taxa of Foc-susceptible samples. b Different metabolic pathways of the root exudates of two cucumber cultivars. Each point represents a different metabolic pathway, and the size of each point represents the degree of change in each metabolic pathway. c Heatmap analysis of 10 compounds selected by PCA, random forest classification, and pathway enrichment analyses; these compounds were significantly (t-test, p<0.05) different in terms of their relative abundance between the root exudates of the two cucumber cultivars.
To determine whether these ten compounds or core differential pathways influenced the assembly of rhizosphere microbial communities and variation between the two cultivars, we employed the FishTaco framework for microbial communities to predict the functional information within the microbial communities and to determine the causes of the shifts in rhizosphere microorganisms. Marked upregulation of both protein synthesis and secretion and bacterial chemotaxis were observed (Wilcoxon test: p < 0.05) for the Foc-susceptible cultivar (FSC), which may be related to the rapid response of soil microorganisms to the roots and corresponded to the enrichment of OTUs from members of the Comamonadaceae and Oxalobacteraceae families (Supplementary Fig. 1). Many amino acids and small-molecule fatty acids were also more abundant in the rhizosphere of the FSC, including the citric acid cycle. The results showed that enrichment of the citric acid cycle was mainly attributed to shifts in the relative abundance of the Comamonadaceae family, as this family contains many genes involved in the citric acid cycle (Fig. 3a and Supplementary Fig. 2). Moreover, the pathway enrichment analysis results based on root exudate data showed that the selected small-molecule organic acids of 10 core root exudates were enriched in the root exudates of the Foc-susceptible cucumber (Supplementary Table 6) associated with the citric acid cycle. Therefore, we suspect that the four organic acids may promote recruitment of Comamonadaceae in the rhizosphere of the Foc-susceptible cultivar by enriching compounds in the TCA cycle.
Impacts of selected small-molecule organic acids on Comamonadaceae members
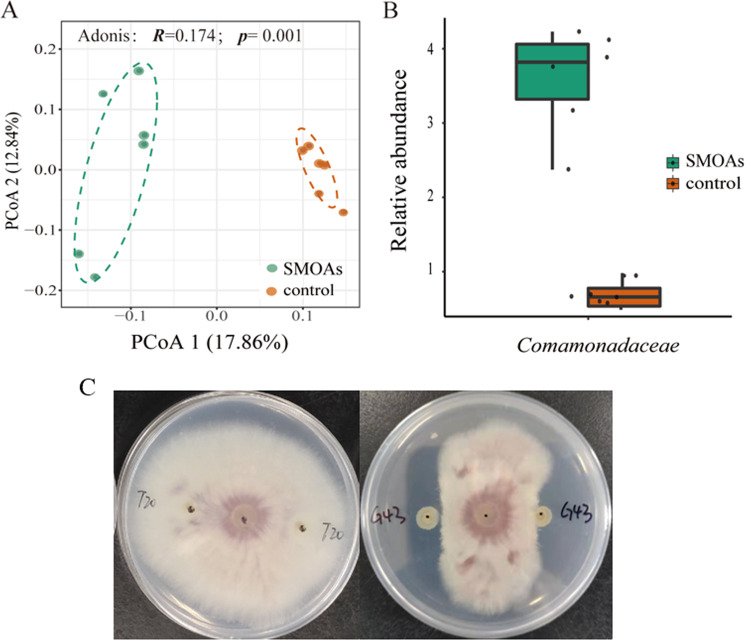
Four small-molecule organic acids that were significantly enriched in the exudates of the Foc-susceptible cultivar were mixed together and repeatedly added to condition the soil. Sequencing results of the communities produced a total of 632,721 sequences, with 15,344–63,539 (35,151 ± 14,705) reads per sample. Higher evenness measures were observed for the rhizosphere soil of the control treatment compared to the SMOA treatment (Fig. 4a, Wilcoxon test: p = 0.031). Principal coordinate analysis (PCoA) showed a clear difference (Adonis: p = 0.003, R = 0.17) in rhizosphere community composition between SMOAs and control (Fig. 4b). Comparative analysis of the microbiome between the SMOAs and control revealed a distinct differential abundances of specific family groups (Supplementary Table 9), including higher relative abundance of the family Comamonadaceae in the soil after the application of the four small-molecule organic acids (Fig. 4b).
Fig. 4. Diversity analysis of bacterial communities in soil treated with different compounds.
a Principal coordinate analysis (PCoA) with Bray-Curtis dissimilarity of the microbial community in soil treated with four small-molecule organic acids (SMOAs) and the control at the OTU level. The R-value and p-value were evaluated via the Adonis test. b Relative abundance (%) of Comamonadaceae enriched in the soil treated with SMOAs (t-test: p = 0.001). The horizontal bars within boxes represent the medians. The tops and bottoms of the boxes represent the 75th and 25th quartiles, respectively. All the outliers were plotted as individual points. c Inhibitory effects of Comamonadaceae strain G43 on F. oxysporum.
In total, we obtained 14 different isolates (Supplementary Table 11) from the SMOAs conditioned and control soils, which belong to Bacillaceae, Pseudomonadaceae, Comamonadaceae, Xanthomonadgaceae, Sphingomonadaceae, Burkholderiaceae, Alcaligenaceae, Oxalobacteraceae, and Rhizobiaceae. We then found that Comamonadaceae strain G43 had a strong inhibitory effect on the growth of Foc (Fig. 4c). Interestingly, more single colonies were isolated, similar to strain G43, from the SMOA-conditioned soils than from the control soils (Supplementary Table 11). Further alignment indicated that strain G43 was the same as Comamonadaceae strain G11.
Discussion
The rhizosphere microbiome is the first line of soil pathogen defense and plays a vital role in the prevention of pathogen invasion. Among the reported mechanisms, the evenness and richness of the rhizosphere microbiome are central players in providing stability and resilience to stress and invasion2,20. High evenness was observed in the Foc-susceptible rhizosphere, which could be attributed to a relatively high variety of exudates supporting more microbial niches. If these niches are left nonstimulated/uninhabited in the Foc-resistant rhizosphere, this could provide a window for successful invasion by other pathogens21. In this study, the higher evenness found in the Foc-susceptible rhizosphere compared with the Foc-resistant rhizosphere may result from sufficient coevolution between the host plant and soil microbiome. In other words, the soil microbial metabolic capacity for the resources from FRC roots has not developed21. It has been shown that traditional cultivars tend to maintain relatively high evenness of rhizosphere microbiomes17,22 and relatively strong interactions between plants and the environment21. For instance, mycorrhizal associations have been shown to be increased in older wheat cultivars compared to modern landraces14. Similarly, Germida et al.23 found that ancient landraces could recruit a more diverse rhizosphere bacterial community than could modern cultivars. We also found higher abundances of Comamonadaceae and Xanthomonadaceae in the Foc-susceptible cultivar rhizosphere community than in the Foc-resistant one, both of whose members have been reported to be abundant in disease-suppressive soils24–26. Xanthomonadaceae genera falling within this family, such as Pseudoxanthomonas, Lysobacter, and Stenotrophomonas, have been used as biocontrol agents of several pathogens, including Fusarium oxysporum27–30. It has been widely observed that fluorescent Pseudomonas species produce the antifungal compound 2,4-diacetylphloroglucinol (DAPG) to resist pathogens5. All these beneficial genera were more abundant for the Foc-susceptible cultivar than for the Foc-resistant cultivar, and five strains of Comamonadaceae (strains G11 and FM2), Pseudomonas (strains M8 and G2), and Stenotrophomonas (strain G47) showed strong inhibitory effects on the growth of pathogens in vitro (Fig. 1e), which may contribute to the resistance of other pathogens and natural attenuation of Fusarium oxysporum. Previous research has shown that crops sensitive to pathogens tend to form disease-suppressive soils more readily than do resistant varieties when under continuous pathogen attack5; this phenomenon was supported by our results, as more beneficial bacterial groups were recruited in the rhizosphere of the susceptible crop than in that of the resistant crop. However, some researchers have reported that, compared with susceptible varieties, resistant varieties can recruit more beneficial microorganisms31. Recently, Mendes et al.32 investigated the composition of the rhizosphere bacterial community of common bean cultivars with different levels of resistance to the fungal pathogen F. oxysporum and found that beneficial bacteria, such as Bacillaceae, Pseudomonadaceae, Solibacteraceae, and Cytophagaceae, were abundant in the rhizobacterial community of the FRC cultivar. On the other hand, our study showed that the FSC cultivar was more enriched in beneficial rhizosphere microbes. This is probably due to the different crops and cultivars used and the mechanism by which resistance breeding influences plant physiology. More work needs to be done with other crop species.
Root exudates are important for regulating and controlling the composition and function of rhizosphere microorganisms. Previous research has shown that plant species, even different genotypes of the same species, may vary in terms of their rhizosphere microbiome composition and root exudates19. In our experiment, the root exudate profiles varied between the FSC and FRC. Interestingly, four small-molecule organic acids (citric acid, fumaric acid, succinic acid, and pyruvic acid) were observed to be the main driving force for the separation of the two exudate patterns and were produced in higher quantities by the FSC compared with the FRC. These small-molecule organic acids reportedly impacted specific beneficial bacterial groups, such as crop growth-promoting rhizobacteria. For example, malic acid and citric acid could chemotactically attract Pseudomonas fluorescens WCS365, which could competently colonize tomato roots. Infection of Pseudomonas syringae pv. into Arabidopsis could induce root secretion of malic acid and thus promote colonization and biofilm formation on the root surface by strain FB177,33. Similarly, watermelon roots could secrete citric acid and malic acid to induce root colonization of the plant growth-promoting rhizobacterial strain Paenibacillus polymyxa SQR-2134. In our experiment, the four small-molecule organic acids added to the soil enriched the relative abundance of Comamonadaceae, an important beneficial bacterial group, and Comamonadaceae strain G43, whose sequence is the same as that of G11, showed a strong inhibitory effect on the growth of Foc. These results showed that FSC enriched more beneficial bacteria by regulating root exudates.
Indeed, plants employ complex defense strategies to protect themselves from infection by pathogens. Some physical structures and chemical components of plants show antidisease effects, such as cell wall keratin, wax deposition, lignin, special pores, water holes, and lenticels and induction of various pathogenesis-related (PR) proteins, such as chitinase and glucanase35. Additionally, defense responses are induced by plants; these responses mainly include the release of various reactive oxygen species, the expression of defense genes and the development of the hypersensitive response (HR)36,37. In recent years, a consensus has gradually been reached in which rhizosphere microorganisms help plants resist pathogens via the secretion of antimicrobial substances, forming biofilms, and via competition for space and nutrients by occupying pathogen niches38.
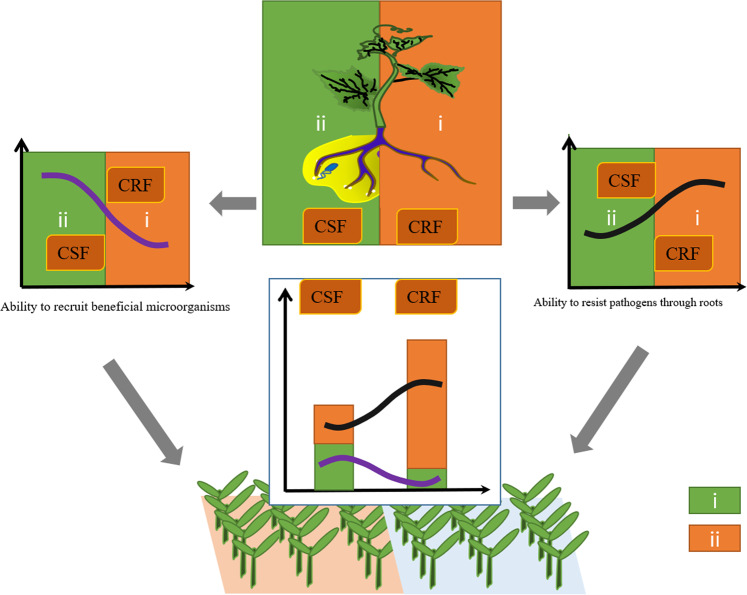
Direct and indirect (via microbial associations) plant pathogen defense strategies coexist. We divided the concept of plant responses to soil-borne pathogens into two aspects: (i) plant resistance, in which plants act as executors by improving physical structures and secreting various new chemical components; and (ii) rhizosphere resistance, in which rhizosphere microorganisms recruited by plants act as executors to confront pathogens by perceiving them, activating the plant immune system, and secreting various effective antimicrobial chemicals (Fig. 5). In this study, two cucumber cultivars displaying different Fusarium wilt resistance were used to assess the level of these two different strategies. The fundamental data indicated that the FRC showed stronger disease resistance than did the FRC (<15% vs >60% of disease incidence), especially plant resistance (Supplementary Table 12), while stronger rhizosphere resistance was found in the FSC than the FRC. In recent years, breeders have been focusing on the role of microorganisms in plant disease resistance. Owing to the complexity of microbial community-plant interactions and uncertain influences of root exudates on rhizosphere microorganisms, compared with strategy i, strategy ii has received less attention by plant breeders. However, improved tools and reduced costs associated with microbiome analyses can add new standards for plant breeding programs. For example, plant phenotypes that target beneficial rhizosphere microbiomes can be evaluated by profiling root exudates or by using community compositional profiles (i.e., diversity and evenness metrics) as criteria to evaluate a cultivar.
Fig. 5. The disease resistance of Fusarium wilt by crops is normally divided into two strategies: strategy i (green) and strategy ii (orange).
These strategies are illustrated here, and the resistance level of the two strategies in the Foc-resistant cucumber and Foc-susceptible cucumber (FSC) are shown. In strategy i (plant resistance), plants act as executors by improving physical structures and secreting various new chemical components. In strategy ii (rhizosphere resistance), rhizosphere microorganisms recruited by plants act as executors to confront pathogens by perceiving pathogens, activating the plant immune system, and secreting various effective antimicrobial chemicals.
Conclusion
In this experiment, using high-throughput sequencing, we determined the rhizosphere microbial communities of two cucumber cultivars with different resistance to Fusarium wilt. The evenness of the rhizosphere microbial community of the Foc-susceptible cucumber cultivar was higher than that of the Foc-resistant cultivar; specifically, the relative abundance of Comamonadaceae and Xanthomonadaceae was greater in the Foc-susceptible cultivar than in the Foc-resistant one. At a lower taxonomic level, higher abundances of the genera Pseudoxanthomonas, Lysobacter, Stenotrophomonas, Pseudomonas, Methylibium, Hydrogenophaga, and Rubrivivax were found in the rhizosphere soil of the Foc-susceptible cultivar compared with the resistant cultivar. As an important medium in the rhizosphere microbial community-plant interaction, different root exudation patterns were found between Foc-resistant and Foc-susceptible cucumber cultivars. The small-molecule organic acids pyruvate acid, citric acid, succinic acid, and fumarate in the cucumber root exudates of susceptible cultivars could recruit Comamonadaceae, which has been shown to be abundant in disease-suppressive soils in many studies. Taken together, our results indicate that the susceptible cucumber cultivar could enrich beneficial microbes (rhizosphere resistance) to compensate for the weakness of the “plant resistance” to pathogens; this overall process may be caused by regulation of the root exudate profile.
Materials and Methods
Soil sampling, plant material, and rhizosphere sampling in the pot experiment
A pot experiment was conducted to evaluate the rhizosphere bacterial communities of two cucumber cultivars with different resistance levels to Fusarium wilt disease. The soil was collected from Baimao town, Changshu city, China (31°35′36.19″N, 120°54′54.93″E, ~300 km away from Nanjing) with no history of cucumber cultivation. The topsoil (20 cm) was air dried, sieved (2-mm sieve) to remove plant debris and rocks, homogenized and stored in plastic bags at room temperature. The seeds of two cucumber cultivars, Foc-resistant cucumber (FRC) Lifeng (disease incidence <15%) and Foc-susceptible cucumber (FSC) B80 (disease incidence >60%), were provided by the Vegetable Research Institute, Guangdong Academy of Agricultural Sciences, China. Before planting, the cucumber seeds were surface sterilized with 75% ethanol for 30 s followed by 5% NaClO for 5 min. The sterilized seeds were placed in Petri dishes with wet autoclaved filter paper in a growth chamber (25 °C, 70% relative humidity in the dark). After 2 days of pregermination, the seedlings were transferred to 200 mL pots (5 cm × 8 cm × 5 cm) filled with 150 g of soil (one seedling per pot). There were 18 seedlings of each cultivar (6 replicates and 3 pots per replication) potted, for a total of 36 pots; the pots were then randomly placed in a growth chamber (28/26 °C day/night cycle, 70% relative humidity, and 180 μmol light m−2 s−1) and irrigated as needed. The plants were killed at the early flowering stage (30 days after planting), and roots with closely attached soil were harvested from the pot after removing the loosely attached soil by shaking. Three cucumber plants of each replicate were pooled into one sample. In total, 12 rhizosphere soil samples were obtained (2 cucumber cultivars × 6 rhizosphere soil samples) and stored at −70 °C for further microbiota analysis.
DNA extraction, 16S rRNA gene amplification and amplicon sequencing
Before DNA extraction, 0.5 g of roots with tightly adhered soil was placed into a 2-mL centrifuge tube containing 1 mL of phosphate-buffered saline solution and several sterilized glass beads, after which the mixture was vortexed at maximum speed for 15 min. The suspension (without root materials) was then transferred to a new 2 mL centrifuge tube and centrifuged for 30 min at 15,000 rpm. The supernatant was discarded, and the precipitate was used for DNA extraction. Total DNA was extracted from the precipitate using a Power Lyzer PowerSoil DNA Isolation Kit (Qiagen, Germany) according to the manufacturer’s protocol. The DNA quality and quantity were measured via a 1.2% agarose gel and a NanoDrop 1000 spectrophotometer (Thermo Scientific, USA).
For taxonomical profiling of the bacterial communities, PCR of the 12 DNA samples that targeted the 16S rRNA gene (V4 region) was performed. The primers 515F/806R (F: GTGYCAGCMGCCGCGGTAA; R: GGACTACNVGGGTWTCTAAT) were used for PCR39, with an amplicon length of ~292 bp. For amplification, 50 μL reaction mixtures consisted of 1 μL of each primer (10 μM), 25 μL of sterilized ultrapure water, 25 μL of 2× Premix Taq (Takara Biotechnology, Dalian Co., Ltd., China), and 3 μL of DNA (20 ng/μL). A Bio-Rad S1000 (Bio-Rad Laboratory, CA) instrument was used to perform PCR amplification with the following amplification procedure: 95 °C for 5 min; 30 cycles of 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 30 s; and then 72 °C for 10 min. The PCR products were run on a 1.2% agarose gel and The DNA marker used was DNA Marker (100–2000 bp; B500350 Sangon Biotech (Shanghai) Co., Ltd.), and those with clear bands between 290 and 310 bp were combined for sequencing. The PCR products were then mixed and sequenced on an Illumina HiSeq 2500 platform with the same sequencing information as reported previously40.
The sequences were assigned to each of samples based on their unique barcode. All the clean reads were trimmed to a minimum length of 200 and had a Phred score of at least 20 by using the split_libraries_fastq.py script (QIIME 1.9.0)41. The reads were clustered into OTUs using the UPARSE strategy42 by dereplication with USEARCH 10. All the reads were mapped to their representative sequences using the usearch_global method (USEARCH 10). The OTU table was converted to BIOM format 1.3.1 using BIOM convert for downstream analysis in QIIME 1.9.0. The Greengenes database (V.13.5) was used for taxonomic annotations with the representative sequences. Summary information of the represented taxonomic groups was generated by the use of the summarize_taxa.py script.
Root exudate collection and GC-TOF-MS detection
The seeds of both cultivars were surface sterilized as mentioned above and then placed in tissue culture bottles (400 mL) containing 100 mL of MS agar media43 in a growth chamber (28/26 °C day/night temperature cycle, 70% relative humidity, and 180 μmol light m−2 s−1) for seven days. Afterward, the sterile seedlings were carefully transferred to conical bottles (one seedling per bottle) containing sterile water and allowed to grow for another seven days. The sterility of the seedlings was tested by plating 100 μL of water from each conical bottle onto an LB44 plate and incubating at 30 °C for 3 days. The contaminated seedlings were discarded. Each cultivar was grown as 3 replicates, with 1 replicate including 3 seedlings, that is, 2 cucumber cultivars × 9 individual seedlings, resulting in 18 samples. All the bottles were fully randomized during the experiment. For cucumber growth, we transferred the sterile cucumber seedlings into sterile Hoagland media under gentle shaking (50 rpm) for 2 h each day on a shaker. To collect root exudates, the seedlings were placed in sterile water for three days, and the container was gently shaken as described above. Finally, nine samples of exudates for each cultivar were obtained, stored immediately at −80 °C, and then dried with a lyophilizer (LGJ- 18 S Beijing Songyuanhuaxing Technology Develop Co., Ltd., China).
For extraction, the lyophilized root exudates (with a dose equal to the amount collected from one cucumber seedling) were put into 2 mL EP tubes and then extracted with 1 mL of extraction liquid (Vmethanol:Vwater = 3:1), after which 10 μL of adonitol (0.5 mg/mL stock in water) was added as an internal standard; the contents was then mixed for 30 s by vortexing. The mixtures were homogenized in a ball mill for 4 min at 45 Hz, ultrasound treated for 5 min (while incubating in ice water), and centrifuged for 15 min at 13,000 rpm and 4 °C, after which the supernatant (0.75 mL) was transferred to a new 2 mL GC/MS glass vial. After completely drying in a vacuum concentrator without heating, 40 μL of methoxy amination hydrochloride (20 mg/mL in pyridine) was added; the sample was then incubated for 30 min at 80 °C, after which 50 μL of the BSTFA (bis(trimethylsilyl) trifluoroacetamide) reagent (1% TMCS (trimethylchlorosilane), v/v) was added, followed by incubation for 1.5 h at 70 °C. The GC-TOF-MS analysis and raw peak analysis were performed as reported by Li et al.45.
Impacts of four small-molecule organic acids present in the root exudates on the soil microbiome
A soil application experiment was conducted to test the effects of four select small-molecule organic acids (SMOAs; pyruvic acid, citric acid, fumaric acid, and succinic acid) on the soil microbiome. These four compounds were abundant in the FSC root exudate samples. The soil used in the experiment was the same soil as that mentioned above. Organic acid solutions in water were prepared such that they contained each of the selected compounds in equal amounts (2.5 mM citric acid, 2.5 mM pyruvic acid, 2.5 mM succinic acid, and 2.5 mM fumaric acid) and at a final total concentration of 10 mM. Before adding the compound mixture, 15 g of soil was placed into each well of six-well plates. The plates were then preincubated in a growth chamber at 30 °C for 1 week to allow the soil microbiome to acclimate for distinguishing seedling rhizospheres from the bulk soil. Each well then received 1.5 ml of compound mixture solution twice a week for 8½ weeks (17 total applications) in a growth chamber at 30 °C40. Sterile ultrapure water was added to each well as a control. Each treatment consisted of 18 wells in three plates. All the plates were randomly arranged during the incubation period. Finally, soils in the 18 wells of each treatment were collected, and three wells were pooled together into one sample. In total, 12 samples for the two treatments (2 treatments × 6 soil samples) were obtained and stored at −80 °C.
For taxonomical profiling of the bacterial communities, 12 samples targeting the V3–V4 region of the 16S rRNA gene were analyzed. The amplification was conducted with the primers 341F/806R (F: CCTAYGGGRBGCASCAG; R: GGACTACHVGGGTWTCTAAT), with an amplicon size of 465 bp. The PCR amplification and 16 S rRNA sequencing were the same as those described above.
Before sequence processing, target sequences were extracted from the raw sequences based on matches to the 515F/806R primers, assuring the same region of the 16 S rRNA gene as that of the rhizosphere samples. Afterward, sequence processing was performed in the same manner as that described above.
Isolation, identification, and in vitro anti-Foc assays
Strains were isolated from the rhizosphere soil of Foc-susceptible and Foc-resistant cucumber varieties by the dilution plate technique. A 0.5-g aliquot of roots with tightly adhering soil was placed into a 2-mL centrifuge tube containing 1 mL of sterile water and then vortexed at maximum speed for 15 min. Then, 100 μL of soil suspension was used for dilution. Specifically, 100 μL of the soil suspension was pipetted into a dilution tube containing 0.9 mL of sterile water. The tube was subsequently vortexed for ~10 s. After vertexing, 0.1 mL of this solution was removed and placed into a second dilution tube containing 0.9 mL of sterile water. TSA agar was used for all plate media.
Strains were also isolated from SMOA-conditioned and control soils by the dilution plate technique. The soil (0.5 g) was mixed with 5 mL of autoclaved water and placed on an orbital shaker for 30 min, after which 100 μL of soil suspension was used for dilution as described above.
We selected plates with fewer than 100 single colonies, and there was a total of three plates used for each treatment. Then, single colonies were selected and purified twice. In total, 238 single colonies from the FSC rhizosphere and 238 from the FRC rhizosphere were isolated. Each of 189 single colonies was isolated from the SMOA-conditioned soil and that of the control treatment.
DNA extraction, amplification, and sequencing were performed as described by Zhang et al.46. The sequences were quality filtered and demultiplexed according to their barcode, and the taxonomy of the sequences was classified using the Greengenes database (V.13.5) as described above.
The antagonistic activity of the isolates against F. oxysporum was evaluated with the method reported by Bordoloi et al.47. Briefly, a 4-mm agar plug of F. oxysporum was placed in the middle of a PDA plate, and the strain was inoculated between the edge of the plate and the plug. The zone of inhibition was measured after incubation at 28 °C for 5 days.
Statistical methods
For downstream analyses after sequence processing, to describe the rhizosphere community structure, a minimum number of sequences was extracted randomly for each sample to calculate the Shannon index estimated via QIIME by the alpha_diversity.py script. A nonparametric t-test was used to determine if the Shannon indices differed between the FRC and FSC. Before calculation of the beta diversity, the cumulative-sum scaling (CSS) method48 was used to standardize the OTU profiles by the normalize_table.py script, and Bray-Curtis similarity matrices were prepared using the beta_diversity.py script. Adonis was used to determine whether the beta diversity differed between the two cultivars. Principal coordinate analysis (PCoA) plots were generated from Bray–Curtis similarity matrices created using the R package ggplot2. To determine the percent change in taxa, we used t-tests for all family- or genus-level taxa with relative abundances >0.001 to measure the significant difference in these abundances between the two cultivars, with the P-values corrected according to the Benjamini–Hochberg method. For functional predictions, the UCLUST method was used to select closed-reference operational taxonomic units49 at 97% sequence similarity using the Greengenes database (V. 13.5). Functional inferences according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database were made using PICRUSt50. Downstream FishTaco analyses were performed according to the methods of David et al.51. Briefly, the top 99 phylotypes with the maximum relative abundance across our OTU table from PICRUSt and normalized with MUSICC were selected. Then, a precomputed OTU-KO table from the PICRUSt analysis, output from MUSICC, and OTU relative abundance table was prepared for input into the FishTaco frame. Multitaxon mode was selected for each pairwise comparison between two cultivar samples. Finally, the R package ggplot was used to visualize the function-contribution distribution.
For root exudate analyses, principal component analysis (PCA)52 was used to visualize the root exudate structure of the two cultivars using the R package vegan. The p-values were corrected by the Benjamini–Hochberg FDR procedure for multiple comparisons53. To identify the exudate compounds with the greatest contribution to the classification, a random forest approach was applied, and the default parameters in the R implementation of the algorithm (R package RandomForest, ntree = 1000) were used. Enrichment analysis of the metabolic pathways was performed and plotted using the online platform MetaboAnalyst54 (http://www.metaboanalyst.ca/faces/home.xhtml). All the plots were created using the R package ggplot2.
Supplementary information
Acknowledgements
This study was financially supported by the Natural Science Foundation of Jiangsu Province (BK20170724), the Natural Science Foundation of China (31902107), the Special Fund for Agro-scientific Research in the Public Interest - Integrated Management Technology of Crop Wilt Disease (No. 201503110), the Innovative Research Team Development Plan of the Ministry of Education of China (Grant No. IRT_17R56), and the Fundamental Research Funds for the Central Universities (Grant Nos. KYT201802, KYXK2020010, and KJQN202017).
Author contributions
T.W. and J.Y. performed all the experiments; J.Y. and Q.H. designed the study and wrote the majority of the manuscript; X.H. and Y.L. provided critical comments on the study and helped write the paper; and Q.S. participated in the design of the study, provided comments, and edited the manuscript.
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive of the BIG Data Center, Chinese Academy of Sciences, under accession code CRA002328.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-00380-3).
References
- 1.Ratnaparkhe MB, Santra DK, Tullu A, Muehlbauer FJ. Inheritance of inter-simple-sequence-repeat polymorphisms and linkage with a fusarium wilt resistance gene in chickpea. Theor. Appl. Genet. 1998;96:348–353. doi: 10.1007/s001220050747. [DOI] [PubMed] [Google Scholar]
- 2.Bakker PA, Berendsen RL, Doornbos RF, Wintermans PC, Pieterse CM. The rhizosphere revisited: root microbiomics. Front. Plant Sci. 2013;4:165. doi: 10.3389/fpls.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berendsen RL, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018;12:1496–1507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwak M-J, et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018;36:1100–1109. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- 5.Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 2002;40:309–348. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 6.Ulloa M, Hutmacher RB, Davis RM, Wright SD, Marsh B. Breeding for fusarium wilt race 4 resistance in cotton under field and greenhouse conditions. J. Cotton Sci. 2006;10:114–127. [Google Scholar]
- 7.De WS, et al. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 2002;15:1173. doi: 10.1094/MPMI.2002.15.11.1173. [DOI] [PubMed] [Google Scholar]
- 8.Zhang N, et al. Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil. 2014;374:689–700. doi: 10.1007/s11104-013-1915-6. [DOI] [Google Scholar]
- 9.Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. Application of natural blends of phytochemicals derived from the root exudates of arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 2013;288:4502–4512. doi: 10.1074/jbc.M112.433300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jousset A, et al. Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol. Plant Microbe Interact. 2011;24:352–358. doi: 10.1094/MPMI-09-10-0208. [DOI] [PubMed] [Google Scholar]
- 11.Huang AC, et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science. 2019;364:546–546. doi: 10.1126/science.aau6389. [DOI] [PubMed] [Google Scholar]
- 12.Badri DV, et al. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 2009;151:2006–2017. doi: 10.1104/pp.109.147462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitter B, et al. A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 2017;8:11. doi: 10.3389/fmicb.2017.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hetrick, B. A. D., Wilson, G. W. T. & Cox, T. S. Mycorrhizal dependence of modern wheat cultivars and ancestors: a synthesis. Can. J. Bot.71, 512–518 (1993).
- 15.Smith KP, Goodman RM. Host variation for interactions with beneficial plant-associated microbes. Annu. Rev. Phytopathol. 1999;37:473–491. doi: 10.1146/annurev.phyto.37.1.473. [DOI] [PubMed] [Google Scholar]
- 16.Engelhard M, Hurek T, Reinhold‐Hurek B. Preferential occurrence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ. Microbiol. 2000;2:131–141. doi: 10.1046/j.1462-2920.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- 17.Karasov TL, et al. Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe. 2018;24:168–179 e164. doi: 10.1016/j.chom.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escudero-Martinez C, Bulgarelli D. Tracing the evolutionary routes of plant–microbiota interactions. Curr. Opin. Microbiol. 2019;49:34–40. doi: 10.1016/j.mib.2019.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Jaramillo JE, Mendes R, Raaijmakers JM. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016;90:635–644. doi: 10.1007/s11103-015-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 21.Bakker MG, Manter DK, Sheflin AM, Weir TL, Vivanco JM. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil. 2012;360:1–13. doi: 10.1007/s11104-012-1361-x. [DOI] [Google Scholar]
- 22.Wissuwa M, Mazzola M, Picard C. Novel approaches in plant breeding for rhizosphere-related traits. Plant Soil. 2008;321:409–430. doi: 10.1007/s11104-008-9693-2. [DOI] [Google Scholar]
- 23.Germida J, Siciliano S. Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fertil. Soils. 2001;33:410–415. doi: 10.1007/s003740100343. [DOI] [Google Scholar]
- 24.Proença DN, et al. The microbiome of endophytic, wood colonizing bacteria from pine trees as affected by pine wilt disease. Sci. Rep. 2017;7:4205. doi: 10.1038/s41598-017-04141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, et al. Declined soil suppressiveness to Fusarium oxysporum by rhizosphere microflora of cotton in soil sickness. Biol. Fertil. Soils. 2015;51:935–946. doi: 10.1007/s00374-015-1038-8. [DOI] [Google Scholar]
- 26.Jack, A. L. et al. Mining and learning from metagenomes plus Workshop on bioinformatic tools. in Thünen Symposium on Soil Metagenomics (DEU, Braunschweig, DEU, 2013).
- 27.Jochum CC, Osborne LE, Yuen GY. Fusarium head blight biological control with Lysobacter enzymogenes strain C3. Biol. Control. 2006;39:336–344. doi: 10.1016/j.biocontrol.2006.05.004. [DOI] [Google Scholar]
- 28.Koberl M, Dita M, Martinuz A, Staver C, Berg G. Members of Gammaproteobacteria as indicator species of healthy banana plants on Fusarium wilt-infested fields in Central America. Sci. Rep. 2017;7:45318. doi: 10.1038/srep45318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figueroa-Lopez AM, et al. Rhizospheric bacteria of maize with potential for biocontrol of Fusarium verticillioides. Springerplus. 2016;5:330. doi: 10.1186/s40064-016-1780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu L, et al. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biol. Biochem. 2017;104:39–48. doi: 10.1016/j.soilbio.2016.10.008. [DOI] [Google Scholar]
- 31.Yao H, Wu F. Soil microbial community structure in cucumber rhizosphere of different resistance cultivars to fusarium wilt. FEMS Microbiol. Ecol. 2010;72:456–463. doi: 10.1111/j.1574-6941.2010.00859.x. [DOI] [PubMed] [Google Scholar]
- 32.Mendes LW, Mendes R, Raaijmakers JM, Tsai SM. Breeding for soil-borne pathogen resistance impacts active rhizosphere microbiome of common bean. ISME J. 2018;12:3038–3042. doi: 10.1038/s41396-018-0234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudrappa T, Czymmek KJ, Pare PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling N, Raza W, Ma J, Huang Q, Shen Q. Identification and role of organic acids in watermelon root exudates for recruiting Paenibacillus polymyxa SQR-21 in the rhizosphere. Eur. J. Soil Biol. 2011;47:374–379. doi: 10.1016/j.ejsobi.2011.08.009. [DOI] [Google Scholar]
- 35.Beardmore J, Ride J, Granger J. Cellular lignification as a factor in the hypersensitive resistance of wheat to stem rust. Physiol. Plant Pathol. 1983;22:209–220. doi: 10.1016/S0048-4059(83)81010-8. [DOI] [Google Scholar]
- 36.Zhang NW, Lindhout P, Niks RE, Jeuken MJW. Genetic dissection ofLactuca salignanonhost resistance to downy mildew at various lettuce developmental stages. Plant Pathol. 2009;58:923–932. doi: 10.1111/j.1365-3059.2009.02066.x. [DOI] [Google Scholar]
- 37.Guo C, et al. Cloning of novel rice blast resistance genes from two rapidly evolving NBS-LRR gene families in rice. Plant Mol. Biol. 2015;90:95–105. doi: 10.1007/s11103-015-0398-7. [DOI] [PubMed] [Google Scholar]
- 38.van Dam NM, Bouwmeester HJ, etc. Metabolomics in the Rhizosphere: tapping into belowground chemical communication. Trends Plant Sci. 2016;21:256–265. doi: 10.1016/j.tplants.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Walters, W. et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems.10.1128/mSystems.00009-15 (2016). [DOI] [PMC free article] [PubMed]
- 40.Yuan J, et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome. 2018;6:156. doi: 10.1186/s40168-018-0537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 43.Skoog F, Murashige T. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 44.Sezonov G, Joseleau-Petit D, d’Ari R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, et al. Insight into the characteristics and new mechanism of nicosulfuron biodegradation by a Pseudomonas sp. LAM1902. J. Agric. Food Chem. 2020;68:826–837. doi: 10.1021/acs.jafc.9b06897. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, et al. NRT1. 1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019;37:676. doi: 10.1038/s41587-019-0104-4. [DOI] [PubMed] [Google Scholar]
- 47.Bordoloi GN, et al. Isolation and structure elucidation of a new antifungal and antibacterial antibiotic produced by Streptomyces sp. 201. Bioscience. Biotechnology. 2001;65:1856–1858. doi: 10.1271/bbb.65.1856. [DOI] [PubMed] [Google Scholar]
- 48.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 50.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Biosci. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casero D, et al. Space-type radiation induces multimodal responses in the mouse gut microbiome and metabolome. Microbiome. 2017;5:105. doi: 10.1186/s40168-017-0325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worley B, Halouska S, Powers R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal. Biochem. 2013;433:102–104. doi: 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 54.Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive of the BIG Data Center, Chinese Academy of Sciences, under accession code CRA002328.