Abstract
TC10-like (TCL) is a small GTPase that has been implicated in carcinogenesis. Elevated TCL expression has been observed in many different types of cancers although the underlying epigenetic mechanism is poorly understood. Here we report that TCL up-regulation was associated with high malignancy in both human colorectal cancer biopsy specimens and in cultured colorectal cancer cells. Hypoxia, a pro-metastatic stimulus, up-regulated TCL expression in HT-29 cells. Further studies revealed that myocardin-related transcription factor A (MRTF-A) promoted migration and invasion of HT-29 cells in a TCL-dependent manner. MRTF-A directly bound to the proximal TCL promoter in response to hypoxia to activate TCL transcription. Chromatin immunoprecipitation (ChIP) assay showed that hypoxia stimulation specifically enhanced acetylation of histone H4K16 surrounding the TCL promoter, which was abolished by MRTF-A depletion or inhibition. Mechanistically, MRTF-A interacted with and recruited the H4K16 acetyltransferase hMOF to the TCL promoter to cooperatively regulate TCL transcription. hMOF depletion or inhibition attenuated hypoxia-induced TCL expression and migration/invasion of HT-29 cells. In conclusion, our data identify a novel MRTF-A-hMOF-TCL axis that contributes to colorectal cancer metastasis.
Subject terms: Epigenetics, Histone post-translational modifications, Metastasis, Cell migration
Introduction
Colorectal carcinoma (CRC) represents one of the leading causes for cancer-related deaths worldwide1. With over 1 million new cases each year, CRC is the third most commonly diagnosed malignancy globally. Despite the development of sophisticated screening techniques that detect early polyps and the use of chemotherapeutic drugs to treat patients under the guidelines of precision medicine, CRC remains the third most lethal form of cancers to which close to 700,000 patients succumb yearly2. The prognosis for those with late-stage and highly malignant CRC is especially dim due to widespread metastasis3. These highly malignant types of CRC are characterized by aggressive migratory and invasive behaviors. A wide range of pro-metastatic stimuli can promote migration and invasion of CRC cells. For instance, hyper-proliferation of cancer cells limits the availability of oxygen supply to create a hypoxic micro-environment. Hypoxia in turn promotes cancer cell migration and invasion via multiple different mechanisms. Hypoxia can stimulate epithelial-mesenchymal transition (EMT) of CRC cells, a process in which cancer cells discard epithelial markers and acquire mesenchymal-like characteristics thus becoming more capable of migration and invasion4–6. Hypoxia can also induce the expression and release of metalloproteinases (MMPs) from CRC cells to remodel extracellular matrix and facilitate migration/invasion7. In addition, hypoxia, via hypoxia-inducible factor 1 (HIF-1α), enhances the canonical Wnt signaling to sustain the stemness of CRC cells and greatly augment migration/invasion8. Another potential mechanism by which hypoxia promotes CRC malignancy is suggested by the observation that PGC-1a, up-regulated by low oxygen content, regulates mitochondrial function to fuel the aberrant migration and invasion of CRC cells.
TC10-like (TCL), also known as RhoJ, is a member of the small GTPase superfamily that shares significant homology with Cdc429. Early characterization of its function revealed that TCL plays a crucial role in adipogenesis likely by activating PPARγ10 and by regulating mitotic clonal expansion11. More recently, it has been demonstrated by independent investigations that TCL may regulate cell mobility. For instance, Heath and colleagues have observed that TCL is located to the focal adhesions in endothelial cells12. Pro-angiogenic stimuli activates TCL, which in turn down-regulates formation of stress fibers and actomyosin contractility to facilitate cell migration. Mechanistically, TCL interacts with the scaffolding protein GIT–PIX to disassemble focal adhesion allowing the cells to move forward13. The observation that TCL activity is positively associated with cell motility has since been confirmed in cornea epithelial cells14 and cancer cells15,16.
Mounting evidence suggests that TCL expression is elevated and correlated with poor prognosis in patients with malignant types of cancers15–18. We have previously reported that TCL mediates hypoxia-induced endothelial-mesenchymal transition by activating the H3K4 methyltransferase WDR519. It remains unknown whether TCL expression levels are correlated with CRC malignancy and, if so, how TCL expression is regulated. Here we provide evidence to show that the histone acetyltransferase hMOF, recruited by the transcriptional modulator MRTF-A, mediates hypoxia-induced TCL expression to promote CRC cell migration and invasion. Therefore, targeting this novel MRTF-A-hMOF-TCL axis may yield novel therapeutic solutions against colorectal cancer metastasis.
Results
High TCL expression correlates with CRC malignancy
We first evaluated the correlation between TCL expression levels and colorectal cancer malignancies in a small cohort of patients. As shown in Fig. 1a, b, TCL expression was significantly up-regulated as CRC progressed from a low-malignant type (grade I&II) to a high-malignant type (grade III&IV). We also compared TCL expression levels in a panel of CRC cell lines with varying metastatic abilities. TCL expression, at both mRNA (Fig. 1c) and protein (Fig. 1d) levels, were much higher in SW480 cells and Caco-2 cells than in HT-29 cells and HCT116 cells. Exposure to hypoxia, a well-established pro-metastatic stimulus, significantly up-regulated TCL expression in HT-29 cells (Fig. 1e, f) and HCT116 (Fig. S1) cells. More important, Kaplan–Meier analysis using data from the TCGA database with the help of a bioinformatics web server (http://gepia.cancer-pku.cn) revealed that high TCL expression was associated with poorer disease-free survival in patients with CRC (Fig. 1g). Together, these data suggest that high TCL expression may be correlated with CRC malignancy.
Fig. 1. High TCL expression correlates with CRC malignancy.
a, b Gene expression levels in human CRC specimens are examined by qPCR and western blotting. c, d TCL expression levels in a panel of CRC cells were examined by qPCR and western. e, f HT-29 cells were exposed to 1% O2 and harvested at indicated time points. TCL expression levels were examined by qPCR and western. Data represent averages of three independent experiments and error bars represent SEM. g Kaplan–Meier plot of survival in patients with high and low TCL expression.
MRTF-A contributes to CRC cell migration and invasion by regulating TCL expression
Myocardin-related transcription factor A (MRTF-A) is a transcriptional modulator that has been implicated in carcinogenesis. Similar to TCL, MRTF-A levels were also higher in more malignant forms of human CRC specimens (Fig. 1b). Over-expression of a constitutively active (CA) form of MRTF-A promoted the migration and invasion of TCL-low HT-29 cells (Fig. 2a, b) and HCT116 cells (Fig. S2A, B). Of interest, depletion of endogenous TCL blocked the augmentation of cell migration/invasion by MRTF-A. In addition, MRTF-A knockdown weakened migration/invasion of HT-29 cells (Fig. 2c, d) and HCT116 cells (Fig. S2C, D) exposed to hypoxia stimulation whereas forced expression of exogenous TCL restored cell migration/invasion. On the contrary, depletion of MRTF-A or TCL with siRNAs similarly attenuated the migration (Fig. S3A) and invasion (Fig. S3B) of SW480 cells and Caco-2 cells.
Fig. 2. MRTF-A contributes to CRC cell migration and invasion by regulating TCL expression.
a A constitutively active (CA) MRTF-A was transfected into HT-29 cells with or without siRNA targeting TCL. Cell migration was measured by scratch wound healing assay and quantified by Image Pro as described in Methods. b A constitutively active (CA) MRTF-A was transfected into HT-29 cells with or without siRNA targeting TCL. Cell migration was measured by transwell assay and quantified by Image Pro as described in Methods. c HT-29 cells were transfected with siRNA targeting MRTF-A or scrambled siRNA (SCR) in the presence or absence of TCL followed by treatment with 1% O2 for 48 h. Cell migration was measured by scratch wound healing assay and quantified by Image Pro as described in Methods. d HT-29 cells were transfected with siRNA targeting MRTF-A or scrambled siRNA (SCR) in the presence or absence of TCL followed by treatment with 1% O2 for 48 h. Cell invasion was measured by transwell assay and quantified by Image Pro as described in Methods. e, f HT-29 cells were transfected with siRNA targeting MRTF-A or scrambled siRNA (SCR) followed by treatment with 1% O2 for 48 h. Gene expression levels were examined by qPCR and western. g, h HT-29 cells were treated with 1% O2 in the presence or absence of CCG-1423 for 48 h. Gene expression levels were examined by qPCR and western. Data represent averages of three independent experiments and error bars represent SEM.
These data suggest that MRTF-A may contribute to CRC cell migration and invasion by regulating TCL expression. In support of this notion, MRTF-A depletion by two separate pairs of siRNAs markedly suppressed TCL induction by hypoxia in HT-29 cells (Fig. 2e, f) and HCT116 cells (Fig. S2E, F). In the TCL-high SW480 (Fig. S3C, D) and Caco-2 (Fig. S3E, F) cells, MRTF-A knockdown similarly repressed basal TCL expression. Further, treatment with CCG-1423, a small-molecule inhibitor of MRTF-A, also attenuated TCL induction by hypoxia (Fig. 2g, h).
MRTF-A directly regulates TCL transcription
Next, we assessed the possibility that MRTF-A may regulate TCL expression at the transcription level. To this end, a luciferase reporter gene fused to the minimal TCL promoter20 (−100/+142) was transfected into HEK293 cells. Over-expression of MRTF-A dose-dependently activated the TCL promoter (Fig. 3a). On the contrary, a dominant-negative MRTF-A that lacks the trans-activation domain21 repressed the TCL promoter (Fig. 3b). In keeping with these observations, inhibition of MRTF-A activity by CCG-1423 also down-regulated the TCL promoter activity (Fig. 3c). Chromatin immunoprecipitation (ChIP) assay confirmed that hypoxia stimulation promoted the binding of MRTF-A to the proximal TCL promoter (−132/+25) but not the distal TCL promoter (−1018/−872), which contains no known TF binding motifs, in HT-29 cells (Fig. 3d). These data combined suggest that MRTF-A may directly bind to the TCL promoter to activate transcription.
Fig. 3. MRTF-A directly regulates TCL transcription.
a HEK293 cells were transfected with a TCL promoter-luciferase construct and increasing doses of MRTF-A. Luciferase activities were normalized by protein concentration and GFP fluorescence and expressed as relative luciferase unit compared to the control group. b HEK293 cells were transfected with a TCL promoter-luciferase construct and increasing doses of dominant-negative (DN) MRTF-A. Luciferase activities were normalized by protein concentration and GFP fluorescence and expressed as relative luciferase unit compared to the control group. c HEK293 cells were transfected with a TCL promoter-luciferase construct and increasing doses of CCG-1423. Luciferase activities were normalized by protein concentration and GFP fluorescence and expressed as relative luciferase unit compared to the control group. d HT-29 cells were exposed to 1% O2 and harvested at indicated time points. ChIP assays were performed with anti-MRTF-A or IgG. Data represent averages of three independent experiments and error bars represent SEM.
MRTF-A modulates histone H4K16 acetylation surrounding the TCL promoter
When HT-29 cells were exposed to hypoxia, there were changes in histone acetylation surrounding the TCL promoter. Abundant H3 acetylation was already associated with the proximal TCL promoter under normoxic conditions and hypoxia did not alter H3 acetylation (Fig. 4a). On the contrary, H4 acetylation was significantly up-regulated by hypoxia stimulation surrounding the proximal, but not the distal, TCL promoter (Fig. 4b). Further analyses revealed that among the four major lysines within the H4 tail, acetylation levels of K5 (Fig. 4c), K8 (Fig. 4d), and K12 (Fig. 4e) were not appreciably changed by hypoxia surrounding the TCL promoter. H4K16 acetylation, on the other hand, was significantly higher after than before hypoxia exposure (Fig. 4f). Both MRTF-A knockdown by siRNAs (Fig. 4g) and MRTF-A inhibition (Fig. 4h) led to diminished accumulation of H4K16 acetylation surrounding the TCL promoter. These data suggest that H4K16 acetylation may be the rate-limiting step in hypoxia-induced TCL transcription and that MRTF-A may play an essential role in this process.
Fig. 4. MRTF-A modulates histone H4K16 acetylation surrounding the TCL promoter.
a–f HT-29 cells were exposed to 1% O2 and harvested at indicated time points. ChIP assays were performed with anti-acetyl H3 (a), anti-acetyl H4 (b), anti-acetyl H4K5 (c), anti-acetyl H4K8 (d), anti-acetyl H4K12 (e), and anti-acetyl H4K16 (f). g HT-29 cells were transfected with siRNA targeting MRTF-A or scrambled siRNA (SCR) followed by treatment with 1% O2 for 48 h. ChIP assays were performed with anti-acetyl H4K16. h HT-29 cells were treated with 1% O2 in the presence or absence of CCG-1423 for 48 h. ChIP assays were performed with anti-acetyl H4K16. Data represent averages of three independent experiments and error bars represent SEM.
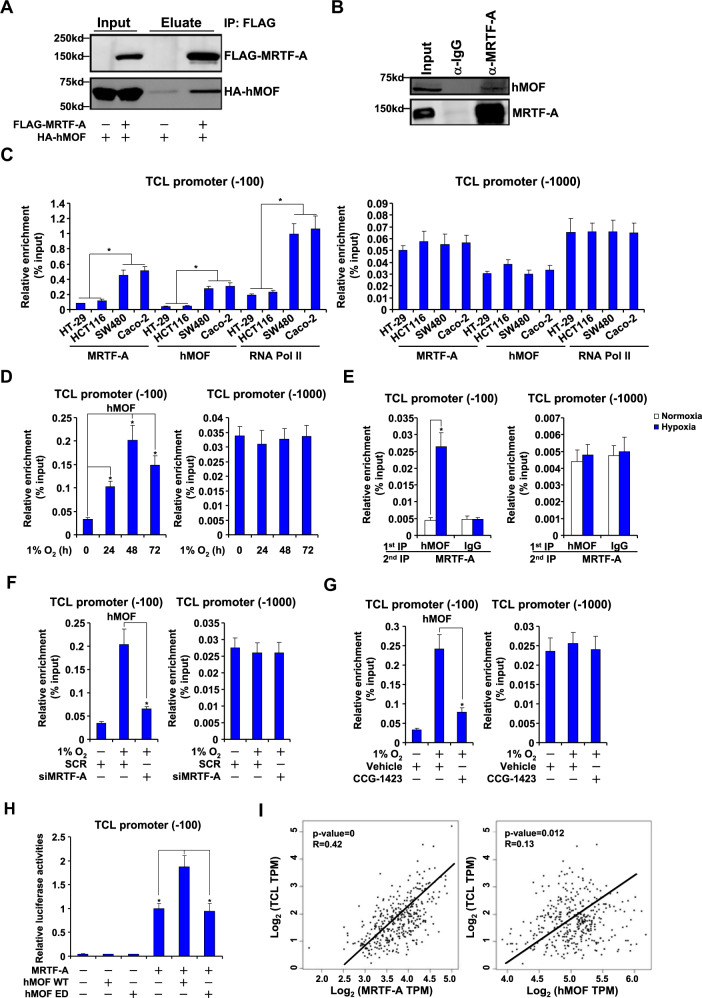
MRTF-A interacts with hMOF to regulate TCL transcription in CRC cells
hMOF is the dedicated H4K16 acetyltransferase in mammals. Based on the observation that MRTF-A deficiency was associated with reduced H4K16 acetylation surrounding the TCL promoter, we hypothesized that an interplay between MRTF-A and hMOF may be accountable. Several lines of evidence supported this hypothesis. First, when exogenous FLAG-MRTF-A and HA-hMOF were over-expressed in HEK293 cells, co-immunoprecipitation assay showed that an anti-FLAG antibody precipitated both MRTF-A and hMOF (Fig. 5a). In addition, co-immunoprecipitation assay also showed that endogenous MRTF-A and hMOF interacted with each other in HT-29 cells (Fig. 5b). Second, ChIP assay showed that MRTF-A bound to the TCL promoter with higher affinity in SW480 cells and Caco-2 cells than in HT-29 cells and HCT116 cells; hMOF displayed a similar binding pattern as MRTF-A (Fig. 5c). Of interest, higher occupancies of RNA polymerase II (Pol II) were detected in SW480 cells and Caco-2 cells than in HT-29 cells and HCT116 cells (Fig. 5c), confirming altered expression of TCL in these cells likely stems from differential transcription rate. In addition, hypoxia exposure stimulated the recruitment of Pol II to the TCL promoter in HT-29 cells (Fig. S4). MRTF-A knockdown suppressed the binding of RNA Pol II to the TCL promoter in SW480 cells and Caco-2 cells (Fig. S5) and in hypoxia-exposed HT-29 cells (Fig. S4) suggesting that MRTF-A may contribute to the regulation of TCL expression by influencing RNA polymerase II access to the promoter and transcription initiation.
Fig. 5. MRTF-A interacts with hMOF to regulate TCL transcription in CRC cells.
a HEK293 cells were transfected with indicated expression constructs. Immunoprecipitation was performed with anti-FLAG or anti-HA. b Nuclear lysates were extracted from HT-29 cells and immunoprecipitation was performed with indicated antibodies. c Nuclear lysates were extracted from a panel of different cells and ChIP assay was performed with anti-MRTF-A or anti-hMOF. d HT-29 cells were exposed to 1% O2 and harvested at indicated time points. ChIP assay was performed with indicated antibodies. e HT-29 cells were exposed to 1% O2 for 48 h. Re-ChIP assay was performed with indicated antibodies. f HT-29 cells were transfected with siRNA targeting MRTF-A or scrambled siRNA (SCR) followed by treatment with 1% O2 for 48 h. ChIP assay was performed with anti-hMOF. g HT-29 cells were treated with 1% O2 in the presence or absence of CCG-1423 for 48 h. ChIP assay was performed with anti-hMOF. h HEK293 cells were transfected with a TCL promoter-luciferase construct, MRTF-A, and/or hMOF. Luciferase activities were normalized by protein concentration and GFP fluorescence and expressed as relative luciferase unit compared to the control group. Data represent averages of three independent experiments and error bars represent SEM. i Expression data of MRTF-A, hMOF and TCL were extracted from the TCGA database to draw the scatter plot. Pearson correlation co-efficient was calculated.
Hypoxia treatment promoted the binding of hMOF on the TCL promoter in HT-29 cells with a similar kinetics as MRTF-A (Fig. 5d). More importantly, hypoxia enhanced the interaction between MRTF-A and hMOF on the TCL promoter in HT-29 cells (Fig. 5e) and HCT116 cells (Fig. S6). Third, either MRTF-A depletion (Fig. 5f) or MRTF-A inhibition (Fig. 5g) abrogated hMOF binding to the TCL promoter induced by hypoxia. Fourth, co-expression of hMOF with MRTF-A activated the TCL promoter more strongly than over-expression of MRTF-A alone (Fig. 5h). Finally, using data extracted from the TCGA database, a positive correlation was detected between TCL and MRTF-A or hMOF in patients with colorectal cancer (Fig. 5i). Combined, these data suggest that MRTF-A may rely on hMOF to regulate TCL transcription in CRC cells.
hMOF contributes to CRC malignancy by regulating TCL transcription
Two strategies were exploited to verify whether hMOF may be essential for hypoxia-induced TCL expression in CRC cells. First, siRNAs were used to knock down hMOF expression, which was accompanied by attenuation of hypoxia-induced TCL expression (Fig. 6a, b). ChIP assay showed that hMOF knockdown blocked the accumulation of H4K16 acetylation surrounding the TCL promoter following hypoxia stimulation (Fig. 6c). We next determined whether hMOF may be functionally relevant in CRC malignancy. Quantitative PCR showed that hMOF expression was elevated in highly malignant CRC specimens compared to those with a lesser malignant grade (Fig. 1c). Wound healing and transwell assays confirmed that hMOF depletion weakened migration (Fig. 6d) and invasion (Fig. 6e) of HT-29 cells exposed to hypoxia.
Fig. 6. hMOF contributes to CRC malignancy by regulating TCL transcription.
a–e HT-29 cells were transfected with siRNA targeting hMOF or scrambled siRNA (SCR) followed by treatment with 1% O2 for 48 h. Gene expression levels were examined by qPCR (a) and western (b). ChIP assay was performed with anti-acetyl H4K16 (c). Cell migration (d) and invasion (e) were measured by scratch wound healing assay and transwell assay, respectively. f–j HT-29 cells were treated with 1% O2 in the presence or absence of MG149 for 48 h. Gene expression levels were examined by qPCR (f) and western (g). ChIP assay was performed with anti-acetyl H4K16 (h). Cell migration (i) and invasion (j) were measured by scratch wound healing assay and transwell assay, respectively. Data represent averages of three independent experiments and error bars represent SEM. k A schematic model.
Alternatively, we used a small-molecule hMOF inhibitor MG149 to treat HT-29 cells. MG149 treatment not only repressed the induction of TCL expression by hypoxia (Fig. 6f, g) and reduced the accumulation of H4K16 acetylation surrounding the TCL promoter (Fig. 6j), but mitigated the aggressive migratory and invasive behaviors of hypoxia-treated HT-29 cells (Fig. 6h, i). Collectively, these data clearly demonstrate that hMOF contributes to CRC malignancy by regulating TCL transcription.
Discussion
The past decade has witnessed the birth of cancer epigenomics along with great expansion in both scope and depth of our understanding with regard to the epigenetic regulation of cancer development and progression22. Here we present evidence to show that the histone H4K16 acetyltransferase hMOF, through its interaction with MRTF-A, activates the transcription of the small GTPase TCL to promote colorectal cancer cell migration and invasion (Fig. 6k). Mounting evidence suggests that hMOF may play a critical role in cancer biology. For instance, Jaganathan et al. observed that augmented proliferation of prostate cancer cells can be accounted for, at least in part, by hMOF potentiating NF-κB and androgen receptor (AR)-dependent transcription23. It has been reported by Chen et al. that low hMOF expression is associated with better prognosis in patients with non-small cell lung cancer24. Mechanistically, hMOF directly acetylates a non-histone transcription factor Nrf2 to up-regulate anti-drug genes. Alternatively, Zhao et al. have demonstrated that hMOF activates the transcription of Skp2, an E3 ubiquitin ligase that drives cell cycling, to promote NSCLC tumorigenesis25. Several independent studies have pointed to a role for hMOF in facilitating DNA repair in cancer cells to avoid growth arrest and apoptosis26–28. Consistent with our results as summarized here, Giampieri et al. have shown that hMOF, along with a panel of other genes, can be used to predict relapse in high-risk CRC patients29. Recently, genomewide gene expression analysis in mouse embryonic fibroblast (MEF) cells suggests that hMOF regulates the transcription of genes involved in stress response30, but it remains to be determined whether hMOF directly binds to the promoters to regulate transcription or whether hMOF regulates transcription through influencing the acetylation levels of H4K16 or non-histone factors. Future studies exploiting the ChIP-seq technique would provide more insights on the regulatory role of hMOF in CRC pathogenesis.
We show here that TCL promotes CRC cell migration and invasion (Fig. 2) and that TCL levels can be used as a predictor of CRC malignancies (Fig. 1). The mechanism whereby TCL regulates CRC metastasis remains an open question. Hypoxia can stimulate colorectal cancer cell migration and invasion by promoting angiogenesis31, epithelial-mesenchymal transition (EMT)4, and cancer stem cell (CSC) self-renewal32. It has been previously demonstrated that TCL blockade leads to failed angiogenesis during breast cancer metastasis18. We have previously shown19 that TCL is essential for hypoxia-induced endothelial-mesenchymal transition (EndMT), a pathophysiological process that shares many similar regulatory molecules with EMT, by regulating the activity of TWIST1 and SNAIL, two transcription factors with well-documented roles in promoting CRC metastasis33. There have also been indications that TCL might be involved in the regulation of stemness in different cells34–36. Additional investigations are warranted to sort out these lingering issues.
There is increasing evidence to suggest that MRTF-A plays a key role in carcinogenesis including lung cancer37, breast cancer38, and hepatocellular cancer39,40. Here we show that MRTF-A contributes to hypoxia-induced CRC cell migration and invasion by recruiting hMOF to activate TCL transcription. Of note, MRTF-A has been shown to interact with a plethora of epigenetic factors to promote carcinogenesis. For instance, an interaction between MRTF-A and the histone H3K4 methyltransferase complex COMPASS results in activation of MMP9 transcription and augmented migration/invasion of lung cancer cells37. Jehanno et al. have observed that a dynamic association between MRTF-A activity and H3K9 methylation status may account for breast cancer metastasis41. In addition, recruitment of histone acetyltransferase CBP/p300 by MRTF-A has been found to be a rate-limiting step in metastasis-related transcriptional events in cancer cells38,42,43. The chromatin remodeling protein BRG1 has been demonstrated to mediate MRTF-A-dependent trans-activation of MMP2 gene in ovarian cancer cells44. Crosstalk between the various MRTF-A-interacting epigenetic factors has been noted. BRG1, for example, is associated with both an H3K4 methyltransferase activity and an H3K9 demethylase activity likely by interacting with COMPASS45,46 and JMJD1A47, respectively. MRTF-A appears to play a brokering role central to the communications between these factors45. Whether hMOF can form a dialog with other epigenetic factors to cooperatively up-regulate TCL transcription and CRC cell migration/invasion awaits further investigation.
In summary, our data suggest that an MRTF-A-hMOF-TCL axis may contribute to colorectal cancer metastasis. It should be noted that these data are largely based on cell culture experiments and a rather small number of human specimens. Future studies should aim to validate our finding by using large cohorts of human data and by exploiting multiple clinically-relevant animal models. Small-molecule inhibitors targeting MRTF-A or hMOF have been found effective in the intervention of a host of diseases in animal models48,49. On other hand, specific TCL inhibitors are not available. Our findings therefore provide a strong rationale for the development of novel TCL-targeting reagents for the treatment of malignant colorectal cancers.
Materials and methods
Human colorectal cancer samples
All human studies were reviewed and approved by the intramural Nanjing Medical University Committee on Ethical Conduct of Studies with Human Subjects. Colorectal cancer tissues were collected, under informed consent, from surgical resection specimens of patients who had not undergone radiotherapy or chemotherapy in the Affiliated Hospital of Nantong University. Diagnoses of all cases were confirmed by histological examination. Tumor differentiation was graded by the Edmondson grading system. Samples were processed essentially as previously described50,51.
Cell culture, plasmids, transient transfection, and reporter assay
Human colorectal cancer cells HT-29, HCT116, SW480, and Caco-2 have been previously described52. The cells were re-authenticated using a fingerprint method every 6 months in the laboratory. Where indicated, hypoxia (1% O2) was achieved by a mixture of ultra-high purity gases (5% CO2, 10% H2, 85% N2) in a 37 oC incubator (Thermo Fisher). MRTF-A expression constructs53,54, hMOF expression constructs48, TCL expression constructs12, and TCL promoter-luciferase constructs20 have been previously described. For transient transfection, cells were plated in 12-well culture dishes (~60,000 cells/well). The next day, equal amounts (0.1 μg) of reporter construct and effector construct were transfected into each well. DNA content was normalized by the addition of an empty vector (pcDNA3). For monitoring transfection efficiency and for normalizing luciferase activity, 0.02 μg of GFP construct was transfected into each well. Raw luciferase activities were divided by both protein concentration and GFP fluorescence. Data are expressed as relative luciferase unit compared to the control group arbitrarily set as 1.
Protein extraction, immunoprecipitation, and western blot
Whole-cell lysates were obtained by re-suspending cell pellets in RIPA buffer (50 mM Tris pH7.4, 150 mM NaCl, 1% Triton X-100) with freshly added protease inhibitor (Roche) as previously described55–59. Specific antibodies or pre-immune IgGs (P.I.I.) were added to and incubated with cell lysates overnight before being absorbed by Protein A/G-plus Agarose beads (Santa Cruz). Precipitated immune complex was released by boiling with 1X SDS electrophoresis sample buffer. Western blot analyses were performed with anti-TCL (Abcam, ab105311), anti-FLAG (Sigma, F3165), anti-HA (Invitrogen, 26183), anti-hMOF (Bethyl Laboratories, A300-992A), anti-MRTF-A (Santa Cruz, sc-32909), and anti-β-actin (Sigma, A2228) antibodies. All experiments were repeated three times.
RNA isolation and real-time PCR
RNA was extracted with the RNeasy RNA isolation kit (Qiagen) as previously described60–64. On-membrane DNase digestion was performed during RNA extraction to eliminate contamination of genomic DNA. Reverse transcriptase reactions were performed using a SuperScript First-strand Synthesis System (Invitrogen). SYBRgreen real-time PCR reactions were performed on an ABI Prism 7500 system with the following primers: TCL, 5′-CGGCTGCAATGGACATGAG-3′ and 5′-GGCACGTATTCCTCTGGGAAG-3′; MRTF-A, 5′-ACCGTGACCAATAAGAATGC-3′ and 5′-CCGCTCTGAATGAGAATGTC-3′; hMOF, 5′- GAAGGAGCATGAGGCGATCA-3′ and 5′-TTTCGTAGTTCCCGATGTGGAT-3′. Ct values of target genes were normalized to the Ct values of a housekeekping control gene (18s, 5′-CGCGGTTCTATTTTGTTGGT-3′ and 5′-TCGTCTTCGAAACTCCGACT-3′) using the ΔΔCt method65 and expressed as relative mRNA expression levels compared to the control group which is arbitrarily set as 1. All experiments were performed in triplicate wells and repeated three times.
Scratch-wound healing/migration assay
Cells were re-suspended in serum-free media. When the cells reached confluence, scratch wound was created by using a sterile micropipette tip. Cell migration was measured 24 h after the creation of the wound and calculated by Image Pro. Data were expressed as % migration compared to control arbitrarily set as 100%.
Boyden chamber invasion assay
24-well inserts (Costar) with 10 μg/ml Matrigel (Sigma) were used for invasion assays. Cells were re-suspended in serum-free media and plated into the upper chamber with the lower chamber filled with complete media. Following exposure to indicated stimuli, the cells on the upper chamber were removed. Invaded cells were stained with 0.1% crystal violet and counted. Data were expressed as % invasion compared to control arbitrarily set as 100%.
Chromatin immunoprecipitation (ChIP)
Chromatin Immunoprecipitation (ChIP) assays were performed essentially as described before47,66–80. In brief, chromatin in control and treated cells were cross-linked with 1% formaldehyde for 8 min at room temperature, and then sequentially washed with ice-cold phosphate-buffered saline, Solution I (10 mM HEPES, pH 7.5, 10 mM EDTA, 0.5 mM EGTA, 0.75% Triton X-100), and Solution II (10 mM HEPES, pH 7.5, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). Cells were then incubated in lysis buffer (150 mM NaCl, 25 mM Tris pH 7.5, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate) supplemented with protease inhibitor tablet and PMSF. DNA was fragmented into 200–500 bp pieces, verified by agarose electrophoresis, using a Branson 250 sonicator. Aliquots of lysates containing 200 μg of protein were used for each immunoprecipitation reaction with anti-acetyl H3 (Millipore, 06-599), anti-acetyl H4 (Millipore, 06-866), anti-acetyl H4K5 (Millipore, 07-327), anti-acetyl H4K8 (Millipore, 07-328), anti-acetyl H4K12 (Millipore, 07-595), anti-acetyl H4K16 (Millipore, 07-329), anti-hMOF (Bethyl Laboratories, A300-992A), anti-MRTF-A (Santa Cruz, sc-32909), or pre-immune IgG followed by adsorption to protein A/G PLUS-agarose beads (Santa Cruz Biotechnology). Precipitated DNA-protein complexes were washed sequentially with RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), high salt buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), LiCl buffer (50 mM Tris, pH 8.0, 250 mM LiCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mM EDTA), and TE buffer (10 mM Tris, 1 mM EDTA pH 8.0). DNA-protein cross-link was reversed by heating the samples to 65 °C overnight. Proteins were digested with proteinase K (Sigma) at 37 °C for 2 h, and DNA was phenol/chloroform-extracted and precipitated by 100% ethanol. For re-ChIP, immune complexes were eluted with the elution buffer (1% SDS, 100 mM NaCO3), diluted with the re-ChIP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris pH 8.1), and subject to immunoprecipitation with a second antibody of interest. Dried DNA was dissolved in 50 μl of deionized distilled water, and 5 μl was used for amplification by real-time PCR with the following primers: TCL proximal promoter, 5′-AGTGGGACCCCTAGTGTTTTC-3′ and 5′-AGGAAATCATGGGTTTCCTG-3′; TCL distal promoter, 5′-GGGTTCCTATAAATACGGACTGC-3′ and 5′-CTGGCACTGCACAAGAAGA-3′. A total of 10% of the starting material is also included as the input. Data are then normalized to the input and expressed as % recovery relative the input. All experiments were performed in triplicate wells and repeated three times.
Statistical analysis
Sample sizes reflected the minimal number needed for statistical significance based on power analysis and prior experience. One-way ANOVA with post-hoc Scheffe analyses were performed using an SPSS package. Unless otherwise specified, P values smaller than 0.05 were considered statistically significant.
Supplementary information
Acknowledgements
This work was supported, in part, by grants from the National Natural Science Foundation of China (81872360, 81700554, and 81702799), from the Nanjing Municipal Administration of Health and Human Services (YKK17061), from the Fundamental Research Funds for Central Universities (021414380323), and from Nanjing Medical Science and Technology Development Foundation (QRX17004 and ZKX17013).
Author contributions
Y.F. and X.F. conceived the project; B.C., Z.F., L.S., J.C., Y.F., X.F., and Y.X. designed experiments; B.C., Z.F., L.S., and J.C. performed experiments and collected and analyzed data; Y.X. wrote the manuscript; Z.F., L.S., and X.F. provided funding.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Baoyu Chen, Zhiwen Fan, Lina Sun, Junliang Chen
Contributor Information
Yifei Feng, Email: fengyifeinjmu@163.com.
Xiangshan Fan, Email: fxs23@hotmail.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41389-020-00269-9).
References
- 1.Fakih MG. Metastatic colorectal cancer: current state and future directions. J. Clin. Oncol. 2015;33:1809–1824. doi: 10.1200/JCO.2014.59.7633. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Riihimaki M, Hemminki A, Sundquist J, Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci. Rep. 2016;6:29765. doi: 10.1038/srep29765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi BJ, Park SA, Lee SY, Cha YN, Surh YJ. Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: A potential role of Sox9. Sci. Rep. 2017;7:15918. doi: 10.1038/s41598-017-15139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hongo K, et al. Hypoxia enhances colon cancer migration and invasion through promotion of epithelial-mesenchymal transition. J. Surgical Res. 2013;182:75–84. doi: 10.1016/j.jss.2012.08.034. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, et al. HIF-1alpha promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PLoS ONE. 2015;10:e0129603. doi: 10.1371/journal.pone.0129603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu S, et al. Hypoxia promotes colorectal cancer cell migration and invasion in a SIRT1-dependent manner. Cancer Cell Int. 2019;19:116. doi: 10.1186/s12935-019-0819-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santoyo-Ramos P, Likhatcheva M, Garcia-Zepeda EA, Castaneda-Patlan MC, Robles-Flores M. Hypoxia-inducible factors modulate the stemness and malignancy of colon cancer cells by playing opposite roles in canonical Wnt signaling. PLoS ONE. 2014;9:e112580. doi: 10.1371/journal.pone.0112580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vignal E, et al. Characterization of TCL, a new GTPase of the rho family related to TC10 andCcdc42. J. Biol. Chem. 2000;275:36457–36464. doi: 10.1074/jbc.M003487200. [DOI] [PubMed] [Google Scholar]
- 10.Nishizuka M, Arimoto E, Tsuchiya T, Nishihara T, Imagawa M. Crucial role of TCL/TC10beta L, a subfamily of Rho GTPase, in adipocyte differentiation. J. Biol. Chem. 2003;278:15279–15284. doi: 10.1074/jbc.M211479200. [DOI] [PubMed] [Google Scholar]
- 11.Kawaji A, Nishizuka M, Osada S, Imagawa M. TC10-like/TC10betaLong regulates adipogenesis by controlling mitotic clonal expansion. Biol. Pharm. Bull. 2010;33:404–409. doi: 10.1248/bpb.33.404. [DOI] [PubMed] [Google Scholar]
- 12.Kaur S, et al. RhoJ/TCL regulates endothelial motility and tube formation and modulates actomyosin contractility and focal adhesion numbers. Arteriosclerosis Thrombosis Vasc. Biol. 2011;31:657–664. doi: 10.1161/ATVBAHA.110.216341. [DOI] [PubMed] [Google Scholar]
- 13.Wilson E, et al. RhoJ interacts with the GIT-PIX complex and regulates focal adhesion disassembly. J. Cell Sci. 2014;127:3039–3051. doi: 10.1242/jcs.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou A, et al. Rho GTPases and regulation of cell migration and polarization in human corneal epithelial cells. PLoS ONE. 2013;8:e77107. doi: 10.1371/journal.pone.0077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho H, et al. RhoJ modulates melanoma invasion by altering actin cytoskeletal dynamics. Pigment Cell Melanoma Res. 2013;26:218–225. doi: 10.1111/pcmr.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C, et al. Rho GTPase RhoJ is associated with gastric cancer progression and metastasis. J. Cancer. 2016;7:1550–1556. doi: 10.7150/jca.15578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho H, et al. RhoJ regulates melanoma chemoresistance by suppressing pathways that sense DNA damage. Cancer Res. 2012;72:5516–5528. doi: 10.1158/0008-5472.CAN-12-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim C, et al. Vascular RhoJ is an effective and selective target for tumor angiogenesis and vascular disruption. Cancer Cell. 2014;25:102–117. doi: 10.1016/j.ccr.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Chen J, Sun L, Xu Y. RhoJ promotes hypoxia induced endothelial-to-mesenchymal transition by activating WDR5 expression. J. Cell. Biochem. 2018;119:3384–3393. doi: 10.1002/jcb.26505. [DOI] [PubMed] [Google Scholar]
- 20.Yuan L, et al. RhoJ is an endothelial cell-restricted Rho GTPase that mediates vascular morphogenesis and is regulated by the transcription factor ERG. Blood. 2011;118:1145–1153. doi: 10.1182/blood-2010-10-315275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cen B, et al. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 23.Jaganathan A, et al. Coactivator MYST1 regulates nuclear factor-kappaB and androgen receptor functions during proliferation of prostate cancer cells. Mol. Endocrinol. 2014;28:872–885. doi: 10.1210/me.2014-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, et al. The histone acetylranseferase hMOF acetylates Nrf2 and regulates anti-drug responses in human non-small cell lung cancer. Br. J. Pharm. 2014;171:3196–3211. doi: 10.1111/bph.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao L, Wang DL, Liu Y, Chen S, Sun FL. Histone acetyltransferase hMOF promotes S phase entry and tumorigenesis in lung cancer. Cell. Signal. 2013;25:1689–1698. doi: 10.1016/j.cellsig.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Li N, Tian GW, Tang LR, Li G. hMOF reduction enhances radiosensitivity through the homologous recombination pathway in non-small-cell lung cancer. Onco. Targets Ther. 2019;12:3065–3075. doi: 10.2147/OTT.S192568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh DK, et al. MOF Suppresses Replication Stress and Contributes to Resolution of Stalled Replication Forks. Mol. Cell. Biol. 2018;38:e00484–17. doi: 10.1128/MCB.00484-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong J, et al. Acetylation of hMOF modulates H4K16ac to regulate DNA repair genes in response to oxidative stress. Int J. Biol. Sci. 2017;13:923–934. doi: 10.7150/ijbs.17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giampieri R, et al. Cancer stem cell gene profile as predictor of relapse in high risk stage II and stage III, radically resected colon cancer patients. PLoS ONE. 2013;8:e72843. doi: 10.1371/journal.pone.0072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheikh BN, et al. MOF maintains transcriptional programs regulating cellular stress response. Oncogene. 2016;35:2698–2710. doi: 10.1038/onc.2015.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zong S, et al. Identification of hypoxia-regulated angiogenic genes in colorectal cancer. Biochemical Biophysical Res. Commun. 2017;493:461–467. doi: 10.1016/j.bbrc.2017.08.169. [DOI] [PubMed] [Google Scholar]
- 32.Ullmann P, et al. Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett. 2019;450:32–41. doi: 10.1016/j.canlet.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 33.Fan XJ, et al. Snail promotes lymph node metastasis and Twist enhances tumor deposit formation through epithelial-mesenchymal transition in colorectal cancer. Hum. Pathol. 2013;44:173–180. doi: 10.1016/j.humpath.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 34.Angelos MG, Abrahante JE, Blum RH, Kaufman DS. Single cell resolution of human hematoendothelial cells defines transcriptional signatures of hemogenic endothelium. Stem Cells. 2018;36:206–217. doi: 10.1002/stem.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JJ, et al. Discovery of consensus gene signature and intermodular connectivity defining self-renewal of human embryonic stem cells. Stem Cells. 2014;32:1468–1479. doi: 10.1002/stem.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peffer ME, et al. Caveolin-1 regulates genomic action of the glucocorticoid receptor in neural stem cells. Mol. Cell. Biol. 2014;34:2611–2623. doi: 10.1128/MCB.01121-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng X, et al. MKL1 potentiates lung cancer cell migration and invasion by epigenetically activating MMP9 transcription. Oncogene. 2015;34:5570–5581. doi: 10.1038/onc.2015.14. [DOI] [PubMed] [Google Scholar]
- 38.He H, et al. Transcriptional factors p300 and MRTF-A synergistically enhance the expression of migration-related genes in MCF-7 breast cancer cells. Biochemical Biophysical Res. Commun. 2015;467:813–820. doi: 10.1016/j.bbrc.2015.10.060. [DOI] [PubMed] [Google Scholar]
- 39.Hermanns C, et al. The novel MKL target gene myoferlin modulates expansion and senescence of hepatocellular carcinoma. Oncogene. 2017;36:3464–3476. doi: 10.1038/onc.2016.496. [DOI] [PubMed] [Google Scholar]
- 40.Muehlich S, et al. The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene. 2012;31:3913–3923. doi: 10.1038/onc.2011.560. [DOI] [PubMed] [Google Scholar]
- 41.Jehanno C, et al. Envisioning metastasis as a transdifferentiation phenomenon clarifies discordant results on cancer. Breast Dis. 2016;36:47–59. doi: 10.3233/BD-150210. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, et al. Histone acetyltransferase p300 promotes MKL1-mediated transactivation of catechol-O-methyltransferase gene. Acta Biochim Biophys. Sin. 2013;45:1002–1010. doi: 10.1093/abbs/gmt108. [DOI] [PubMed] [Google Scholar]
- 43.Shu XZ, et al. Histone acetyltransferase p300 promotes MRTF-A-mediates transactivation of VE-cadherin gene in human umbilical vein endothelial cells. Gene. 2015;563:17–23. doi: 10.1016/j.gene.2015.02.076. [DOI] [PubMed] [Google Scholar]
- 44.Xu W, Xu H, Fang M, Wu X, Xu Y. MKL1 links epigenetic activation of MMP2 to ovarian cancer cell migration and invasion. Biochemical Biophysical Res. Commun. 2017;487:500–508. doi: 10.1016/j.bbrc.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Weng X, et al. A crosstalk between chromatin remodeling and histone H3K4 methyltransferase complexes in endothelial cells regulates angiotensin II-induced cardiac hypertrophy. J. Mol. Cell. Cardiol. 2015;82:48–58. doi: 10.1016/j.yjmcc.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, et al. MRTF-A steers an epigenetic complex to activate endothelin-induced pro-inflammatory transcription in vascular smooth muscle cells. Nucleic Acids Res. 2014;42:10460–10472. doi: 10.1093/nar/gku776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao J, et al. Angiotensin II induced CSF1 transcription is mediated by a crosstalk between different epigenetic factors in vascular endothelial cells. Biochimica et. Biophysica Acta Gene Regul. Mech. 2019;1862:1–11. doi: 10.1016/j.bbagrm.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Yu L, et al. Megakaryocytic leukemia 1 (MKL1) bridges epigenetic activation of NADPH oxidase in macrophages to cardiac ischemia-reperfusion injury. Circulation. 2018;138:2820–2836. doi: 10.1161/CIRCULATIONAHA.118.035377. [DOI] [PubMed] [Google Scholar]
- 49.Zhang B, et al. Role of macrophage migration inhibitory factor in the proliferation of smooth muscle cell in pulmonary hypertension. Mediators Inflamm. 2012;2012:840737. doi: 10.1155/2012/840737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen B, et al. MKL1 mediates TGF-β induced RhoJ transcription to promote breast cancer cell migration and invasion. Front. Cell Dev. Biol. 2020;8:832. doi: 10.3389/fcell.2020.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun L, et al. A SUMOylation-dependent pathway regulates SIRT1 transcription and lung cancer metastasis. J. Natl Cancer Inst. 2013;105:887–898. doi: 10.1093/jnci/djt118. [DOI] [PubMed] [Google Scholar]
- 52.Sun LN, et al. SIRT1 suppresses colorectal cancer metastasis by transcriptional repression of miR-15b-5p. Cancer Lett. 2017;409:104–115. doi: 10.1016/j.canlet.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y, Li Z, Guo J, Xu Y. Deacetylation of MRTF-A by SIRT1 defies senescence induced down-regulation of collagen type I in fibroblast cells. Biochimica et. Biophysica Acta Mol. Basis Dis. 2020;1866:165723. doi: 10.1016/j.bbadis.2020.165723. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, et al. An interplay between MRTF-A and the histone acetyltransferase TIP60 mediates hypoxia-reoxygenation induced iNOS transcription in macrophages. Front. Cell Dev. Biol. 2020;8:484. doi: 10.3389/fcell.2020.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, et al. Dual roles of chromatin remodeling protein BRG1 in angiotensin II-induced endothelial-mesenchymal transition. Cell Death Dis. 2020;11:549. doi: 10.1038/s41419-020-02744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z, et al. BRG1 stimulates endothelial derived alarmin MRP8 to promote macrophage infiltration in an animal model of cardiac hypertrophy. Front. Cell Dev. Biol. 2020;8:569. doi: 10.3389/fcell.2020.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lv F, et al. CDKN2a/p16 antagonizes hepatic stellate cell activation and liver fibrosis by modulating ROS levels. Front. Cell Dev. Biol. 2020;8:176. doi: 10.3389/fcell.2020.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao L, et al. Histone deacetylase 11 contributes to renal fibrosis by repressing KLF15 transcription. Front. Cell Dev. Biol. 2020;8:235. doi: 10.3389/fcell.2020.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao L, Liu L, Zhang T, Wu X, Xu Y. MKL1 mediates TGF-beta-induced CTGF transcription to promote renal fibrosis. J. Cell. Physiol. 2020;235:4790–4803. doi: 10.1002/jcp.29356. [DOI] [PubMed] [Google Scholar]
- 60.Chen B, et al. BRG1 activates PR65A transcription to regulate NO bioavailability in vascular endothelial cell. Front. Cell Dev. Biol. 2020;8:774. doi: 10.3389/fcell.2020.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong W, et al. Activation of TWIST transcription by chromatin remodeling protein BRG1 contributes to liver fibrosis in mice. Front. Cell Dev. Biol. 2020;8:340. doi: 10.3389/fcell.2020.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan Z, et al. Brahma related gene 1 (Brg1) regulates cellular cholesterol synthesis by acting as a Co-factor for SREBP2. Front. Cell Dev. Biol. 2020;8:259. doi: 10.3389/fcell.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li N, et al. Transcriptional activation of matricellular protein Spondin2 (SPON2) by BRG1 in vascular endothelial cells promotes macrophage chemotaxis. Front. Cell Dev. Biol. 2020;8:794. doi: 10.3389/fcell.2020.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Q, et al. Peli1 induction impairs cardiac microvascular endothelium through Hsp90 dissociation from IRE1alpha. Biochimica et. Biophysica Acta Mol. Basis Dis. 2019;1865:2606–2617. doi: 10.1016/j.bbadis.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 65.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 66.Fan Z, et al. An interaction between MKL1, BRG1, and C/EBPbeta mediates palmitate induced CRP transcription in hepatocytes. Biochimica et. Biophysica Acta Gene Regul. Mech. 2019;1862:194412. doi: 10.1016/j.bbagrm.2019.194412. [DOI] [PubMed] [Google Scholar]
- 67.Kong M, et al. Serum response factor (SRF) promotes ROS generation and hepatic stellate cell activation by epigenetically stimulating NCF1/2 transcription. Redox Biol. 2019;26:101302. doi: 10.1016/j.redox.2019.101302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kong M, et al. Ablation of serum response factor in hepatic stellate cells attenuates liver fibrosis. J. Mol. Med. 2019;97:1521–1533. doi: 10.1007/s00109-019-01831-8. [DOI] [PubMed] [Google Scholar]
- 69.Li Z, et al. MKL1 promotes endothelial-to-mesenchymal transition and liver fibrosis by activating TWIST1 transcription. Cell Death Dis. 2019;10:899. doi: 10.1038/s41419-019-2101-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, et al. The chromatin remodeler Brg1 integrates ROS production and endothelial-mesenchymal transition to promote liver fibrosis in mice. Front Dev. Cell Biol. 2019;7:245. doi: 10.3389/fcell.2019.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Z, et al. A non-autonomous role of MKL1 in the activation of hepatic stellate cells. Biochimica et. Biophysica Acta Gene Regul. Mech. 2019;1862:609–618. doi: 10.1016/j.bbagrm.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 72.Li Z, et al. Activation of galectin-3 (LGALS3) transcription by injurious stimuli in the liver is commonly mediated by BRG1. Front. Cell Dev. Biol. 2019;7:310. doi: 10.3389/fcell.2019.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z, Xia J, Fang M, Xu Y. Epigenetic regulation of lung cancer cell proliferation and migration by the chromatin remodeling protein BRG1. Oncogenesis. 2019;8:66. doi: 10.1038/s41389-019-0174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu L, et al. BRG1 regulates endothelial-derived IL-33 to promote ischemia-reperfusion induced renal injury and fibrosis in mice. Biochimica et. Biophysica Acta Mol. Basis Dis. 2019;1865:2551–2561. doi: 10.1016/j.bbadis.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 75.Liu L, Mao L, Xu Y, Wu X. Endothelial-specific deletion of Brahma-related gene 1 (BRG1) assuages unilateral ureteral obstruction induced renal injury in mice. Biochemical Biophysical Res. Commun. 2019;517:244–252. doi: 10.1016/j.bbrc.2019.07.077. [DOI] [PubMed] [Google Scholar]
- 76.Lu Y, et al. A cAbl-MRTF-A feedback loop contributes to hepatic stellate cell activation. Front. Cell Dev. Biol. 2019;7:243. doi: 10.3389/fcell.2019.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weng X, et al. Class II transactivator (CIITA) mediates IFN-gamma induced eNOS repression by enlisting SUV39H1. Biochimica et. Biophysica acta Gene Regul. Mech. 2019;1862:163–172. doi: 10.1016/j.bbagrm.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Yang Y, Liu L, Fang M, Bai H, Xu Y. The chromatin remodeling protein BRM regulates the transcription of tight junction proteins: Implication in breast cancer metastasis. Biochimica et. Biophysica Acta Gene Regul. Mech. 2019;1862:547–556. doi: 10.1016/j.bbagrm.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 79.Yang Y, et al. The chromatin remodeling protein BRG1 links ELOVL3 trans-activation to prostate cancer metastasis. Biochimica et. Biophysica Acta Gene Regul. Mech. 2019;1862:834–845. doi: 10.1016/j.bbagrm.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, et al. An interaction between BRG1 and histone modifying enzymes mediates lipopolysaccharide-induced proinflammatory cytokines in vascular endothelial cells. J. Cell. Biochem. 2019;120:13216–13225. doi: 10.1002/jcb.28595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.