Abstract
In this study, we examined the association between antimicrobial resistance, CRISPR/Cas systems and virulence with phage susceptibility in Acinetobacter baumannii and investigated draft genomes of phage susceptible multidrug resistant A. baumannii strains from Thailand. We investigated 230 A. baumannii strains using 17 lytic A. baumannii phages and the phage susceptibility was 46.5% (107/230). Phage susceptibility was also associated with resistance to numerous antibiotics (p-value < 0.05). We also found association between biofilm formation and the presence of ompA gene among phage susceptible A. baumannii strains (p-value < 0.05). A. baumannii isolates carrying cas5 or combinations of two or three other cas genes, showed a significant increase in phage resistance. Whole-genome sequences of seven phage susceptible A. baumannii isolates revealed that six groups of antibiotic resistance genes were carried by all seven phage susceptible A. baumannii. All strains carried biofilm associated genes and two strains harbored complete prophages, acquired copper tolerance genes, and CRISPR-associated (cas) genes. In conclusion, our data exhibits an association between virulence determinants and biofilm formation among phage susceptible A. baumannii strains. These data help to understand the bacterial co-evolution with phages.
Subject terms: Genetics, Microbiology
Introduction
Acinetobacter baumannii is a major cause of opportunistic infection, especially among immunocompromised patients. The emergence of multidrug-resistant A. baumannii (MDR-AB) and even the extensively drug-resistant A. baumannii (XDR-AB) has been increasing worldwide, and especially in Thailand and Nepal1–3. Thus, alternative treatments against A. baumannii infection is urgently needed. Bacteriophages (phages) are good candidates, specifically killing host bacteria resulting in minimal impact on bacterial normal flora and with no known critical side effects4. To use phages for therapy, it is important to identify broad host range phages that kill the highest possible number of strains of bacterial species. In addition, it is crucial to understand the host-phage susceptibility mechanism. One of the important mechanisms impacting host range specificity is phage adsorption5. This is a crucial step in the infection process, which represents the initial contact between virus and its host and requires phage receptors; outer membrane proteins (OMPs), lipopolysaccharides and teichoic acids5. Several phages use the outer membrane protein OmpA as a receptor to infect Gram negative bacteria6,7. Alterations to this molecule result in a decrease of bacterial virulence in phage resistance strains8. Previous studies reported a positive correlation of antibiotic resistance in A. baumannii with phage susceptibility9,10. Phage susceptibility represents an evolutionary trade-off in A. baumannii strains that were selected for antibiotic resistance, particularly in hospital environments with high antibiotic use10. However, the mechanism and genetic basis of phage susceptibility in A. baumannii is not completely understood. In this study, we aimed to determine the association between antimicrobial resistance and virulence with phage susceptibility in a large collection of A. baumannii strains and to investigate draft genomes of phage susceptible of MDR-AB strains from Thailand to identify antibiotic resistance genes and virulence genes.
Results
Characterization and antimicrobial susceptibility profiles
Among 230 A. baumannii isolates, the resistance to various antibiotics was as follows; amikacin (53.47%), cefotaxime (80.43%), ceftazidime (83.04%), ceftriaxone (83.91%), cefepime (73.91%), ciprofloxacin (85.22%), gentamicin (63.48%), imipenem (82.17%), meropenem (81.74%), trimethoprim/sulfamethoxazole (65.65%), tetracycline (61.74%), cefoperazone/sulbactam (28.70%), piperacillin/tazobactam (82.60%). Among all A. baumannii isolates, 86.52% (199/230) were MDR-AB, 83.49% (192/230) were carbapenem resistant A. baumannii (CR-AB) and 12.17% (28/230) were XDR-AB. A total of 28 (12.17%) isolates were non MDR-AB. All isolates were sensitive to colistin and tigecycline.
Phage susceptibility among MDR-AB, CR-AB, XDR-AB and non MDR-AB
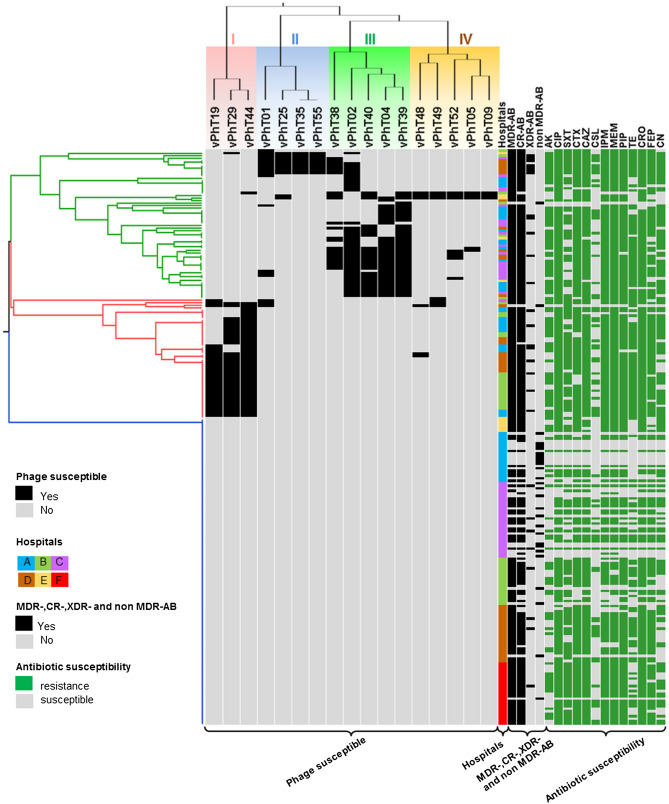
We grouped the 17 phages (Table 1) into four clusters (I, II, III, and IV), depending on their ability to infect 230 A. baumannii isolates (Fig. 1). The vPhT19, vPhT29 and vPhT44 phages belonged to cluster I and infected between 13.91 and 20.43% of bacterial hosts. The phages vPhT01, vPhT25, vPhT35 and vPhT55 were grouped into cluster II, showing between 3.91 and 7.83% infectivity. Phages vPhT02, vPhT04, and vPhT39 showed infectivity against 14.78–18.26% of A. baumannii isolates and belonged to cluster III. Cluster IV contained five phages (vPhT05, vPhT09, vPhT48, vPhT49 and vPhT52), infecting approximately 1.30–3.48% of A. baumannii. Overall phage susceptibility of the 230 A. baumannii strains was 46.5% (107/230). The bacteria were divided into three groups. Group 1 (Fig. 1; green) were the bacteria susceptible to a variety of phages and were mostly infected by phages in cluster II, III and IV. In Group 2 (Fig. 1; red) were the bacteria mostly susceptible to phages in cluster I. In Group 3 (Fig. 1; blue) were the phage resistant bacteria. Among the 199 MDR-AB isolates, 105 (52.76%) were infected specifically by at least one phage. Most of the CR-AB (53.64%) and XDR-AB (71.43%) were infected specifically by at least one phage. Only two of twenty-eight of non MDR-AB (7.14%) were phage susceptible strains (Fig. 1). Of the 230 A. baumannii strains, 205 were isolated from Thailand and 25 were from Nepal. We found 52.2% (107/205) of A. baumannii isolates from Thailand were phage susceptible (Supplementary Table S1). Interestingly, we observed that all 17 phages isolated from Thailand did not infect the 25 Nepalese A. baumannii clinical isolates.
Table 1.
List of bacteriophages used in this study.
| Previous bacteriophage nomenclature | Bacteriophages (full name; abbreviated name) | Source of isolate | A. baumannii host strain | References |
|---|---|---|---|---|
| ØABP-01; PAB01 | vB_AbaP_PhT01; vPhT01 | Waste water HE | A1589a | Kitti33 |
| ØABP-02; PAB02 | vB_AbaM_PhT02; vPhT02 | Waste water HE | A1389a, AB183b | Kitti9,33 |
| ØABP-04; PAB04 | vB_AbaM_PhT04; vPhT04 | Waste water HE | A1522a, AB22b | Kitti33 |
| ØABP-05; PAB05 | vB_AbaX_PhT05; vPhT05 | Waste water HE | A1521a | Kitti9 |
| ØABP-09; PAB09 | vB_AbaX_PhT09; vPhT09 | Waste water HE | A1589a, AB20b | Kitti33 |
| ØABP-19; PAB19 | vB_AbaP_PhT19; vPhT19 | Waste water HE | A1589a | Kitti9,33 |
| ØABP-25; PAB25 | vB_AbaX_PhT25; vPhT25 | Waste water HE | A1589a | Kitti33 |
| ØABP-29; PAB29 | vB_AbaP_PhT29; vPhT29 | Waste water HG | A1589a, AB20b | Kitti9,33 |
| ØABP-35; PAB35 | vB_AbaX_PhT35; vPhT35 | Waste water HG | A1589a | Kitti33 |
| ØABP-38; PAB38 | vB_AbaX_PhT38; vPhT38 | Waste water HE | A1589a | Kitti33 |
| ØABP-39; PAB39 | vB_AbaP_PhT39; vPhT39 | Waste water HE | A1511a, AB22b | Kitti9,33 |
| ØABP-40; PAB40 | vB_AbaX_PhT40; vPhT40 | Waste water HE | A1522a | Kitti33 |
| ØABP-44; PAB44 | vB_AbaM_PhT44; vPhT44 | Waste water HE | ATCC19606a, AB20b | Kitti9,33 |
| ØABP-48; PAB48 | vB_AbaX_PhT48; vPhT48 | Waste water HG | A1522a | Kitti33 |
| ØABP-49; PAB49 | vB_AbaX_PhT49; vPhT49 | Waste water HG | A1521a | Kitti33 |
| ØABP-52; PAB52 | vB_AbaX_PhT52; vPhT52 | Waste water HG | A1522a | Kitti33 |
| ØABP-55; PAB55 | vB_AbaX_PhT55; vPhT55 | Waste water HG | A1589a | Kitti33 |
All phages were isolated in 2010 from Buddhachinaraj hospital (HE) and Bang Rakam hospital (HG), Phitsanulok, Thailand.
aHost strains for phage isolation.
bHost strains for phage propagation.
Figure 1.
UPGMA dendrogram based on phage susceptibility patterns among 230 A. baumannii clinical isolates.) Tree branches represent three bacterial groups of phage susceptibility (Group1: green, Group2 red, Group 3 blue), phage susceptible is displayed as yes (black) and no (gray). Hospitals A-E located in Central, Lower Northern, Northern, East and Northern regions of Thailand, respectively. Hospital F, located in Nepal. Classifications of MDR-, CR-, XDR- and non MDR-AB were presented as yes (black) and no (gray). Antibiotic sensitivity is presented as resistant (green) and susceptible (gray). Antibiotics are abbreviated as follows: AK amikacin, CIP ciprofloxacin, SXT trimethoprim/sulfamethoxazole, CTX cefotaxime, CAZ ceftazidime, CSL cefoperazone/sulbactam, IPM imipenem, MEM meropenem, PIP piperacillin/tazobactam, TE tetracycline, CRO ceftriaxone, FEP cefepime, CN gentamicin.
Association between antibiotics, drug resistance patterns, biofilm formation, REP-PCR typing, copper tolerance, and phage susceptibility
Comparisons of phage susceptibility between antibiotics and drug resistance patterns are shown in Table 2. Phage susceptibility was positively associated with resistance to amikacin, ciprofloxacin, cefotaxime, ceftazidime, cefoperazone/sulbactam, imipenem, meropenem, piperacillin/tazobactam, tetracycline, ceftriaxone, cefepime and gentamicin (p-value < 0.05). We found the association of phage susceptibility and drug resistance across the MDR-AB, CR-AB, XDR-AB and non MDR-AB strains (p-value < 0.05). An analysis of phage susceptibility and biofilm formation also showed association (Table 3). The most common virulence gene associated with biofilm formation, ompA, was detected in 94.39% (101/107) of phage susceptible strains (Supplementary Table S1). We found the association of phage susceptibility and present of ompA gene (p-value < 0.05) (Table3). A repetitive element palindromic-PCR (REP-PCR) analysis of all isolates characterized the phage susceptible A. baumannii into 15 REP-types, while phage resistant A. baumannii belonged to 22 REP-types. The majority REP-types that presented in bacteriophage susceptible strains were R16 (59/107, 55.14%), R4 (18/107, 16.82%) and R24 (9/107, 8.41%), while the majority REP-types that presented in phage resistant strains were R16 (28/123, 22.76%), R12 (23/123, 18.70%) and R1 (10/123, 8.13%) (Supplementary Table S2). Our results showed the relationships between phage susceptibility and the presence of copper tolerance phenotype and genotype (Table 3).
Table 2.
Association between antibiotics, drug resistance patterns and phage susceptibility.
| Antibiotics/drug resistance patterns | No. of phage susceptible isolates | p-values* | |
|---|---|---|---|
| Drug resistant | Drug susceptible | ||
| Antibiotics | |||
| Amikacin (AK) | 80/123 (65.04%) | 27/107 (25.23%) | < 0.001 |
| Ciprofloxacin (CIP) | 104/196 (53.06%) | 3/34 (8.82%) | < 0.001 |
| Trimethoprim/sulfamethoxazole (SXT) | 75/151 (49.67%) | 32/79 (40.51%) | 0.1859 |
| Cefotaxime (CTX) | 96/185 (51.89%) | 11/45 (24.44%) | 0.0009 |
| Ceftazidime (CAZ) | 100/191 (52.35%) | 7/39 (17.95%) | 0.0001 |
| Cefoperazone/sulbactam (CSL) | 38/66 (57.57%) | 69/164 (42.07%) | 0.0330 |
| Imipenem (IPM) | 103/189 (54.49%) | 4/41 (9.76%) | < 0.001 |
| Meropenem (MEM) | 104/188 (55.32%) | 3/42 (7.14%) | < 0.001 |
| Piperacillin/tazobactam (PIP) | 101/190 (53.16%) | 6/40 (15.00%) | < 0.001 |
| Tetracycline (TE) | 89/142 (62.67%) | 18/88 (20.45%) | < 0.001 |
| Ceftriaxone (CRO) | 101/193(52.33%) | 6/37 (16.22%) | 0.0001 |
| Cefepime (FEP) | 89/170 (52.35%) | 18/60 (30.00%) | 0.0028 |
| Gentamicin (CN) | 83/146 (56.85%) | 24/84 (28.57%) | < 0.001 |
| Drug resistance patterns | |||
| MDR-AB | 105/199 (52.76%) | 2/31 (6.45%) | < 0.001 |
| CR-AB | 103/192 (53.64%) | 4/38 (10.53%) | < 0.001 |
| XDR-AB | 20/28 (71.43%) | 87/202 (43.07%) | 0.0048 |
| Non MDR-AB | 2/28 (7.14%) | 105/202 (51.98%) | < 0.001 |
*p-values less than 0.05 were considered as a statistically significant difference (Fisher's exact test). Bold font indicates statistically significant difference between two groups.
Table 3.
Association between biofilm formation, ompA gene and copper tolerance among phage susceptible A. baumannii strains.
| Biofilm formation and copper tolerance | No. of phage susceptible isolates | p-values* | |
|---|---|---|---|
| Positive (Yes) | Negative (No) | ||
| Biofilm formation | |||
| Biofilm formation phenotype | 88/173 (50.87%) | 19/57 (33.33%) | 0.0214 |
| ompA gene | 101/198 (51.01%) | 6/32 (18.75%) | 0.0007 |
| Copper tolerance | |||
| Copper tolerance phenotype | 9/53 (16.98%) | 98/177 (55.37%) | < 0.001 |
| copRS gene | 9/46 (19.56%) | 98/184 (53.26%) | < 0.001 |
*p-values less than 0.05 were considered as a statistically significant difference (Fisher's exact test) and are shown in bold.
Association between CRISPR-associated (cas) genes and phage susceptibility
Across the 230 strains, amplicons of cas1 (506 bp), cas2 (196 bp) and cas3 (850 bp) genes were present in 32 (13.91%), 2 (0.87%) and 30 (13.04%) isolates, respectively. The cas5 gene was found in 34 (14.78%) isolates, while cas6 was detected in 7 (3.04%) isolates. The cas9 gene was not found among any of the A. baumannii isolates. Overall, a total of 44 of the 230 strains were found to contain at least one cas gene. The majority (31/44, 70.5%) of the cas positive strains were classified as phage resistant strains. The correlation between the presence of cas genes and phage susceptibility was statistically determined (p-value < 0.05), showing that only cas5 gene was associated with A. baumannii phage resistance (Table 4). Phage resistant A. baumannii isolates were more than twice as likely to carry two or more cas genes than phage susceptible isolates (resistant; 23/123 (18.70%), susceptible; 9/107 (8.41%),) (p-value < 0.05) (Table 4).
Table 4.
Comparisons of CRISPR-associated (cas) genes between phage susceptible and resistant A. baumannii.
| Characteristics | No. of isolates | p-values* | |
|---|---|---|---|
| Phage susceptible (n = 107) |
Phage resistant (n = 123) |
||
| cas1 positive | 11/107 (10.28%) | 21/123 (17.07%) | 0.1376 |
| cas2 positive | 0/107 (0.00%) | 2/123 (1.67%) | NC |
| cas3 positive | 9/107 (8.41%) | 21/123 (17.07%) | 0.0517 |
| cas5 positive | 8/107 (7.47%) | 26/123 (21.14%) | 0.0036 |
| cas6 positive | 2/107 (1.87%) | 5/123 (4.06%) | NC |
| cas9 positive | 0/107 (0.00%) | 0/123 (0.00%) | NC |
| Positive for two or three cas-types | 9/107 (8.41%) | 23/123 (18.70%) | 0.0245 |
Bold font indicates statistically significant difference between two groups.
NC not comparable.
*p-values less than 0.05 were considered as statistically significant difference (Fisher's exact test).
Genomic sequence analysis of phage susceptible A. baumannii
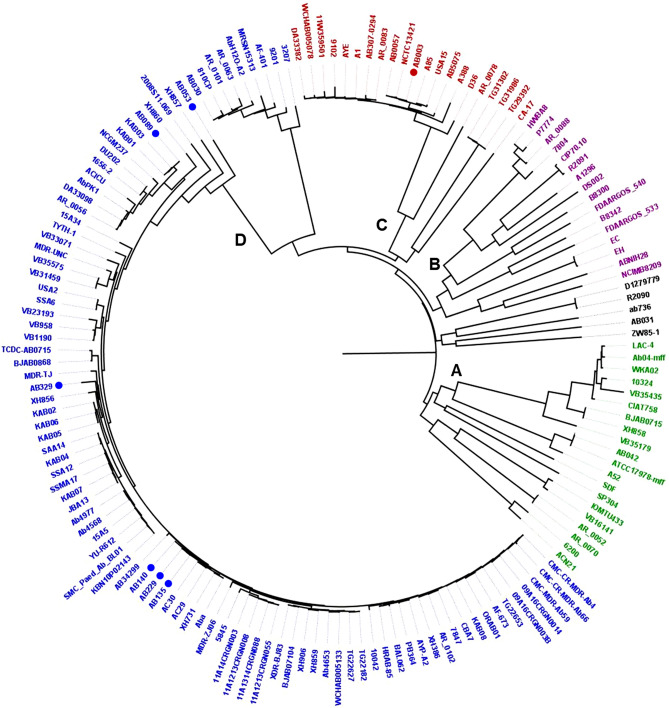
The whole genome of seven phage susceptible A. baumannii strains (AB003, AB053, AB089, AB135, AB140, AB229 and AB329) was sequenced and the result of the genomic sequence analyses are shown in Fig. 2 and Table 5. Antibiotic resistance genes carried by all phage susceptible A. baumannii strains were classified into six groups: sulphonamide (sul1, sul2), tetracycline (tet(B)), β-lactam (blaADC, blaOXA, blaTEM, blaCARB), aminoglycoside (aac(6′)-Ib3, aadA1, ant(2′')-Ia, aph(3′′)-Ib, aph(3′)-Ia, aph(3′)-IIa, aph(3′)-VIa aph(6)-Id, armA), macrolide (mph(E), msr(E)), and phenicol (catB8). Strain AB003, which is highly phage susceptible (9/17, 53%), carried antibiotic resistance genes in two groups: β-lactam and aminoglycoside. Tetracycline, β-lactam, aminoglycoside, and macrolide resistance genes were detected in most phage susceptible A. baumannii (6/7). Multilocus sequence typing (MLST) types 1, 2, 98, and 129 (Pasteur) were investigated in phage susceptible A. baumannii. Two strains (AB003 and AB053) carried one complete prophage each and had acquired copper tolerance genes. Two complete predicted prophages contained genome size ranging from 31.3 to 40.6 kb and GC contents ranging from 39.10 to 40.01%. Incomplete prophage regions were detected in all seven strains. All seven strains carried biofilm associated genes including ompA, adeRS, csuE, gacS, csuCD, bap, and bfmS genes. Plasmid groups, GR1 (1/7), GR2 (7/7), GR6 (4/7), and pRAY (1/7) were detected (Table 5). CRISPR-associated (cas) genes were only detected in two strains, AB003 and AB329, when searched by CRISPRCasFinder. The creation of a whole-genome SNP-based phylogenetic tree using the seven A. baumannii genomes and 149 published A. baumannii genomes indicated that these bacteria are divided into four large clusters, which are designated as clusters A, B, C, and D (Fig. 2). Strain AB003 was classified into cluster C. Other phage susceptible A. baumannii strains from this study were grouped into cluster D (Fig. 2).
Figure 2.
Whole-genome SNP-based phylogenetic tree. A phylogenetic tree base on WGS analysis showing the relationship between seven phage susceptible A. baumannii and 149 published A. baumannii genomes. Colours indicate four major clusters (A = green, B = purple, C = red, and D = blue). Seven phage susceptible A. baumannii strains are marked with dots.
Table 5.
Genome features, acquired antimicrobial resistance genes, putative prophages, copper tolerance, virulence genes, and CRISPR-associated (cas) genes identified in whole genome sequences of phage susceptible A. baumannii.
| Strain ID/Genome characteristics | AB003 | AB053 | AB089 | AB135 | AB140 | AB229 | AB329 |
|---|---|---|---|---|---|---|---|
| Hospital/year isolated | HE/2006 | HA/2013 | HA/2013 | HB/2014 | HB/2014 | HB/2014 | HD/2015 |
| Specimens | Sputum | Sputum | Sputum | Pus | Sputum | Urine | Sputum |
| Phage susceptibility | (9/17) | (2/17) | (3/17) | (3/17) | (4/17) | (3/17) | (6/17) |
| Genome features | |||||||
| Number of assembled contigs | 57 | 142 | 174 | 99 | 77 | 333 | 256 |
| Average genome size (bp) | 4,050,570 | 4,108,360 | 3,897,375 | 4,036,789 | 3,990,813 | 4,070,648 | 3,943,615 |
| GC content (%) | 38.8 | 38.9 | 39.0 | 38.9 | 39.8 | 39.1 | 39.0 |
| Number of CDSs | 3,965 | 4,059 | 3,808 | 3,955 | 3,875 | 4,134 | 3,923 |
| Number of RNAs | 70 | 69 | 70 | 72 | 73 | 68 | 68 |
| MLST/Rep type | ST1/R34 | ST129/R15 | ST2/R4 | ST2/R4 | ST2/R4 | ST2/R4 | ST98/R4 |
| Acquired antimicrobial resistance genes | |||||||
| Sulfonamide resistance | ND | sul1, sul1 | sul2 | sul2 | ND | sul2 | sul2 |
| Tetracycline resistance | ND | tet(B) | tet(B) | tet(B) | tet(B) | tet(B) | tet(B) |
| Beta-lactam resistance | blaADC-25, blaOXA-23, blaOXA-69, blaTEM-181 | blaADC-25, blaOXA-23, blaOXA-66 | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | blaADC-25, blaCARB-16/49/5, blaOXA-23, blaOXA-66, blaTEM-1D | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D | blaADC-25, blaOXA-23, blaOXA-66, blaTEM-1D |
| Aminoglycoside resistance | aph(3′)-IIa | aac(6′)-Ib3, aadA1, ant(2′')-Ia, aph(3′')-Ib, aph(3′)-Ia, aph(6)-Id, armA | aph(3′')-Ib, aph(3′)-Ia, aph(6)-Id, armA | aph(3′')-Ib, aph(3′)-Ia, aph(6)-Id, armA | aph(3′')-Ib, aph(3′)-VIa, aph(6)-Id, armA | aph(3′')-Ib, aph(3′)-Ia, aph(6)-Id, armA | aph(3′')-Ib, aph(3′)-Ia, aph(6)-Id, armA |
| Macrolide resistance | ND | mph(E), msr(E) | mph(E), msr(E) | mph(E), msr(E) | mph(E), msr(E) | mph(E), msr(E) | mph(E), msr(E) |
| Phenicol resistance | ND | catB8 | ND | ND | ND | ND | ND |
| Virulence genes | |||||||
| Iron acquisition | entE, zur | entE, zur | entE, zur | entE, zur | entE, zur | entE, zur | entE, zur |
| Biofilm formation | ompA, adeRS, csuE, gacS, csuCD, bfmS | ompA, adeRS, gacS, bfmS | ompA, adeRS, csuE, gacS, csuCD, bap, bfmS | ompA, adeRS, csuE, gacS, csuCD, bap, bfmS | ompA, csuE, gacS, csuCD, bap, bfmS | ompA, adeRS, csuE, gacS, csuCD, bap, bfmS | ompA, adeRS, csuE, gacS, csuCD, bfmS |
| Type V, VI and IV secretion systems | hcp, traU, traC | ata, hcp | ata, hcp | ata, hcp, traU, traC | ata, hcp, traU, traC | ata, hcp, traU, traC | ata, hcp, traU, traC |
| Other systems | ostA, ompF, rstA | ostA, ompF, rstA | ostA, ompF, rstA | ostA, ompF, rstA | ostA, ompF, rstA | ostA, ompF, rstA | ostA, ompF, rstA |
| Copper tolerance (genotype/phenotype) | copRS+/tolerance | copRS+/tolerance | copRS−/susceptible | copRS−/susceptible | copRS−/susceptible | copRS−/susceptible | copRS−/susceptible |
| Number of prophages (complete/incomplete) | 9 (1/8) | 8 (1/7) | 4 (0/4) | 10 (0/10) | 5 (0/5) | 8 (0/8) | 6 (0/6) |
| Plasmid replicon typing (GR) | GR2, GR6 | GR1, GR2, pRAY | GR2 | GR2, GR6 | GR2, GR6 | GR2, GR6 | GR2 |
| CRISPR-associated (cas) genes | CAS-TypeI (cas6, cas3-cas2, cas1) | Negative | Negative | Negative | Negative | Negative | CAS-TypeI (cas3) |
| Accession no/Bioproject | PRJEB32181 | JABCNM000000000 | JABCNJ000000000 | JABCNK000000000 | JABCNI000000000 | JABCNL000000000 | JABCNH000000000 |
| Reference | Thummeepak et al.35 | This study | This study | This study | This study | This study | This study |
ND not detected.
Discussion
In Thailand, the incidence of MDR-AB, CR-AB and XDR-AB infection has increased in the past decade3,11. Tigecycline and colistin are still last resort drugs of choice for treatment of multidrug-resistant A. baumannii. However, cases of colistin resistance in A. baumannii was reported in Thailand and can cause serious problems in treatment outcome12. Phage therapy is one potential candidate for the treatment of multidrug resistant bacteria. We found that 53.5% of A. baumannii strains isolated from six hospitals in Thailand and Nepal were resistant to all 17 phages tested. This is close to what was found previously by Thawal et al. in 2012, where approximately 51% of A. baumannii strains tested in this study were found to be phage resistant13. We found that A. baumannii strains were more sensitive to phages from the same geographic area, since the Thai phages could not lyse A. baumannii strains isolated from Nepal. This is also consistent with previously published investigation, where one study has reported that phage-host specificity was limited to specific geographic areas14. Host strain AB003, which was highly susceptible to bacteriophages, was isolated from the same hospital (HE), where the phages of this study were isolated. Most of the 26 non MDR-AB (92.8%) strains were classified as phage resistance. Our finding is consistent with previous studies by Kitti et al. and Chen et al. that showed that antibiotic-resistant A. baumannii clinical isolates have higher susceptibility to phages than antibiotic-sensitive strains9,10. The increase in phage susceptibility may be a result of the high antibiotic usage in the hospital environment and co-evolution of phages and bacteria10. Chen et al. proposed that the antibiotic-resistant A. baumannii strains that remained in the hospital could be more easily infected by phages, because of the adaptations for multiple antibiotics resistance, especially in medical environments that have high antibiotic use since the phage resistant strains of A. baumannii displayed diverse RAPD-PCR types, when compared with phage susceptible strains10. Our study found that phage resistant strains of A. baumannii can be classified into more than 20 different REP-types, while 80% of phage susceptible strains can be grouped into just three similar REP-types (R4, R16 and R24). The phage resistant strains were derived from various sources and did not have a common stressor or environmental selector meaning they presented a diversity of REP-type patterns. In contrast, the phage susceptible strains that can infect by at least one phage may be caused by the selective pressure of adopting resistance to multiple antibiotics and limitation of phage immunity.
Bacterial biofilm formation is one strategy mediating protection against phage lysis. However, our study showed positive association between biofilm formation and phage susceptibility. Interestingly, some studies also revealed that phages can induce and strengthen biofilms and it has been shown that biofilm composition and maturation had an impact on susceptibility towards phage infection15. Quorum sensing system, OmpA and OmpC are critical host factors for phage infection7,16,17. The receptor for Shigella flexneri phage Sf6 and E. coli phage M1 is the outer membrane protein A (OmpA). In this study, 94.39% (101/107) of phage susceptible strains A. baumannii harbored the ompA gene. We found positive association between the present of OmpA in phage susceptible strains (Table3). These data imply that OmpA could also be a receptor for A. baumannii phage adsorption.
Bacteria have numerous strategies to prevent bacteriophage infection and cell lysis. The CRISPR/Cas systems are one of the phage immunity systems that are present in bacteria and enabling the organisms to respond to and eliminate invading genetic material18. In 2016 Lin et al. showed that Klebsiella pneumoniae isolated from the hospital environment have a decreased level of phage immunity and a reduction in CRISPR-Cas activity compared to strains isolated from outside the hospital environment19. PCR with cas-specific primers showed that 19% of the A. baumannii isolates had a CRISPR-Cas system. The majority (70.5%) of the cas positive strains were classified as phage resistant strains. The positive correlation between the presence of cas5 or combinations of two or three different cas genes and phage resistant strains was statistically determined (p-value < 0.05). This data supports a hypothesis that CRISPR/Cas systems are one of the important phage resistance mechanisms in A. baumannii. We analyzed whole genome sequence of Acinetobacter spp. deposited on National Center for Biotechnology Information (NCBI) database showed that these contain six cas genes including cas1, cas2, cas3, cas5, cas6 and cas920,21. Cas1 and Cas2 proteins recognize invading phage nucleic acids and insert them into the CRISPR/Cas array as a spacer in order to transcribe into pre-CRISPR-RNA (crRNA)22, whereas, the Cas5 protein has RNase activity, resulting in inhibition of the transcription machinery in phages18. The cas6 gene encodes a type I-F CRISPR-associated endoribonuclease Cas6/Csy4 protein, which has an important role in the cleavage of the repeat sequences to yield mature CRISPR-RNAs (crRNAs) that complementary pair with invading phage nucleic acid bases, causing destruction of the target phage DNA23.
Phage resistance may be related to lower virulence, making the resistant bacteria less virulent than non-phage-resistant strains24. We detected antibiotic resistance, CRISPR-associated (cas) genes and virulence genes among A. baumannii isolates using PCR and found association between phage susceptibility with antibiotic resistance, CRISPR-associated (cas) genes and virulence genes. Whole genome sequencing of virulence genes involving biofilm formation revealed that the seven phage susceptible A. baumannii in this study harbored various virulence genes linked to the ability to form biofilms, iron acquisition and bacterial secretion systems. cas genes (cas1, cas3-cas2, and cas6) were found in two of seven the phage susceptible A. baumannii genomes (AB003 and AB329). Antibiotic resistance genes involved sulphonamide, tetracycline, β-lactam, aminoglycoside, macrolide and phenicol resistance were found in seven phage susceptible A. baumannii genomes. In a previous study, we identified the acquired copper tolerance genes, copRS that respond to copper toxicity in the genome of A. baumannii25. These genes were identified in two of the seven phage susceptible strains and both strains also showed the copper tolerance phenotype. The correlation between phage susceptibility and regulating heavy metal toxicity is in agreement with the findings of Zhang et al., where plasmid-borne cadmium resistant determinants were associated with the susceptibility of Listeria monocytogenes to phages26. Antibiotic resistance genes and heavy metal tolerance genes can be disseminated within the microbial population by horizontal gene transfer mechanisms using plasmids and phages. Among 19 groups of plasmids identified in A. baumannii, in this study, we found plasmids GR1, GR2, GR6, and pRAY in phage susceptible A. baumannii. Plasmid GR6, linked to the dissemination via horizontal gene transfer by conjugation was detected in four strains of phage susceptible A. baumannii27. In contrast, phages are important genetic vehicles for transferring genetic information between bacteria via transduction28. All seven phage susceptible A. baumannii strains in our analysis carried prophage associated sequences on their chromosomes. However, only two strains harbored complete prophages. Prophages are directly related to genome diversity, evolution and strains variation as well as an association with the presence of antibiotic resistance genes and virulence genes29,30. In addition, prophages are responsible for gene disruption or translocation to phenotypic changes in their host and can introduction of pathogenicity determinants that contribute positively to bacterial fitness29,31.
An analysis of the clonal relationship of the seven phage susceptible A. baumannii genomes from this study with 149 previously published A. baumannii genomes (Fig. 2.) revealed that strain AB003, isolated from hospital HE, was closely related to strains AB0057 (CP001182), NCTC13421 (LS483472), A85 (CP021782) and USA15 (CP020595). The three strains isolated from hospital HB (AB140, AB229 and AB135) all belonged to the same ST type and are very closely related with Malaysian and Chinese isolates AC30 (CP007577), AC29 (CP007535), XH731 (CP021321) and Aba (CP030083). The presence of closely related bacterial strains collected from Thailand, Malaysia and China might be descriptive that these organisms shared ancestry and emerged at the same time, caused by rapid dissemination of genetic material.
In conclusion, we found that phage susceptibility was associated with antibiotic resistance and virulence among A. baumannii strains. Moreover, in silico analysis showed that seven strains of phage susceptible A. baumannii carried at least six antibiotic resistance genes which included sulphonamide, tetracycline, β-lactam, aminoglycoside, macrolide, phenicol and several virulence genes involved biofilm formation. Thus, the data from this study can be used as essential information about phage therapy in the future.
Materials and methods
Bacteria and phages used in this study
We used 230 A. baumannii isolates collected from inpatient units of five hospitals in Thailand (HA-HE) and one hospital in Nepal (HF) as described by Niumsup et al., Joshi et al., and Leungtongkam et al.2,3,32 (Supplementary Table S1). Seventeen A. baumannii phages used in this study were isolated from wastewater treatment plants of two hospitals (HE and HG) in Phitsanulok province, Thailand9,33 (Table 1). A. baumannii strains used as the host for phage propagation are shown in Table 1. The protocol was approved by Naresaun University Institutional Biosafety Committee (NUIBC) (No. NUIBC GM 62-06-22).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (2017) with fifteen antibiotics; amikacin, cefepime, cefotaxime, cefoperazone/sulbactam, ceftazidime, ceftriaxone, ciprofloxacin, colistin, gentamicin, imipenem, meropenem, piperacillin/tazobactam, tetracycline, tigecycline and trimethoprim/sulfamethoxazole. Escherichia coli ATCC 25922 was used as a quality control strain. All isolates were defined as being MDR-AB, when there was resistance to more than three antibiotic classes, as carbapenem- resistant A. baumannii (CR-AB), when there was resistance to carbapenems and as XDR-AB, when there was resistance to all antimicrobial agents tested except the polymyxin colistin, and tigecycline. Non MDR-AB classified as bacteria that non-resistant or less resistant than the three antibiotic group34.
Phages susceptibility of A. baumannii
Assessment of phage susceptibility was determined by spot test on all A. baumannii isolates. A colony of bacteria was suspended in 0.85% (w/v) NaCl to the equivalent of a 0.5 McFarland standard (1 × 108 CFU/ml). The suspensions were swabbed on to Trypticase Soy Agar (TSA). Phage suspensions (2 µl at 1 × 108 PFU/ml) were dropped into the bacterial lawn. Then, plates were incubated at 37 °C for 8 h. The result of spot test being clearance at the location of phage inoculation when the host was sensitive to phage (Supplementary Fig. S1). All experiments were performed in duplicate.
Biofilm formation, detection of ompA gene, repetitive element palindromic-PCR (REP-PCR) and copper tolerance
The genetic diversity among phage susceptible A. baumannii isolates was studied using REP-PCR typing as described previously by Leungtongkam et al.3. Detection of ompA gene and biofilm formation were performed as described by Thummeepak et al.35. The copper tolerance phenotype and copper-related genes (copRS) studies were conducted by using Minimum Inhibitory Concentration (MIC) and PCR methods as described previously25.
Detection of CRISPR-associated (cas) genes
The bacterial protein sequences and GenBank nucleotide sequences of well-identified cas genes, detected in six example Acinetobacter spp., were used as templates to design the specific primers using Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Supplementary Table S3). The genomic DNA of all isolates in this study was extracted by a previously described boiling method36 for use as a template in a multiplex-PCR method with cas specific primers (Supplementary Table S3) to detect CRISPR-associated (cas) genes. Conditions for multiplex-1 was the following: 95 °C for 5 min, and then 35 cycles at 94 °C for 30 s, at 62 °C for 40 s, and 72 °C for 50 s, followed by a final extension at 72 °C for 5 min. Multiplex-2 condition was one cycle of 95 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 58 °C for 40 s, and 72 °C for 50 s and finally, one cycle of 72 °C for 5 min. The PCR product was visualized by agarose gel electrophoresis, stained with ethidium bromide in a UV transilluminator.
Whole genome sequencing of A. baumannii
We selected seven phage susceptible A. baumannii strains (Group1 and Group2) that were susceptible to phages from all four clusters, namely AB003 (cluster III), AB053 (cluster IV), AB089 (cluster I), AB135 (cluster I), AB140 (cluster II), AB229 (cluster I) and AB329 (cluster IV) to study the whole genome sequence. Genomic DNA of A. baumannii strains were isolated using HiYield Genomic DNA Mini Kit (RBC Bioscience, New Taipei, Taiwan). Extracted DNA was quantified using a Qubit DNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies, CA, USA). Genomes were sequenced by the Illumina Miseq platform (250 bp paired end). DNA libraries were constructed by Nextera XT DNA Library Preparation Kit according to the manufacturers’ instructions. Reads were trimmed and assembled by using Sickle v1.3337 and SPAdes genome assembler v3.6.038 with default settings, respectively. After assembled contigs, annotation was conducted with RAST pipeline using default parameters39. Single nucleotide polymorphisms (SNPs) phylogenetic analysis was conducted by using CSI Phylogeny v1.4 with default options40 (with reference strain SDF, NC_010400.1). Phylogenetic tree image was visualized and edited by FigTree v1.4.4 (https://tree.bio.ed.ac.uk/software/figtree/). Antibiotic resistance genes in draft genomes were detected by ResFinder tool41. CRISPR-associated (cas) genes within bacterial genomes were detected by using CRISPRCasFinder programs and the prediction of prophage regions genomes were performed by using PHASTER42,43. Prophage hits with status “questionable” were scored as incomplete prophages. The virulence genes were examined by BlastN algorithm using collection of virulence genes as queries.
Statistical analysis
The Fisher’s exact test was used to analyze phage susceptibility associated with the drug resistance patterns, biofilm formation, ompA gene, copper tolerance and CRISPR-associated (cas) genes. Data with a p-value < 0.05 were classed as statistically significant.
Nucleotide sequence accession number
The nucleotide sequences of seven A. baumannii have been deposited to the GenBank database under accession numbers PRJEB32181 (AB003), JABCNM000000000 (AB053), JABCNJ000000000 (AB089), JABCNK000000000 (AB135), JABCNI000000000 (AB140), JABCNL000000000 (AB229), and JABCNH000000000 (AB329).
Supplementary information
Acknowledgements
The authors would like to thank the staffs of six hospitals for collecting the bacterial samples.
Author contributions
S.S., K.T., and J.W. conceived and designed the experiments; U.L., R.T., and T.K. conducted the experiments; U.L., R.T., S.S. A.P.S., K.M.S., and A.D.M. interpretation of data; R.T., U.L., A.D.M., S.S. performed the data and bioinformatic analysis; U.L. and S.S. wrote the manuscript; S.S., A.P.S., K.M.S., A.D.M., and E.M.W edited the manuscript, funding acquisition was done by A.P.S. and S.S. with E.M.W and A.D.M. as collaborators; All authors read and approved the manuscript.
Funding
This work was supported by grants of the Newton Fund Institutional Links to SS (RDG61W0003) and APS, an Institutional Links grant, ID 332371796, under the UK—Thailand Research and Innovation Partnership Fund. The grant is funded by the UK Department for Business, Energy and Industrial Strategy and delivered by the British Council. For further information, please visit www.newtonfund.ac.uk. RT and UL were supported by The Royal Golden Jubilee Ph.D. Program (PHD/0031/2558 and PHD/0227/2560).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-73123-y.
References
- 1.Tal-Jasper R, et al. Clinical and epidemiological significance of carbapenem resistance in Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 2016;60:3127–3131. doi: 10.1128/AAC.02656-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joshi PR, et al. Co-existence of blaOXA-23 and blaNDM-1 genes of Acinetobacter baumannii isolated from Nepal: antimicrobial resistance and clinical significance. Antimicrob. Resist. Infect. Control. 2017;6:1–7. doi: 10.1186/s13756-017-0180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leungtongkam U, et al. Dissemination of blaOXA-23, blaOXA-24, blaOXA-58, and blaNDM-1 genes of Acinetobacter baumannii isolates from four tertiary hospitals in Thailand. Microb. Drug Resist. 2018;24:55–62. doi: 10.1089/mdr.2016.0248. [DOI] [PubMed] [Google Scholar]
- 4.Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011;1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertozzi Silva J, Storms Z, Sauvageau D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016;363:fnw002. doi: 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- 6.Morona R, Krämer C, Henning U. Bacteriophage receptor area of outer membrane protein OmpA of Escherichia coli K-12. J. Bacteriol. 1985;164:539–543. doi: 10.1128/JB.164.2.539-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parent KN, et al. OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol. Microbiol. 2014;92:47–60. doi: 10.1111/mmi.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azam AH, Tanji Y. Bacteriophage-host arm race: An update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 2019;103:2121–2131. doi: 10.1007/s00253-019-09629-x. [DOI] [PubMed] [Google Scholar]
- 9.Kitti T, Thummeepak R, Leungtongkam U, Kunthalert D, Sitthisak S. Efficacy of Acinetobacter baumannii bacteriophage cocktail on Acinetobacter baumannii growth. Afr. J. Microbiol. Res. 2015;9:2159–2165. [Google Scholar]
- 10.Chen LK, et al. Clinical Antibiotic-resistant Acinetobacter baumannii strains with higher susceptibility to environmental phages than antibiotic-sensitive strains. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inchai J, et al. Risk factors of multidrug-resistant, extensively drug-resistant and pandrug-resistant Acinetobacter baumannii ventilator-associated pneumonia in a Medical Intensive Care Unit of University Hospital in Thailand. J. Infect. Chemother. 2015;21:570–574. doi: 10.1016/j.jiac.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Naksena P, Tribuddharat C, Thaipisuttikul I. Colistin resistance in Acinetobacter baumannii isolated in Siriraj Hospital is associated with mutations in pmrCAB operon. Am. J. Med. 2012;64:A21–A21. [Google Scholar]
- 13.Thawal ND, Yele AB, Sahu PK, Chopade BA. Effect of a novel podophage AB7-IBB2 on Acinetobacter baumannii biofilm. Curr. Microbiol. 2012;65:66–72. doi: 10.1007/s00284-012-0127-2. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, et al. Characterization and genome annotation of a newly detected bacteriophage infecting multidrug-resistant Acinetobacter baumannii. Arch. Virol. 2019;164:1527–1533. doi: 10.1007/s00705-019-04213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen MF, Svenningsen SL, Røder HL, Middelboe M, Burmølle M. Big Impact of the Tiny: Bacteriophage-Bacteria Interactions in Biofilms. Trends Microbiol. 2019;27:739–752. doi: 10.1016/j.tim.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Hashemolhosseine S, Holmes Z, Mutschler B, Henning U. Alterations of receptor specificities of coliphages of the T2 family. J. Mol. Biol. 1994;240:105–110. doi: 10.1006/jmbi.1994.1424. [DOI] [PubMed] [Google Scholar]
- 17.Saucedo-Mora MA, et al. Selection of functional quorum sensing systems by lysogenic bacteriophages in Pseudomonas aeruginosa. Front. Microbiol. 2017;8:1669. doi: 10.3389/fmicb.2017.01669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makarova KS, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015;13:722. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin TL, et al. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci. Rep. 2016;6:31644. doi: 10.1038/srep31644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahl JW, et al. Phylogenetic and genomic diversity in isolates from the globally distributed Acinetobacter baumannii ST25 lineage. Sci. Rep. 2015;5:15188. doi: 10.1038/srep15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicoloff H, Hjort K, Levin BR, Andersson DI. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 2019;4:504–514. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell KL. Phages fight back: inactivation of the CRISPR-Cas bacterial immune system by anti-CRISPR proteins. PLOS Pathog. 2016;12:e1005282. doi: 10.1371/journal.ppat.1005282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiedenheft B, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.León M, Bastías R. Virulence reduction in bacteriophage resistant bacteria. Front. Microbiol. 2015;6:343. doi: 10.3389/fmicb.2015.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thummeepak R, et al. Essential gene clusters involved in copper tolerance identified in Acinetobacter baumannii clinical and environmental isolates. Pathogens. 2020;9:60. doi: 10.3390/pathogens9010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, et al. Plasmid-borne cadmium resistant determinants are associated with the susceptibility of Listeria monocytogenes to bacteriophage. Microbiol. Res. 2015;172:1–6. doi: 10.1016/j.micres.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Leungtongkam U, et al. Acquisition and transfer of antibiotic resistance genes in association with conjugative plasmid or class 1 integrons of Acinetobacter baumannii. PLoS ONE. 2018;13:e0208468. doi: 10.1371/journal.pone.0208468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balcazar JL. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLOS Pathog. 2014;10:e100429. doi: 10.1371/journal.ppat.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortier LC, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4:354–365. doi: 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brueggemann AB, et al. Pneumococcal prophages are diverse, but not without structure or history. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niumsup PR, Boonkerd N, Tansawai U, Tiloklurs M. Carbapenem-resistant Acinetobacter baumannii producing OXA-23 in Thailand. Jpn. J. Infect. Dis. 2009;62:152–154. [PubMed] [Google Scholar]
- 33.Kitti T, et al. Characterization and detection of endolysin gene from three Acinetobacter baumannii bacteriophages isolated from sewage water. Indian J. Microbiol. 2014;54:383–388. doi: 10.1007/s12088-014-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 35.Thummeepak R, Kongthai P, Leungtongkam U, Sitthisak S. Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int. Microbiol. 2016;19:121–129. doi: 10.2436/20.1501.01.270. [DOI] [PubMed] [Google Scholar]
- 36.Dashti AA, Jadaon MM, Abdulsamad AM, Dashti HM. Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med J. 2009;41:117–122. [Google Scholar]
- 37.Joshi, N. A. & Fass, J. N. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software]. https://github.com/najoshi/sickle (2011).
- 38.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genom. 2008;9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zankari EH, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couvin D, et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arndt D, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.