Abstract

This work presents a non-targeted high-resolution mass spectrometry inter-laboratory study for the detection of new chemical markers responsible of soft refined oils addition to extra virgin olive oils. Refined oils (soft deodorized and soft deacidified) were prepared on a laboratory scale starting from low-quality olive oils and analyzed together with a set of pure extra virgin olive oil (EVOO) samples and with mixtures of adulterated and pure EVOO at different percentages. The same analytical workflow was applied in two different laboratories equipped with two types of instrumentation (Q-Orbitrap and Q-TOF); a group of discriminant molecules was selected, and a tentative identification of compounds was also proposed. In summary, 12 molecules were identified as markers of this specific adulteration, and seven of them were selected as discriminative in both the laboratories, with a similar trend throughout the samples (i.e., propylene glycol 1 stearate). The results obtained in the two laboratories are comparable, concretely demonstrating the inter-laboratory repeatability of non-targeted studies. As a confirmation, the same markers were detected also in “in-house” mixtures and in suspect commercial deodorized mixtures, reinforcing the robustness of the results obtained and proving that, thanks to these molecules, mixtures containing at least 40% of adulterated oils can be detected.
Introduction
Extra virgin olive oil (EVOO) is one of the most important and expensive edible oils, and therefore, it is also one of the most adulterated food commodities over the global market.1
The International Olive Council has clearly defined the different categories of olive oils2 and, together with the European Union, the limits of the specific chemical parameters able to protect EVOO against potential adulterations with other edible oils.3
Typical frauds like the addition of other type of oils (i.e., seed oils, pomace oils), the presence of re-esterified oils, or the creation of mixtures with refined oils can be easily detected with standard methods. For this reason, the fraudsters are now focused on developing more sophisticated adulterations (like, indeed, the use of soft refined oils or the use of oils with tailored composition) that would allow for the creation of mixtures that cannot be discovered with regular methods; in addition, another relevant authenticity issue is the false declaration of oils’ geographical provenance.4
Although official recognized methods for the detection of these frauds do not exist up to now, the literature suggests different approaches, like gas chromatography,5 nuclear magnetic resonance,6 isotopic fingerprint studies,7,8 or liquid chromatography coupled with chemometrics,9 and the results presented are encouraging.
The use of soft deodorized (<100 °C) or soft deacidified virgin or lampante olive oils can be considered one of the most critical point within this scenario: the products obtained at the end of the soft-refinement processes cannot be considered “virgin” according to the current legislation, so any mixture of EVOO with soft refined oils is considered a fraud.4
As clearly described by Aparicio-Ruiz et al., this adulteration process is able to remove unpleasant volatile components or reduce the acidity of the oils without any macroscopic changes in the other chemical parameters.10 The lack of changes in the chemical bulk of the EVOO during soft refinement is actually the reason that makes the detection of soft-refined oil addition particularly challenging with the official analyses established by the European Union.
The content of Fatty acid alkyl esters,11 lignans’ profile,12 amount of pyropheopytin “A”,13 and kinetic of diacyl glycerol isomerization14 were proposed in the past as possible tools to detect this fraud.
In addition, also the study of the trans- and cis-phytol isomer distribution15 is a possibility presented in the literature, together with the evaluation of the global volatile profile,16 but a robust analytical solution is not available up to now, especially when the aim is to detect mixtures of soft refined oils with pure EVOO.10
Within this scenario, non-targeted analyses by LC-HRMS could represent an interesting approach for the identification of specific compounds responsible for the fraudulent process.17
Untargeted LC-HRMS methods were recently applied for the detection of defective olive oils18 or for the assessment of the geographical origin of the product,19 but, to the best of our knowledge, this is the first proposal of application for the detection of soft refined oil additions to extra virgin olive oils.
Due to the intrinsic nature of these non-targeted approaches, one critical point is their reliability when applied across different laboratories;20 although ring-tests are highly recommended, they have not been largely applied yet for food protection purposes. Only some attempts of “metabo-ring tests” were reported recently in the literature in other fields.21,22
On the basis of what was described above, this work presents a non-targeted liquid chromatography–high-resolution mass spectrometry inter-laboratory study between two laboratories (equipped with two different types of mass spectrometers, a Q-Orbitrap and a Q-TOF) for the detection of new chemical markers able to identify the addition of soft deodorized and soft deacidified low-quality virgin or lampante olive oils to EVOO.
“In-house” soft refined oils were created and analyzed together with a group of pure extra virgin olive oils; in addition, different mixtures of pure EVOO and adulterated oils were included in the sample set.
After a robust data elaboration, the markers selected as a discriminant in the two laboratories were compared with the aim to assess the reproducibility of the inter-laboratory study, and a tentative identification of compounds was performed.
Results and Discussion
Despite the fact that the main constituents of the extra virgin olive oil matrix are lipophilic molecules, the results presented in this work are related to the polar fraction of the oils, and this is coherent with the recent trends presented in the literature in which the volatile and the phenolic fractions of virgin olive oils are studied, for both quality and authenticity purposes.23,24
In a preliminary phase of this research, the nonpolar fraction of the oils was also extracted and studied, but the results obtained were not encouraging. For this reason, the attention was totally focused on the polar component that, on the contrary, also provided interesting results at the early stage of this study.
As an example, the total ion chromatograms (TIC) obtained in positive ionization mode of a pure EVOO sample and of different mixtures are presented in the Supporting Information (Section 1, Figure S1): without the extraction of any specific masses, the TIC itself shows that there are differences in the nonvolatile profile through the pure EVOO and the adulterated mixtures (especially between 3 and 5 min of the chromatographic run). These visual differences encouraged the subsequent data elaboration steps.
Multivariate Studies
After peak alignment, blank subtraction, and isotope merging, in Laboratory #1, 3024 features were extracted in positive mode and 1019 in negative, while in Laboratory #2, the software selected 5612 and 2800 features in positive and negative modes, respectively.
The preliminary PCA models highlighted a tight clustering of the QCs and a substantial overlay of the replicate samples for both the laboratories, certifying the goodness of the analytical procedure performed in both the laboratories (see Figures S2 and S3, Section 1, Supporting Information).
Therefore, the PCA models were obtained without the inclusion of QC samples and using the average values of the replicate extracts. In this case, a partial separation of the EVOO samples from the non-authentic ones was achieved.
The ESI+ score plot is reported in Figure 1, while the ESI– score plot can be found in the Supporting Information (Section 1, Figure S4).
Figure 1.
ESI + PCA score plots of the samples (X axis, PC1; Y axis, PC2). (A) Laboratory #1; number of PCs, 8; explained variance of each PC: PC1 33.1%, PC2 13.1%, PC3 8.7%, PC4 6.9%, PC5 6.2%, PC6 4.0%, PC7 3.0%, PC8 2.4%. (B) Laboratory #2; number of PCs, 7; explained variance of each PC: PC1 9.7%, PC2 8.9%, PC3 8.7%, PC4 8.5%, PC5 8.1%, PC6 7.2%, PC7 6.7%. Green dots: EVOO samples; blue dots: OTHER samples.
The clustering of the two groups dramatically increase when we move to supervised PLS-DA models, as presented in Figure 2 for the ESI+ mode and in Figure S5 (Supporting Information) for the ESI– mode. The variance of the x and y variables explained by the model (R2X(cum) and R2Y(cum)) and the cumulative predicted variation in the Y matrix (Q2(cum)) are reported in the plots as well.
Figure 2.
ESI + PLS-DA score plots of the samples (X axis, Component 1; Y axis, Component 2). (A) Laboratory #1; R2X(cum) = 0.564; R2Y(cum) = 0.964; Q2(cum) = 0.903. (B) Laboratory #2; R2X(cum) = 0.208; R2Y(cum) = 0.991; Q2(cum) = 0.902. Green dots: EVOO samples; blue dots: OTHER samples.
In order to validate these results, new predictive PLS-DA models were created without six samples that were used as validation set: three EVOO samples (CP_30, CP_31, and CP_32) selected with a Kennard-Stone algorithm, and three “NOT EVOO” samples (DEO3, DEO_DEA2, and Mix D) selected with the aim to predict both pure adulterated samples and mixtures.
The affinity of these samples to the two groups was predicted by the models and, as presented in Tables S1 and S2, Section 1, Supporting Information, all the six oils were correctly classified as EVOO or NOT EVOO.
This global differentiation of the groups encouraged the subsequent univariate study, aimed at the selection and identification of the molecules responsible for the fraudulent processes.
Compound Selection and Identification
Different one-way ANOVA tests were applied on the data obtained in Laboratory #1, comparing the EVOO group with the DEA, the DEO, and the DEO_DEA groups: the molecules with a p-value lower than 0.05 were selected for subsequent studies.
The compounds selected in Laboratory #1 were searched in the data set from Laboratory #2, evaluating that in these studies, their p-value was also lower than 0.05.
Subsequently, a tentative identification of compounds was executed for the features that were visible also in the mixtures, taking into account that only these could represent a realistic tool for fraud detection.
The summary of the results obtained is detailed in Tables 1 and 2; when more than one feature is related to the same compound, this means that features are in-source fragments of the precursor ion.
Table 1. Comparison of the Adulterations Markers Detected in Positive Ionization Modea.
| Laboratory
#1 |
Laboratory #2 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| compound ID | formula | m/z [M + H]+ | Δmass (ppm) | RT [min] | p-value DEA | p-value DEO | p-value DEO_DEA | formula | m/z [M + H]+ | Δmass (ppm) | RT [min] | p-value DEA | p-value DEO | p-value DEO_DEA |
| 1 | C21H42O3 | 343.3207 | 1.5 | 6.06 | 2.68 × 10–4 | 2.00 × 10–6 | 1.32 × 10–7 | N.D. | ||||||
| C18H36O3 | 301.2737 | 1.7 | 6.06 | 6.29 × 10–5 | 1.05 × 10–7 | 6.41 × 10–9 | C18H36O3 | 301.2707 | 11.9 | 3.99 | 1.30 × 10–49 | 1.87 × 10–23 | 5.48 × 10–53 | |
| C18H34O2 | 283.2632 | 1.8 | 6.06 | 2.68 × 10–4 | 2.00 × 10–6 | 1.32 × 10–7 | C18H34O2 | 283.2625 | 4.2 | 3.99 | 2.07 × 10–24 | 1.61 × 10–22 | 2.26 × 10–28 | |
| C18H32O | 265.2526 | 1.9 | 6.06 | 6.29 × 10–5 | 1.05 × 10–7 | 6.41 × 10–9 | C18H30 | 247.2407 | 7.7 | 3.99 | 3.24 × 10–33 | 2.25 × 10–24 | 6.72 × 10–33 | |
| 2 | C10H12O2 | 165.0909 | 4.2 | 3.65 | 6.57 × 10–6 | 4.06 × 10–5 | 1.24 × 10–7 | N.D. | ||||||
| 3 | C11H15NO3 | 210.1128 | 0.9 | 2.10 | 2.16 × 10–10 | 1.72 × 10–8 | 1.00 × 10–15 | N.D. | ||||||
| 4 | C10H16O2 | 169.1223 | 3.0 | 4.57 | 4.53 × 10–7 | 3.28 × 10–1 | 1.00 × 10–15 | N.D. | ||||||
| 5 | C12H16O5 | 241.107 | 2.5 | 2.58 | 1.13 × 10–2 | 2.28 × 10–5 | 4.82 × 10–5 | N.D. | ||||||
| 6 | C21H40O3 | 341.3049 | 2.1 | 6.19 | 1.36 × 10–4 | 1.27 × 10–1 | 1.16 × 10–6 | N.D. | ||||||
| C18H34O3 | 299.2579 | 2.3 | 6.19 | 1.89 × 10–4 | 1.35 × 10–2 | 1.28 × 10–7 | N.D. | |||||||
N.D. = Not Detected
Table 2. Comparison of the Adulterations Markers Detected in Negative Ionization Mode.
| Laboratory #1 |
Laboratory #2 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| compound ID | formula | m/z [M – H]− | Δmass (ppm) | RT [min] | p-value DEA | p-value DEO | p-value DEO_DEA | formula | m/z [M – H]− | Δmass (ppm) | RT [min] | p-value DEA | p-value DEO | p-value DEO_DEA |
| 7 | C23H29ClO4 | 403.1682 | 1.5 | 5.67 | 2.03 × 10–10 | 1.00 × 10–15 | 1.17 × 10–8 | C23H29ClO4 | 403.1663 | 3.2 | 3.74 | 5.19 × 10–37 | 1.52 × 10–37 | 1.22 × 10–20 |
| 8 | C23H27ClO4 | 401.1534 | 3.7 | 5.60 | 1.00 × 10–15 | 1.00 × 10–15 | 1.09 × 10–5 | C23H27ClO4 | 401.151 | 2.2 | 3.67 | 7.33 × 10–56 | 7.16 × 10–43 | n.a. |
| 9 | C25H33ClO6 | 463.1903 | 3.5 | 5.35 | 9.10 × 10–10 | 1.00 × 10–15 | 1.07 × 10–7 | C25H33ClO6 | 463.182 | 14.5 | 3.49 | 2.70 × 10–24 | 2.26 × 10–22 | 1.04 × 10–19 |
| 10 | C25H31ClO6 | 461.1748 | 3.9 | 5.41 | 1.00 × 10–15 | 1.00 × 10–15 | 1.00 × 10–15 | C25H31ClO6 | 461.1723 | 1.5 | 3.53 | 4.33 × 10–21 | 2.67 × 10–13 | 6.85 × 10–24 |
| 11 | C23H33ClO5 | 423.1955 | 4.0 | 5.40 | 1.00 × 10–15 | 1.00 × 10–15 | 1.00 × 10–15 | C23H33ClO5 | 423.1927 | 2.6 | 3.52 | 2.06 × 10–42 | 2.75 × 10–50 | 2.69 × 10–24 |
| 12 | C23H32O4 | 371.2231 | 2.4 | 5.50 | 1.29 × 10–5 | 3.34 × 10–9 | 8.05 × 10–6 | C23H32O4 | 371.222 | 0.5 | 3.58 | 3.59 × 10–26 | 4.23 × 10–28 | 1.21 × 10–15 |
Table 3 presents, as example, the “Compound 1” supposed name, together with the identification level25 and with its mean area values in the different groups.
Table 3. Compound 1 Description and Comparison of Its Mean Area Values (+/– Standard Error) through the Groupsa.
Yellow bar, EVOO; red bar, DEO; green bar, DEA; blue bar, DEO + DEA samples.
The complete list of the supposed names, identification levels, and mean area values in the groups for all the selected compounds is presented in the Supporting Information (Section 2, Table S3).
As far as we know, this is the first time that these molecules are connected to the soft deodorization of low quality olive oils. However, as reported in the subsequent paragraph, an explanation of their presence is detailed.
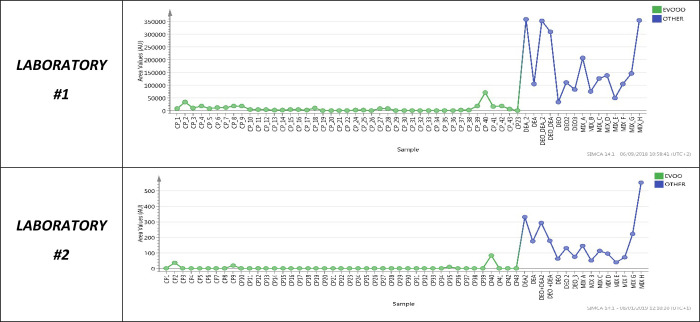
All the selected molecules were detected at significant levels in the eight mixtures (the six ones created in the laboratory and the two suspect commercial blends). As an example, the “Variables trends plot” chart of “Compound 12” is given in Figure 3. Other examples of “Variables trends plot” charts can be found in the Supporting Information (Section 2, Figure S6).
Figure 3.
Variable trend plots of Compound 12. Green dots: area values of the marker in the EVOO group; blue dots: area values of the marker in the OTHER group.
Markers Interpretation
All the 12 selected compounds were searched also in the virgin and lampante olive oils used for the refinement processes, and they were all detected. Accordingly, it is likely that these molecules are characteristic of oils obtained with low-quality olives.
Moreover, this information certifies that these compounds are not affected by the adulteration procedures. This fact turns them into a set of robust markers because they are not originated during refinement and thus subjected to possible fluctuation according to the process applied, but they clearly indicate the use of low-quality raw materials.
This result is consistent with other markers presented in the past, like the Fatty acid alkyl esters that were not affected by the soft deodorization procedure.11
A chemical interpretation of these features is not always easy to perform; for example, it seems that compounds 4 and 6 are produced by the degradation of the oxidized fatty acids.
The reason why these compounds are present mostly in the adulterated samples is not totally clear, but a hypothesis was developed: virgin and especially lampante olive oils have a high amount of oxidized fatty acids, and this should justify the presence of more products of this chemical process.26
Moreover, oxidation generates peroxides and hydroperoxides that subsequently bring to the creation of conjugated dienes with a loss of water; for this reason, for example, the oleic acid is transformed into the conjugated linolenic acid.27−29 This process is catalyzed by thermal and physical stresses that happen in virgin and lampante olive oils more than in extra virgin olive oils.
Our hypothesis is that, in a minor percentage, the oxidized fatty acids could also degrade to unsaturated hydroxyl acids instead of moving to the formation of a conjugated diene with a loss of water. This justifies, for example, the presence of the propyl-12-hydroxy-9-octadecenoate (named “Compound 6” in the tables) as one of the markers.
Compounds 2, 3, and 5 are part of the metabolic pathway of phenolic compound30 formation and are probably connected to a lower quality of the olive used for the creation of the oil.
For “Compound 1”, another less likely hypothesis is that it could be used as emulsifier to increase the extraction yield from low-quality olives that were used to the produce lampante or virgin olive oils.
The identification process of the 12 selected molecules will be undoubtedly reinforced in the future in order to increase the identification level of some of these markers.
In any case, these features are present exclusively (i.e., “Compound 10” and “Compound 11”) or mostly (i.e., “Compound 1” and “Compound 2”) in the adulterated samples and were identified also in the mixtures, allowing for the detection of fraudulent samples that would be wrongly considered “legal” according to the official legislation.
Moreover, the fact that these molecules were also detected in the commercial mixtures (not prepared in the laboratory) reinforce the results obtained, avoiding the risk that these data are affected by some mistakes performed during the in-house adulteration processes.
If compared with the other works presented in the literature, the results obtained seems encouraging, taking into account that the compounds presented in this study were also detected in different mixtures.
All the works presented in the past, indeed, studied pure samples,12-15 sometimes also using deodorization conditions stronger than the ones used in this work.11
Evaluation of the Inter-Laboratory Study
The whole analytical process (from sample preparation to markers interpretation) was performed in two different laboratories with two different HRMS platforms in order to evaluate the inter-laboratory applicability of a non-targeted mass spectrometry study.
The amount of features detected in Laboratory #2 is higher than the ones obtained in Laboratory #1.
This is partially explained with the fact that MarkerView software treated all the ionization adducts (i.e. ammonium, sodium, etc.) of the same compound as separated features; on the contrary, Compound Discoverer software merged all the adducts as unique features. This strongly contributed to the discrepancy in the total amount of compounds between the two laboratories. In addition, the area threshold used for Compound Discoverer and MarkerView software are not the same due to the intrinsic differences in the instruments, and this may have affected the number of features detected. The multivariate approaches (for both unsupervised PCA and supervised PLS-DA models) showed comparable results, showing that in both the laboratories the fingerprints detected were fit for the purpose of this work.
Moving to the univariate analysis, seven compounds were selected as discriminative in both the laboratories, with a similar trend throughout the samples.
“Compound 1” was detected in Laboratory #2 only as the in-source fragmentations feature and not “as is”. This is probably due to the different ESI ionization sources present in the mass spectrometers.
Except for this discrepancy, for each of the common m/z values, the results obtained are comparable not only for the information provided with the “Full scan” experiment but also for the MS/MS spectra (see the Supporting Information, Section 3, Figures S7 to S13).
These results put in evidence a similar behavior of the molecules in the two instrumentations despite the differences in the elution methods (due to the different columns lengths) and even though two different approaches were applied in the mass spectrometers for compound fragmentation and for accurate mass measurement. The results obtained in the two laboratories are similar despite the use of two different mass spectrometers (a Q-Orbitrap and a Q-TOF): it must be underlined that the good agreement was obtained with freshly prepared samples and not injecting the same extracts in different platforms.
In addition, according to our results, it seems that the negative ionization mode is more reproducible through different instruments than the positive one, taking into account that both the laboratories were able to select the same discriminant markers using this polarity.
In summary, the results obtained within the inter-laboratory ring test and the two analytical processes are comparable on the whole, demonstrating the goodness of the untargeted approach for the detection of chemical markers introduced by the addition of soft refined oils (soft deodorized and soft deacidified) to extra virgin olive oils. Seven compounds related to soft refined oils additions were selected as markers and tentatively identified. These markers are a valuable tool for unmasking this specific fraud, considering that they were detected in raw materials and in the final products, remaining unaffected by soft-refining. In addition, their robustness was proven independently over two laboratories and confirmed by the application to samples from the market.
In this work, the minimum adulteration percentage of the mixtures was 40%: although it may seem to be a high value, we know that it is a real condition because, as also reported in the literature, all the methods presented in the past, including the official methods, were not able to detect additions of 50% of soft refined oils.9 Lowest percentages of addition did not provide robust results and for this reason were not included in this work.
In any case, the markers selected in our study were detected in the two commercial mixtures: we do not know the percentage of additions in these two samples, but we know that these samples were commercialized as “extra virgin olive oil”; this means that they were considered pure EVOO by the market. According to the results obtained, the presence of at least one of these compounds should be enough to consider a sample as a “suspect”; however, an overall judgment concerning the number of markers required for considering a sample as a “suspect” should be performed only after the development of a quantitative target method that could be able to set up concentration limits for each molecule.
For this reason, further efforts will be performed for a better clarification of the chemical structures, leading subsequently to the development of a target method for a quantitative evaluation.
Materials and Methods
Chemicals and Sample Description
Methanol, ammonium formate, sodium hydroxyde, and sodium sulfate were purchased from VWR International, Ltd. (Poole, United Kingdom).
Isopropyl alcohol and formic acid were purchased from Sigma Aldrich (St. Louis, MO).
Water was purified using a Milli-Q system (Millipore, Bedford, MA).
Pure EVOO samples (43) were collected trying to include the highest variability in their characteristic (i.e., coming from different suppliers, related to different production years, with different storage conditions, etc.). Forty of them were produced with Italian olives and three with a mixture of olives coming from different countries of the European Union.
Moreover, three “soft deodorized”, two “soft deacidifed”, and two “soft deacidified and then deodorized” samples were prepared in a laboratory scale starting from virgin and lampante olive oils that were provided for us specifically for this research purpose from a trusted supplier and that were classified as “virgin” or “lampante” after performing the official chemical and sensorial analyses.
These adulterated samples were analyzed for the official EVOO parameters (fatty acids, aliphatic alcohols, sterols, triglycerides, and hydrocarbons);2 subsequently, according to the results obtained, six mixtures were prepared at different percentages of adulteration (with the aim to create frauded samples that could be in compliance with the current legislation) and were analyzed together with two suspect commercial blends with deodorized oils obtained from the bulk market.
The sample list is detailed in the Supporting Information (Section 4, Table S4).
Soft Deodorization
The process was executed partially according to a previous work presented in the literature.10
Defective oil (400 mL) was introduced into a round-bottom flask with a triple neck containing a magnetic stir bar.
Two necks were used to connect the vacuum tubing and a dropping funnel containing Milli-Q grade water. A thermometer was inserted into the third neck in order to constantly monitor the temperature of the solution.
This apparatus was placed on a magnetic stirring hotplate (VELP Scientifica, Italy) operating at 300 rpm.
When the oil reached a temperature of 60 °C, 0.2 mL of water was added to the oil in order to start the distillation. Thereafter, the temperature was raised to 100 °C and maintained for 60 min. Once the initial water was evaporated, 0.2 mL more were added to carry on the distillation process, repeating this addition during the process. At the end of this procedure, the sample was cooled down to room temperature and stored at 4 °C before the analyses.
Three different samples were prepared following this procedure that were labeled as “DEO”, “DEO2”, and “DEO3”.
Soft Deacidification
This process was executed on two defective olive oils with an acidity of 1.4 and 1.0%.
600 mL of defective oil were weighed and transferred into a beaker containing a magnetic stir bar and put on a magnetic stirrer working at room temperature.
A specific amount (in mL) of a 50% (w/w) sodium hydroxide solution was added followed by a double amount (in mL) of a 10% (w/w) sodium sulfate solution, according to the following formula:
 |
where A is the starting acidity (free fatty acids, FFA) of the oil (% oleic acid), B is the target % FFA of the oil (0.4% in our work), C is the oil weight in grams, D is the sodium hydroxide molecular weight (40 g/mol); E is the oleic acid molecular weight (282 g/mol), F is the concentration of the sodium hydroxide solution, and G is the density of the 50% (w/w) sodium hydroxide solution (1.52 g/mL).
After 5 min of stirring, the oil was centrifuged at 5000 rpm, and 200 mL of the supernatant (the neutralized oil) was stored at 4 °C before the analyses.
The remaining 400 mL of the neutralized oil were subjected to the soft deodorization process described above.
At the end, we created two “soft deacidified” samples (named DEA and DEA2) and two “soft deacidified and then deodorized” samples (named DEO_DEA and DEO_DEA2).
Sample Extraction
The extraction procedure was based on a previous work of Vaclavik et al.31 Both the laboratories applied the same extraction procedure: 1 mL of sample was transferred into a PTFE cuvette (15 mL), and 3 mL of a mixture methanol:water (80:20, v/v) were added for isolation of the polar fraction. The sample was then shaken for 20 min at 240 RPM using a horizontal laboratory shaker (IKA Labortechnik, Germany) and then centrifuged at 10,000 rpm (12,768 RCF) for 5 min (Hettich, Germany).
One milliliter of the hydroalcoholic layer was transferred into an amber glass vial and stored at −20 °C until LC-HRMS analysis.
For the evaluation of method reproducibility, 20% of the samples were double extracted.
The same procedure was also performed into empty tubes in which all the steps were executed without the oil addition. These samples were labeled as “extraction blank”.
Additionally, a quality control (QC) sample was prepared by mixing 10 μL of each extract sample (extra virgin olive oils, adulterated samples, and mixtures). As detailed below in the text, this sample will be periodically injected during the analytical sequences in order to check the goodness of the measurements, being sure that no retention time drifts or problems in signals intensity eventually occurred.
LC-HRMS Analysis
In Laboratory #1, HPLC analysis was performed with a Dionex UltiMate 3000 UHPLC (Thermo Fisher Scientific, Inc., Waltham, MA) equipped with a BEH C18 150 × 2.1 mm, 1.7 μm particle size analytical column (Waters, Milford, MA, USA) maintained at 45 °C. Gradient elution was performed using formic acid (FA) and ammonium formate (AF) as mobile phase modifiers with a constant flow rate of 0.3 mL/min.
Mobile phase A was constituted by a 5 mM AF and 0.1% FA solution of a mixture of water and methanol (95:5 v/v), while mobile phase B was constituted by a 5 mM AF and 0.1% FA solution of a mixture of isopropyl alcohol, methanol, and water (65:30:5, v/v/v).
Gradient conditions are the following: after 1 min with 75% of mobile phase A and 25% of mobile phase B, the percentage of solvent B increased to 80% in 2.5 min, then to 100% in 4 min and then was maintained at this percentage for 3.5 min before column re-equilibration (5 min).
The autosampler was maintained at 5 °C, and the injection volume was 4 μL.
Mass spectrometry detection was performed with a benchtop Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) equipped with a heated (H) electrospray ionization (ESI) interface (Thermo Fisher Scientific,Waltam, MA). Two analytical sequences (one with positive and one with negative ionization mode) were executed with a “full scan data-dependent fragmentation” experiment.
The resolution of the full scan experiment was 70,000 full width at half-maximum (FWHM) (m/z 200) and was 17,500 FWHM (m/z 200) for the MS/MS experiment.
All the other ionization source and mass spectrometer conditions are the same as described in a previous work of the Barilla research group.32
Samples were randomly injected, while the QC sample was injected at the beginning and at the end of the sequence and every 10 sample injections.
In Laboratory #2, HPLC analysis was performed with a Dionex UltiMate 3000 UHPLC (Thermo Fisher Scientific, Inc., Waltham, MA) equipped with a BEH C18 100 × 2.1 mm, 1.7 μm particle size analytical column (Waters, Milford, MA, USA) maintained at 45 °C.
HPLC and chromatographic conditions were the same as used in Laboratory #1 with the exception of the gradient steps, which are the following: after 0.5 min with 75% of mobile phase A and 25% of mobile phase B, the percentage of solvent B increased to 80% in 1.5 min, then to 100% in 3 min and then was maintained at this percentage for 2.5 min before column re-equilibration (2.5 min).
Mass spectrometry detection was performed with TripleTOF 6600 quadrupole time-of-flight (TOF) mass spectrometer (SCIEX) equipped with a DuoSpray source with separated ESI and APCI ion sources. ESI was used for the sample measurement, and APCI was used for exact mass calibration of the TripleTOF instrument.
Two analytical sequences (one with positive and one with negative ionization mode) were executed with a TOF-MS and information-dependent acquisition (IDA) experiments in order to simultaneously collect the full scan spectrum and the product ion spectra of the most abundant ions.
The achieved resolving power was >40,000 FWHM (m/z 829.5393 for positive ionization mode and m/z 933.637 for negative ionization mode), while >25,000 FWHM (m/z 811.5288 for positive ionization mode and m/z 933.637 for negative ionization mode) for the MS/MS experiments.
All the other ionization source and mass spectrometer conditions applied to the TripleTOF 6600 are the same as described in a previous work of Hurkova et al.33
Samples were randomly injected, while the QC sample was injected at the beginning and at the end of the sequence and every 10 sample injections.
Data Treatments and Statistics
Laboratory #1
UHPLC-HRMS raw data were acquired using Xcalibur software (version 3.0 Thermo Fisher Scientific,Waltam, MA); peaks alignment, extraction blanks subtraction, and features extraction were performed using Compound Discoverer software (version 2.1 Thermo Fisher Scientific,Waltam, MA) directly connected to Chemspider and m/z CLOUD databases and able to perform in silico fragmentations; the mass range inspected was between 100 and 1000 m/z from 0.5 to 11 min of the chromatographic runs.
The values of the critical parameters for features extractions and identification are the same as described in a previous work.32
Laboratory #2
UHPLC-HRMS raw data were acquired using Analyst 1.7.1 TF (SCIEX), and the qualitative analysis was performed using PeakView 2.2 (SCIEX) equipped with MasterView and Formula Finder and directly linked to the ChemSpider database.
MarkerView software (version 1.2.1, SCIEX) was used for data processing; the mass range inspected was between 100 and 1000 m/z from 0.6 to 8 min of the chromatographic runs.
Peak alignment was carried out with a tolerance of 0.2 min for the retention time and of 0.01 Da for the m/z values, while MS/MS spectra were studied using METLIN (https://metlin.scripps.edu/landing_page.php?pgcontent=mainPage )34 online database.
In both the laboratories, the resulting two data matrixes (for positive and negative ionization modes), containing the area values for all the features, were exported and processed with SIMCA software (version 14.1 Umetrics, Umea, Sweden) for chemometric data elaboration.
Data were log transformed and Pareto scaled, and then a preliminary Principal Component Analysis (PCA) was executed in order to check the clusterization of the samples and the QC positioning in the score plot.
Subsequently, QCs were removed, and the replicate samples were averaged; then, explorative principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) models were created comparing the group of the pure EVOO samples against the group “OTHER” containing the adulterated samples.
In this work, different kinds of adulterations are studied (DEO, DEA, and DEO_DEA), and the amount of samples is different in each group; for these reasons, multivariate models were used only as preliminary approaches for a global evaluation, but markers selection was executed with univariate models.
For each data matrix, different one way ANOVA tests were applied using Microsoft Excel 2016 professional plus software (Microsoft Corporation) comparing the EVOO group with the pure DEA, the pure DEO, and the pure DEO_DEA samples.
The molecules with a p-value lower than 0.05 were selected, and a tentative identification of compounds was executed, following the workflow described in a previous paper32 and referring to the Standard Initiative on Metabolomics35 and subsequent improvements25 for identification ranking.
The presence of the most interesting features was evaluated in the mixture samples in order to check their value as discriminant markers in real fraudulent situations.
Acknowledgments
Authors would like to thank Raffaele Maranzoni and Evelyne Sasso from “Coppini Arte Olearia” for the invaluable support in EVOO authentic sample collection. This work was supported by the European Union, Seventh Framework Program (FP7/2007–2013), under grant agreement 613688 (Ensuring the Integrity of the European food chain FoodIntegrity). This work has received funding from the Czech National Agency for Agriculture Research (NAZV-QJ1530272), the Operational Programme Prague Competitiveness (CZ.2.16/3.1.00/21537 and CZ.2.16/3.1.00/24503), and by the National Program of Sustainability I (NPU I LO1601). This research has also received funding from the European Union Horizon 2020 research and innovation program under grant agreement no. 692195 (MultiCoop).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c00346.
TIC chromatograms in ESI+ ionization mode of different samples, PCA scores plots, PLS-DA scores plots, predicted group affinity tables, compounds description and comparison through the groups, variable trend plots, MS/MS spectra, and sample list (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- de Lange E. Report on the food crisis, fraud in the food chain and the control thereof (2013/2091(INI)); European Parliament, 2013.
- I. O. C. IOC , COI/T.15/NC No 3/Rev. 14 - Trade standard applying to Olive Oils and Olive-Pomace Oils; International olive council, coi, Madrid, Spain, 2019. [Google Scholar]
- Commission Delegated Regulation (EU) 2019/1604 of 27 September 2019 amending Regulation (EEC) No 2568/91 on the characteristics of olive oil and olive-residue oil and on the relevant methods of analysis, Official Journal Of the European Union, n. L250, p. 14, 2019. [Google Scholar]
- Garcia-Gonzalez D.; Aparicio D.; Aparicio-Ruiz R.. Olive oil in FoodIntegrity Handbook: A guide to food authenticity issues and analytical solutions, Nantes, 2018, pp. 336–357. [Google Scholar]
- Melucci D.; Bendini A.; Tesini F.; Barbieri S.; Zappi A.; Vichi S.; Conte L.; Gallina Toschi T. Rapid direct analysis to discriminate geographic origin of extra virgin olive oils by flash gas chromatography electronic nose and chemometrics. Food Chem. 2016, 204, 263–273. 10.1016/j.foodchem.2016.02.131. [DOI] [PubMed] [Google Scholar]
- Alonso-Salces R. M.; Moreno-Rojas J. M.; Holland M. V.; Reniero F.; Guillou C.; Héberger K. Virgin Olive Oil Authentication by Multivariate Analyses of 1H NMR Fingerprints and δ13C and δ2H Data. J. Agric. Food Chem. 2010, 58, 5586–5596. 10.1021/jf903989b. [DOI] [PubMed] [Google Scholar]
- Faberi A.; Marianella R.; Fuselli F.; La Mantia A.; Ciardiello F.; Montesano C.; Mascini M.; Sergi M.; Compagnone D. Fatty acid composition and δ13 C of bulk and individual fatty acids as marker for authenticating Italian PDO/PGI extra virgin olive oils by means of isotopic ratio mass spectrometry. J. Mass Spectrom. 2014, 49, 840–849. 10.1002/jms.3399. [DOI] [PubMed] [Google Scholar]
- Bontempo L.; Paolini M.; Franceschi P.; Ziller L.; García-González D. L.; Camin F. Characterisation and attempted differentiation of European and extra-European olive oils using stable isotope ratio analysis. Food Chem. 2019, 276, 782–789. 10.1016/j.foodchem.2018.10.077. [DOI] [PubMed] [Google Scholar]
- Vera D. N.; Jiménez-Carvelo A. M.; Cuadros-Rodríguez L.; Ruisánchez I.; Pilar Callao M. Authentication of the geographical origin of extra-virgin olive oil of the Arbequina cultivar by chromatographic fingerprinting and chemometrics. Talanta 2019, 203, 194–202. 10.1016/j.talanta.2019.05.064. [DOI] [PubMed] [Google Scholar]
- Aparicio-Ruiz R.; Romero I.; García-González D. L.; Oliver-Pozo C.; Aparicio R. Soft-deodorization of virgin olive oil: Study of the changes of quality and chemical composition. Food Chem. 2017, 220, 42–50. 10.1016/j.foodchem.2016.09.176. [DOI] [PubMed] [Google Scholar]
- Pérez-Camieno M. D. C.; Cert A.; Romero-Segura A.; Cert-Trujillo R.; Moreda W. Alkyl esters of fatty acids a useful tool to detect soft deodorized olive oils. J. Agric. Food Chem. 2008, 56, 6740–6744. 10.1021/jf801131b. [DOI] [PubMed] [Google Scholar]
- Cecchi L.; Innocenti M.; Melani F.; Migliorini M.; Conte L.; Mulinacci N. New isobaric lignans from Refined Olive Oils as quality markers for Virgin Olive Oils. Food Chem. 2017, 219, 148–157. 10.1016/j.foodchem.2016.09.132. [DOI] [PubMed] [Google Scholar]
- Serani A.; Piacenti D. Sistema analitico per I’dentificazione di oli deodorati in oli vergini di oliva – Nota 1 – Analisi dei pigmenti clorofilliani in oli vergini di oliva. Riv. Ital. Sostanze Grasse. 2001, 78, 459–463. [Google Scholar]
- Serani A.; Piacenti D.; Staiano G. Sistema analitico per I’dentificazione di oli deodorati in oli vergini di oliva – Nota 2 – Cinetica di isomerizzazione dei digliceridi in oli vergini di oliva. Riv. Ital. Sostanze Grasse. 2001, 78, 525–528. [Google Scholar]
- Vetter W.; Schröder M.; Lehnert K. Differentiation of Refined and Virgin Edible Oils by Means of the trans- and cis-Phytol Isomer Distribution. J. Agric. Food Chem. 2012, 60, 6103–6107. 10.1021/jf301373k. [DOI] [PubMed] [Google Scholar]
- Cerretani L.; Bendini A.; Barbieri S.; Lercker G. Preliminary observations on the change of some chemical characteristics of virgin olive oils subjected to a ″soft deodorization″ process. Riv. Ital. Sostanze Grasse. 2008, 85, 75–82. [Google Scholar]
- Rubert J.; Zachariasova M.; Hajslova J. Advances in high-resolution mass spectrometry based on metabolomics studies for food – a review. Food Addit. Contam., Part A 2015, 32, 1685–1708. 10.1080/19440049.2015.1084539. [DOI] [PubMed] [Google Scholar]
- Kalogiouri N. P.; Alygizakis N. A.; Aalizadeh R.; Thomaidis N. S. Olive oil authenticity studies by target and nontarget LC–QTOF-MS combined with advanced chemometric techniques. Anal. Bioanal. Chem. 2016, 408, 7955–7970. 10.1007/s00216-016-9891-3. [DOI] [PubMed] [Google Scholar]
- Gil-Solsona R.; Raro M.; Sales C.; Lacalle L.; Díaz R.; Ibáñez M.; Beltran J.; Sancho J. V.; Hernández F. J. Metabolomic approach for Extra virgin olive oil origin discrimination making use of ultra-high performance liquid chromatography −Quadrupole time-of-flight mass spectrometry. Food Control. 2016, 70, 350–359. 10.1016/j.foodcont.2016.06.008. [DOI] [Google Scholar]
- Cavanna D.; Righetti L.; Elliott C.; Suman M. The scientific challenges in moving from targeted to non-targeted mass spectrometric methods for food fraud analysis: A proposed validation workflow to bring about a harmonized approach. Trends Food Sci. Technol. 2018, 80, 223–241. 10.1016/j.tifs.2018.08.007. [DOI] [Google Scholar]
- Martin J.-C.; Maillot M.; Mazerolles G.; Verdu A.; Lyan B.; Migné C.; Defoort C.; Canlet C.; Junot C.; Guillou C.; Manach C.; Jabob D.; Bouveresse D. J.-R.; Paris E.; Pujos-Guillot E.; Jourdan F.; Giacomoni F.; Courant F.; Favé G.; Le Gall G.; Chassaigne H.; Tabet J.-C.; Martin J.-F.; Antignac J.-P.; Shintu L.; Defernez M.; Philo M.; Alexandre-Gouaubau M.-C.; Amiot-Carlin M.-J.; Bossis M.; Triba M. N.; Stojilkovic N.; Banzet N.; Molinié R.; Bott R.; Goulitquer S.; Caldarelli S.; Rutledge D. N. Can we trust untargeted metabolomics? Results of the metabo-ring initiative, a large-scale, multi-instrument inter-laboratory study. Metabolomics 2015, 11, 807–821. 10.1007/s11306-014-0740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajka T.; Smilowitz J. T.; Fiehn O. Validating quantitative untargeted lipidomics across nine liquid chromatography–high-resolution mass spectrometry platforms. Anal Chem. 2017, 89, 12360–12368. 10.1021/acs.analchem.7b03404. [DOI] [PubMed] [Google Scholar]
- Quintanilla-Casas B.; Bustamante J.; Guardiola F.; García-González D. L.; Barbieri S.; Bendini A.; Toschi T. G.; Vichi S.; Tres A. Virgin olive oil volatile fingerprint and chemometrics: Towards an instrumental screening tool to grade the sensory quality. LWT- Food Sci. Technol. 2020, 121, 108936. 10.1016/j.lwt.2019.108936. [DOI] [Google Scholar]
- Esposto S.; Selvaggini R.; Taticchi A.; Veneziani G.; Sordini B.; Servili M. Quality evolution of extra-virgin olive oils according to their chemical composition during 22 months of storage under dark conditions. Food Chem. 2020, 311, 126044. 10.1016/j.foodchem.2019.126044. [DOI] [PubMed] [Google Scholar]
- Schymanski E. L.; Jeon J.; Gulde R.; Fenner K.; Ruff M.; Singer H. P.; Hollender J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- Rovellini P.; Cortesi N.; Fedeli E. Ossidazione dei lipidi. Riv. Ital. Sostanze Grasse. 1997, 74, 181–189. [Google Scholar]
- Frankel E. N. Review. Recent advances in lipid oxidation. J. Sci. Food Agric. 1991, 54, 495–511. 10.1002/jsfa.2740540402. [DOI] [Google Scholar]
- Gutiérrez F.; Villafranca M. J.; Castellano J. M. Changes in the main components and quality indices of virgin olive oil during oxidation. J. Am. Oil Chem. Soc. 2002, 79, 669–676. 10.1007/s11746-002-0541-3. [DOI] [Google Scholar]
- Montaño A.; Hernández M.; Garrido I.; Llerena J.; Espinosa F. Fatty Acid and Phenolic Compound Concentrations in Eight Different Monovarietal Virgin Olive Oils from Extremadura and the Relationship with Oxidative Stability. Int. J. Mol. Sci. 2016, 17, 1960. 10.3390/ijms17111960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Xu J.; Gong X.; Yang M.; Zhang C.; Li M. Biosynthesis, Chemistry, and Pharmacology of Polyphenols from Chinese Salvia Species: A Review. Molecules 2019, 24, 155. 10.3390/molecules24010155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaclavik L.; Cajka T.; Hrbek V.; Hajslova J. Ambient mass spectrometry employing direct analysis in real time (DART) ion source for olive oil quality and authenticity assessment. Anal. Chim. Acta 2009, 645, 56–63. 10.1016/j.aca.2009.04.043. [DOI] [PubMed] [Google Scholar]
- Cavanna D.; Catellani D.; Dall’Asta C.; Suman M. Egg product freshness evaluation: a metabolomic approach. J. Mass Spectrom. 2018, 53, 849–861. 10.1002/jms.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurkova K.; Rubert J.; Stranska-Zachariasova M.; Hajslova J. Strategies to Document Adulteration of Food Supplement Based on Sea Buckthorn Oil: a Case Study. Food Anal. Methods. 2017, 10, 1317–1327. 10.1007/s12161-016-0674-4. [DOI] [Google Scholar]
- https://metlin.scripps.edu/landing_page.php?pgcontent=mainPage (last access: 19/08/2019).
- Sumner L. W.; Amberg A.; Barrett D.; Beale M. H.; Beger R.; Daykin C. A.; Fan T. W.-M.; Fiehn O.; Goodacre R.; Griffin J. L.; Hankemeier T.; Hardy N.; Harnly J.; Higashi R.; Kopka J.; Lane A. N.; Lindon J. C.; Marriott P.; Nicholls A. W.; Reily M. D.; Thaden J. J.; Viant M. R. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.